Abstract
Human adenocarcinomas commonly harbor mutations in the KRAS and MYC proto-oncogenes and the TP53 tumor suppressor gene. All three genetic lesions are potentially pro-angiogenic, as they sustain production of vascular endothelial growth factor (VEGF). Yet Kras-transformed mouse colonocytes lacking p53 formed indolent, poorly vascularized tumors, whereas additional transduction with a Myc-encoding retrovirus promoted vigorous vascularization and growth. In addition, VEGF levels were unaffected by Myc, but enhanced neovascularization correlated with downregulation of anti-angiogenic thrombospondin-1 (Tsp1) and related proteins, such as connective tissue growth factor (CTGF). Both Tsp1 and CTGF are predicted targets for repression by the miR-17-92 microRNA cluster, which was upregulated in colonocytes coexpressing K-Ras and c-Myc. Indeed, miR-17-92 knockdown with antisense 2′-O-methyl oligoribonucleotides partly restored Tsp1 and CTGF expression; in addition, transduction of Ras-only cells with a miR-17-92–encoding retrovirus reduced Tsp1 and CTGF levels. Notably, miR-17-92–transduced cells formed larger, better-perfused tumors. These findings establish a role for microRNAs in non–cell-autonomous Myc-induced tumor phenotypes.
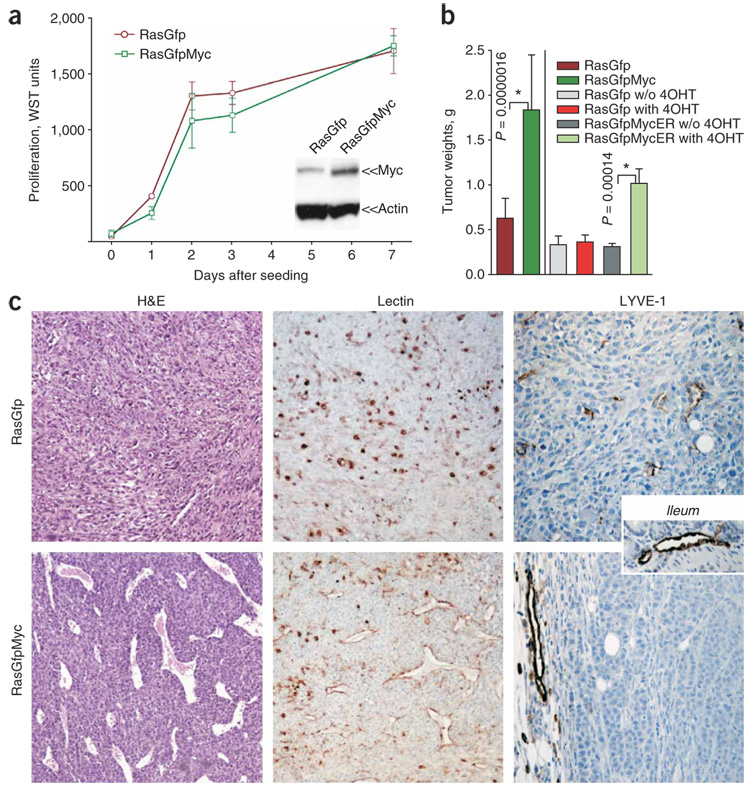
The role of c-Myc in neovascularization of one-hit neoplasms is well established1–4 and involves both upregulation of pro-angiogenic VEGF4–6 and downregulation of anti-angiogenic Tsp1 (refs. 7–9). To address the role of Myc in neovascularization of genetically complex tumors, we used p53-null mouse colonocytes, which can be transformed in vitro by low-grade overexpression of either activated K-Ras or Myc10,11. When engrafted into syngeneic mice, subcutaneously or orthotopically (into the cecal wall), Ras-overexpressing cells formed tumors, but their Myc-overexpressing counterparts did not (data not shown). Then we introduced MYC into ras-transformed colonocytes, either constitutively (‘RasGfpMyc’) or in a 4-hydroxytamoxifen (4OHT)-dependent form (‘RasGfpMycER’). We did not observe any increase in cell accumulation in vitro compared with Ras cells expressing GFP alone (‘RasGfp’) (Fig. 1a and data not shown). We then tested whether Myc and Ras cooperate in vivo. Whereas control RasGfp cells formed relatively small tumors, RasGfpMyc neoplasms were on average three times larger (Fig. 1b). The same increase in tumor sizes was observed with RasGfpMycER cells in animals continuously treated with 4OHT (Fig. 1b).
Figure 1. Growth properties of RasGfp versus RasGfpMyc p53-null colonocytes in vivo and in vitro.
(a) Cell accumulation assay performed on RasGfp and RasGfpMyc colonocytes. We assessed the number of viable cells in triplicate plates using the WST reagent. We assessed levels of Myc overexpression by immunoblotting (inset) using mouse β-actin as a loading control. (b) Average weights of subcutaneous tumors formed by RasGfp (bars 1, 3, 4), RasGfpMyc (bar 2) and RasGfpMycER (bars 5, 6) colonocytes. Where indicated, tumor-bearing animals were treated with 4OHT (‘with 4OHT’) or left untreated (‘w/o 4OHT’). Error bars in a and b refer to s.d. (c) Comparative analysis of RasGfp and RasGfp Myc tumors. Left: hematoxylin and eosin staining (H&E). Perfused blood vessels contain numerous red blood cells. Center: staining of endothelial cells with lectin (brown). Right: staining of lymphatic vessels with an antibody against LYVE-1 (brown). A large vessel in the RasGfpMyc LYVE-1 image localizes in the surrounding adipose tissue. The inset depicts staining of normal ileum.
To determine the contribution of Myc to neoplastic growth, we performed histological examination of size-matched tumors. Most notably, Myc-overexpressing tumors possessed much more robust neovascularization. Especially numerous were large-caliber vessels richly perfused with red blood cells (Fig. 1c). Similar differences emerged when the same sections were stained with lectin to visualize endothelial cells. Whereas RasGfp sections contained only solitary lectin-positive cells, the latter surrounded apparent luminal structures in RasGfpMyc neoplasms (Fig. 1c). In contrast, there was no increase in the density of lymphatic vessels, as judged by staining for the lymphatic-specific LYVE-1 marker (Fig. 1c), in spite of the reported propensity of Myc to promote lymphangiogenesis5. To determine whether the effects of Myc on angiogenesis are mediated by hypoxia, we assessed levels of labile hypoxia-induced factor 1α (HIF1α) in lysates from RasGfp and RasGfpMyc tumors. HIF1α is stable only under hypoxic conditions and was undetectable in either RasGfp or RasGfpMyc tumors (Fig. 2a). To determine whether hypoxia-activated genes are elevated in RasGfpMyc tumors, we performed microarray analysis on mRNAs from RasGfp and RasGfpMyc tumors. There was no upregulation, at the mRNA level, of a variety of known hypoxia-activated genes such as Slc2a1 (also known as Glut1) or Vegfa (data not shown). To investigate possible deregulation of VEGF at the protein level, we performed ELISA on tumor cell lysates. We did not observe any difference in VEGF production between RasGfp and RasGfpMyc neoplasms (Fig. 2b).
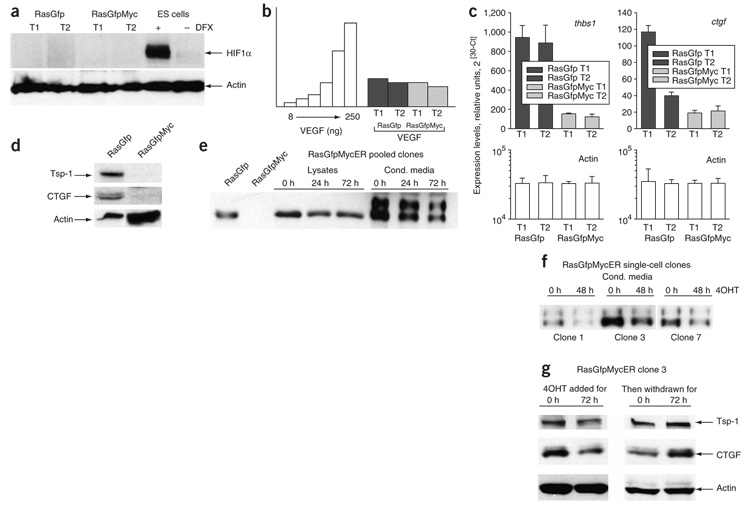
Figure 2. Expression of pro- and anti-angiogenic factors in RasGfp and RasGfpMyc carcinomas.
(a) Lack of detectable HIF1α in RasGfp and RasGfpMyc tumor lysates. Two independent tumors of each type (T1 and T2) were assayed. Mouse ES cells cultured in the presence or absence of hypoxia mimetic desferrioxamine (DFX) were used for comparison. (b) ELISA-based quantification of VEGF A in the same neoplasms. (c) Real-time RT-PCR analysis of thrombospondin-1 (thbs1) and CTGF gene expression in the same neoplasms. Error bars refer to s.d. (d) Immunoblotting analysis of Tsp1 and CTGF expression levels. For CTGF detection, cultured cells were used. (e) Immunoblotting analysis of CTGF expression levels in mass cultures of RasGfpMycER cells treated with 4OHT for indicated number of hours. We analyzed both whole-cell lysates and conditioned medium. RasGfp and RasGfpMyc cells were used for comparison. (f) The same analysis performed on three single-cell RasGfpMycER clones. When we analyzed conditioned medium, we normalized TSR protein levels to cell numbers. (g) Immunoblotting analysis of Tsp1 and CTGF expression levels in RasGfpMycER clone 3. Assayed cells were initially treated with 4OHT for 72 h (left) and then deprived of 4OHT for additional 72 h (right). Trimeric form of Tsp1 was predominantly expressed in these lysates.
Thus, we used microarray data to analyze the effects of Myc on expression of other pro- and anti-angiogenic molecules. We compared the list of differentially expressed genes with the Gene Ontology (GO) database to determine which of 192 known angiogenesis-related genes are subject to regulation by Myc at the mRNA level. No inducers of angiogenesis were significantly upregulated. On the other hand, the list of Myc-downregulated genes included not only thrombospondin-1 but also other proteins with thrombospondin type 1 repeats (TSR): CTGF, spondin-1 (f-spondin), thrombospondin repeat–containing protein 1, clusterin, SPARC, and thrombospondin type I domain–containing protein 6 (Supplementary Table 1 online). The TSR superfamily members SPARC and spondin-1 are known to possess anti-angiogenic properties12, whereas CTGF can either promote or inhibit angiogenesis13, depending on developmental context14.
We validated Tsp1 and CTGF data using real-time quantitative RT-PCR (Fig. 2c) and immunoblotting (Fig. 2d). To confirm that their downmodulation is directly related to the overexpression of Myc, we examined CTGF and Tsp1 protein levels in RasGfpMycER cells. Treatment with 4OHT for 24–72 h resulted in a reduction of CTGF levels in MycER cell lysates and medium conditioned by pooled MycER clones (Fig. 2e). We also confirmed this in several single-cell RasGfpMycER clones with detectable CTGF expression. In all such clones, this protein was downregulated upon 4OHT treatment (Fig. 2f); one clone (#3) was chosen for further analyses. Using this clone, we confirmed that Tsp1 and CTGF are repressed in the presence of activated MycER and return to basal levels upon subsequent removal of 4OHT (Fig. 2g). Although repression was less marked than that observed in RasGfpMyc tumors, it indicated that Tsp1 and CTGF are bona fide Myc effectors.
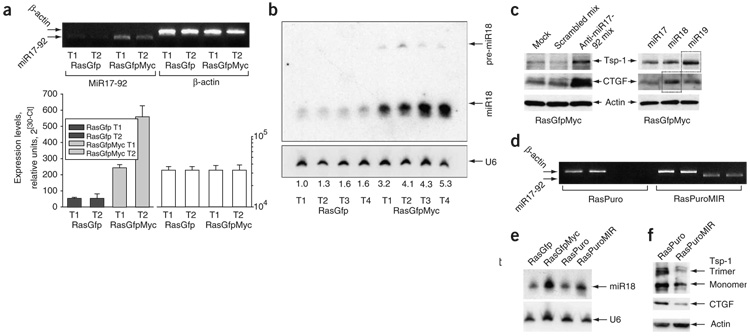
We have demonstrated previously that rather than affecting the thrombospondin-1 promoter, Myc decreases Tsp1 mRNA half-life8. More recently, microRNAs have emerged as important regulators of mRNA stability15, and at least one microRNA cluster (miR-17-92) is directly activated by Myc in human lymphocytes16 and cooperates with Myc during B-lymphomagenesis17. Its role in promoting growth of solid tumors has not been fully elucidated. Using quantitative RT-PCR analysis, we determined that the steady-state levels of its primary transcript are indeed elevated in the presence of overexpressed Myc (Fig. 3a), as are levels of its cleavage products (such as miR-18), as judged by RNA blotting (Fig. 3b). Notably, several members of the TSR superfamily are predicted targets of the miR-17-92 cluster (according to the MiRanda algorithm18) (Supplementary Table 2 online).
Figure 3. miR-17-92 and TSR protein expression in RasGfp and RasGfpMyc cells.
(a) Real-time RT-PCR analysis of the miR-17-92 primary transcript. We tested the same tumors here as in Figure 2. Upper part of panel depicts PCR products quantified in the bar graphs below. Error bars refer to s.d. (b) RNA blot analysis of four RasGfp and four RasGfpMyc tumors. The miR-18 probe detects both pre-miR-18 and mature miR-18 species. U6 RNA was used as a loading control. Numbers below the autoradiogram refer to the increase in miR-18 levels as a multiple of that in RasGfp T1. (c) Immunoblotting analysis of Tsp1 and CTGF expression levels in RasGfpMyc cells transfected with antisense 2′-O-methyl oligoribonucleotides targeting components of the miR-17-92 cluster. Left: cells transfected with mixtures of scrambled or miR-17-92-specific oligoribonucleotides. Mock-transfected cells were used as an additional control. Right: the same cells transfected with oligoribonucleotides targeting individual microRNAs. (d) RT-PCR performed on Ras-only colonocytes transduced with either empty vector (RasPuro) or the miR-17-92–encoding retrovirus (RasPuroMIR). PCR primers were specific for the human miR-17-92 pre-miRNA and did not detect the endogenous mouse transcript. (e) RNA blot analysis of the same cells. RasGfp and RasGfpMyc cells were used for comparison. Other designations are as in b. (f) Immunoblotting analysis of Tsp1 and CTGF expression levels in RasPuro versus RasPuroMIR cells.
We sought to determine whether Myc-induced upregulation of the miR-17-92 cluster is directly responsible for the downregulation of TSR proteins in RasGfpMyc cells. As microRNA function can be inhibited with specific 2′-O-methyl oligoribonucleotides19, we examined TSR protein expression in transiently transfected RasGfpMyc cells. Using a mixture of antisense oligoribonucleotides targeting six microRNAs from the miR-17-92 cluster, we partly restored expression of Tsp1 and CTGF in RasGfpMyc cells (Fig. 3c, left). Transfection of antisense oligonucleotides to individual microRNAs further suggested that within the cluster, miR-19 is primarily responsible for Tsp1 downregulation and miR-18 for CTGF downregulation in response to Myc (Fig. 3c). Antisense oligonucleotides to miR-17 (Fig. 3c), miR-20 and miR-92 (data not shown) did not affect TSR protein levels, consistent with bioinformatic predictions (Supplementary Table 2). We also generated, using retrovirus transduction, Ras cells overexpressing the human miR-17-92 cluster (‘RasPuroMIR’, Fig. 3d), which upon cleavage yields microRNAs that are identical to their mouse counterparts. The level of their overexpression was physiological; that is, comparable to that attained in RasGfpMyc cells (Fig. 3e). As predicted, RasPuroMIR cells produced lower levels of thrombospondin- 1 and especially CTGF, as compared with vector-transduced RasPuro cells (Fig. 3f). In the case of CTGF, we also observed a 90% reduction in mRNA levels, indicative of regulation at the level of mRNA turnover (data not shown).
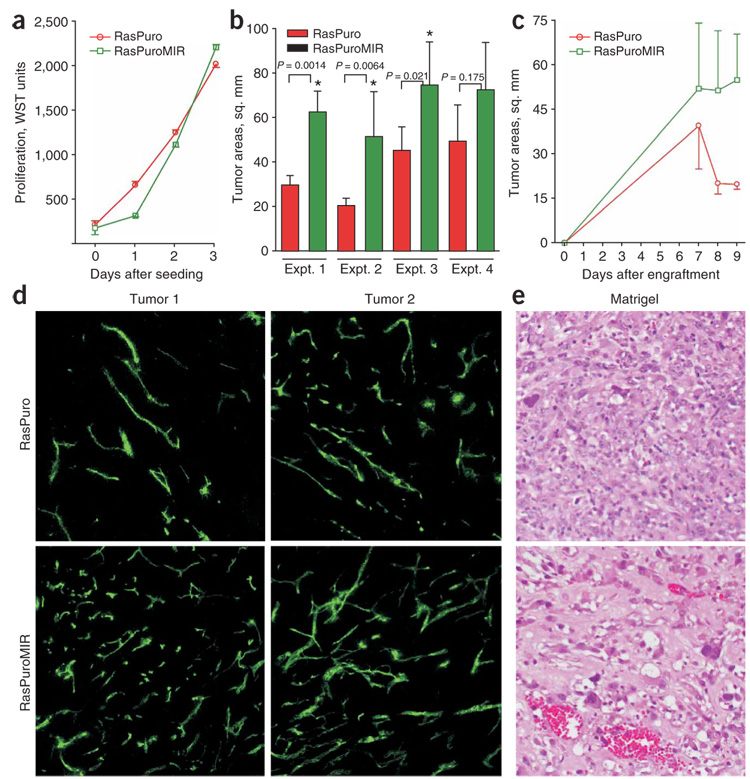
We then asked whether overexpression of miR-17-92 could partly recapitulate Myc-induced phenotypes and confer non–cell-autonomous advantages to RasPuroMIR cells. Using the water-soluble tetrazolium- 1 (WST) assay, we determined that in vitro RasPuro and RasPuroMIR cells grow at a similar rate (Fig. 4a). However, when we implanted cells into C57BL6/NCr mice, RasPuroMIR cells formed tumors that were on average 1.6–2.5 times larger than RasPuro tumors (data from four independent experiments, Fig. 4b). By monitoring tumor kinetics, we determined that the two sets of neoplasms initially grew at similar rates but diverged when they were several millimeters in diameter, a recognized threshold for angiogenic tumors20. At that time point, only RasPuroMIR cells were capable of progressive growth, whereas RasPuro tumors stagnated or even slightly regressed (Fig. 4c, data from experiment 2 in Fig. 4b; similar observations were made in experiments 1, 3 and 4). To examine the effects of miR-17-92 overexpression on tumor vasculature, we injected tumor-bearing mice intravenously with FITC-conjugated lectin and killed them 20 min later. In numerous sections examined using confocal microscopy, RasPuroMIR tumors exhibited a higher density of perfused vessels (Fig. 4d). Moreover, when we embedded the same cells in Matrigel and injected them subcutaneously into syngeneic hosts, RasPuroMIR cells promoted more vigorous neovascularization, as judged by hemoglobin assay (data not shown). In particular, only RasPuroMIR implants contained large-caliber vascular channels reminiscent of RasGfpMyc tumors (Fig. 4e).
Figure 4. The effects of miR-17-92 upregulation in Ras-only cells on neoplastic growth.
(a) Cell accumulation assay performed on RasPuro and RasPuroMIR colonocytes. Numbers of viable cells were assessed using the WST reagent as in Figure 1a. (b) Average sizes of subcutaneous tumors formed by RasPuro and RasPuroMIR colonocytes in syngeneic mice. * indicates statistical significance (P < 0.05). P values were determined using unpaired Student’s t-test. (c) Kinetics of tumor formation by RasPuro and RasPuroMIR colonocytes from experiment 2 in b. Error bars in a–c represent s.d. (d) Blood perfusion of RasPuro and RasPuroMIR tumors. Green staining corresponds to FITC-conjugated lectin bound to vascular endothelial cells after intravenous injection. Two independent tumors were assayed. Representative 10× sections from each neoplasm are shown. (e) Matrigel neovascularization induced by the same cells. The richly perfused, large-caliber vascular channels in the lower image are typical of RasPuroMIR samples.
Although Myc clearly contributes to angiogenesis in one-hit model neoplasms1–4, its involvement in angiogenesis is uncertain in the case of tumors (for example, colon carcinomas) in which Myc is coactivated with Ras and in which mutations in the TP53 tumor suppressor gene are common. Both activation of Ras and inactivation of p53 are considered pro-angiogenic. Besides being the repressors of thrombospondin-1, H-Ras and K-Ras are known to upregulate VEGF and increase the activity of matrix metalloproteinases (MMP) required for endothelial cell migration (reviewed in ref. 21). Notably, the ability of Ras to promote angiogenesis has been documented in a transgenic tumor setting22. The loss of p53 may result in improved stability of hypoxia-induced factor alpha (HIF1α)23, as well as upregulation of VEGF and downregulation of Tsp1 (ref. 24).We were surprised to find that in our colon cancer model, a combination of mutations in Kras and Trp53 mutations yielded indolent, poorly vascularized tumors. It was not before Myc overexpression and a further decrease in TSR protein levels that robust tumor vasculature developed, greatly boosting overall neoplastic growth. Profound downregulation of Tsp1 and CTGF might stem from both acute and delayed effects of Myc. In short-term experiments with MycER-transduced cells, in which only acute effects are assessed, Tsp1 and CTGF were downregulated 65%–80%. Approximately the same level of repression was apparent in miR-17-92–transduced Ras-cells. Thus, activation of the miR-17-92 cluster can fully account for the acute effects of Myc on TSR protein expression. Additional delayed effects could stem from the propensity of Myc to activate certain metalloproteinases25 and thus indirectly affect extracellular proteins.
Recently, several microRNAs have been ascribed important roles in cancerous growth, but thus far they were implicated exclusively in cell-autonomous processes such as proliferation and death. For example, miR-15 and miR-16 are frequently deleted or downregulated in chronic lymphocytic leukemia, owing to their ability to promote apoptosis, and let-7 loss causes upregulation of Ras and ensuing increase in cell proliferation (reviewed in ref. 26). miR-17-92 is upregulated in many types of human cancers, including colon carcinomas27, but its contribution to neoplastic growth is just beginning to emerge. It is implicated in enhanced cell cycle progression28 and, through targeting of E2F1, in a block to tumor cell apoptosis16. Our data demonstrate that miR-17-92 also affects non–cell-autonomous processes such as tumor neovascularization. Thus, antisense-based microRNA targeting, an emerging therapeutic technology19,29, might be effective even against apoptosis-resistant tumors. Admittedly, activation of the miR-17-92 pathway may not be the sole pro-angiogenic event triggered by Myc. However, even partial restoration of TSR expression could tip the balance between pro- and anti-angiogenic factors in favor of the latter. This shift might afford significant therapeutic benefits in colon carcinomas, which are known to respond well to anti-angiogenic therapies30.
METHODS
Cell lines and tumor production
p53-null colonocytes transformed with retroviruses encoding K-Ras, Myc and MycER have been described previously10,11. To obtain doubly transduced cells, MigR1 retroviral vectors encoding either Myc or the Myc-estrogen receptor fusion were transfected into GP293 cells using Lipofectamine 2000 (Invitrogen) along with plasmids encoding viral proteins: gag-pol (pGP) and VSV-G protein from vesicular stomatitis virus. Viral supernatants were harvested 48–72 h later and added to recipient cells. Polybrene was added to cells to facilitate infection. GFP-positive cells were obtained using FACS.
The mouse stem cell virus–based vector (MSCV) encoding miR-17-92 has been described previously16. MSCV-miR-17-92–transduced cells were obtained using puromycin selection. C57BL6/NCr mice were obtained from the US National Cancer Institute. Transformed colonocytes were implanted either subcutaneously or orthotopically, into the wall of the cecum. For 4-hydroxytamoxifen (4-OHT) treatment, the hormone powder (Sigma) was dispersed, via sonication, in corn oil (Sigma) at the concentration of 10 mg/ml. We administered 4-OHT daily by intraperitoneal injection at the dose of 1 mg per mouse. Cultured cells expressing MycER were exposed to 250 nM 4-OHT dissolved in ethanol. All mouse experiments were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Antisense inhibition of miR-17-92 cluster
2′-O-methyl oligoribonucleotides were synthesized by Integrated DNA Technologies (see sequences in Supplementary Table 3 online). For the analysis of CTGF and Tsp1 protein levels, a mixture of 2′-O-methyl oligoribonucleotides (100 pmol each) targeting individual members of the cluster or 600 pmol of the scrambled oligoribonucleotide were transfected into RasGfpMyc colonocytes growing in six-well dishes (plated at 200,000 cells per well 24 h before transfection) using Lipofectamine 2000. We confirmed transfection efficiency (>95%) using BLOCK-iT Fluorescent Oligo (Invitrogen). Protein lysates were collected 48 h after transfection and analyzed by immunoblotting.
Analyses of tumor specimens and blood vasculature
Tumor sizes were measured using calipers and tumor weights were recorded on the day of tumor excision. Levels of VEGF in protein extracts were determined by ELISA (R&D Systems), using a purified VEGF standard. Alternatively, tissues were fixed in formalin, embedded in paraffin, sectioned and subjected to histological staining or immunohistochemistry. Vascular endothelial cells were stained using the Bandeiraea simplicifolia lectin (BS-1; Sigma). Lymphatics were visualized using a 1:500 dilution of rabbit polyclonal antibody to LYVE-1 (Research Diagnostics), a biotinylated secondary antibody, and avidin-linked peroxidase. Images were captured at 20× magnification using a Nikon Eclipse E600 microscope and a Photometrix CoolSnap camera.
To detect perfused blood vessels, tumor-bearing mice were injected intravenously with 100 µl of 2 mg/ml FITC-conjugated Lycopersicon esculentum lectin (Vector Labs) 20 min before being killed. After excision, tumors were embedded in OCT (Fisher Scientific), frozen in liquid nitrogen, and sectioned into thick (50-µm) sections using a cryostat. Slides were examined using an upright Zeiss Axiovert 200M microscope equipped with a Zeiss LSM510 Vis/UV META confocal system. FITC fluorescence was detected by a 30 mW argon laser system. Images were viewed through a 10× objective, and serial images were acquired at 2-µm intervals using LSM510 META V3.2 software. Images were integrated to create a composite projection of a vessel in three dimensions. The in vivo Matrigel neovascularization assay has been described in detail earlier3.
Microarray and microRNA target analyses
We used total RNA from RasGfp and RasGfpMyc tumors. cDNAs were synthesized using in vitro transcription with biotinylated CTP and UTP. Labeled cDNAs were hybridized to the Mouse Genome 430 2.0 Array chip (Affymetrix) using the University of Pennsylvania Microarray Facility standard protocol (http://www.med.upenn.edu/microarr/Data%20Analysis/Affymetrix/methods.htm). We calculated Affymetrix MAS5 probe set signals and presence/absence flags. The LPE (Local Pooled Error) test for differential expression as implemented in S+ArrayAnalyzer v 1.1 (Insightful Corporation) was applied with 1% Bonferroni multiple testing correction to median interquartile range-normalized MAS 5 signal values. The resulting lists were imported into GeneSpring v 6.1 (Silicon Genetics), filtered for Presence (per Affymetrix MAS5 analysis) in two of two samples in one or more conditions (RasGfp or RasGfpMyc) and then filtered for a change of at least 60%. The comparison with the Gene Ontology database lists was carried out using DAVID 2.0 software, as implemented at http://apps1.niaid.nih.gov/david/. Putative miR-17-92 targets were identified using the MiRanda algorithm18, as implemented at http://www.microrna.org and http://microrna.sanger.ac.uk.
Protein and RNA blotting
For thrombospondin-1 and CTGF expression analysis, we used either cell lysates or conditioned medium. Myc and HIF1α expression was detected in cell lysates. Membranes were probed with antibodies to Tsp1 (Ab-11, Lab Vision), CTGF (L-20, Santa Cruz Biotechnology), Myc (N-262/sc-764, Santa Cruz Biotechnology) and HIF1α (NB 100-105; Novus Biologicals) diluted according to manufacturers’ recommendations. Conditioned medium was loaded on PAGE without dilution. Appropriate secondary antibodies were used in horseradish peroxidase–conjugated forms (Amersham Biosciences). Antibody binding was detected using the enhanced chemiluminescence system (Amersham). When indicated, we used a monoclonal antibody reactive with mouse β-actin (Sigma) to confirm equal loading. Detection of mouse miR-17-92 RNAs using RNA blotting was performed as described in ref. 16.
Real-time PCR
Total RNAs were isolated using TRI Reagent (Sigma) and treated with a TURBO DNA-free kit (Ambion). cDNAs were prepared from 2 µg RNA using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). The nucleotide sequences of primers used are provided in Supplementary Table 3. Amplifications were performed using Smartcycler (Cepheid). Typical conditions were as follows: 95 °C for 150 s (one cycle), then 95 °C for 10 s and 60 °C for 30 s (30 cycles). All reactions were performed in duplicates or triplicates to ensure accuracy of quantification.
Supplementary Material
Note: Supplementary information is available on the Nature Genetics website.
ACKNOWLEDGMENTS
We are indebted to C. Dang (Johns Hopkins University) for fostering collaborations between the investigators. We are grateful to C. Simon and C.-J. Hu (University of Pennsylvania) for providing HIF1α-overexpressing cell lysates for protein blot analysis and J. Tobias (University of Pennsylvania) for his help with microarray data analysis. We thank W. El-Deiry, A. Rustgi, and S. Fuchs (University of Pennsylvania) for stimulating discussions and comments on the manuscript. M. Goldschmidt and J. Burns (University of Pennsylvania) are acknowledged for their help with histopathological analyses. This work was supported by US National Institutes of Health grants CA 097932 and DK 050306 and a University of Pennsylvania Research Foundation grant to A.T.-T.
Footnotes
Accession codes. Microarray data described herein have been deposited to the ArrayExpress (http://www.ebi.ac.uk/arrayexpress) database under accession number E-MEXP-757.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Pelengaris S, Littlewood T, Khan M, Elia G, Evan GI. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol. Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 2.Brandvold KA, Neiman P, Ruddell A. Angiogenesis is an early event in the generation of myc-induced lymphomas. Oncogene. 2000;19:2780–2785. doi: 10.1038/sj.onc.1203589. [DOI] [PubMed] [Google Scholar]
- 3.Ngo C, et al. An in vivo function for the transforming myc protein: elicitation of the angiogenic phenotype. Cell Growth Differ. 2000;11:201–210. [PMC free article] [PubMed] [Google Scholar]
- 4.Baudino TA, et al. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 2002;16:2530–2543. doi: 10.1101/gad.1024602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruddell A, Mezquita P, Brandvold KA, Farr A, Iritani BM. B lymphocyte-specific c-Myc expression stimulates early and functional expansion of the vasculature and lymphatics during lymphomagenesis. Am. J. Pathol. 2003;163:2233–2245. doi: 10.1016/S0002-9440(10)63581-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knies-Bamforth UE, Fox SB, Poulsom R, Evan GI, Harris AL. c-Myc interacts with hypoxia to induce angiogenesis in vivo by a vascular endothelial growth factor-dependent mechanism. Cancer Res. 2004;64:6563–6570. doi: 10.1158/0008-5472.CAN-03-3176. [DOI] [PubMed] [Google Scholar]
- 7.Tikhonenko AT, Black DJ, Linial ML. Viral Myc oncoproteins in infected fibroblasts down-modulate thrombospondin-1, a possible tumor suppressor gene. J. Biol. Chem. 1996;271:30741–30747. doi: 10.1074/jbc.271.48.30741. [DOI] [PubMed] [Google Scholar]
- 8.Janz A, Sevignani C, Kenyon K, Ngo C, Thomas-Tikhonenko A. Activation of the Myc oncoprotein leads to increased turnover of thrombospondin-1 mRNA. Nucleic Acids Res. 2000;28:2268–2275. doi: 10.1093/nar/28.11.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watnick RS, Cheng YN, Rangarajan A, Ince TA, Weinberg RA. Ras modulates Myc activity to repress thrombospondin-1 expression and increase tumor angiogenesis. Cancer Cell. 2003;3:219–231. doi: 10.1016/s1535-6108(03)00030-8. [DOI] [PubMed] [Google Scholar]
- 10.Sevignani C, et al. Tumorigenic conversion of p53-deficient colon epithelial cells by an activated Ki-Ras gene. J. Clin. Invest. 1998;101:1572–1580. doi: 10.1172/JCI919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas-Tikhonenko A, et al. Myc-transformed epithelial cells down-regulate clusterin which inhibits their growth in vitro and carcinogenesis in vivo. Cancer Res. 2004;64:3126–3136. doi: 10.1158/0008-5472.can-03-1953. [DOI] [PubMed] [Google Scholar]
- 12.Tucker RP. The thrombospondin type 1 repeat superfamily. Int. J. Biochem. Cell Biol. 2004;36:969–974. doi: 10.1016/j.biocel.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 13.Inoki I, et al. Connective tissue growth factor binds vascular endothelial growth factor (VEGF) and inhibits VEGF-induced angiogenesis. FASEB J. 2002;16:219–221. doi: 10.1096/fj.01-0332fje. [DOI] [PubMed] [Google Scholar]
- 14.Perbal B. CCN proteins: multifunctional signalling regulators. Lancet. 2004;363:62–64. doi: 10.1016/S0140-6736(03)15172-0. [DOI] [PubMed] [Google Scholar]
- 15.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 17.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.John B, et al. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerbel RS, Folkman J. Clinical translation of angiogenesis inhibitors. Nat. Rev. Cancer. 2002;2:727–739. doi: 10.1038/nrc905. [DOI] [PubMed] [Google Scholar]
- 21.Rak J, Yu JL, Kerbel RS, Coomber BL. What do oncogenic mutations have to do with angiogenesis/vascular dependence of tumors? Cancer Res. 2002;62:1931–1934. [PubMed] [Google Scholar]
- 22.Chin L, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 23.Ravi R, et al. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1 alpha. Genes Dev. 2000;14:34–44. [PMC free article] [PubMed] [Google Scholar]
- 24.Dameron KM, Volpert OV, Tainsky MA, Bouck N. Control of angiogenesis in fibroblasts by p53 regulation of thrombospondin-1. Science. 1994;265:1582–1584. doi: 10.1126/science.7521539. [DOI] [PubMed] [Google Scholar]
- 25.Himelstein BP, Lee EJ, Sato H, Seiki M, Muschel RJ. Transcriptional activation of the matrix metalloproteinase-9 gene in an H-ras and v-myc transformed rat embryo cell line. Oncogene. 1997;14:1995–1998. doi: 10.1038/sj.onc.1201012. [DOI] [PubMed] [Google Scholar]
- 26.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 27.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayashita Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 29.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 30.Hurwitz H, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N. Engl. J. Med. 2004;350:2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Genetics website.