Summary
The Ca2+-independent membrane interactions of the soluble C2 domains from synaptotagmin 1 (syt1) were characterized using a combination of site-directed spin labeling (SDSL) and vesicle sedimentation. The second C2 domain of syt1, C2B, binds to membranes containing phosphatidylserine (PS) and phosphatidylcholine (PC) in a Ca2+-independent manner with a lipid partition coefficient of approximately 3.0 × 102 M−1. A soluble fragment containing the first and second C2 domains of syt1, C2A and C2B, has a similar affinity, but C2A alone has no detectable affinity to PC/PS bilayers in the absence of Ca2+. Although the Ca2+-independent membrane affinity of C2B is modest, it indicates that this domain will never be free in solution within the cell. SDSL was used to obtain bilayer depth restraints, and a simulated annealing routine was used to generate a model for the membrane docking of C2B in the absence of Ca2+. In this model, the polybasic strand of C2B forms the membrane binding surface for the domain; however, this face of C2B does not penetrate the bilayer, but is localized within the aqueous double-layer when C2B is bound. This double-layer location indicates that C2B interacts in a purely electrostatic manner with the bilayer interface. In the presence of Ca2+, the membrane affinity of C2B is increased approximately 20 fold, and the domain rotates so that the Ca2+ binding loops of C2B insert into the bilayer. This Ca2+-triggered conformational change may act as a switch to modulate the accessibility of the polybasic face of C2B, and control interactions of syt1 with other components of the fusion machinery.
Keywords: Site-directed spin labeling, EPR spectroscopy, membrane fusion, protein-membrane interactions, calcium-binding protein
Introduction
Neuronal exocytosis is a highly-regulated Ca2+-triggered process involving the membrane fusion of synaptic vesicles with the presynaptic plasma membrane. This fusion process is mediated and regulated by a large number of proteins, including the SNARE proteins, which constitute the core of the conserved membrane fusion machinery 1; 2; 3. There is strong evidence that the synaptic vesicle protein synaptotagmin 1 (syt1) acts as the Ca2+ sensor for this process 4; 5; 6; 7. Synaptotagmin 1 consists of two tandem cytosolic C2 domains, C2A and C2B, that are anchored to the synaptic vesicle by a single transmembrane helical pass (see Fig. 1). However, the mechanisms by which syt1 triggers membrane fusion are not understood. It is not known whether syt1 acts by directly binding to and regulating the SNAREs 8, whether it acts by binding membranes and creating curvature strain within the bilayer 9, or whether individual 10 or tandem C2 domains act to bridge across the vesicle and the plasma membrane, thereby driving these two membranes closer together 11.
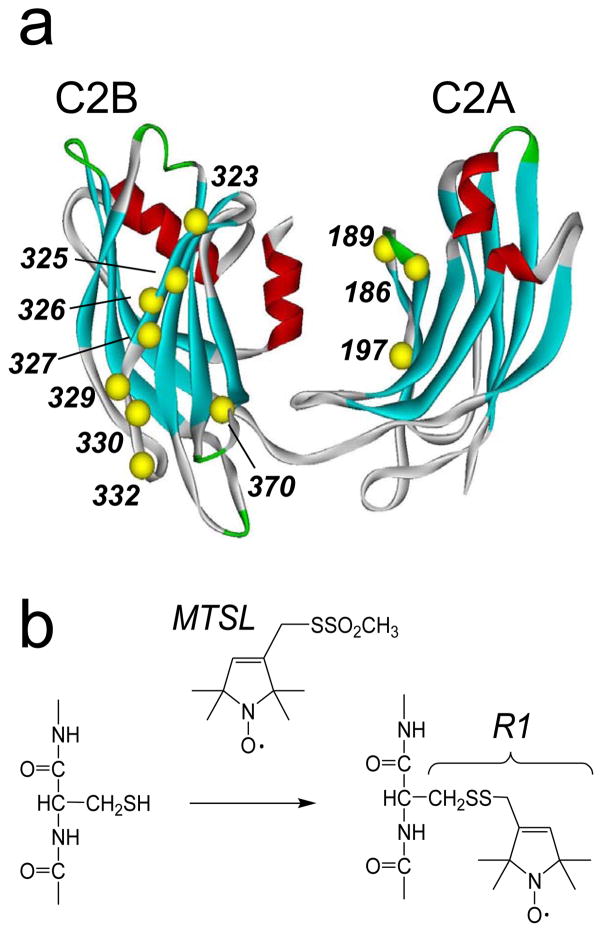
Figure 1.
a) Model of the water-soluble fragment of synaptotagmin 1, syt1C2AB, showing the labeled sites and labeling scheme. The first domain, C2A includes residues 140–263; the second, C2B, includes 273–418. b) The spin labeled side chain, R1, was incorporated into 11 positions within syt1C2AB (Ca positions in yellow), and into the corresponding positions in syt1C2A and syt1C2B, using the sulfhydryl reactive methanethiosulfonate spin label (see Methods). The C2AB model shown was built from models for the isolated C2A(PDB:1BYN) and C2B(PDB:1K5W) domains, that were connected with a linker using InsightII.
Each C2 domain in syt1 is composed of an 8 stranded β-sandwich, where the flexible loops that connect the β-strands at one end bind to Ca2+. When bound to Ca2+, the C2 domains of syt1 penetrate into lipid bilayers that contain anionic phospholipids 12; 13; 14. Site-directed spin labeling (SDSL) demonstrated that the Ca2+-binding loops in C2A and C2B in a soluble construct of syt1 (syt1C2AB) penetrate approximately 10Å below the phosphate plane in 3:1 POPC:POPS membranes 15. This position is 6–7 Å deeper than that seen when the isolated C2 domains are examined 16; 17, indicating that the tandem domains act synergistically to enhance their mutual penetration. Synaptotagmin 1 also interacts with the SNAREs, and there is evidence that syt1 binds simultaneously to phospholipids and the SNARE complex through interactions of its C2 domains 18.
Long-range Coulombic interactions are a major driving force for the Ca2+-dependent membrane binding of the syt1 C2 domains. When bound to Ca2+, the Ca2+-binding loops of C2A attain a positive electrostatic potential, thereby becoming attracted to membrane surfaces containing anionic phospholipids 19. Both C2A and C2B contain a region in their fourth β-strand, which contains a number of positively charged lysine residues. This polybasic region is more pronounced in C2B with eight, almost consecutive, highly conserved positive residues. This region of C2B is implicated in binding membranes as well as the SNARE proteins syntaxin and SNAP-25 18; 20.
The polybasic region of Synaptotagmin 1 C2B has been shown to interact with anionic phospholipid membranes. It is reported to associate with membranes containing PIP2 in a Ca2+-independent manner, and this interaction is proposed to pre-dock C2A-C2B to the plasma membrane surface 21. There are indications that PIP2 may not be necessary to achieve Ca2+-independent binding, and both Ca2+-dependent and Ca2+-independent binding to liposomes are reported for syt1C2AB 9. However, both the Ca2+-dependent and Ca2+-independent binding of syt1C2AB are strongly influenced by ionic strength, and in another study, the Ca2+-independent phospholipid binding is not detected without PIP2 under conditions of normal ionic strength 22. Fluorescence experiments indicate that polybasic region, residues (K326, K327) in syt1C2AB insert into the interfacial environment of phospholipid head groups in a Ca2+-dependent manner, leading to the proposal that the C2B domain may simultaneously bind two membrane surfaces with this positively charged region and with the calcium binding loops 10. However, measurements made using SDSL, do not provide an indication of membrane penetration for the polybasic region of C2B when syt1 is bound in a Ca2+-dependent manner 15; 17.
In this work, we determined the membrane binding affinities for syt1C2A, syt1C2B and syt1C2AB in the absence of Ca2+ to lipid bilayers composed of 3:1 POPC:POPS using a well-established equilibrium vesicle sedimentation assay. The syt1C2A domain alone does not associate with bilayers containing acidic lipid in the absence of Ca2+, however, syt1C2B and syt1C2AB bind with similar affinities. Site-directed spin labeling indicates that in the absence of Ca2+, residues in the polybasic face of C2B reside in the aqueous double-layer at the bilayer interface but do not penetrate into the bilayer interior, and a model for the membrane docking of C2B was determined from EPR-derived membrane depth parameters using an established simulated annealing protocol. Although the membrane affinities for syt1C2B and syt1C2AB are weak in the absence of Ca2+, they indicate that the C2B domain will never be free in the cytosol, but will always be either SNARE or membrane associated. Upon Ca2+ addition, the affinity of syt1C2B to the membrane interface increases by 20 fold and the domain rotates so that it now interacts through its Ca2+-binding region 17. We discuss the possibility that this Ca2+ triggered change in membrane docking may function as a conformational switch to regulate the exposure of the polybasic face of C2B.
Results
Syt1C2AB and syt1C2B bind to POPC:POPS membranes in the absence of Ca2+
Figure 2 presents membrane binding data obtained by vesicle sedimentation for syt1C2A, syt1C2B and syt1C2AB to POPC:POPS (75:25) bilayers. The partition coefficients are summarized in Table 1 and represent equilibrium partitioning data obtained under conditions where the syt1 C2 domains are dilute on the membrane surface. In the absence of Ca2+, the data in Fig. 2a indicate that there is no membrane binding for syt1C2A, up to the highest lipid concentrations tested (15 mM total lipid, 7.5 mM accessible lipid). In contrast to C2A, the C2B domain binds to POPC:POPS bilayers in the absence of Ca2+ with a partition coefficient of 290 M−1 (Fig. 2b). This is a modest binding affinity and it indicates that approximately half of the C2B will be membrane bound at an accessible lipid concentration of 3 mM POPC:POPS,. The data for syt1C2AB are similar (Fig. 2c), and yield an affinity for the tandem C2 domain fragment of approximately, 160 M−1 in the absence of Ca2+. Although these affinities are modest, they indicate that the C2B domain of syt1 will never be found free in the cytosol within the cell (see Discussion).
Figure 2.
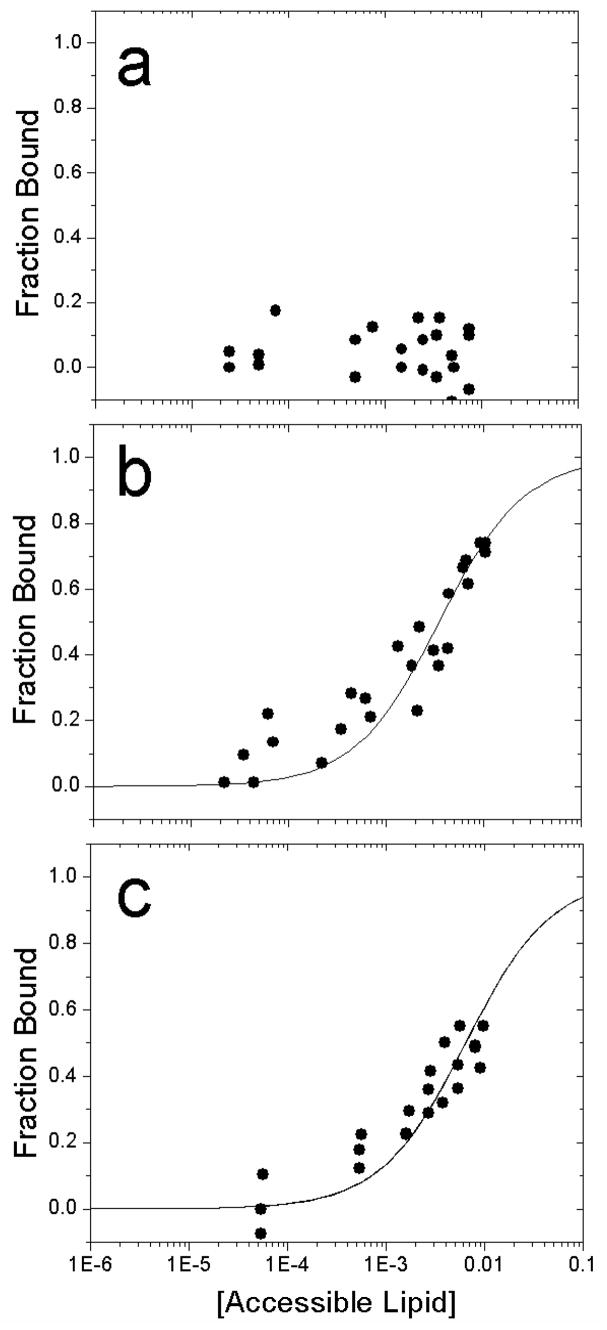
Ca2+-independent binding of a) syt1C2A, b) syt1C2B and c) syt1C2AB fragments of synaptotagmin 1 to membranes composed of POPC:POPS (3:1), using a vesicle sedimentation assay (see Methods). The lines are fits to Eq. 2 and the values of K are given in Table 1. No binding of syt1C2A to POPC:POPS is detected in the absence of Ca2+.
Table 1.
Molar Partition Coefficients, K, for the Ca2+-dependent and Ca2+-independent membrane affinity of the syt1 C2 domains.†
| POPC:POPS (3:1) | Syt1C2A | Syt1C2B | Syt1C2AB |
|---|---|---|---|
| [Ca2+]=1 mM | 1.1 (± 0.1) × 103 | 6.9 (± 0.1) × 103 | 5.3 (± 1.0) × 103 |
| No Ca2+ | Not detected | 2.9 (± 0.3) × 102 | 1.2 (± 0.1) × 102 |
Values of K are give in units of M−1. The errors represent the uncertainty obtained from the non-linear regression procedure used in the fitting the data to K. Samples in the Ca2+-free state contained 2 to 5 mM EGTA. The reciprocal of K corresponds to the accessible molar lipid concentration at which 50% of the protein is membrane bound.
The membrane affinities of C2A and C2B are not additive in the presence of Ca2+
The partition coefficients obtained in the absence of Ca2+ may be compared those obtained in the presence of Ca2+. Shown in Fig. 3 are the binding data obtained by sedimentation for syt1C2A, syt1C2B and syt1C2AB bound to POPC:POPS bilayers in the presence of Ca2+. The affinity of C2B is approximately 20 fold higher than that observed in the absence of Ca2+, and C2A binds in the presence of Ca2+, with an affinity that is slightly lower than that of C2B (see Table 1).
Figure 3.
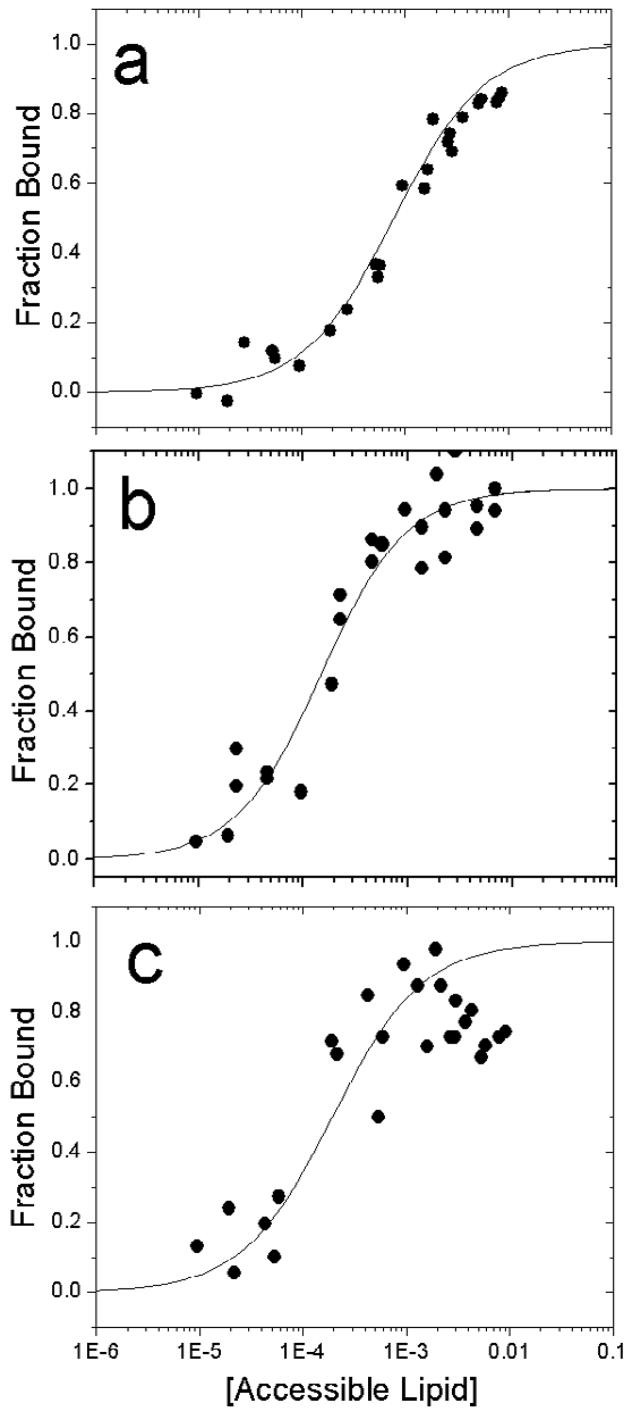
Ca2+-dependent binding of a) syt1C2A, b) syt1C2B and c) syt1C2AB fragments of synaptotagmin 1 to membranes composed of POPC:POPS (3:1), using a vesicle sedimentation assay (see Methods). The lines are fits to Eq. 2 and the values of K are given in Table 1.
Remarkably, the binding affinity of the tandem fragment containing both C2 domains, syt1C2AB, is intermediate between those of C2A and C2B alone (Fig. 3c), indicating that the free energy of interaction for the two C2 domains to PC:PS bilayers is not additive. The reason for this non-cooperativity is not clear, but it might be the result of a demixing of acidic lipids, or the effects of the C2 domains on bilayer curvature strain (see Discussion).
EPR spectra are consistent with the C2A and C2B domain structures
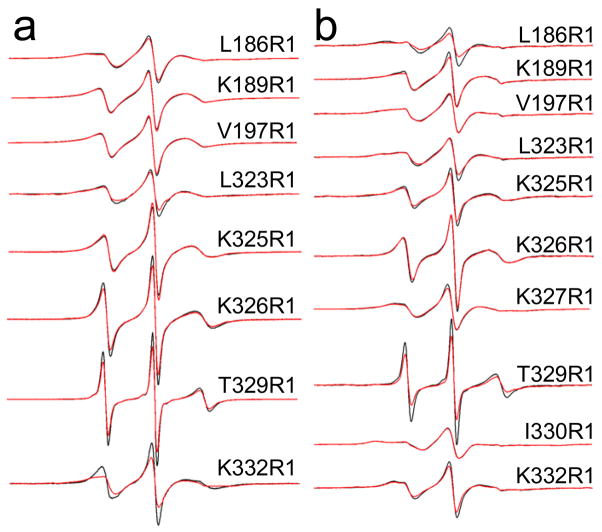
The spin labeled side chain R1 (Fig. 1b) was incorporated one at a time into sites within both C2A and C2B (Fig. 1a). Figure 4 shows X-band EPR spectra in the absence and presence of POPC:POPS bilayers for R1 mutants in the Ca2+-free state from syt1C2AB and from the same sites in either syt1C2A or syt1C2B. These EPR spectra reflect spin labels having a range of motion. For example, the narrow lineshapes obtained from T329R1 are characteristic of a label attached to a flexible loop and undergoing relatively fast (τ < 2 ns) isotropic motion. This residue is located in a twisted portion of β-strand 4, which is solvent exposed. Some of the more immobile spin labels include V197R1 and I330R1. At these sites, the spin-labeled side chain may interact with a nearby tyrosine, as observed previously for specific sites in T4 lysozyme 23. Many of the spectra display EPR lineshapes that are typical of those seen in β-sheets 24; and in general, similar lineshapes are seen whether the spin label is incorporated into the tandem syt1C2AB or in the isolated syt1C2A or syt1C2B domains.
Figure 4.
X-band EPR spectra obtained from single R1 substitutions in the absence of Ca2+ for labeled sites in a) syt1C2A and syt1C2B, and in b) syt1C2AB. Aqueous spectra are shown in black, spectra of R1 mutants in the presence of POPC:POPS lipid vesicles are shown in red. For syt1C2B and syt1C2AB, the protein is completely bound or membrane-associated at the concentrations of lipid used (25 mM). For syt1C2A (sites 186 and 189), sedimentation data (Figure 2) indicate the domain does not bind to POPC:POPS. Spectra are normalized against the total spin number so that the amplitudes provide an approximate measure of the motional averaging of the R1 side-chain. The spectra are all 100 Gauss scans.
In the presence of POPC:POPS bilayers, most of the EPR spectra are unchanged (see Fig. 4); however, at a few sites slight decreases in the normalized amplitudes are observed. For example, T329R1 undergoes a decrease in normalized amplitude, indicating that rate of motion is slowed in the presence of bilayers. This site is near the second Ca2+-binding loop in C2B, and this change may indicate membrane proximity or slight changes in the local structure or dynamics of this strand in the presence of membranes. L186R1, which is located near a basic region on C2A, also undergoes a decrease in motion, but only when this labeled site is examined in the tandem syt1C2AB fragment. As discussed below (see Discussion), this region of C2A may make contact with PC:PS bilayers in the absence of Ca2+, but only when tethered to C2B.
The polybasic region of C2B is membrane-associated in the absence of Ca2+
Continuous wave power saturation was performed on the EPR spectra from the eight spin-labeled mutants in the C2B portion of C2AB in the presence of POPC:POPS bilayers to obtain a membrane depth parameter, Φ (see Methods). Table 2 lists these parameters in the presence or absence of Ca2+, along with depth parameters obtained for several sites on C2B studied as an isolated domain. Phi values of −2.0 to −2.5 are associated with labels having aqueous exposure and lying more than 5 Å on the aqueous side of the membrane interface 16. Values greater than this, but still negative, indicate that the R1 side chains are localized in the interface region of the bilayer. In the presence of Ca2+, all sites on C2B (either in syt1C2B or syt1C2AB) display phi values in the aqueous range, indicating that this face of C2B does not approach within 5Å of the phosphates on the bilayer surface. However, in the absence of Ca2+, the phi values obtained from certain labeled sites (for example, K327R1, T329R1, and K332R1) indicate that this surface of C2B approaches to the membrane interface. The labeled site N370R1 is not in the polybasic region of C2B, but was also power saturated in order to provide orientation information for the positioning of the domain.
Table 2.
Depth Parameters, F, for C2AB or isolated C2B.†
| mutant, Ca2+ | C2AB | C2B | mutant, EGTA | C2AB | C2B |
|---|---|---|---|---|---|
| L323R1 | −2.3±0.2 | −2.1±0.1 | L323R1 | −2.0±0.1 | −1.6±0.1 |
| K325R1 | −2.0±0.2 | −2.9±0.1 | K325R1 | −2.1±0.2 | −2.1±0.1 |
| K326R1 | −2.1±0.2 | −2.5±0.1 | K326R1 | −1.9±0.3 | −1.8±0.1 |
| K327R1 | −2.2±0.2 | K327R1 | −1.7±0.2 | ||
| T329R1 | −2.1±0.1 | −2.1±0.1 | T329R1 | −1.4±0.4 | |
| I330R1 | −2.3±0.4 | I330R1 | −2.2±0.1 | ||
| K332R1 | −2.4±0.5 | −2.3±0.1 | K332R1 | −1.7±0.1 | −1.3±0.1 |
| N370R1 | −2.3±0.3 | N370R1 | −2.0±0.2 |
Depth parameters for C2AB are the average of 2 to 3 measurements, and errors represent standard deviations for the data. Previous work has shown that the depth parameters are in the range of approximately −2.0 to −2.5 for labels in the aqueous phase 45.
The C2B domain is localized in the aqueous double layer but does not penetrate the bilayer interior in the absence of Ca2+
The depth parameters shown in Table 2 were used to determine the membrane-associated position of C2B for syt1C2AB. As described in Materials and Methods, the experimental Φ values for labeled residues in the C2B polybasic region were used to estimate a distance from the plane of the lipid phosphates using a calibration curve obtained previously (Eq. 3) 15. The depth parameter for residue I330R1 was not used since it is believed that the side chain at this site may be sterically restricted and might not provide an accurate depth measurement.
Models for the orientation and position of docked C2B (for the data obtained for syt1C2AB) at the POPC:POPS interface were generated using XPLOR-NIH as described in Methods, using the distances and distance ranges given in Table 3. For labels with values of Φ more negative than −1.9 or −2.0, a significant error is associated with the distance estimate. In these cases, the labels do not approach closer than 5Å to the level of the lipid phosphates, but might assume a wide range of positions in the aqueous phase. As a result, they were allowed to assume a wide distance distribution in the simulated annealing, but not allowed to penetrate the bilayer. Shown in Fig. 5 are the 10 lowest energy models that resulted from simulated annealing. Figure 5b shows the lowest energy structure, which nearly represents an averaged structure. In these models, the backbone of C2B at the polybasic region sits roughly parallel to the bilayer surface and ranges from 5–12 Å away from the phospholipid phosphate plane. In the majority of these structures, the charged protein side chains do not penetrate the lipid bilayer interface, but appear to be positioned within the aqueous double layer. The localization of these charged side-chains in the aqueous double-layer is consistent with EPR spectra and depth data form this region, which show no evidence for membrane penetration of the polybasic face of C2B.
Table 3.
Distances from the R1 side chain to lipid phospholipid phosphates.†
| mutant | ϕ | distance (Å) | Range (Å) |
|---|---|---|---|
| L323R1 | −2.0 | −5.4 | −4.0 < x < −7.3 |
| K325R1 | −2.1 | −7.3 | −4.0 < x < −21.1 |
| K326R1 | −1.9 | −4.0 | −1.3 < x < −10.6 |
| K327R1 | −1.7 | −2.1 | −0.6 < x < −4.0 |
| T329R1 | −1.4 | 0.0 | +2.0 < x < −3.0 |
| K332R1 | −1.7 | −2.1 | −1.3 < x < −3.0 |
| N370R1 | −2.0 | −5.4 | −3.0 < x < −10.6 |
Depth parameters and distances for the Ca2+-free C2B(C2A) R1 side chains to a plane defined by the lipid phosphates, see Materials and Methods. Negative distances indicate labels that lie on the aqueous side of the membrane-solution interface. The distance range represents the uncertainty in the position of the spin labeled side chain (x) based upon the uncertainty in the experimental measurement of Φ.
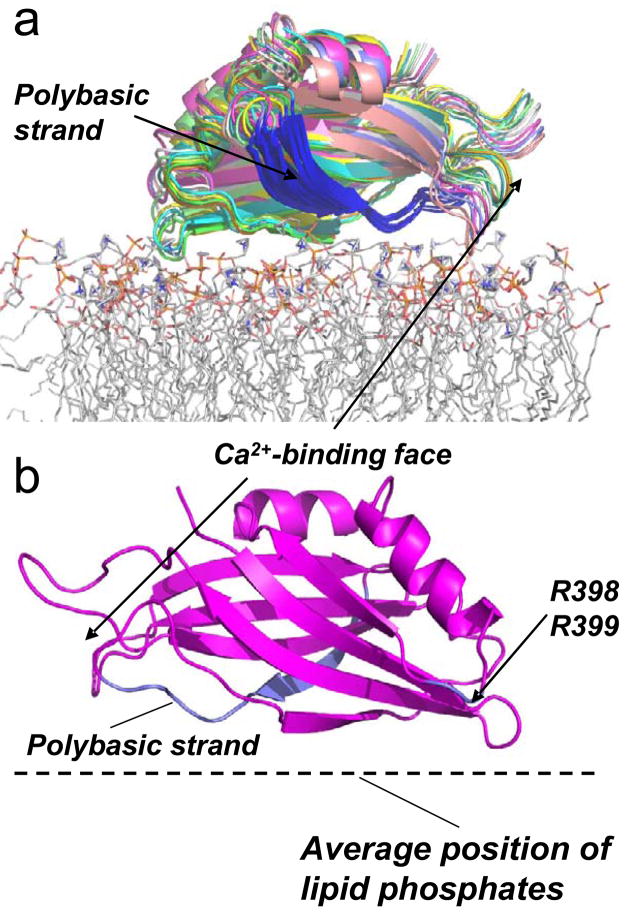
Figure 5.
a) The result of simulated annealing showing models for the 10 lowest energy structures for the Ca2+-independent docking of synaptotagmin C2B domain at the membrane interface. The position was generated for C2B using the distance data obtained for syt1C2AB at the POPC:POPS interface (Table 3) and the Xplor-NIH docking routine (see Methods). The membrane associated structure was produced using the PDB structure file for C2B, PDB ID:1K5W. The highly positively charged (polybasic) region of C2B is shown in blue. The protein is shown docked to a POPC bilayer obtained by a molecular dynamics simulation. b) One structure for C2B docked to the membrane interface is shown for clarity. The positively charged clusters that appear to interact electrostatically with the interface are highlighted in blue, and include the polybasic face and the two arginine residues at positions 398 and 399. The protein orientation is rotated 180° on the bilayer surface relative to the orientation shown in A).
The variation in the orientation and position of the models generated by simulated annealing provides an indication of the uncertainty in these models. The distance restraints used have several sources of error, including the uncertainty in the depth parameter, the actual populations of rotameric states of the spin labeled side-chain, R1, and the assumption that the solution NMR structure represents the membrane associated structure. Not every surface exposed site on C2B was labeled, and it is conceivable that sites on C2B, which have not been identified, might penetrate into bilayer in the Ca2+ free state. However, membrane penetration of the domain is not consistent with the data obtained and the structures generated in Figure 5, and it was not possible to align the domain to place other likely binding sites (hydrophobic side chains) into the interface and maintain consistency with the data in Tables 2 and 3.
Discussion
In the work described here, we compare the Ca2+-dependent and Ca2+-independent binding modes of syt1. The Ca2+-dependent binding of the C2 domains of syt1 has been extensively studied, but the Ca2+-indepenent binding mode is not well-characterized. As shown using vesicle sedimentation (Table 1), only the C2B domain or the C2AB fragment exhibited a measurable Ca2+-independent binding. This membrane interaction is not strong, which may account for the conflicting reports in the literature that C2B either does or does not bind to PC:PS bilayers in the absence of Ca2+. Although weak, the affinity is sufficient so that 50% of C2B will be bound when the accessible lipid concentration is on the order of a few mM. Within the cell this affinity suggests that C2B will be associated with a membrane interface if it is not bound to a protein target.
Synaptotagmin 1 is tethered to the vesicle membrane through a single helical membrane pass within the cell, a constraint that will raise the effective lipid concentration experienced by the C2B domain to a very high level. This effective concentration may be estimated using a simple geometric model 25. If it is assumed that C2B is tethered 100Å away from the vesicle surface, and can freely diffuse over a hemisphere around this tether point, C2B will experience an effective lipid concentration in excess of 350 mM. This exceeds the membrane binding affinity listed in Table 1 for the soluble fragment of C2B (or C2AB) in the absence of Ca2+ by two orders of magnitude. It is not known how closely the membrane partition coefficients presented in Table 1 resemble those within the cell. The PS content on the inner membrane plasma membrane or the synaptic vesicle membrane is difficult to determine. Even when the lipid content of a membrane is known (as it is for the synaptic vesicle 26), PS may be asymmetrically distributed, and a conservative estimate for the PS concentration on the inner plasma membrane lies in the range of 15 to 30 mol% 27. Nevertheless, even at the lower estimates for the PS content, the effective lipid concentration is likely to greatly exceed the C2B affinity. Thus, in the context of the cell, the membrane tethered C2B domain unlikely to diffuse in the cytoplasm in the absence of Ca2+, but will be bound to the membrane interface if it is not associated with protein targets, such as the SNAREs.
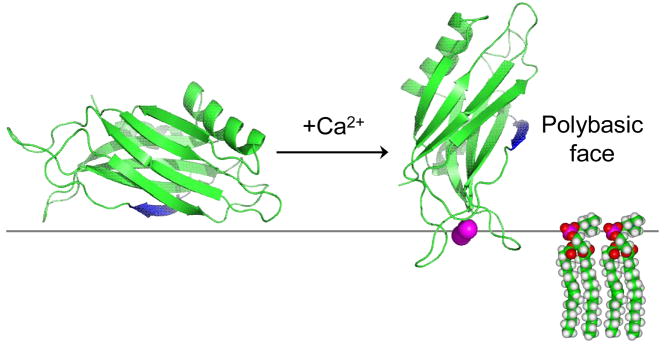
As shown in Figs. 5 and 6, different surfaces of C2B are involved in Ca2+-dependent and Ca2+-independent binding. In the Ca2+-dependent binding mode, C2B penetrates the bilayer interface, and hydrophobic side-chains within the 1st and 3rd Ca2+-binding loops penetrate the interface 15; 17. However, in the Ca2+-independent binding mode, there is no penetration of C2B into the bilayer interior, and the domain remains within the double layer displaced a few Angstroms from the membrane phosphates. This purely electrostatic interaction has also been observed for highly basic peptides that lack aromatic or hydrophobic residues. For example, peptides based upon lysine which lack aromatic residues bind but do not penetrate the membrane interface 28, and a peptide derived from the effector domain of the myristoylated alanine-rich C-kinase substrate (MARCKS) protein binds but fails to penetrate bilayers when its five phenylalanine residues are replaced with alanine 29. As discussed previously, the localization of these peptides within the double layer, several Angstroms on the aqueous side of the lipid headgroup phosphates, is a result of a repulsive dehydration force that is experienced as charged residues approach the membrane interface 30. In the absence of a hydrophobic interaction, charged protein segments or domains will associate with the interface but will not penetrate into the bilayer interior.
Figure 6.
Models for Ca2+-dependent and Ca2+-independent modes of membrane binding of syt1C2B. In the absence of Ca2+, the bilayer “protects” C2B from interactions with other components of the fusion machinery. The binding of Ca2+ to the domain alters the surface of C2B that interacts with the bilayer interface, and promotes the insertion of the Ca2+ binding loops into the bilayer. The Ca2+ bound orientation allows the polybasic face and other positively charged segments of C2B to interact with the SNAREs or with other bilayer surfaces.
Previous work demonstrated that C2B bound to phospholipids bilayers containing PI(4,5)P2 in the absence of Ca2+21. The work presented here, indicates that PS alone is sufficient to promote the Ca2+-independent binding of C2B. PI(4,5)P2 is a highly negatively charged lipid, and the polybasic face of C2B also appears to mediate the interaction with this lipid. In general, low levels of PI(4,5)P2 drive the electrostatic membrane binding of polybasic protein domains with enhanced affinity compared to monovalent PS 31. The interaction of C2B with PI(4,5)P2 may function to direct this domain to the target membrane in the absence of Ca2+ so that syt1 assumes a trans-configuration where C2A and C2B bind opposite membrane surfaces 11; 21. The C2B domain may also act to sequester PI(4,5)P2 as seen for other polybasic membrane binding proteins 32, and as observed for the juxta-membrane region of syntaxin 33.
The polybasic face of C2B plays an important role in fusion. The ability of syt1 to accelerate SNARE mediated fusion appears to be mediated by the polybasic region of C2B, which facilitates Ca2+-independent priming of synaptic vesicles at release sites 20; 34. Exactly how C2B carries out these functions is not known. The polybasic face of C2B has been shown to interact with an acidic C-terminal region of SNAP-25, and interact simultaneously with membranes and SNAREs in the presence of Ca2+18. However, it is not clear whether the C2B domain interacts with membrane surfaces or with membrane bound SNAREs in the absence of calcium. The polybasic face of C2B clearly has the capacity to interact either SNAREs or with membranes, and the orientation change depicted in Fig. 6 may act as a switch, so that the polybasic face of C2B is protected from interaction with SNAREs until it associates with Ca2+.
Previous work using fluorescence spectroscopy indicated that polybasic region of the C2B domain, residues K326 and K327, associated with the membrane interface in the presence of Ca2+. In this work, it was proposed that the C2B domain of syt1C2AB functioned to simultaneously bind two membranes through both its Ca2+-binding loops and its polybasic face. Such an association with two membranes might assist fusion by driving the vesicle and target membranes closer together, and might act to stabilize highly curved membrane structures that occurred during an intermediate stage of fusion 10. Under the conditions of the EPR experiments carried out here, we do not see evidence for a Ca2+-dependent membrane association of the polybasic face (Table 2), and only the calcium binding loops appear to associate 17. Conceivably, the different findings may be due to different experimental conditions, or the fact that the EPR measurements are only sensitive to labels that come within 5Å of the lipid phosphates. It has recently been proposed that R398 and R399 may bind membranes and allow C2B to bridge across two lipid surfaces 35. Although we have not tested these sites directly, these sites do appear to be localized close to the membrane surface in the absence of Ca2+ (see Fig. 5b). Ca2+ binding will increase the exposure of this region, and might act to facilitate interactions with opposing bilayer surfaces.
In the absence of Ca2+, we were not able to detect membrane binding by C2A in a vesicle sedimentation assay, indicating that the partition coefficient must be less than 25 M−1. We also were unable to detect binding of C2A to PC:PS bilayers using fluorescence correlation spectroscopy (data not shown). However, C2A may interact with membrane surfaces composed of PC:PS when tethered to C2B. As shown in Fig. 4b, L186R1 on C2AB exhibits a change in lineshape in the presence of lipid, and the depth parameter, Φ, at this site increased by 0.5 compared to that in the absence of bilayers, indicating that this site approaches within a few Angstroms of the interface. Why would C2A associate with bilayers when tethered to C2B, but not associate as a separate domain? As indicated above, when a domain is tethered to the bilayer surface, the effective local concentration of lipid is raised dramatically. We estimate that when C2B is bound to the bilayer surface, C2A may experience a local lipid concentration in excess of 0.8M.
In the presence of Ca2+, the binding data obtained here with vesicle sedimentation indicate that the membrane affinity for the tandem syt1C2AB domain is similar to the affinity of either syt1C2A or syt1C2B alone. This is an unexpected result. Both domains are able to simultaneously bind membranes 15; 36 and the linker connecting C2A and C2B is flexible 37, indicating that the interaction of the two domains with a common surface should not be restricted. In this case, the free energy of membrane binding for the tandem fragment should greatly exceed that of either isolated domain. Using a simple ball-and-string model described previously 25 and assuming a 4 nm separation between C2A and C2B, the affinity of the tandem domain should be at least a 1000 fold greater than that of either domain alone. Several phenomena could explain the weaker than expected affinity of syt1C2AB. For example, C2 domains may promote the demixing of acidic lipids 38 and the enhanced demixing of lipids by the tandem syt1C2AB might explain this result. The C2 domains of synaptotagmin 1 may alter membrane curvature strain, and this strain may be generated at the expense of binding free energy for syt1C2AB. Currently, we are testing several ideas that might explain the non-cooperative binding behavior for the syt1C2AB membrane interaction.
In summary, we investigated the membrane binding of the C2 domains of synaptotagmin 1 in the absence of Ca2+. The C2AB and C2B domains bind weakly in the absence of Ca2+, and C2A alone fails to bind membranes. It is important to consider the affinities of the individual domains in the context of the intact protein, and in the case of C2B the fact that this protein is tethered to the vesicle bilayer dramatically raises the effective lipid concentration presented to the domain. As a result, within the presynaptic cell, the C2B domain will never reside free in the cytosol, but will either be associated with membranes or with SNAREs. A novel approach using depth restraints derived from site-directed spin labeling and simulated annealing was used to generate a model for C2B that is bound to membranes in a Ca2+-independent manner. This model indicates that C2B associates electrostatically with bilayers through its polybasic face. When the domain binds Ca2+, the orientation of C2B on the membrane surface changes to expose the polybasic face. This change in orientation may act as a switch to trigger interactions between synaptotagmin 1 and the SNARE complex.
Materials and Methods
DNA Modification
DNA of rat syt1 (P21707), was obtained from Dr. Carl Creutz (Pharmacology Department, University of Virginia) in the pGEX-KG vector encoding amino acid residues 96–265 (syt1C2A), 249–421 (syt1C2B), and 96–421 (syt1C2AB) 39. All DNA modifications followed published protocols 40. The single native cysteine residue at position 277 was mutated to alanine by typical polymerase chain reaction (PCR) strategies. Either two-step PCR or a QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA), were used to produce single-cysteine mutants of C2A, C2B, or C2AB. Individual cysteine residues were introduced at positions in the isolated or tandem C2A domain: 186, 189, 197, 256 and in the isolated or tandem C2B domain: 269, 323, 325, 326, 327, 329, 330, 332, 370, 415. All cysteine substitutions were confirmed by DNA sequencing.
Protein Expression and Purification
For the synaptotagmin 1 C2A, C2B, and C2AB cysteine mutants, protein expression and purification were carried out as described previously 15; 16; 17. For C2B, the procedure was modified slightly so that the affinity-purified and benzonase-treated protein was exchanged into a buffer of 100mM NaCl, 50mM MOPS, 5mM EDTA, pH6.5. The protein was then loaded onto two 5ml HiTrap SP HP prepacked columns (Amersham Bioscience), and eluted with a gradient running from 100 mM to 700 mM NaCl over 10 column volumes. The UV spectrum of the purified protein had a maximum absorption at 278 nm indicating that it was free of nucleic acid contamination.
Spin Labeling syt1 C2 Domains
The protein was spin-labeled as described previously 17 using the sulfhydryl reactive spin label, (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate (MTSL; Toronto Research Chemicals, North York, ON, Canada) at a 1:3:30 mole ratio of protein:DTT:MTSL. Excess free spin-label was removed by passing the sample through a HiPrep 26/10 desalting column (Amersham Biosciences). The spin-labeled protein was concentrated to 20–300 μM using Centripreps, Centricons, and Microcons (Millipore).
Large Unilamellar Vesicles
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS) (Avanti Polar Lipids, Alabaster, AL) were used to prepare large unilamellar vesicles (LUVs) with a POPC:POPS ratio of 3:1 as described previously 16. For Ca2+-free experiments, the LUVs were prepared in buffer that lacked CaCl2. Sucrose-loaded large unilamellar vesicles (LUVs) were prepared as described previously 41. Briefly, appropriate aliquots of lipids (75% POPC:25% POPS) were mixed, dried under a stream of nitrogen and vacuum desiccated overnight. The resulting film was rehydrated in sucrose buffer (176mM sucrose, 1mM MOPS, pH=7.0), vortexed thoroughly, cycled through 5 freeze-thaw cycles, and extruded through 0.1 μm polycarbonate filters using a hand-held Mini-Extruder (Avanti Polar Lipids, Alabaster, AL). To remove the external sucrose, the LUVs were diluted into 5 volumes of buffer without sucrose (100mM KCl, 1mM MOPS, pH 7.0), centrifuged for an hour at 100,000 × g, 25 °C and resuspended with fresh buffer. For syt1 in its Ca2+-bound state, the buffer contained 1 mM CaCl2.
Sedimentation assay to determine Vesicle Binding
A vesicle sedimentation binding assay described previously 41 was used to determine the membrane affinity of each domain of synaptotagmin 1 individually and when present in the tandem fragment. Briefly, protein was added to final concentrations of 5–10 μM with sucrose-loaded LUVs at lipid concentrations ranging from 0.05 mM to 15 mM, and centrifuged at 100,000 × g for 1 hour to pellet the LUVs. The lipid concentrations were confirmed by phosphate analysis 42. Each domain studied contained a single cysteine residue (sytC2A, T256C; syt1C2B, A415C; syt1C2AB, E269C), which enabled cysteine and protein levels to be detected in the supernatant spectrophotometrically by the addition of 200μM of 4, 4′dithiodipyridine 43; 44. Equivalent binding affinities were obtained by detecting the tryptophan fluorescence from these domains. In a few experiments, the binding affinity was determined at protein concentrations of 1 μM. Similar affinities were obtained in these cases indicating that the protein was sufficiently dilute on the membrane surface. For each lipid concentration, at least three measurements of the fraction of bound protein, fb, were made and used to determine a molar partition coefficient, K, which is defined as:
| [1] |
Here, [P] is the concentration of protein in the bulk aqueous phase, [P]m is the molar concentration of protein bound to the membrane and [L] is the molar concentration of accessible lipid (typically one half of the total lipid concentration) 41. If the protein concentrations are sufficiently dilute ([lipid] ≫ [protein]), the fraction of protein bound fb, will be given by:
| [2] |
The data were fit to Eq. 2 using OriginPro 7.5 to yield a value of K, which has units of M−1.
EPR Spectroscopy
EPR spectra were recorded at room temperature using a Varian E-line Century series spectrometer fitted with a microwave preamplifier and an X-band loop-gap resonator (Medical Advances, Milwaukee, WI). Aqueous syt1 protein concentrations ranged from 20–300 μM. For Ca2+-free conditions, 5 mM EGTA was present in the protein sample. Membrane-associated or membrane-bound protein spectra were obtained under these same conditions but with the addition of LUVs (25 mM total lipid) to ensure complete membrane binding and low surface densities of either syt1C2B or syt1C2AB. Continuous-wave power saturation experiments were performed on the syt1 mutants using a Bruker EMX spectrometer as described elsewhere 45 and used to calculate a depth parameter, F, based upon the saturation behavior in samples containing oxygen and Ni(II)EDDA 46. Ni(II)EDDA was used at a concentration of 10 mM, but the ΔP1/2 values were scaled to an effective concentration of 20 mM as described previously 45.
Membrane Docking of the sytI C2B domain
The distance (x) of the spin label from a plane defined by the lipid phosphates was determined from the depth parameters (Φ) using the following expression:
| [3] |
This is an empirically derived expression where A and D set the bulk values of Φ in water and hydrocarbon, C defines the position of the reflection point of the curve, and B determines the slope of the curve 16; 45. Point-to-plane distances were determined by using the calibration curve obtained previously, where A, B, C, and D have values of 3.4, 0.11, 8.56, and 1.1, respectively 15.
Spin label depths (x in Eqn. 3) were used as input for standard XPLOR-NIH restrained simulated annealing and energy minimization protocols 47; 48 to determine the position of C2B relative to the phospholipid bilayer. Starting structures for simulated annealing were obtained by mutating in silico positions 323, 325, 326, 327, 329, 332, and 370 of the high-resolution structure of C2B (residues 272–419) (PDB ID: 1K5W) to Cys and attaching MTSL groups. The MTSL groups were then allowed to undergo molecular dynamics and energy minimization in order to generate likely MTSL conformations. The relative coordinates of the R1 side chains were then fixed and the C2B domain was then oriented relative to the plane by using restrained simulated annealing and energy minimization. The python XPLOR-NIH plane distance potential (planeDistPot) was used to restrain the distances between the spin label nitrogen atoms and the phospholipid phosphate plane. The uncertainty in these distances were set according to the distance range derived from the empirically determined calibration curve (Eq. 3).
The forcefield for R1 used in the simulated annealing routinely produces rotameric forms of R1 that are more extended and not as defined that those observed by crystallography 49. Although we did not take this approach in the present case, the variation in the structures obtained might be narrowed by setting side chain rotamers to the most favorable conformations based upon crystallography.
Acknowledgments
We would like to thank Charles D. Schwieters (Center for Information Technology, NIH) for help with the implementation of XPLOR in this calculation. This work was supported by grants from the National Institutes of Health, NIGMS, GM 062305 and GM 072694.
Abbreviations used
- EPR
electron paramagnetic resonance spectroscopy
- LUV
large unilamellar vesicle
- MOPS
3-(N-morpholino) propanesulfonic acid
- MTSL
(1-oxy-2,2,5,6-tetramethylpyrroline-3-methyl) methanethiosulfonate
- PI(4
5)P2, phosphatidylinositol 4–5 bisphosphate
- POPC
palmitoyloleoylphosphatidycholine
- POPS
palmitoyloleoylphosphatidylserine
- R1
spin-labeled side chain produced by derivatization of a cysteine with MTSL
- SDSL
site-directed spin labeling
- SNARE
soluble N-ethylmaleimide–sensitive factor attachment receptor
- syt1
synaptotagmin 1
- syt1C2AB
soluble fragment of syaptotagmin 1 containing the C2A and C2B domains
- syt1C2A
C2A domain of synaptotagmin 1
- syt1C2B
C2B domains of synaptotagmin 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 2.Jahn R, Lang T, Suhof TC. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 3.Sudhof TC. The synaptic vesicle cycle. Ann Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 4.Chapman ER. How does synaptotagmin trigger neurotransmitter release? Annu Rev Biochem. 2008;77:615–41. doi: 10.1146/annurev.biochem.77.062005.101135. [DOI] [PubMed] [Google Scholar]
- 5.Rizo J, Chen X, Arac D. Unraveling the mechanisms of synaptotagmin and SNARE function in neurotransmitter release. Trends Cell Biol. 2006;16:339–50. doi: 10.1016/j.tcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Jackson MB, Chapman ER. Fusion pores and fusion machines in Ca2+-triggered exocytosis. Annu Rev Biophys Biomol Struct. 2006;35:135–60. doi: 10.1146/annurev.biophys.35.040405.101958. [DOI] [PubMed] [Google Scholar]
- 7.Jahn R, Scheller RH. SNAREs [mdash] engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 8.Bhalla A, Chicka MC, Tucker WC, Chapman ER. Ca(2+)-synaptotagmin directly regulates t-SNARE function during reconstituted membrane fusion. Nat Struct Mol Biol. 2006;13:323–30. doi: 10.1038/nsmb1076. [DOI] [PubMed] [Google Scholar]
- 9.Martens S, Kozlov MM, McMahon HT. How synaptotagmin promotes membrane fusion. Science. 2007;316:1205–1208. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- 10.Arac D, Chen X, Khant HA, Ubach J, Ludtke SJ, Kikkawa M, Johnson AE, Chiu W, Sudhof TC, Rizo J. Close membrane-membrane proximity induced by Ca2+-dependent multivalent binding of synaptotagmin-1 to phospholipids. Nat Struct Mol Biol. 2006;13:209–217. doi: 10.1038/nsmb1056. [DOI] [PubMed] [Google Scholar]
- 11.Stein A, Radhakrishnan A, Riedel D, Fasshauer D, Jahn R. Synaptotagmin activates membrane fusion through a Ca2+-dependent trans interaction with phospholipids. Nat Struct Mol Biol. 2007;14:904–911. doi: 10.1038/nsmb1305. [DOI] [PubMed] [Google Scholar]
- 12.Brose N, Petrenko AG, Sudhof TC, Jahn R. Synaptotagmin: a calcium sensor on the synaptic vesicle surface. Science. 1992;256:1021–1025. doi: 10.1126/science.1589771. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez I, Arac D, Ubach J, Gerber SH, Shin O, Gao Y, Anderson RG, Sudhof TC, Rizo J. Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine. Neuron. 2001;32:1057–69. doi: 10.1016/s0896-6273(01)00548-7. [DOI] [PubMed] [Google Scholar]
- 14.Sutton RB, Davletov BA, Berghuis AM, Sudhof TC, Sprang SR. Structure of the first C2 domain of synaptotagmin I: A novel Ca2+/phospholipid-binding fold. Cell. 1995;80:929–938. doi: 10.1016/0092-8674(95)90296-1. [DOI] [PubMed] [Google Scholar]
- 15.Herrick DZ, Sterbling S, Rasch KA, Hinderliter A, Cafiso DS. Position of synaptotagmin I at the membrane interface: cooperative interactions of tandem C2 domains. Biochemistry. 2006;45:9668–9674. doi: 10.1021/bi060874j. [DOI] [PubMed] [Google Scholar]
- 16.Frazier AA, Roller CR, Havelka JJ, Hinderliter A, Cafiso DS. Membrane-bound orientation and position of the synaptotagmin I C2A domain by site-directed spin labeling. Biochemistry. 2003;42:96–105. doi: 10.1021/bi0268145. [DOI] [PubMed] [Google Scholar]
- 17.Rufener E, Frazier AA, Wieser CM, Hinderliter A, Cafiso DS. Membrane-bound orientation and position of the synaptotagmin C2B domain determined by site-directed spin labeling. Biochemistry. 2005;44:18–28. doi: 10.1021/bi048370d. [DOI] [PubMed] [Google Scholar]
- 18.Dai H, Shen N, Arac D, Rizo J. A quaternary SNARE-synaptotagmin-Ca2+-phospholipid complex in neurotransmitter release. J Mol Biol. 2007;367:848–63. doi: 10.1016/j.jmb.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray D, Honig B. Electrostatic control of the membrane targeting of C2 domains. Mol Cell. 2002;9:145–154. doi: 10.1016/s1097-2765(01)00426-9. [DOI] [PubMed] [Google Scholar]
- 20.Rickman C, Archer DA, Meunier FA, Craxton M, Fukuda M, Burgoyne RD, Davletov BA. Synaptotagmin interaction with the syntaxin/SNAP-25 dimer Is mediated by an evolutionarily conserved motif and Is sensitive to inositol hexakisphosphate. J Biol Chem. 2004;279:12574–12579. doi: 10.1074/jbc.M310710200. [DOI] [PubMed] [Google Scholar]
- 21.Bai J, Tucker WC, Chapman ER. PIP2 increases the speed of response of synaptotagmin and steers its membrane-penetration activity toward the plasma membrane. Nat Struct Mol Biol. 2004;11:36–44. doi: 10.1038/nsmb709. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Shin OH, Rhee JS, Arac D, Rah JC, Rizo J, Sudhof T, Rosenmund C. Phosphatidylinositol phosphates as Co-activators of Ca2+ binding to C2 domains of synaptotagmin 1. J Biol Chem. 2006;281:15845–15852. doi: 10.1074/jbc.M600888200. [DOI] [PubMed] [Google Scholar]
- 23.Langen R, Oh KJ, Cascio D, Hubbell WL. Crystal structures of spin labeled T4 lysozyme mutants: implications for the Interpretation of EPR spectra in terms of structure. Biochemistry. 2000;39:8396–8405. doi: 10.1021/bi000604f. [DOI] [PubMed] [Google Scholar]
- 24.Lietzow MA, Hubbell WL. Motion of spin label side chains in cellular retinol-binding protein: correlation with structure and nearest-neighbor interactions in an antiparallel beta-sheet. Biochemistry. 2004;43:3137–3151. doi: 10.1021/bi0360962. [DOI] [PubMed] [Google Scholar]
- 25.Buser CA, Sigal CT, Resh MD, McLaughlin S. Membrane binding of myristylated peptides corresponding to the NH2 terminus of src. Biochemistry. 1994;33:13093–101. doi: 10.1021/bi00248a019. [DOI] [PubMed] [Google Scholar]
- 26.Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, Urlaub H, Schenck S, Brugger B, Ringler P, Muller SA, Rammner B, Grater F, Hub JS, De Groot BL, Mieskes G, Moriyama Y, Klingauf J, Grubmuller H, Heuser J, Wieland F, Jahn R. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–46. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 27.Gambhir A, Hangyas-Mihalyne G, Zaitseva I, Cafiso DS, Wang J, Murray D, Pentyala SN, Smith SO, McLaughlin S. Electrostatic sequestration of PIP2 on phospholipid membranes by basic/aromatic regions of proteins. Biophys J. 2004;86:2188–207. doi: 10.1016/S0006-3495(04)74278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Victor KG, Cafiso DS. Location and dynamics of basic peptides at the membrane interface: electron paramagnetic resonance spectroscopy of tetramethyl-piperidine-N-oxyl-4-amino-4-carboxylic acid-labeled peptides. Biophys J. 2001;81:2241–2250. doi: 10.1016/S0006-3495(01)75871-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Victor K, Jacob J, Cafiso DS. Interactions controlling the membrane binding of basic protein domains: phenylalanine and the attachment of the myristoylated alanine-rich C-kinase substrate protein to interfaces. Biochemistry. 1999;38:12527–12536. doi: 10.1021/bi990847b. [DOI] [PubMed] [Google Scholar]
- 30.Ben-Tal N, Honig B, Peitzsch RM, Denisov G, McLaughlin S. Binding of small basic peptides to membranes containing acidic lipids: theoretical models and experimental results. Biophys J. 1996;71:561–75. doi: 10.1016/S0006-3495(96)79280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLaughlin S, Wang J, Gambhir A, Murray D. PIP(2) and proteins: interactions, organization, and information flow. Annu Rev Biophys Biomol Struct. 2002;31:151–75. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- 32.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–11. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 33.James DJ, Khodthong C, Kowalchyk JA, Martin TF. Phosphatidylinositol 4,5-bisphosphate regulates SNARE-dependent membrane fusion. J Cell Biol. 2008;182:355–66. doi: 10.1083/jcb.200801056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loewen CA, Lee SM, Shin YK, Reist NE. C2B polylysine motif of synaptotagmin facilitates a Ca2+-independent stage of synaptic vesicle priming in vivo. Mol Biol Cell. 2006;17:5211–5226. doi: 10.1091/mbc.E06-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue M, Ma C, Craig TK, Rosenmund C, Rizo J. The Janus-faced nature of the C(2)B domain is fundamental for synaptotagmin-1 function. Nat Struct Mol Biol. 2008;15:1160–8. doi: 10.1038/nsmb.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hui E, Bai J, Chapman ER. Ca2+-triggered simultaneous membrane penetration of the tandem C2-domains of synaptotagmin I. Biophys J. 2006;91:1767–77. doi: 10.1529/biophysj.105.080325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang H, Cafiso DS. Conformation and Membrane Position of the Region Linking the Two C2 Domains in Synaptotagmin 1 by Site-Directed Spin Labeling. Biochemistry. 2008 doi: 10.1021/bi801470m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hinderliter A, Biltonen RL, Almeida PF. Lipid modulation of protein-induced membrane domains as a mechanism for controlling signal transduction. Biochemistry. 2004;43:7102–10. doi: 10.1021/bi036334t. [DOI] [PubMed] [Google Scholar]
- 39.Damer CK, Creutz CE. Synergistic membrane interactions of the two C2 domains of synaptotagmin. J Biol Chem. 1994;269:31115–31123. [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Press; Plainview, N.Y: 1989. [Google Scholar]
- 41.Buser CA, McLaughlin S. Ultracentrifugation technique for measuring the binding of peptides and proteins to sucrose-loaded phospholipid vesicles. Methods Mol Biol. 1998;84:267–81. doi: 10.1385/0-89603-488-7:267. [DOI] [PubMed] [Google Scholar]
- 42.Ames BN. Assay of inorganic phosphate, total phosphate and phosphatases. Meth Enzymol. 1966;8:115–118. [Google Scholar]
- 43.Grassetti DR, Murray JF., Jr Determination of sulfhydryl groups with 2,2′- or 4,4′-dithiodipyridine. Arch Biochem Biophys. 1967;119:41–49. doi: 10.1016/0003-9861(67)90426-2. [DOI] [PubMed] [Google Scholar]
- 44.Liao H, Ellena J, Liu L, Szabo G, Cafiso D, Castle D. Secretory carrier membrane protein SCAMP2 and phosphatidylinositol 4,5-bisphosphate interactions in the regulation of dense core vesicle exocytosis. Biochemistry. 2007;46:10909–20. doi: 10.1021/bi701121j. [DOI] [PubMed] [Google Scholar]
- 45.Frazier AA, Wisner MA, Malmberg NJ, Victor KG, Fanucci GE, Nalefski EA, Falke JJ, Cafiso DS. Membrane orientation and position of the C2 domain from cPLA2 by site-directed spin labeling. Biochemistry. 2002;41:6282–6292. doi: 10.1021/bi0160821. [DOI] [PubMed] [Google Scholar]
- 46.Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwieters CD, Kuszewski JJ, Clore GM. Using xplor-NIH for NMR molecular structure determination. Prog Nuclear Magn Reson Spec. 2006;48:47–62. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 48.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 49.Guo Z, Cascio D, Hideg K, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: solvent-exposed sites in helix B of T4 lysozyme. Protein Sci. 2008;17:228–39. doi: 10.1110/ps.073174008. [DOI] [PMC free article] [PubMed] [Google Scholar]