Abstract
The Notch receptor and its ligands are key components in a core metazoan signalling pathway which regulates the spatial patterning, timing and outcome of many cell-fate decisions. Ligands contain a disulphide-rich Delta/Serrate/LAG-2 (DSL) domain required for Notch trans-activation or cis-inhibition. Here we report the first X-ray structure of a functional fragment of a Notch ligand, the DSL-EGF3 domains of human Jagged-1 (J-1DSL-EGF3). The structure identifies a highly conserved face of the DSL domain and we show, by functional analysis of Drosophila ligand mutants, that this surface is required for both cis- and trans-regulatory interactions with Notch. We also identify, using NMR, a surface of Notch-1 involved in J-1DSL-EGF3 binding. Our data imply that cis- and trans-regulation may occur through formation of structurally distinct complexes which, unexpectedly, involve the same surfaces on both ligand and receptor.
The Notch receptor is part of a core signalling pathway which has been highly conserved in all metazoan species and is required at multiple stages of development 1,2. In addition to its role in regulating cell-fate decisions during early life, the Notch receptor is required for maintenance and differentiation of mammalian neural and haemopoietic stem cell populations in the adult organism 3,4. Furthermore it has been shown to play a key role in the development and regulation of the immune system including the induction of T-cell tolerance 5. Notch pathway dysfunction is associated with both acquired and inherited disease states in humans 6.
Notch is a transmembrane protein which undergoes proteolytic processing by furin during its trafficking within the secretory pathway, and is subsequently presented at the cell surface as a non-covalently associated heterodimer 7-9. Proximal to the membrane are three Lin-12-Notch repeats (LNRs) and a heterodimerisation domain. These comprise the extracellular negative regulatory region (NRR), which is important for maintaining the receptor in the “off” state. A recent atomic structure of this region demonstrates that a tumor necrosis factor alpha converting enzyme (TACE) cleavage site is buried by interdomain interactions, and it has been proposed that significant conformational changes in this region occur upon Notch activation to expose the protease site 10. The latter may occur, following ligand binding, as a result of endocytosis of the Notch-ligand complex by the ligand-expressing cell, which removes the extracellular moiety of the Notch heterodimer 11,12. The remaining, membrane-tethered, Notch fragment on the signal-receiving cell then undergoes two distinct intramembrane proteolytic steps catalysed by TACE and γ-secretase. Proteolysis results in the release of a soluble intracellular fragment of Notch 13,14, which subsequently translocates to the nucleus and binds to a DNA-binding protein of the CBF1/ Suppressor of Hairless/ Lag-1 (CSL) family and its co-activator Mastermind, thus relieving repression of Hairy/Enhancer-of-split (HES) gene expression 15. In addition to promoting Notch activation through trans-interactions with the receptor expressed on adjacent cells, Notch ligands can also form cis-inhibitory interactions with Notch expressed in the same cell, limiting the zone of Notch activity 16-20.
In Drosophila there is a single Notch receptor, whereas in mammals the signalling pathway is more complex, with four Notch receptors (Notch1-4). The major part of the extracellular region of Notch comprises up to 36 EGF domains, many of which contain a calcium-binding (cb) consensus sequence 21. EGF domains 11 and 12 are known to be essential for ligand binding, as is calcium 22. The solution structure of human Notch-1 EGFs 11-13 (N-111-13) previously demonstrated a rod-like conformation for the N-111-12 region, with both calcium co-ordination and hydrophobic packing interactions contributing to the extended organisation of domains 23. It is likely that the rod-shaped organisation facilitates the formation of a binding surface for protein-protein interactions, as seen in other proteins with tandem repeats of cbEGF domains 24.
All Notch ligands contain a variable number of EGF domain repeats and an N-terminal Delta/Serrate/Lag-2 (DSL) domain. Two ligand families can be distinguished by the presence or absence of a cysteine-rich domain. This gives rise to the Serrate/Jagged ligand family and the Delta/Delta-like ligand family, respectively 21. In Drosophila there is only one ligand of each class (Serrate and Delta), while in mammals there are two Serrate class ligands (Jagged-1 and Jagged-2) and three Delta-like ligands (Dll1, Dll3 and Dll4). Site-directed mutagenesis and deletion analysis was used to demonstrate that the DSL domain confers specificity of binding to Notch, and that C-terminal linkage of two EGF domains appears to facilitate binding 18,25,26. Although cellular and in vivo studies have provided considerable insight into the downstream consequences of signalling, and some advances have been made in determining the structure of the Notch receptor, there are no high-resolution structural data currently available for the ligands, or for the Notch-ligand complex, to explain how receptor binding and activation occurs. Furthermore, the molecular basis for the productive (trans-activation) and non-productive (cis-inhibition) interactions is not understood.
In this study we identify functional fragments of human Notch-1 (N-1) and Jagged-1 (J-1) which interact in a calcium-dependent manner. We describe the first high resolution structure of the DSL-EGF3 fragment of human J-1 (J-1DSL-EGF3) and the first crystal structure for the ligand-binding region of N-1 (N-111-13). These data, together with NMR titration of 15N-labelled N-111-13 with J-1DSL-EGF3, allow definition of the face of N-1 involved in ligand binding. An interaction surface on the DSL domain of J-1 is predicted based on our structure and tested using mutagenesis and functional analysis of the DSL domain in the Drosophila homologue of J-1, Serrate. These experiments confirm the importance of this DSL surface in both the activation and cis-inhibition of Notch. When these data are combined, using in silico docking of the two protein structures, anti-parallel and parallel models for the complex are generated. This suggests that, despite our finding that the same region of J-1 is involved in cis- and trans-regulatory activities, the physical basis for these functions may be formation of two, structurally distinct, complexes. The results presented therefore provide the first atomic structure of a Notch ligand, a model for the early molecular events associated with Notch-Jagged interaction and a basis for the design of selective inhibitors of Notch activity.
RESULTS
Jagged-1DSL-EGF3 interacts with Notch-111-13
To increase our understanding of the molecular basis of the human Notch-Jagged recognition event we designed truncated fragments of N-1 and J-1, encompassing the domains that are implicated in binding. These fragments correspond to cbEGFs 11-13 of N-1 (N-111-13), and the DSL and first three EGF domains of J-1 (J-1DSL-EGF3). The N-terminus of the DSL domain was defined based on sequence homology between Jagged and Delta-like proteins as in the Swiss-Prot database 27. Correct folding of N-111-13 has previously been demonstrated by determination of the solution structure of this construct 23. We have now used a similar overexpression and refolding strategy to generate J-1DSL-EGF3 and a N-111-13 construct modified by the addition of a BirA-tag at the C-terminus. Addition of the tag was found to increase the efficiency of the refolding of EGF13. We have used pull-down experiments (Fig. 1a, b) and surface plasmon resonance (SPR) (Figure 1c, d, e) to demonstrate a specific, Ca2+-dependent interaction between these constructs, a property of the cell-surface interaction mediated by the full-length proteins.
Figure 1. Specific recognition of N-111-13 by J-1DSL-EGF3.
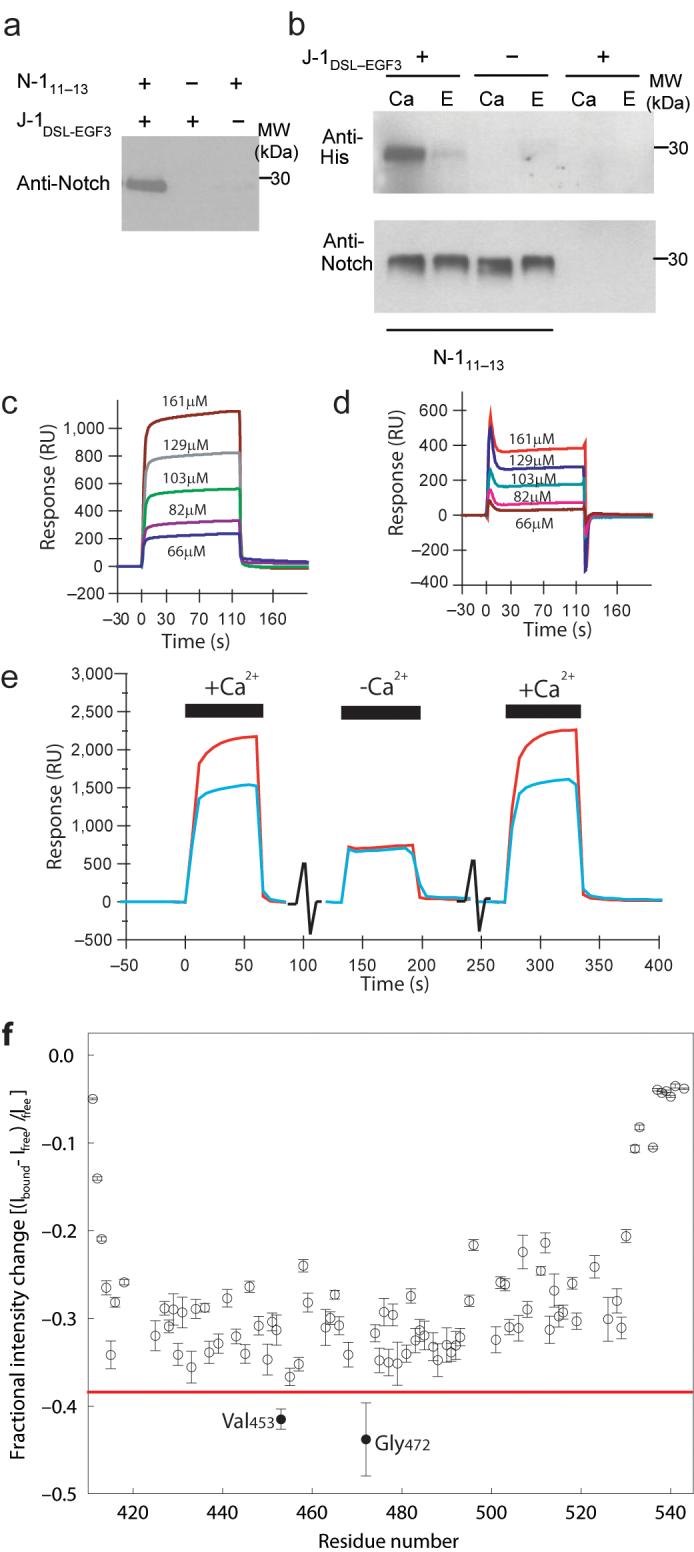
(a) Western blot analysis to demonstrate the pull down of N-111-13 with His-tagged J-1DSL-EGF3 on capture with Ni-NTA Magnetic Agarose Beads. Anti-Notch antibody was used for detection. (b) The calcium dependence of the interaction was demonstrated with biotinylated N-111-13 immobilised on Dynabeads M-270 Streptavidin (lanes 1-4). Lanes 5 and 6 were control beads without N-111-13. Detection of His-tagged J-1DSL-EGF3 by anti-RGS.His HRP conjugate showed the interaction of N-111-13 with J-1DSL-EGF3 occurred in the presence of calcium (Ca) and was inhibited by EDTA (E). Anti-Notch antibody confirmed equivalent amounts of N-111-13 were eluted from the beads (lower panel). (c) SPR measurements were taken with 7085RU of J-1DSL-EGF3 coupled to the chip surface and N-111-13 (without the BirA-tag) injected over the chip surface at the concentrations indicated in Tris buffered saline supplemented with 1mM Ca2+. (d) Trace from (c) corrected relative to reference cell. (e) To demonstrate the dependence on Ca2+ of the specific interaction three sequential injections of N-111-13 at 110μM were made over the same J-1DSL-EGF3 surface. The first and last injections were carried out in the buffer described above, the second injection contained an additional 10mM EGTA to chelate available Ca2+. Interaction of N-111-13 with the J-1DSL-EGF3 coupled surface and with the reference surface are shown by the red and blue traces respectively. (f) Addition of unlabelled J-1DSL-EGF3 (0.15mM) to 15N-labelled N-111-13 (0.5mM) leads to a loss of intensity of NMR peaks following formation of the large complex between the two proteins. The red line is a visual aid to highlight the greater intensity changes for V453 and G472.
NMR studies of the Notch-Jagged interaction
Complex formation between N-111-13 and J-1DSL-EGF3 was investigated, at the residue-specific level, using NMR spectroscopy. Stepwise addition of unlabelled J-1DSL-EGF3 to a solution of 15N-labelled N-111-13 led to a gradual decrease in the intensity of most peaks in 1H-15N HSQC spectra of N-111-13. At J-1DSL-EGF3: N-111-13 ratios above ~0.35 precipitation of J-1DSL-EGF3 is observed, therefore the HSQC spectrum of fully-bound N-111-13 cannot be obtained. No measurable changes in chemical shifts were seen in the HSQC spectra, nor were any new peaks arising from the N-111-13:J-1DSL-EGF3 complex observed, most likely due to exchange broadening under the conditions studied. The fractional intensity data plotted in Figure 1f are consistent with the participation of ~30% of the N-111-13 molecules in a higher molecular weight complex with J-1DSL-EGF3. The remaining ~70% intensity arises from unbound N-111-13. Very little or no change in intensity was observed for the N- and C-termini of N-111-13 (residues 411-413 and 532-543, Fig. 1f). These residues undergo significant motions on a picosecond timescale 23 and the linewidths of their peaks are determined, to a large extent, by these independent fast motions. The absence of any change in intensity for these peaks indicates that the dynamics of these N- and C-terminal residues are not affected by the interaction of J-1DSL-EGF3 with the EGF domains of N-111-13. In contrast, V453 and G472 display a larger decrease in peak intensity than the majority of residues (Fig. 1f). These larger changes arise from exchange contributions for residues of N-111-13 with the greatest chemical shift differences between the free and bound states; thus V453 and G472 represent the strongest candidates for residues that lie in or close to the binding site for the J-1DSL-EGF3 ligand 28. These residues are located adjacent to one another on one face of EGF12, within the region where the major ligand-binding site is predicted to reside 22,29.
Structure of J-1DSL-EGF3 and N-111-13
In order to dissect further the J-1DSL-EGF3/N-111-13 interactions, we determined the X-ray crystallographic structure of each construct. The structure of J-1DSL-EGF3 was elucidated at a resolution of 2.5 Å (Fig. 2a, Supplementary Fig. 1) and reveals an extended, linear arrangement of the domains with overall dimensions of 120 × 20 × 20 Å. EGF3 has a classical EGF-like fold, closely related at a structural level to the first EGF domain from human factor IX (1edm, r.m.s.d. = 1.0 Å over 35/37 Cα atoms), despite the absence of Ca2+-binding residues. EGF1 and EGF2 exhibit a highly-truncated version of the EGF-fold with no canonical secondary structure and more distant structural homologies to other EGF-like domains (EGF1, 1l3y-Integrin EGF-like domain, r.m.s.d. = 2.1 Å over 30/31 Cα atoms; EGF2, 1xdt-Heparin binding EGF, r.m.s.d. = 1.9 Å over 26/31 Cα atoms). The largest truncation in these two domains is in the loop between cysteine 3 (C3) and cysteine 4 (C4), a feature which is also present in the EGF domain from the β2 integrin subunit (1l3y).
Figure 2. J-1DSL-EGF3 & N-111-13 architecture.
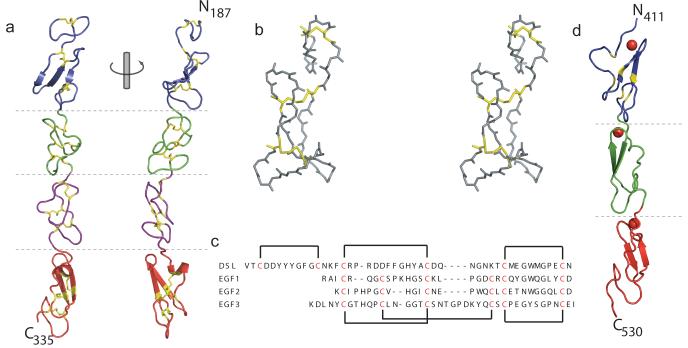
(a) The overall structure of J-1DSL-EGF3 is shown in a cartoon representation coloured from blue at the N-terminus (residue 187) to red at the C-terminus (residue 335). Disulphide bonds are shown in yellow stick representations and two views differing by a rotation of 90° about the long axis are shown. (b) Stereo view of the DSL domain fold. (c) Sequences of the four J-1DSL-EGF3 domains with disulphide bond pairings indicated. (d) Crystallographic structure of N-111-13 shown as in panel (a). The bound Ca2+ in each EGF domain is shown in a space-filling representation. All structural figures were generated with PyMol (http://www.pymol.org) 54.
The DSL domain has a distinct fold (Fig. 2b), with no structural homologues in the PDB with an r.m.s.d < 2.5 Å. It does however have clear structural similarities to an EGF domain, with a double-stranded anti-parallel β-sheet immediately preceding the characteristic loop between the final disulphide-bonded cysteines. However, while the three EGF domains have the classic pattern of disulphide bonding (C1-C3, C2-C4, C5-C6), the DSL domain, which also contains six cysteine residues, adopts a fold with a different disulphide bonded pattern (C1-C2, C3-C4, C5-C6). Intriguingly, the loop between the disulphide bonded C1 and C2 of the DSL superimposes on the loop from C5 to C6 of the same domain, with an r.m.s.d. of 0.25 Å (Supplementary Fig. 2a), while the stretch of residues from C3 to C6 aligns well with the EGF domains (Fig. 2c and Supplementary Fig. 2b & 2c). Based on the structure, we therefore hypothesise that the DSL domain may have evolved from a truncation of two tandem, short EGF domains. The sequence immediately upstream of the DSL is not conserved between members of the Notch ligand family, suggesting structural divergence away from the EGF-like fold beyond this point.
The structure of N-111-13 was solved by molecular replacement using the structure of the first EGF domain from human factor IX (1edm) as the search model (see Experimental Procedures). Like J-1DSL-EGF3, the molecule adopts an extended, gently curving structure (Fig. 2d) with dimensions of 100 × 24 × 20 Å. The model is in overall agreement with the earlier NMR structure obtained for EGF11 and EGF12 in N-111-13 (r.m.s.d. = 3.3 Å, Supplementary Fig. 3) 23. The relative orientation of EGF13 in the NMR model could not be accurately determined, most likely due to the lower stability of EGF13 in the absence of a C-terminal tag. Despite this, NOEs were assigned between Y482 in EGF12 and I509 in EGF13, and these residues are within 3.5 Å of each other in the crystal structure.
A single DSL face mediates both trans- and cis-regulation.
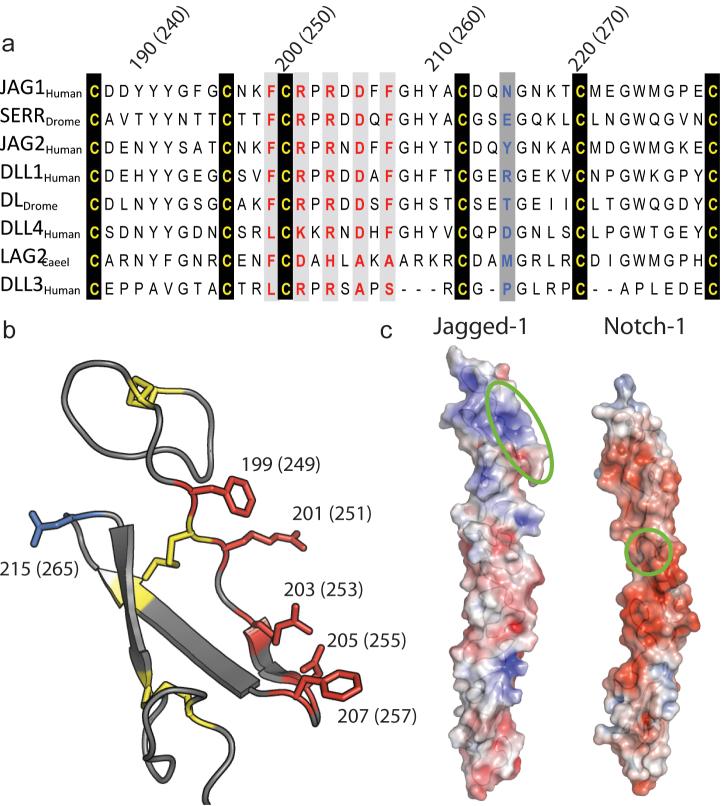
Sequence alignment of DSL domains from both Jagged/Serrate and Delta-like ligand families identifies a series of highly conserved residues (Fig. 3a). Some of these perform a structural role (Supplementary Results), but others (F199, R201, R203, D205, F207) are exposed on one face of the DSL domain and form a putative Notch binding site (Fig. 3b). Calculation of the electrostatic surface potential of the J-1DSL-EGF3 structure reveals that this region is positively charged and therefore complementary to the binding face on N-111-13 predicted by NMR (Fig. 3c). On this basis, we constructed a series of site-directed DSL domain mutants and tested the ability of each to interact with Notch in a well-established in vivo model of Notch activity, the Drosophila wing imaginal disc30. Alanine substitutions were introduced within the Drosophila homologue of J-1, Serrate, at equivalent residues (F249, R251, R253, D255 and F257) to abrogate function. In addition to mutants containing a single substitution, two additional constructs were made creating double (F249A, F257A) and triple (R251A, R253A, D255A) Serrate mutants. Finally, a non-conserved, surface exposed residue within the DSL domain, which maps to the uncharged face of the structure and is not predicted to participate in Notch binding (Fig. 3b), was substituted by alanine (E265A) to act as a positive control; this corresponds to residue N215 in the human J-1 DSL domain.
Figure 3. Predicting surfaces involved in binding and recognition.
Analysis of (a) an alignment of Jagged/Delta family DSL domains representing a variety of species (H. sapiens Jagged-1, residues 187-229; D. melanogaster Serrate, residues 237-279; H. sapiens Jagged-2, residues 198-240; H. sapiens Delta-like 1, residues 179-221; D. melanogaster Delta, residues 184-226; H. sapiens Delta-like 4, residues 175-217; C. elegans LAG-2, residues 124-166; H. sapiens Delta-like 3, residues 178-215) and (b) the DSL structure, reveals a series of highly-conserved, but surface-exposed residues. Residues which are conserved and predicted to form a Notch binding face are coloured red, cysteines are coloured yellow, while a non-conserved residue on the opposite face is coloured blue. (c) Electrostatic surface potential of J-1DSL-EGF3 and N-111-13 plotted at +/- 4 kT/e using APBS 55. Note the positively charged patch (blue) within the DSL domain of Jagged-1 and the negatively charged surface (red) of Notch. Highlighted by green bands are the surfaces predicted by sequence/structure analysis (J-1DSL-EGF3) and NMR studies (N-111-13) to be involved in binding.
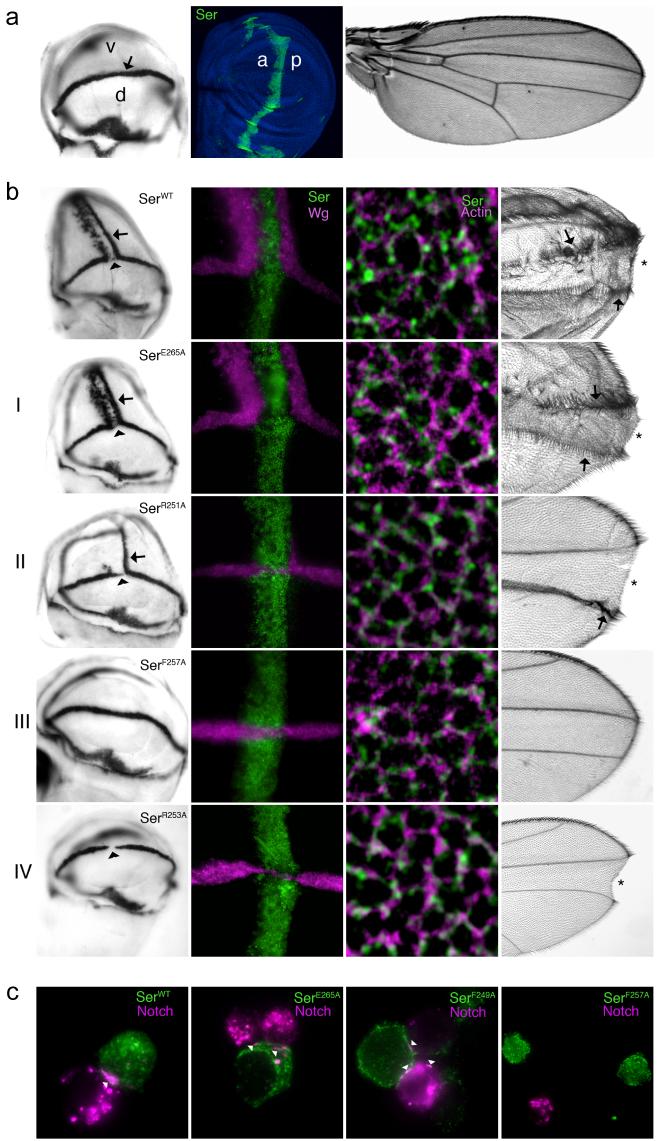
Serrate wild-type (WT) and mutant constructs were expressed in 18°C fly cultures along the anterior-posterior compartment boundary of the third instar Drosophila imaginal wing disc (Methods). The ability of each construct to induce Notch signalling was assayed through the ectopic expression of wingless, a reporter gene for Notch activity (Fig. 4a), and by the ability of each construct to induce ectopic wing margin or to suppress the normal margin of the adult wing. Consistent with previous studies, trans-activation of Notch was induced in ventral cells which lay immediately posterior to the stripe of WT Serrate over-expression, but was inhibited within the stripe of WT Serrate over-expression, through cis-interactions between ligand and receptor (Fig. 4b). The latter blocked both ectopic Notch signalling and the endogenous Notch activity present at the dorsal-ventral (D-V) boundary of the wing disc.
Figure 4. Functional analysis of Serrate DSL mutants in Drosophila wing disc and S2 cells reveals residues important for trans-activation and cis-inhibition.
(a) Left panel, wingless mRNA expression in wild-type 3rd instar wing disc, which on the dorsal (d)-ventral (v) boundary (arrow) acts as a reporter for Notch activity. Central panel shows wing disc over-expressing a Serrate construct along the anterior (a)-posterior (p) compartment boundary; cell nuclei (blue), and anti-Serrate (green). Right panel, wild-type adult wing. (b) Over-expression of wild type and mutant Serrate constructs illustrating four different phenotypic classes. Far left panels, wingless mRNA expression in imaginal wing discs. Ectopic expression is marked with an arrow and suppression of endogenous D-V boundary expression is marked with an arrowhead. Centre left panels, immunofluorescence staining of Serrate (green) and Wingless (purple). Centre right panels, localisation of expressed Serrate construct (green) with actin (purple) at the adherens junction of wing disc epithelial cells. Far right panels, adult wing phenotypes. Ectopic margins are indicated by arrows and notching of endogenous wing margin by asterisks. The results presented are from fly cultures maintained at 18°C, except for the data representative of Class IV which were obtained from a 29°C fly culture. (c) Cell aggregation assay of the Notch-binding potential of different full-length Serrate constructs. The Serrate construct used is labelled in each panel. Panels show merged immunofluoresence images of Serrate (green) and Notch (purple) expression. Cell junctions with clustered Serrate and Notch are indicated by arrowheads.
Four different classes of Serrate mutants could be distinguished after analysis of wingless expression. Representative images for each class are shown in Figure 4b and images for all mutants can be viewed in Supplementary Figure 4. E265A gave a similar result to the over-expressed WT Serrate construct, consistent with its location in the DSL structure on the opposite face of the putative Notch binding surface (Class I). D255A and R251A were able to trans-activate Notch in a similar pattern to over-expressed wild-type Serrate but had weaker signalling activity. The ability of these mutants to suppress Notch signalling in the stripe of Serrate over-expressing cells was also reduced, although still clearly discernable in the form of D-V boundary thinning and adult wing notches (Class II). Expression of the F257A mutant had no effect on the wingless reporter gene, neither activating Notch signalling in ventral cells, nor repressing endogenous Notch activity at the D-V boundary, indicating a key role for this residue in mediating both cis-inhibitory and trans-activating interactions with Notch (Class III). Finally F249A and R253A did not induce trans-activation of Notch or cis-inhibition of Notch activity at the D-V boundary at 18°C, but when expression levels were elevated in 29°C cultures both mutants were able to mimic the cis-inhibitory effect of wild type Serrate at the D-V boundary (Class IV). The combination mutants (F249A, F257A) and (R251A, R253A, D255A) also belonged to Class III, and were unable to induce trans-activation of Notch, or cis-inhibit Notch (Table 1). As expected, the adult wing phenotypes of each mutant class correlated with the ability of Serrate to activate Notch ectopically or inhibit Notch at the D-V boundary (Fig. 4b). Immunostaining revealed that all Serrate mutant constructs were expressed and properly localised at the adherens junction (Fig. 4b and Supplementary Fig. 5). Comparison of expression levels showed that availability of Serrate was not limiting for the observed affects of different constructs on wingless expression (Supplementary Fig. 6). Thus substitution of all amino acid residues predicted by structural data to participate in Notch binding gave functional effects consistent with this. Furthermore, effects of the Serrate mutants on both cis- and trans-interactions were observed, indicating that the Notch binding site identified on the DSL domain is involved in both processes.
Table 1.
Activity of Serrate mutants expressed in the Drosophila 3rd instar wing imaginal disc
| Serrate mutant* | 18°C | 29°C | ||
|---|---|---|---|---|
| INH | TA | INH | TA | |
| WT | +++ | +++ | lethal | |
| Triple R251A+R253A+D255A | 0 | 0 | 0 | 0 |
| Double F249A+F257A | 0 | 0 | nd | nd |
| F249A (F199) | 0 | 0 | ++ | (+) |
| R253A (R203) | 0 | 0 | ++ | 0 |
| D255A (D205) | + | ++ | nd | nd |
| F257A (F207) | 0 | 0 | 0 | 0 |
| E265A (N215) | +++ | +++ | lethal | |
| R251A (R201) | ++ | ++ | nd | nd |
The number in parentheses refers to the equivalent residue in the human Jagged-1 protein sequence. INH- defined as the ability to inhibit endogenous D/V boundary formation, TA-trans-activation of Notch signalling.
Roles for conserved DSL residues in Notch signalling
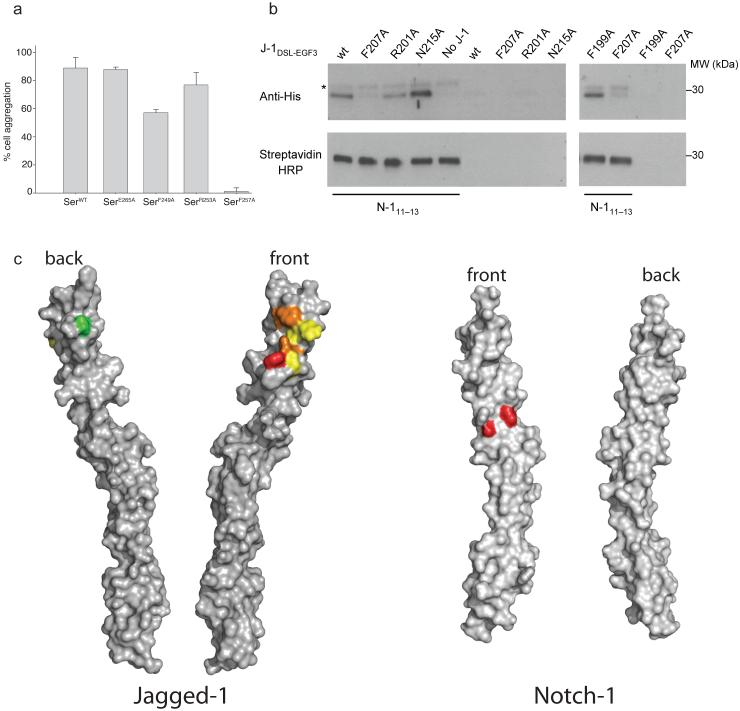
Cell-aggregation assays were performed with cells transfected with either Serrate or Notch (Fig. 4c & 5a) to compare the Notch-binding ability of different Serrate mutants shown to be defective in Notch trans-activation. Immunofluorescence staining showed that F257A was unable to mediate cell aggregation with Notch-expressing cells, unlike WT Serrate or E265A, the control mutant. This suggested that the explanation for the inability of F257A to trans-activate Notch in the wing disc is due to a major defect in Notch binding. Class IV mutants F249A and R253A, however, were each able to promote cellular adhesion and the clustering of ligand and receptor at sites of cellular contact. Therefore, the loss of trans-activation observed in vivo with F249A and R253A is not explained by loss of Notch binding. Since F249A and R253A can still bind to Notch in trans (giving rise to a non-signalling mode), this may contribute to their observed ability in vivo to inhibit endogenous Notch signalling at the D-V boundary. The cell-aggregation assays have been further validated using pull-down assays with purified forms of the J-1DSL-EGF3 mutant proteins (Fig. 5b). In these assays the mutant constructs were expressed and refolded (as previously performed for the wild-type protein, see Methods). These proteins were then assayed for the ability to be pulled-down by N-111-13 coupled to beads via a C-terminal biotin tag (see Methods). The results of the pull-down experiments correlated well with the phenotypes observed for the equivalent Serrate mutations in vivo and in the cell aggregation assays. The N215A (Serrate E265A) control mutant showed WT binding, while F207A (Serrate F257A) was unable to bind the N-111-13 beads. R201A (Serrate equivalent R253A) and F199A (Serrate F249A) had somewhat intermediate phenotypes showing reduced, but not abolished, binding in agreement with the other assays.
Figure 5. Quantification of mutant phenotypes and mapping onto structures.
(a) Graph shows % of total Notch expressing cells bound to cells that express different Serrate constructs. Error bars represent standard deviation from quadruplicate experiments. (b) Western blot analyses of representative experiments to demonstrate the pull down of J-1DSL-EGF3 wild-type and mutant constructs by biotinylated N-111-13 immobilised on Dynabeads M-270 Streptavidin. Detection by anti-RGS.His HRP conjugate (upper panels) demonstrated interaction of N-111-13 with J-1DSL-EGF3 wild-type, F199A, R201A and N215A but not F207A. A faint upper band (indicated by an asterisk) is attributed to a trace of His-tagged N-111-13. Streptavidin-HRP conjugate was used to show equivalent amounts of N-111-13 were eluted from the beads for each mutant (lower panels). (c) Surface representations of both J-1DSL-EGF3 and N-111-13 are shown in two orientations related by 180 degree rotation about the long axis. The J-1DSL-EGF3 is coloured by mutant phenotype class (class 1 - green, class II - yellow, class III - red, class IV - orange) whilst the residues implicated in J-1DSL-EGF3 binding by NMR are coloured red on the surface of N-111-13. We have termed the faces implicated in binding the “front” view on each molecule.
DISCUSSION
Our multi-disciplinary studies have identified for the first time the structure and potential evolutionary origins of a DSL domain, which is common to all Notch ligands. In addition, an extended organisation of domains within the J-1DSL-EGF3 ligand fragment is identified, suggesting the possibility of numerous interactions along its surface. Targeted disruption of a conserved, putative Notch binding site in the DSL domain caused both defective cis-inhibition and trans-activation of Notch in the Drosophila wing, demonstrating that the same surface of the DSL domain is essential for both types of interaction. These data, combined with NMR studies of N-111-13 which demonstrated a J-1 interaction site at the centre of N-111-13, identify a candidate interaction surface on both molecules for the Notch-ligand complex (Figure 5c).
Although we were able to determine structures for functional fragments J-1DSL-EGF3 and the N-111-13, we were unable to find crystallisation conditions compatible with complex formation. We therefore performed in silico docking of J-1DSL-EGF3 and the N-111-13 structures (Methods) using restraints from our NMR binding studies (V453 and G472) to drive the docking and then using the J-1DSL-EGF3 phenotypes to validate the docked models. Interestingly, the docking predicts two models for the N-1/J-1 interface (Supplementary Figure 7). Both of these models involve interactions along the long axes of the two molecules, but one has the two molecules in a parallel arrangement, while the other is anti-parallel. In both models the DSL domain plays the most prominent role in binding, but EGF1 and EGF2 also participate in interface formation. This is consistent with the findings of Shimizu et al.26, where deletion analyses demonstrated that the DSL domain was indispensible for Notch binding, but EGF1 and EGF2 were also required to confer native-like affinity for Notch. The existence of two binding modes could contribute to the exchange broadening observed in the NMR titrations and complicate attempts to crystallise the complex.
That two proteins could interact in completely different ways using the same surfaces is highly unexpected. However, in strong support of this is the observation that all of the J-1DSL-EGF3 which lead to distinct phenotypes when expressed as Serrate mutants in the Drosophila are buried in the docked models, despite the fact that they were not used to drive the docking. Critical to both model complexes is the highly conserved phenylalanine residue (F207/257) which completely abrogates both trans-activation and cis-inhibition when mutated. This residue packs against the hydrophobic surface formed by V453 and G472 of EGF12 from N-111-13 and acts as a pivot; with the N-111-13 considered as a fixed object, the two models are related by a 163° rotation of the J-1DSL-EGF3 centred on F207. Ring current shifts arising from the aromatic ring may explain in part the larger changes in peak intensity for V453 and G472 observed in the NMR spectrum; these residues could have appreciably different chemical shifts in the free and bound states which would result in significant chemical exchange broadening. Interestingly, the differential outcomes on signal activation obtained from our mutational analysis can be interpreted in the context of the docked models (Supplementary Figure 7). Residues F207, F199, R203, homologous to those shown to be essential for ligand-dependent Notch activation by Serrate (F257, F249, R253), are extensively buried in both models. Residues R201 and D205 (R251 and D255 in Serrate), which have a more limited effect, have relatively minor contacts to the N-111-13. Given the known cell-surface locations of ligand and receptor, and the knowledge that both J-1DSL-EGF3 and N-111-13 form extended structures, we suggest that the parallel complex is a working model for the cis-inhibitory interaction and the antiparallel complex a model for the trans-activating complex of ligand and receptor (Supplementary Figure 7).
Different molecular mechanisms underlying the defective trans-activation observed in vivo for Serrate mutants F257A, F249A and R253A were revealed by cell-aggregation assays. F257A was unable to mediate adhesion to Notch expressing cells, while both F249A (Figure 4c) and R253A (Figure 5a) supported binding. The ability of Serrate F249A and R253A mutants to bind to Notch-expressing cells in aggregation assays, despite the absence of trans-activation in the wing disc experiments, suggests that binding alone is not sufficient for promoting signal initiation. Ligand-dependent activation of Notch has been proposed to involve ligand-dependent clustering of Notch-ligand complexes at the cellular surface, followed by initiation of a pulling force through the interaction of the Notch and ligand cytoplasmic domains with the endocytic machinery that removes the Notch extracellular domain, which is transendocytosed with ligand into the signal sending cell 12. This process then elicits a conformational change in the Notch regulatory region (NRR) region 10. One possible mechanism promoting trans-activation may be a requirement for an interaction of a sufficiently high affinity to resist the tensional force necessary for activation of the NRR. If correct, then the F257A mutant (F207 in J-1) has such a low affinity for Notch that no binding is observed, while the F249A (F199 in J-1) and R253A (R203 in J-1) mutants may allow the complex to form and be sufficiently tight to allow cell aggregation and initiation of receptor clustering, but insufficient to complete the subsequent activation step (Supplementary Figure 7). Alternatively, the interactions mediated by F199/249 and R203/253 may be required to initiate the final activation step by transmitting a conformational change to the cytoplasmic domains of ligand and/or receptor. An interesting possibility is that such a conformational change might be required to initiate the endocytosis of Serrate and hence the trans-endocytosis of Notch. Future studies will analyse in detail the endocytic cycle of the different mutant Serrate constructs.
Genetic mutations in human J-1 are associated with severe disease. Some substitutions affect residues predicted by our study to form part of a Notch-ligand interface. For example, R203K, identified in a patient with extrahepatic biliary atresia 31, substitutes the highly conserved arginine residue demonstrated in our functional studies (R253 in Serrate) to play a critical role in transactivation. Similarly, an R252G substitution identified in a patient with Alagille syndrome 32, replaces an arginine involved in a salt bridge at the ligand-receptor interface in both the parallel and anti-parallel docking models, and is therefore likely to destabilise the interface (Supplementary Figure 7).
Signalling via the Notch receptor has been demonstrated to be modulated by Fringe extension of O-fucosylation sites within Notch 33,34. The residue in EGF12 of N-111-13 which has been suggested to be O-fucosylated and subsequently modified by Fringe (T466) is on the opposite face of Notch to that implicated in ligand binding in our model. We therefore predict that O-fucosylation of EGF12 of Notch is not directly involved in J-1DSL-EGF3 binding, in agreement with the calcium-dependent specific interaction observed with our unmodified fragments. Furthermore, it has recently been demonstrated that Notch-ligand interactions are not inhibited by the addition of exogenous sugars, also suggesting that the glycosylated residues do not directly contact the ligand 35. It is also of note that recent work 36 has implicated a novel glucosylase in regulation of Notch activity. Our complexes involve the only potential site of glucosylation in domain 12 and may therefore suggest a role for this modification in modulating ligand interactions.
In summary, we have identified a Notch binding site on the DSL domain from human Jagged-1 which participates in an interface responsible for regulating both cis- and trans-interactions. These data therefore provide the first structural insight into the early events of Notch activation and extend our general understanding of the different ways in which protein-protein complexes may form. Understanding the structural basis of cis- and trans-interactions and the molecular events leading to Notch activation will enable rational approaches to the design of antagonists or agonists of Notch activity, with possible applications in anti-cancer therapeutics, immune system modulation and the control of cell-fate decisions of stem cells and their progeny in vitro or in vivo.
METHODS
Protein Expression and Refolding
We carried out expression, purification and refolding of N-111-13 as described previously 37 with modifications (Supplementary Data) to allow addition of a C-terminal BirA-tag. Biotinylation of N-111-13 and 15N-labelling of EGF11-13 were performed as described previously 23,37. The J-1DSL-EGF3 fragment was expressed in Escherichia coli NM554 as a His-tag fusion protein and purified from cell lysate by Ni2+-affinity chromatography. Native and Selenomethionine (SeMet) material was refolded in vitro and purified as described previously 37 with modifications as described in Supplementary Methods. Missense mutations to generate F199A, R201A, F207A and N215A in the J-1DSL-EGF3 construct were introduced into pQE30 recombinant plasmid containing the cDNA sequence of the wild-type construct by a PCR-based site-directed mutagenesis method with Pfu DNA polymerase. Generation of the desired mutation was confirmed by DNA sequencing. The mutant constructs were purified as described for the wild-type J-1DSL-EGF3. Masses were confirmed using Electrospray Ionisation Mass Spectral analysis (ESI-MS). Correct folding of the functionally dead F207A mutant was confirmed with NMR (data not shown).
Notch antibody
Mouse monoclonal antibodies were generated against purified antigen hN-111-14.38 and affinity purified.
Pull down assays
N-111-13 (12 μg) and J-1DSL-EGF3 (3 μg) were incubated in 10 mM Tris pH 7.5, 150 mM NaCl, 1 mM CaCl2, 20 mM imidazole. The 6xHis-tagged J-1DSL-EGF3 fragment was captured on Ni-NTA Magnetic Agarose Beads (Qiagen) and washed. Proteins were eluted from the beads, separated using SDS PAGE and immunostained using the Notch antibody (Fig. 1a). N-111-13 was also shown to interact with J-1DSL-EGF3 previously immobilised on the magnetic beads. The calcium dependence of the Notch/Jagged interaction was demonstrated by immobilisation of biotinylated N-111-13 on Dynabeads M-270 Streptavidin (10 μg per 0.5 mg beads) and subsequent capture of J-1DSL-EGF3 (8 μg per 100 μl) in buffer (10 mM Tris pH 7.5, 150 mM NaCl) containing 10 mM CaCl2 or 5 mM EDTA. (Figure 1b). Assaying of binding of J-1DSL-EGF3 mutants (Fig. 5b) was performed as described above except the co-eluted N-111-13 was detected with streptavidin-HRP conjugate.
Surface Plasmon Resonance
Surface plasmon resonance (SPR) experiments were performed as described in Supplementary Data. Briefly, J-1DSL-EGF3 fragment was immobilized on the chip surface and interaction data were collected by injecting N-111-13 over the coupled chip surface. Results were identical in the absence (Fig. 1) or presence (data not shown) of the C-terminal BirA-tag. Traces were corrected for refractive index changes by subtraction of a control trace simultaneously recorded. Unless otherwise stated, all experiments were carried out in Tris buffered saline, pH 7.5, supplemented with 1mM Ca2+.
NMR Spectroscopy
NMR data were collected for 0.5 mM 15N-labelled N-111-13 at pH 6.1 with 15 mM Ca2+ in the absence and in the presence of 0.15 mM unlabelled J-1DSL-EGF3 (see Supplementary Data). The fractional intensity change [(Ibound - Ifree)/Ifree] between peaks in the absence (Ifree) and presence (Ibound) of J-1DSL-EGF3 was calculated from peak heights (I); errors were estimated from baseline noise. The fractional intensities in Figure 1f are the average of three experiments.
J-1DSL-EGF3 Crystallisation and Structure Determination
J-1DSL-EGF3 (residues 187-335) was crystallised in sitting drops by the vapour diffusion technique at 4.6 mg ml-1. using 0.2 μl of protein and 0.2 μl of mother liquor. Sparse matrix screening and optimisation led to rod-shaped crystals, with approximate dimensions 100 × 50 × 20 μm, in 8.5% (w/v) PEG 4000, 100mM Imidazole/Malate, pH 7.0. Native and SeMet datasets were collected and processed as described in Supplementary Data and Table 2. The structure was phased using autoSHARP 39 and SHARP 40, built with Xfit 41, and refined using Buster-TNT 42 (Table 1).
Table 2.
Data collection, phasing and refinement statistics for MAD (SeMet) structures
| Notch-Native 1 | Jagged-Native 1 | Jagged-Native 2 | Jagged-SeMet 1 | Jagged-SeMet 3 | |||
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| Space group | P3121 | C2 | C2 | C2 | C2 | ||
| Cell dimensions | |||||||
| a, b, c (Å) | 28.0, 28.0, 281.7 | 61.0, 102.3, 62.7 | 60.6, 102.6, 61.2 | 61.2, 102.8, 60.7 | 61.0, 102.7, 59.9 | ||
| α, β, γ (°) | 90, 90, 120 | 90, 114.3, 90 | 90, 114.2, 90 | 90, 113.7, 90 | 90, 113.3, 90 | ||
| Peak 1 | Peak 2 | Remote | Peak | ||||
| Wavelength | 1.542 | 0.9340 | 0.9800 | 0.9788 | 0.9788 | 0.9537 | 0.9793 |
| Resolution (Å) | 24.3-2.6 (2.7-2.6) | 57.1-2.5 (2.7-2.5) | 55.8-3.1 (3.3-3.1) | 56.0-2.8 (3.0-2.8) | 51.4-2.7 (2.9-2.7) | 51.6-2.9 (3.1-2.9) | 51.4-3.1 (3.3-3.1) |
| R merge | 6.2 (15.3) | 9.2 (42.2) | 10.3 (31.6) | 13.5 (31.9) | 11.6 (32.0) | 11.6 (30.1) | 12.5 (32.2) |
| I / σI | 25.2 (5.3) | 13.1 (3.3) | 5.6 (2.3) | 4.6 (2.1) | 5.6 (2.1) | 5.3 (2.1) | 5.0 (2.0) |
| Completeness (%) | 93.3 (93.3) | 98.6 (98.6) | 98.1 (98.1) | 99.8 (99.2) | 92.4 (92.8) | 98.2 (98.6) | 98.2 (98.7) |
| Redundancy (Ano) | 8.2 | 3.7 | 3.4 | (5.2) | (3.0) | (2.8) | (2.8) |
| Refinement | |||||||
| Resolution (Å) | 24.3-2.6 (2.7-2.6) | 48.5-2.5 (2.7-2.5) | |||||
| No. | 4080 | 11961 | |||||
| reflections | |||||||
| Rwork / Rfree | 23.9 (29.1)/24.3 (36.3) | 22.0 (26.3)/23.9 (30.8) | |||||
| No. atoms | |||||||
| Protein | 897 | 2229 | |||||
| Ligand/ion | 8 | 9 | |||||
| Water | 22 | 208 | |||||
| B-factors | |||||||
| Protein | 34.3 | 32.3 | |||||
| Ligand/ion | 33.9 | 36.4 | |||||
| Water | 38.4 | 67.3 | |||||
| R.m.s deviations | |||||||
| Bond | 0.005 | 0.007 | |||||
| lengths (Å) | |||||||
| Bond | 0.68 | 0.72 | |||||
| angles (°) | |||||||
Each dataset was collected using a single crystal.
Values in parentheses are for highest-resolution shell.
N-111-13 Crystallisation and Structure Determination
N-111-13 was crystallised in sitting drops by the vapour diffusion technique at 15.7 mg ml-1. using 0.2 μl of protein and 0.2 μl of mother liquor. Sparse matrix screening and optimisation led to hexahedral crystals, with approximate dimensions 300 × 100 × 100 μm, in 30% (w/v) PEG-5000-MME, 100mM Na acetate, pH 5.7. Native data were collected and processed as described in Supplementary Data. The structure was solved by molecular replacement (CCP4-MOLREP 43) using 3 copies of an EGF-like domain from factor IX (PDB 1EDM44) and refined to give the model described in Supplementary Methods and Table 2.
Serrate Site-Directed Mutagenesis and Functional analysis in Drosophila
Site-directed mutants of Serrate were constructed by four-primer PCR mutagenesis, using the plasmid pBKS+SerFL 45 as a template (primers in Supplementary Data). EcoRI fragments from Serrate clones were then excised and ligated into EcoRI-digested pUAST vector 46 for the construction of Drosophila lines. Transgenic Drosophila lines were constructed using standard methods by BestGene Inc. (Chino Hills, California). Flies were cultured on standard yeast/agar/cornmeal media. For Serrate expression experiments the Ptc-Gal4 line 47 was crossed to each of the UAS-Serrate constructs in either 18°C or 29°C fly culture as indicated in the text. In situ hybridisation was performed using a digoxigenin-labelled antisense cRNA wg probe and stained with alkaline phosphatase conjugated anti-digoxigenin antibody as previously described 48.
Antibodies For Functional Analyses
Rabbit polyclonal anti-Serrate 49 was used at 1:200 dilution. Mouse monoclonal anti-Wingless (4D4) and anti-Notch (C17.9C6) (DSHB, Iowa University) were used at 1:50 and 1:200. Anti-rabbit RRX or anti-mouse CY5 secondary antibodies (Jackson Immunoresearch Laboratories) were used at 1:600.
Immunocytochemistry
Late third instar larval discs were dissected in phosphate buffered saline (PBS) + 0.1% Tween20 (PBS-Tw) and fixed in 4% formaldehyde/PBS for 20 min at room temperature (RT). The discs were immunostained in PBS + 0.5% Triton X-100, 4% normal donkey serum. For actin labelling FITC-labelled phalloidin (Sigma) was subsequently incubated with the tissue at 1μM concentration for 2 hours at room temperature, the tissues were then rinsed 5 times with PBS-Tw. Preps were transferred into Vectashield mounting medium with DAPI (Vector Laboratories). Images were captured using a cooled digital camera (Hamamatsu) mounted on a Zeiss Axiovert microscope and processed on an Apple Macintosh computer using Improvision Openlab and Adobe PhotoShop software. For images of the adherens junctions deconvolution was performed using 3 nearest neighbours from optical sections obtained with a Z spacing of 0.5μm.
Cell Culture
Drosophila S2 cells (Invitrogen) were transiently transfected as previously described 50. For Serrate expression S2 cells were co-transfected with wild type or mutant pUAST-Ser constructs and pMT-Gal4 51, and induced with 1mM CuSO4 for 16-20 hours prior to mixing with Notch transfected cells. For Notch expression, S2 cells were transfected with H/N 52. H/N transfected cells were mixed with Serrate expressing cells and Notch expression was induced by heat shock of 40 minutes at 42°C. After induction cells were mixed in 1.5ml eppendorf tubes on a rotating platform for 4 hours before transferring the cell suspension to cover slips coated in 0.1% poly-L-lysine (Sigma) for immunostaining.
In silico docking of N-111-13 and J-1DSL-EGF3
Docking was carried out using the program HADDOCK 2.0 53, driving the docking via constraints based on the NMR peak intensity perturbation data observed for N-111-13 upon complex formation. N-111-13 residues which showed a significant decrease in peak intensity upon complex formation (Val453/Gly472) were defined as “active”. In the absence of NMR data for J-1DSL-EGF3, all residues with a high solvent accessibility (>50%) were selected as “passive” residues. The resultant “active” residues of N-111-13 and “passive” residues of J-1DSL-EGF3 were used to define ambiguous interaction restraints (AIRs) with a 3 Å distance. Following rigid-body docking the top ten structures were analysed and found to belong to two classes. Both of these classes involved interactions along the long axes of the 2 molecules. The two best scoring models represented an anti-parallel orientation of J-1DSL-EGF3 relative to N-111-13 and buried 526 Å2 protein-1. The next eight models on the other hand represented a parallel orientation and buried 857 Å2/protein. All ten models buried all of the residues on J-1DSL-EGF3 which produced phenotypes when mutated in Drosophila.
Supplementary Material
ACKNOWLEDGMENTS
We thanks M. Walsh at BM14, the staff of ID29 at the European Synchrotron Radiation Facility and to Ed Lowe (University of Oxford) for assistance with X-ray data collection. We also thank A. McMichael, S. Hambleton & X. Xu (University of Oxford) for advice and Global Phasing for access to a beta version of Buster-TNT. Robert Fleming, Cedric Wesley and Spyros Artavanis-Tsakonas of Harvard Medical School for Drosophila cDNA constructs and fly stocks. This work was supported by the E.P.A. Cephalosporin fund (#CF065 to SML), the UK MRC (#G0400389 to SML, #G000164 to PAH and MRC Studentship G78/7267 to JC.) the Wellcome Trust (#077082 to SML, #078765 to PAH and SML & #079440 to C.R.) and the BBSRC #C503162/1 and E002285/1 to MB).
Accession codes. Protein Data Bank: Coordinates for N-111-13 have been deposited with accession code 2VJ3 and coordinates for J-1 DSL-EGF3 have been deposited with accession code 2VJ2.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–73. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 3.Milner LA, Kopan R, Martin DI, Bernstein ID. A human homologue of the Drosophila developmental gene, Notch, is expressed in CD34+ hematopoietic precursors. Blood. 1994;83:2057–62. [PubMed] [Google Scholar]
- 4.Nyfeler Y, et al. Jagged1 signals in the postnatal subventricular zone are required for neural stem cell self-renewal. Embo J. 2005;24:3504–15. doi: 10.1038/sj.emboj.7600816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKenzie GJ, et al. Notch signalling in the regulation of peripheral T-cell function. Semin Cell Dev Biol. 2003;14:127–34. doi: 10.1016/s1084-9521(02)00181-7. [DOI] [PubMed] [Google Scholar]
- 6.Hansson EM, Lendahl U, Chapman G. Notch signaling in development and disease. Semin Cancer Biol. 2004;14:320–8. doi: 10.1016/j.semcancer.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 7.Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–91. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 8.Logeat F, et al. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci U S A. 1998;95:8108–12. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez-Irizarry C, et al. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol Cell Biol. 2004;24:9265–73. doi: 10.1128/MCB.24.21.9265-9273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon WR, et al. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- 11.Nichols JT, et al. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J Cell Biol. 2007;176:445–58. doi: 10.1083/jcb.200609014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parks AL, Klueg KM, Stout JR, Muskavitch MA. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127:1373–85. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- 13.Mumm JS, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 14.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–6. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 15.Jarriault S, et al. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–8. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 16.de Celis JF, Bray S. Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development. 1997;124:3241–51. doi: 10.1242/dev.124.17.3241. [DOI] [PubMed] [Google Scholar]
- 17.Franklin JL, et al. Autonomous and non-autonomous regulation of mammalian neurite development by Notch1 and Delta1. Curr Biol. 1999;9:1448–57. doi: 10.1016/s0960-9822(00)80114-1. [DOI] [PubMed] [Google Scholar]
- 18.Glittenberg M, Pitsouli C, Garvey C, Delidakis C, Bray S. Role of conserved intracellular motifs in Serrate signalling, cis-inhibition and endocytosis. Embo J. 2006;25:4697–706. doi: 10.1038/sj.emboj.7601337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Micchelli CA, Rulifson EJ, Blair SS. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development. 1997;124:1485–95. doi: 10.1242/dev.124.8.1485. [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto K, Ohara O, Takagi M, Takeda S, Katsube K. Intracellular cell-autonomous association of Notch and its ligands: a novel mechanism of Notch signal modification. Dev Biol. 2002;241:313–26. doi: 10.1006/dbio.2001.0517. [DOI] [PubMed] [Google Scholar]
- 21.Fleming RJ. Structural conservation of Notch receptors and ligands. Semin Cell Dev Biol. 1998;9:599–607. doi: 10.1006/scdb.1998.0260. [DOI] [PubMed] [Google Scholar]
- 22.Rebay I, et al. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991;67:687–99. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- 23.Hambleton S, et al. Structural and functional properties of the human notch-1 ligand binding region. Structure. 2004;12:2173–83. doi: 10.1016/j.str.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 24.Abbott RJ, et al. Structural and functional characterization of a novel T cell receptor co-regulatory protein complex, CD97-CD55. J Biol Chem. 2007;282:22023–32. doi: 10.1074/jbc.M702588200. [DOI] [PubMed] [Google Scholar]
- 25.Henderson ST, Gao D, Christensen S, Kimble J. Functional domains of LAG-2, a putative signaling ligand for LIN-12 and GLP-1 receptors in Caenorhabditis elegans. Mol Biol Cell. 1997;8:1751–62. doi: 10.1091/mbc.8.9.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu K, et al. Mouse jagged1 physically interacts with notch2 and other notch receptors. Assessment by quantitative methods. J Biol Chem. 1999;274:32961–9. doi: 10.1074/jbc.274.46.32961. [DOI] [PubMed] [Google Scholar]
- 27.Tax FE, Yeargers JJ, Thomas JH. Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with Delta and Serrate of Drosophila. Nature. 1994;368:150–4. doi: 10.1038/368150a0. [DOI] [PubMed] [Google Scholar]
- 28.Salmon D, et al. Solution structure and backbone dynamics of the Trypanosoma cruzi cysteine protease inhibitor chagasin. J Mol Biol. 2006;357:1511–21. doi: 10.1016/j.jmb.2006.01.064. [DOI] [PubMed] [Google Scholar]
- 29.Cordle J, et al. Localization of the Delta-like-1-binding Site in Human Notch-1 and Its Modulation by Calcium Affinity. J Biol Chem. 2008;283:11785–11793. doi: 10.1074/jbc.M708424200. [DOI] [PubMed] [Google Scholar]
- 30.Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates Notch-ligand interactions. Nature. 1997;387:908–12. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- 31.Kohsaka T, et al. The significance of human jagged 1 mutations detected in severe cases of extrahepatic biliary atresia. Hepatology. 2002;36:904–12. doi: 10.1053/jhep.2002.35820. [DOI] [PubMed] [Google Scholar]
- 32.Warthen DM, et al. Jagged1 (JAG1) mutations in Alagille syndrome: increasing the mutation detection rate. Hum Mutat. 2006;27:436–43. doi: 10.1002/humu.20310. [DOI] [PubMed] [Google Scholar]
- 33.Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406:411–5. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- 34.Moloney DJ, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–75. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 35.Xu A, et al. In vitro reconstitution of the modulation of Drosophila notch-ligand binding by fringe. J Biol Chem. 2007;282:35153–62. doi: 10.1074/jbc.M707040200. [DOI] [PubMed] [Google Scholar]
- 36.Acar M, et al. Rumi Is a CAP10 Domain Glycosyltransferase that Modifies Notch and Is Required for Notch Signaling. Cell. 2008;132:247–58. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muranyi A, et al. 1H, 13C, and 15N resonance assignments of human Notch-1 calcium binding EGF domains 11-13. J Biomol NMR. 2004;29:443–4. doi: 10.1023/B:JNMR.0000032521.42723.1a. [DOI] [PubMed] [Google Scholar]
- 38.Hutchinson S, et al. Molecular effects of homocysteine on cbEGF domain structure: insights into the pathogenesis of homocystinuria. J Mol Biol. 2005;346:833–44. doi: 10.1016/j.jmb.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 39.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated Structure Solution with autoSHARP. In: Doublie S, editor. Crystallographic Methods. Humana Press; 2005. [DOI] [PubMed] [Google Scholar]
- 40.La Fortelle E.d.B. G. Maximum-Likelihood Heavy-Atom Parameter Refinement for Multiple Isomorphous Replacement and Multiwavelength Anomalous Diffraction Methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 41.McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–65. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 42.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60:2210–21. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 43.Vagin A, Teplyakov A. MOLREP: an Automated Program for Molecular Replacement. J. Appl. Cryst. 1997;30:1022. [Google Scholar]
- 44.Rao Z, et al. The structure of a Ca(2+)-binding epidermal growth factor-like domain: its role in protein-protein interactions. Cell. 1995;82:131–41. doi: 10.1016/0092-8674(95)90059-4. [DOI] [PubMed] [Google Scholar]
- 45.Fleming RJ, Scottgale TN, Diederich RJ, Artavanis-Tsakonas S. The gene Serrate encodes a putative EGF-like transmembrane protein essential for proper ectodermal development in Drosophila melanogaster. Genes Dev. 1990;4:2188–201. doi: 10.1101/gad.4.12a.2188. [DOI] [PubMed] [Google Scholar]
- 46.Brand AG, Manoukian AS, Perrimon N. Ectopic expression in Drosophila. In: Goldstein LSB, Fyrberg EA, editors. Methods in cell biology. Vol. 44. 1994. pp. 635–54. [DOI] [PubMed] [Google Scholar]
- 47.Speicher SA, Thomas U, Hinz U, Knust E. The Serrate locus of Drosophila and its role in morphogenesis of the wing imaginal discs: control of cell proliferation. Development. 1994;120:535–44. doi: 10.1242/dev.120.3.535. [DOI] [PubMed] [Google Scholar]
- 48.Mazaleyrat SL, et al. Down-regulation of Notch target gene expression by Suppressor of deltex. Dev Biol. 2003;255:363–72. doi: 10.1016/s0012-1606(02)00086-6. [DOI] [PubMed] [Google Scholar]
- 49.Thomas U, Jonsson F, Speicher SA, Knust E. Phenotypic and molecular characterization of SerD, a dominant allele of the Drosophila gene Serrate. Genetics. 1995;139:203–13. doi: 10.1093/genetics/139.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fehon RG, et al. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990;61:523–34. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- 51.Klueg KM, Alvarado D, Muskavitch MA, Duffy JB. Creation of a GAL4/UAS-coupled inducible gene expression system for use in Drosophila cultured cell lines. Genesis. 2002;34:119–22. doi: 10.1002/gene.10148. [DOI] [PubMed] [Google Scholar]
- 52.Lieber T, et al. Single amino acid substitutions in EGF-like elements of Notch and Delta modify Drosophila development and affect cell adhesion in vitro. Neuron. 1992;9:847–59. doi: 10.1016/0896-6273(92)90238-9. [DOI] [PubMed] [Google Scholar]
- 53.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–7. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 54.DeLano WL. The PyMOL Molecular Graphics System. 2002. http://www.pymol.org.
- 55.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.