Abstract
The Est3 protein is a small regulatory subunit of yeast telomerase which is dispensable for enzyme catalysis but essential for telomere replication in vivo. Using structure prediction combined with in vivo characterization, we show here that Est3 consists of a predicted OB (oligo-saccharide/oligo-nucleotide binding) fold. Mutagenesis of predicted surface residues was used to generate a functional map of one surface of Est3, which identified a site that mediates association with the telomerase complex. Surprisingly, the predicted OB-fold of Est3 is structurally similar to the OB-fold of the mammalian TPP1 protein, despite the fact that Est3 and TPP1, as components of telomerase and a telomere capping complex, respectively, perform functionally distinct tasks at chromosome ends. The analysis performed on Est3 may be instructive in generating comparable missense mutations on the surface of the OB-fold domain of TPP1.
The enzyme telomerase elongates chromosome termini in most species. In human tissues, telomerase activity can be highly regulated, both at the level of expression of subunits of the enzyme, as well as by regulatory factors. Even modest alterations in enzyme activity are associated with several human disease syndromes1, highlighting the importance of understanding the mechanistic basis for how telomerase is regulated. In budding yeast, the telomerase enzyme is composed of three proteins, Est1, Est2 and Est3, in a complex with the 1.3 kb TLC1 RNA2–4, which provides a flexible scaffold on which telomerase assembles5. Est2 and TLC1 comprise the catalytic core of the enzyme, while the Est1 and Est3 subunits are regulatory proteins, as evidenced by the dramatically differential effects on telomerase function displayed by est1-Δ and est3-Δ strains, as assessed by in vivo versus in vitro assays. For example, est1-Δ strains exhibit an in vivo telomere replication defect that is indistinguishable from that of strains defective for the catalytic core of the enzyme6, even though the Est1 protein is dispensable for in vitro catalysis by telomerase from both S. cerevisiae and S. pombe7–10. The overall architecture of the telomerase RNP appears to be surprisingly well conserved between the evolutionarily distant budding and fission yeasts10–13, despite the fact that individual subunits have evolved rapidly at the primary sequence level.
A number of studies have established that the primary regulatory role for the budding yeast Est1 protein is to recruit telomerase to its site of action, through an electrostatic interaction with the single-strand telomere DNA binding protein Cdc13. Elucidation of this recruitment activity has relied on analysis of strains bearing separation-of-function mutations in CDC13 and EST1 (cdc13-2 and est1-60, respectively), which has established that telomere maintenance relies on a direct association between the telomere-bound Cdc13 protein and the telomerase regulatory subunit Est114,15. In fact, the catalytic Est2 subunit can be re-localized to non-telomeric locations in the genome, including a site that is not adjacent to a DNA terminus, through a process that is dependent on the Cdc13-Est1 interaction16. Furthermore, Est1 and Est2, but not Cdc13, exhibit preferential association with shorter telomeres17, consistent with the demonstration that telomere elongation is preferentially targeted to the shortest telomeres in a population18. These studies have established that the function of the Est1 and Cdc13 proteins that is impaired by the cdc13-2 and est1-60 mutations is recruitment of the catalytic core of telomerase to short telomeres (rather than to activate the enzyme, as alternatively proposed19).
In contrast to Est1, the Est3 telomerase subunit has been much less well-studied. Like Est1, the Est3 protein also performs a regulatory role, as it is also crucial for telomere replication in vivo but not for catalysis in vitro8. However, the molecular mechanism(s) underlying the Est3-specific regulatory function has not yet been elucidated. Even how Est3 physically associates with the telomerase complex has not been established: conflicting reports have suggested that Est3 interacts with telomerase through an interaction with Est24,20 (JSL and VL, unpublished observations) or Est121,22.
Using a panel of structure prediction programs, we report here that the Est3 protein consists of an OB (oligo-saccharide/oligo-nucleotide binding)-fold, and we have used this structural prediction to construct a functional map of one surface of the Est3 protein. Unexpectedly, the predicted OB-fold of Est3 exhibits structural similarity to an OB-fold domain in the mammalian TPP1 protein, as well as weaker similarity to that of the O. nova TEBP β protein. These two proteins are subunits of a telomere end-binding complex, called POT1/TPP1 in most species and TEBP α-β in the ciliate O. nova23–28. Both genetic and biochemical data argue that the POT1/TPP1 complex functions in role(s) distinct from that of telomerase, by protecting chromosome ends from events that would otherwise be lethal for cells25,29–34. The functional differences between Est3 and TPP1 raise intriguing questions about the potential relationship(s) between structurally similar OB-fold domains present in complexes that perform different tasks at chromosome termini.
RESULTS
The Est3 telomerase protein contains a predicted OB-fold domain
As a first step towards identifying structural homologs of the Est3 protein, we constructed an alignment of 20 Est3 protein sequences, displayed in Fig. 1a. This alignment revealed a very low degree of sequence conservation; for example, the experimentally verified Est3 proteins from S. cerevisiae35 and Candida albicans36 exhibited only 18% identity and 15% similarity. The rapid divergence at the primary sequence level presumably explains why detection of Est3 homologs from more distant species has not yet been successful.
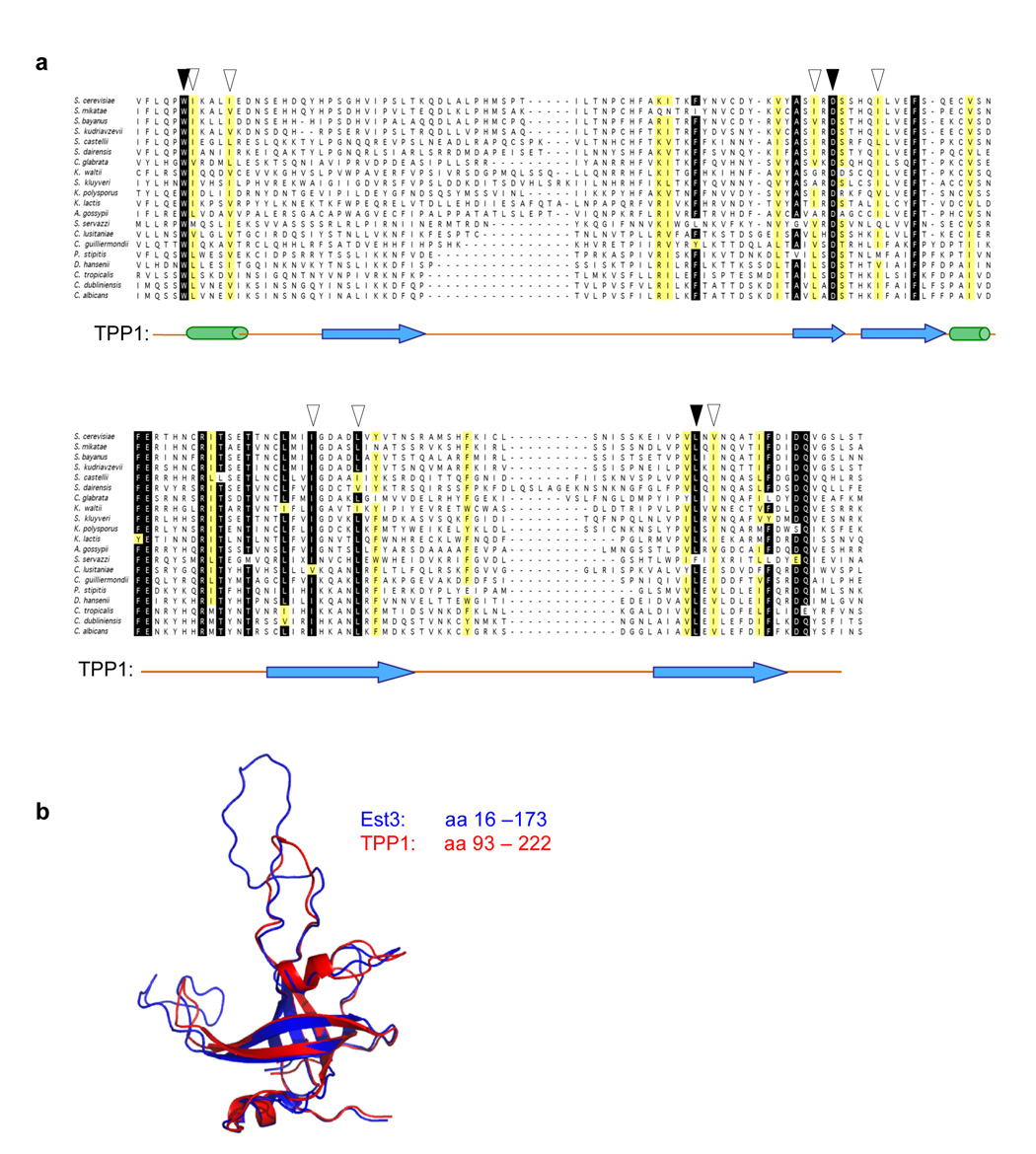
Figure 1.
Est3 is an OB-fold-containing protein with structural similarity to the OB-fold of TPP1. (a) Alignment of the proposed OB-fold domain of the Est3 protein; aa 16 to 173 of the 181 amino acid S. cerevisiae protein is shown. Black arrowheads indicate 3 invariant, or nearly invariant, residues that are common to the Est3 and TPP1 protein families, and white arrowheads indicate an additional 7 amino acids that appear to be conserved between the two proteins (see Supplementary Fig. 1 for an alignment of TPP1). The position of TPP1 secondary structure assignments38 are indicated (α helices and β strands, as cylinders and arrows, respectively), based on the structural comparison between Est3 and TPP1 shown in Fig. 1b; the C-terminal α helix (αC) present in TPP1 could not be detected in Est3. (b) A ribbon representation of the predicted structure of Est3 (aa 16 to 173, indicated in blue), overlaid with the structure of the OB-fold domain of TPP1 (aa 93 to 222, indicated in red).
This alignment was subsequently used in a search of hidden Markov model (HMM) profiles for potential structural homologs in the Protein Data Bank, using the HHpred structure-prediction server37. The top-ranked hit was the OB-fold domain of the human TPP1 protein38, with an E-value of 0.054 (P-value of 3.3 × 10−6) and a probability score of 93.8. Subsequent submission of the S. cerevisiae Est3 sequence to the structure-prediction servers SAM-T0639 and FUGUE40 similarly identified TPP1 as the top-ranked hit, with an E-value of 1.2 × 10−2 and a confidence value above 95%, respectively; SAM-T06 also identified the OB-fold of the O. nova TEPB β protein41, with a score of 8 × 10−1. The S. cerevisiae Est3 protein was also submitted to the I-TASSER server, the highest scoring server at the CASP7 structure prediction competition42. Unlike the above three structure prediction programs, I-TASSER does not rely on global profile-profile searches and instead combines a fragmented structure prediction algorithm with fragment reassembly and ab initio folding of non-aligned regions. The two models for Est3 with the best confidence scores from the I-TASSER submission could be structurally aligned with the OB-fold of TPP1 using DALILITE, with Z-scores of 14.6 and 8.6 respectively (for comparison, TPP1 aligns with the O. nova TEBP β protein with a Z-score of 10.0).
Collectively, the above observations argue that the Est3 subunit of telomerase contains an OB-fold that is structurally similar to that of TPP1. We therefore constructed a 3-dimensional structural model of the S. cerevisiae Est3 protein, based on the HHpred profile-profile comparison. Fig. 1b shows a ribbon representation of the predicted structure of Est3, overlaid with the structure of the OB-fold domain of TPP1. Supplementary Fig. 1 demonstrates the relative position of this domain in Est3 and TPP1: the small Est3 protein consists of just the OB-fold, whereas TPP1 is a larger multi-domain protein.
Based on this structural prediction, an alignment of 16 TPP1 protein sequences was also constructed (Supplementary Fig. 1). TPP1 is similarly very divergent at the primary sequence level, and like Est3, TPP1 cannot be detected in a wide range of eukaryotic species. Due to the limited degree of conservation between these two protein families, it was not possible to construct an alignment composed of both TPP1 and Est3 sequences with high statistical confidence. Consistent with this, a comparison of these two independently constructed alignments revealed a limited number of residues that were conserved across both protein families. Only 3 residues, which were invariant or close to invariant, were common to both alignments (Trp21/Trp98, Asp86/Asp148, and Leu155/Leu204, in Est3 versus TPP1, respectively). Structural alignment of TPP1 and the HHpred-derived model of Est3 demonstrated that these residues share a common structural space (data not shown). An additional 7 amino acid positions, which were primarily hydrophobic, also appeared to be structurally conserved between the two protein families. These 10 residues are indicated on the alignments in Fig. 1a and Supplementary Fig. 1.
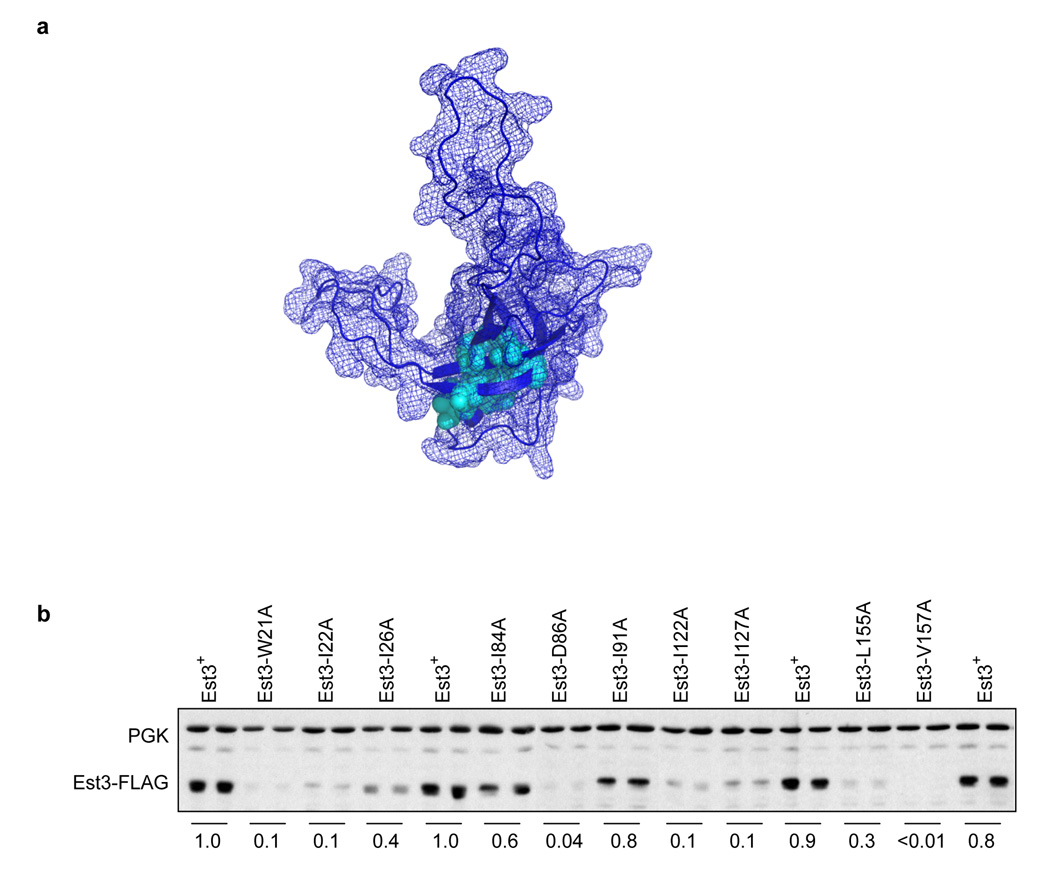
Notably, the 10 amino acids that are in common between Est3 and TPP1 are clustered in the interior of the core of the OB-fold, in both Est3 (Fig. 2a) and TPP138, as calculated by solvent accessibility (data not shown). The proposed internal position of these 10 residues in the Est3 protein suggest that they make critical contributions to protein folding, which predicts that mutations in these residues should destabilize the protein. In agreement with this prediction, mutations in 8 of these 10 internal amino acids to alanine had a dramatic effect on steady-state protein levels, as assessed by western analysis of wild type and mutant Est3-(FLAG)3 derivatives (Fig. 2b), consistent with a disruption in protein folding. This analysis therefore suggests that the limited number of amino acids that are reasonably well conserved between Est3 and TPP1 are primarily involved in protein folding and/or stability of the OB-fold.
Figure 2.
Residues that are conserved between Est3 and TPP1 are located in the core of the OB-fold. (a) Residues common to Est3 and TPP1 (indicated by arrowheads in the alignments shown in Fig. 1a and Supplementary Fig. 1) are depicted in cyan on a wire mesh diagram representing the surface area of the model for Est3, overlaid on a ribbon representation of the OB-fold. (b) Steady-state protein levels of wild type and mutant Est3-(FLAG)3 proteins were assessed by western analysis, by simultaneously probing with anti-FLAG and anti-PGK antibodies. For each mutant construct, the Est3-(FLAG)3 protein signal was quantitated, relative to that of the PGK signal, and normalized relative to wild type Est3-(FLAG)3 protein. The average of two samples prepared in parallel is shown.
Mapping the surface of the Est3 protein by mutagenesis
Since mutations in internal residues are not, in most cases, useful for determining specific function(s) of a protein, we directed our attention to those amino acids that were likely to be on the surface of the Est3 protein, in order to identify a set of missense mutations that impaired Est3 activity due to loss of one or more biochemical property(s) of Est3. Missense mutations were introduced into 29 amino acids that were likely to be on the surface; residues were mutated to alanine and in some cases, to polar and/or charged residues. From an initial in vivo analysis of the effects of this larger set of mutations (data not shown), mutations in 10 amino acids were selected for a more detailed characterization, in parallel with mutations in two highly conserved internal residues (Trp21 and Asp86, Fig. 2).
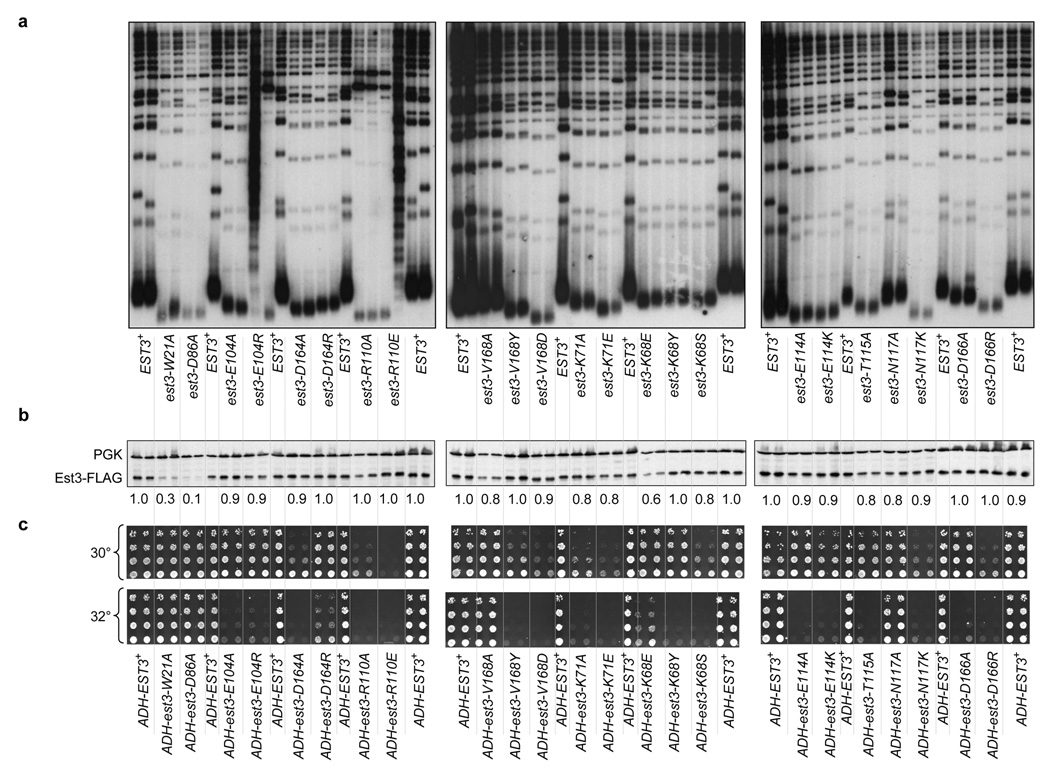
We first examined the impact of these mutations on Est3 function in vivo, using a loss-of-function assay that assessed telomere length (Fig. 3a) and growth phenotypes (Supplementary Fig. 2) at several time points during serial propagation of mutant strains. A subset of mutant alleles exhibited telomere replication defects comparable to that of the est3-Δ null strain: telomeres were extremely short and the strains eventually exhibited the senescence phenotype that is a hallmark of a strain bearing a null mutation in telomerase6. These severely defective alleles included mutations in one of the predicted internal alleles (Asp86) as well two highly conserved potential surface residues (Arg110 and Glu104). Strains expressing mutations in the remaining 8 residues displayed a telomere replication defect that was only slightly less pronounced, with telomere lengths that were 150 to 200 bp shorter (for a subset of the mutations in Trp21, Asn117, Asp166 and Val168) or 100 to 150 bp shorter (for mutations in Lys68, Lys71, Glu114, Thr115 and Asp164) than that of the wild type EST3 strain.
Figure 3.
Detailed phenotypic analysis of mutations in predicted surface residues of Est3. (a) Telomere length analysis: an est3-Δ strain was transformed with single-copy plasmids expressing either a mutant allele or the wild type EST3 gene and propagated for ~80 cell divisions and examined for telomere length, as described previously. The senescence phenotype displayed by a subset of these mutant strains is shown in Supplementary Fig. 2. (b) Steady state levels of wild type and mutant derivatives of Est3-(FLAG)3 proteins were determined as described in Fig. 2c. (c) Dominant negative phenotypes of est3− mutant alleles: high copy (2 µ) plasmids expressing wild type and mutant alleles of EST3, under control of the ADH promoter, were transformed into a yku80-Δ/p YKU80 URA3 strain and subsequently examined for growth by plating 5-fold serial dilutions of equivalent numbers of cells on media that that selects for loss of the YKU80 plasmid. Growth of the yku80-Δ/pADH-est3− strains was examined at both 30° and 32° C, based on our prior observations showing that synthetic growth defects displayed by yku80-Δ strains are enhanced by even a modest increase in temperature (data not shown).
Consistent with the predicted location of these residues on the surface of the Est3 protein, mutations in these 10 residues had little or no effect on protein stability, as monitored by western analysis (Fig. 3b). However, this relatively crude assessment of protein stability cannot rule out more subtle defects in protein folding. We therefore subjected these mutations to an alternative test of protein stability, by examining whether each of these mutant Est3 proteins, when expressed at high levels in an EST3 strain, would be capable of disrupting telomere replication. Such dominant negative phenotypes are often used to infer that the mutant protein in question is functional enough (i.e. properly folded) to perform a subset of the activities of the wild type protein, and hence interfere with the function of the wild type protein. Two assays were used to assess potential dominant negative phenotypes: synthetic lethality in a EST3 yku80-Δ strain (Fig. 3c) and telomere length in an EST3 YKU80 strain (Supplementary Fig. 3). The synthetic lethality assay is based on prior observations demonstrating that a yku80-Δ strain is highly sensitized to defects in telomerase, resulting in severe growth defects or even lethality (43 and data not shown). To assess the validity of this assay, we examined mutations in two internal residues: over-expression of the Est3-W21A and Est3-D86A proteins failed to confer a synthetic lethal phenotype in the EST3 yku80-Δ strain (Fig. 3c), consistent with the fact that both of these mutations reduced protein stability (Fig. 3b). In contrast, most of the mutations in the 10 proposed surface residues conferred some degree of synthetic lethality in the yku80-Δ strain, ranging from very severe to more modest effects on growth (Fig. 3c).
The results from this assay also were supported by a more direct examination of telomere length in an EST3 YKU80 strain, when either wild type or mutant Est3 proteins were over-expressed. Increased expression of the wild type Est3 protein had a very modest effect on telomere length, with a telomere length decline of ≤ 50 bp; in contrast, over-expression of the Est3-W21A mutant protein had no effect on telomere length, further confirming that the W21A mutation creates an unfolded protein (Supplementary Fig. 3). The effects on telomere length in the EST3 YKU80 strain correlated extremely well with the synthetic lethal assay in the EST3 yku80-Δ strain. For example, over-expression of the Est3-D164A mutant protein resulted in both substantial telomere shortening and a severe synthetic growth defect, whereas over-expression of the Est3-D164R protein displayed virtually no dominant negative effects in either assay (Fig. 3c and Supplementary Fig. 3). A stringent interpretation of these results is that the D164A mutation impairs a specific function of Est3, whereas the D164R mutation creates a mutant protein that is at least partially unfolded and thus unable to confer a dominant negative phenotype when over-expressed.
The data in Fig. 3a and Fig. 3c indicated that this panel of est3− mutations could be classified on the basis of their relative behavior in the loss-of-function and dominant negative assays. Mutations in internal residues, such as est3-W21A and est3-D86A, were at one extreme end of the spectrum, with no dominant negative phenotype despite a severe loss-of-function defect, whereas mutations such as est3-R110A and est3-R110E were at the opposite end, with a strong defect in both assays. A graphical representation of the relative effect of these mutations in these two assays (Supplementary Fig. 4) demonstrates that for most mutations, there is an excellent correlation between the severity of the defects in the loss-of-function and dominant negative assays. This is consistent with the premise that these mutations perturb the surface of the protein, which suggests that at least a subset of these may be “separation-of-function” alleles that have specifically lost a single biochemical activity associated with the Est3 telomerase subunit. For a small subset of mutations, however, there is a roughly inverse correlation, suggesting that loss of function in these mutant proteins is potentially due to protein destabilization. For example, the est3-K68E mutation has a loss-of-function defect roughly comparable to that of est3-K68Y but a greatly reduced effect in the dominant negative assay; the Est3-K68E mutant protein also exhibits a modest reduction in protein levels (Fig. 3b). Mutations with little or no dominant negative phenotype (in particular, K68E and D164R) therefore were not included in the analysis described in the subsequent section.
A binding site for telomerase maps to one surface of the OB-fold
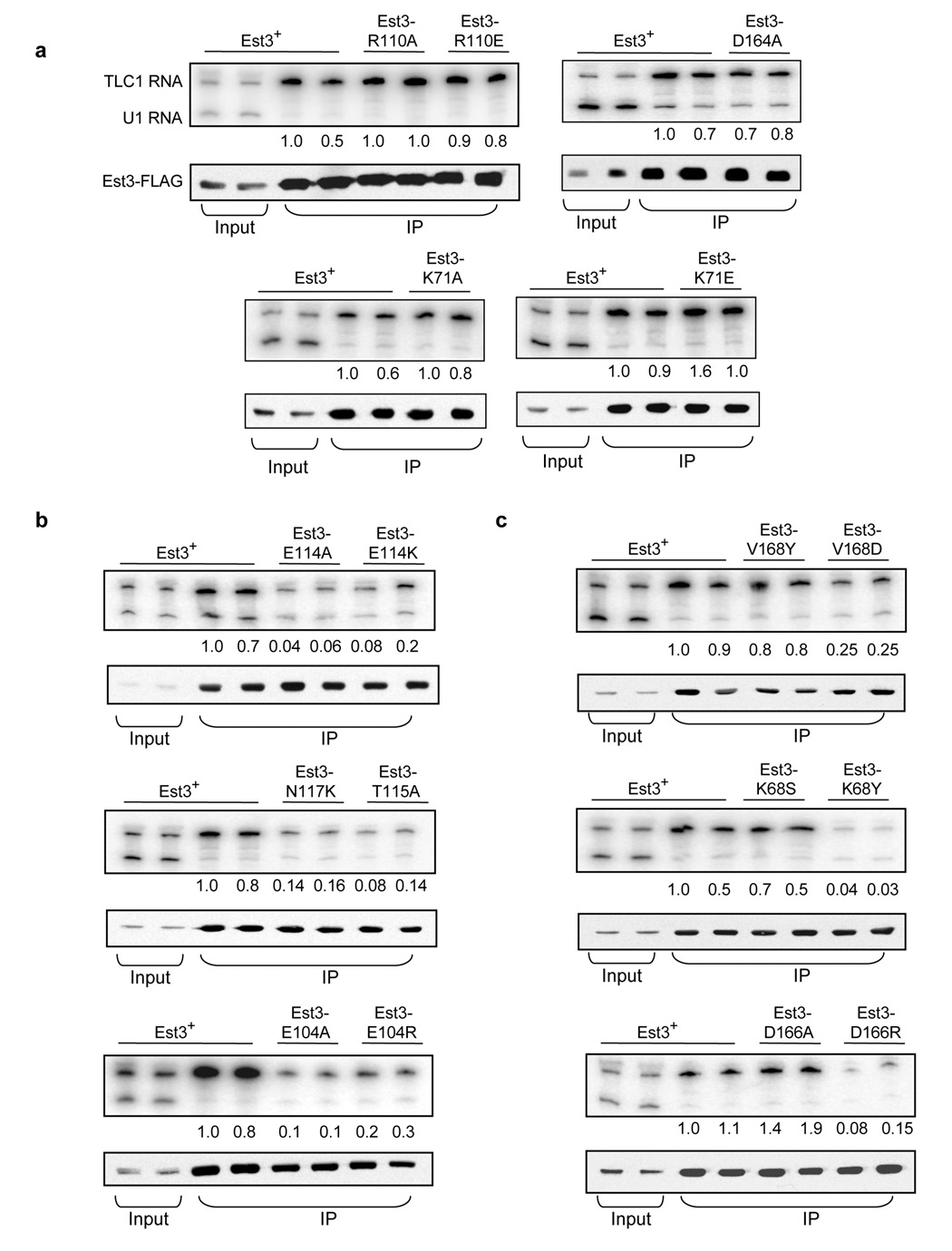
To determine whether this collection of mutations included separation-of-function alleles, the association of each mutant Est3 protein with the telomerase holoenzyme was examined using co-immunoprecipitation of Est3-(FLAG)3 with TLC1. Each mutation was introduced into the Est3-(FLAG)3 construct, and Northern analysis of anti-FLAG immunoprecipitates were analyzed with probes to detect the telomerase TLC1 RNA, as well as the unrelated U1 RNA.
Strikingly, several of the most defective alleles, based on the in vivo analysis shown in Fig. 3, still retained association with telomerase. Mutations in residues Lys71, Arg110 and Asp164 all exhibited wild type levels of co-immunoprecipitation with TLC1 (Fig. 4a), arguing that a property other than interaction with the telomerase RNP is affected. In contrast, mutations in a second set of residues (Glu114, Thr115, Asn117 and Glu104) clearly impaired the ability of the Est3 protein to bind telomerase, with a reduction of 10-fold or more in co-immunoprecipitation with TLC1 (Fig. 4b). It is worth re-emphasizing that mutations in both groups of residues exhibited strong dominant negative phenotypes when over-expressed, indicating that the absence of a dominant negative phenotype was not simply due to a difference in whether the Est3 protein associated with telomerase or not.
Figure 4.
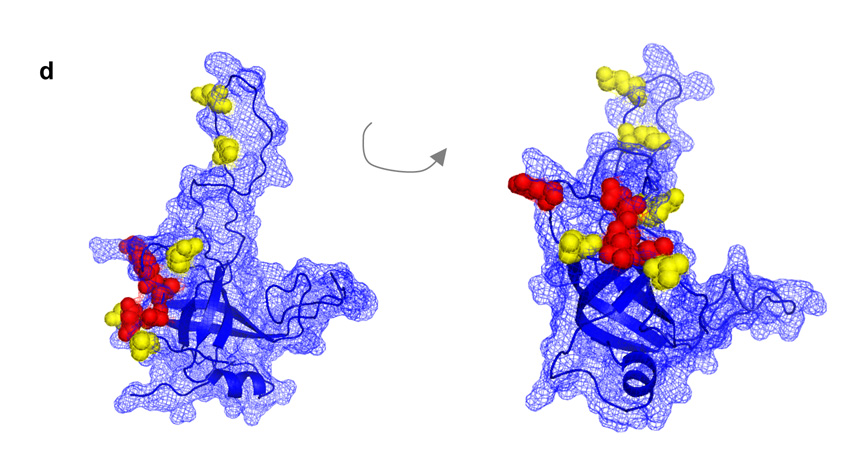
An interaction surface on one face of the OB-fold mediates association of Est3 with the telomerase enzyme. (a) – (c) Association of mutant Est3 proteins with telomerase. Extracts from asynchronous log-phase cultures, expressing mutant or wild type Est3-(FLAG)3 proteins, were immunoprecipitated with anti-FLAG antibody, and immunoprecipitates were analyzed by Northern blotting to detect the TLC1 telomerase RNA or an unrelated (U1) RNA, and by western blotting to detect Est3-(FLAG)3; input samples for the Northern and western analysis are 2% and 10%, respectively. The efficiency of TLC1 co-immunoprecipitation was normalized to Est3-(FLAG)3 protein levels in the immunoprecipitates, and the first wild type sample arbitrarily set to 1.0; absolute efficiency was between 20 and 40%. Representative samples are shown for each set of mutants. (d) Residues that mediate binding of Est3 to telomerase (Glu104, Glu114, Thr115, Asn117, and Asp166) are depicted in red, and residues that perform a separate function (Lys68, Lys71, Arg110, Asp164 and Val168) are depicted in yellow, on a wire mesh model of Est3, overlaid on a ribbon representation of the OB-fold. Note that because this is a predicted structure, the exact relative position of these 10 residues, relative to each other, is not precisely determined.
Three residues (Val168, Lys68 and Asp166) exhibited variable effects in the TLC1 co-immunoprecipitation assay, depending on the mutation (Fig. 4c). For two of these residues (Val168 and Lys68), we propose that their primary role is a function other than mediating binding of the Est3 protein to telomerase, for the following rationale. The est3-V168Y mutation had a reasonably strong in vivo telomere replication defect but the mutant protein nevertheless retained wild type levels of association with telomerase; furthermore, the very modest decrease in telomerase association displayed by the Est3-V168D mutant protein could not account for the severity of the in vivo est3-V168D defect. In a similar manner, the mutant Est3-K68S protein bound telomerase at wild type levels, even though the est3-K68S strain exhibited a more severe in vivo defect than the est3-K68Y strain. In contrast, the est3-D166R strain displayed a far more severe in vivo defect (in both the loss-of-function and dominant negative assays) than the est3-D166A strain, arguing that a primary function of Asp166 is to contribute to the telomerase binding site of Est3.
Fig. 4d maps these two groups of amino acids – those that mediate association of Est3 with telomerase (indicated in red) and those that have a separate distinct function (indicated in yellow) – on the proposed structure of Est3. Notably, four of the residues that are required for binding of Est3 to telomerase (Glu114, Thr115, Asn117, and Asp166) all cluster on one face of the OB-fold, with a fifth residues (E104) immediately adjacent. Thus, this analysis has identified a binding site for telomerase on the proposed surface of Est3 (note, however, that since this is a predicted structure, the specific orientation of these five residues relative to each other cannot be precisely determined). The remaining residues perform an additional function by Est3, as yet to be determined; notably, several of these residues (Arg110, Asp164 and Val168) immediately bracket the telomerase binding site on Est3, suggesting that this interaction face is not solely functioning to bring Est3 to the complex but it is also a site of an additional critical activity mediated by Est3.
DISCUSSION
The predicted 3-dimensional conformation of the Est3 telomerase protein provides a basis for generating separation-of-function mutations in residues that are likely to be on the protein surface. The identification of analogous separation-of-function mutations in CDC1314, and subsequently in EST115 was instrumental in elucidating the regulatory role of the Est1 telomerase subunit. However, the initial mutation (cdc13-2) was a fortuitous discovery, resulting from an extremely labor-intensive mutagenesis screen6. In contrast, using structural predictions to restrict mutagenesis to proposed surface residues offers an alternative, and much more rapid, means of generating this particular class of alleles.
The directed mutagenesis approach has its limitations, however. In particular, our data illustrate the importance of analyzing missense mutations other than just alanine, often the default amino acid for site-directed mutagenesis, due its small size and amphipathic characteristics. For example, changing Val168 or Asn117 to alanine had no phenotypic consequences (Fig. 3); the role of each of these two amino acids in Est3 function was only uncovered with additional mutagenesis. In addition, while the effects of mutations in the 10 predicted surface residues is consistent with the proposed Est3 structure that in Fig. 4c, it is important to emphasize that this is only a model. In particular, the conformation of the extended β1 – β2 and β4 – β5 loops is an open question, as evidenced by the fact the different structure prediction servers handled these two regions differently (data not shown). However, the results from the protein stability and over-expression dominant negative assays increase the likelihood that we have identified functionally important surface residues. Furthermore, the availability of separation-of-function est3− mutations should allow a genetic test of the proposed telomerase interaction surface depicted in Fig. 4c, through the identification of reciprocal mutations in either EST1 or EST2 that suppress this particular class of est3− defects.
The unexpected discovery of a structural relationship between a domain shared by Est3 and TPP1 might suggest that these two proteins are orthologs, i.e. two proteins with common ancestry that are structurally, and presumably functionally, related but separated by speciation. Although this is an intriguing speculation, we would argue against this idea based on what is currently known about these two proteins. Est3, as a 20 kDa subunit of telomerase, and TPP1, as a 58 kDa subunit of an end-binding complex, perform functionally distinct tasks at chromosome ends. The Est3-containing telomerase complex is responsible for enzymatically elongating the G-rich strand of the telomere, whereas the TPP1-POT1 heterodimer is crucial for protecting this telomeric G-rich overhang, once synthesized. Resolving the question of whether these two proteins are orthologs may rely on identification of additional TPP1 and Est3 protein sequences. In particular, identification of TPP1 and Est3 in the same species would definitively argue against the orthology model.
Nevertheless, the identification of structurally similar domains in Est3 and TPP1 may point to one or more biochemical properties shared by these two proteins. A recent study has shown that TPP1 can be co-immunoprecipitated with a TERT-containing complex44, which suggests that the telomerase interaction site on Est3 may correspond to a similar site on TPP1. In fact, although Est3 and TPP1 share extremely limited primary sequence identity overall, three amino acids that are part of the telomere interaction site in Est3 have conserved structural counterparts in TPP1. We are currently testing whether over-expression of TPP1, bearing mutations in these three residues, will have a dominant negative effect on telomere length in human cells, analogous to the effects that we have observed with Est3.
The POT1/TPP1 complex has also been shown to act as a processivity factor for human telomerase in vitro38. Whether Est3, either alone or in combination with other proteins (such as Est1), functions in an analogous manner, is unclear. In C. albicans, null mutations in EST1 or EST3 result in primer-specific defects, as telomerase lacking either of these two subunits is unable to elongate a subset of telomeric primers21,45. We have observed even more severe consequences on telomerase activity in extracts prepared from est1-Δ or est3-Δ strains of a related yeast, S. castellii: loss of either Est1 or Est3 results in an almost completely inactive enzyme, with all primers tested (JSL and VL, unpublished data). This is in sharp contrast to the situation in both S. cerevisiae and the more evolutionarily distant S. pombe, where loss of EST1 function has no apparent effect on telomerase activity7,8,10. These species-specific variations may be due to differences in how the telomerase enzyme is assembled in different yeast species. Lue and colleagues have recently reported that Est1 and Est3 mutually depend on each other for association with the telomerase RNP in C. albicans21, in contrast to what has been observed in S. cerevisiae4. Alternatively, it is possible that complete loss of a regulatory subunit, as the result of a deletion mutation, may destabilize the catalytic core of telomerase more substantially in some species than in others. Thus, whether the effects observed with the S. castellii and C. albicans enzymes will be representative of an evolutionarily conserved role for Est1 or Est3 in enzyme activity will require further study; the separation-of-function mutations described here may be useful in this regard.
Finally, we point out that the 10 residues presented in this study were chosen from a panel of missense mutations introduced into 29 residues, which represents ~50% of the amino acids predicted to reside on the surface of the Est3 protein. However, since this larger set consisted primarily of alanine mutations, it is likely that far less than 50% of the functional surface of Est3 has been probed, as discussed above. Successful construction of a comprehensive map of the functional surface of a protein therefore cannot rely solely on directed mutagenesis. Our study provides one such alternative, by using forward, rather than directed, mutagenesis, and screening for dominant negative phenotypes. Furthermore, it is worth noting that such a strategy, even in the absence of a predicted structure, is likely to target surface residues.
METHODS
Strains and plasmids
The strains used for the loss-of-function and dominant negative assays shown in Fig. 3 were YVL3057 (MATa est3-Δ::LYS2 ura3-52 lys2-801 trp1-Δ1 his3-Δ200 leu2-Δ1/p CEN URA EST3) and YVL3142 (MATa yku80-Δ::KAN ura3-52 lys2-801 trp1-Δ1 his3-Δ200 leu2-Δ1/p CEN URA YKU80), respectively. Supplementary Table 1 lists the plasmids used in this study; the frameshift that is present in the genomic EST3 gene35 has been corrected in these constructs. Following the introduction of missense mutations by site-directed mutagenesis, we sequenced the entire EST3 gene and/or sub-cloned the relevant portion of the mutant est3− allele into an unmutagenized backbone, to eliminate unlinked mutations. The biochemical experiments shown in Fig. 3b and Fig. 4 were performed in the protease-deficient strain JB811 (MATa leu2 trp1 ura3-52 prb− prc− pep4-3).
Sequence alignments
The Est3 sequences shown in the alignment in Fig. 1a were identified by successive BLAST searches, starting with iterative PSI-BLAST searches of the non redundant (NR) database using the S. cerevisiae and C. albicans Est3 protein sequences as seeds. Homologs recovered from these two initial searches with a score of at least 1 × 10−10 were subsequently used to search available fungal genomes from the FGI, JGI and Sanger center, using BLASTP and TBLASTN. TPP1 homologs were identified through BLASTP searches of the NCBI NR database and ENSEMBL genome database. Predicted homologs were considered legitimate if the original and reciprocal BLASTP E-value with experimentally characterized homologs was at least 1 × 10−5. TPP1 sequences were aligned with the EXPRESSO alignment program46. Est3 sequences were aligned using the COMBINE feature of the TCOFFEE server to combine alignments generated by PROMALS and MCOFFEE47,48.
Structure predictions
A full-length alignment of all identified Est3 homologs was used as a seed for structural prediction with the online server HHpred, allowing for more homologous sequences to be used in calculation of the profile than are available in the NR database. Multi-template modeling (HHpred2 method) was used for super-HMM generation, and the 3D model based on the resulting HMM was generated with the program MODELLER49. Predictions from the online servers SAM-T06, FUGUE and I-TASSER were performed using default settings with a single protein sequence submission. The resulting models from prediction servers were subsequently compared with each other, and with TPP1, using the server DALILITE, to insure a similar global topology50. The structural alignments from these comparisons were visualized with PyMol to insure the calculated similarity resided within the core of the predicted OB fold.
Genetic and biochemical analysis of mutant proteins
To monitor loss-of-function phenotypes, plasmids expressing mutant alleles of EST3 (introduced into pVL2537, expressing an untagged version of the Est3 protein under the control of the native EST3 promoter and present on a single copy CEN plasmid) were transformed into freshly derived est3-Δ derivatives of YVL3057, following loss of the EST3 URA3 plasmid present in the strain; telomere length and senescence were assayed as described previously6. Dominant negative phenotypes were determined following transformation of est3− mutations (introduced into pVL1024, a 2 µ plasmid expressing the Est3 protein under the control of the ADH promoter) into YVL3142, and potential synthetic lethal effects were assessed as described previously43. Est3-(FLAG)3 protein levels were determined from whole cell extracts of the JB811 strain transformed with pVL2076, or mutant derivatives of pVL2076, examined on 15% SDS-PAGE gels, which were subsequently probed simultaneously with anti-FLAG (Sigma; 1:5000) and anti-PGK (Invitrogen; 1:5000) antibodies. Co-immunoprecipitation of Est3 with TLC1 was examined in the protease-deficient strain JB811, transformed with plasmids expressing wild type or mutant alleles of Est3-(FLAG)3. The efficiency of co-immunoprecipitation of the wild type Est3-(FLAG)3 protein with TLC1 was ~ 20 to 40%, even in the presence of the genomic wild type EST3 gene, and was not appreciably enhanced in an est3-Δ derivative of JB811 (data not shown); the efficiency of co-immunoprecipitation of the unrelated U1 RNA was ~0.1 to 0.5%. Extract preparation, immunoprecipitation protocols and northern analysis were performed as described previously43; anti-FLAG M2 agarose beads and HRP-conjugated anti-FLAG M2 antibody (Sigma) were used for immunoprecipitation and western analysis, respectively. Northerns were simultaneously probed with a 1.3 kb TLC1 restriction fragment, prepared by random-primed labeling, and a 23-mer oligomer that recognizes the U1 snRNA, prepared by end-labeling.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Leslie Ricks and Erin Ford for outstanding technical assistance. This research was supported by grant AG11728 from the US National Institutes of Health.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Garcia CK, Wright WE, Shay JW. Human diseases of telomerase dysfunction: insights into tissue aging. Nucleic Acids Res. 2007;35:7406–7416. doi: 10.1093/nar/gkm644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 3.Lingner J, et al. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 4.Hughes TR, Evans SK, Weilbaecher RG, Lundblad V. The Est3 protein is a subunit of yeast telomerase. Curr Biol. 2000;10:809–812. doi: 10.1016/s0960-9822(00)00562-5. [DOI] [PubMed] [Google Scholar]
- 5.Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc Natl Acad Sci U S A. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohn M, Blackburn EH. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 8.Lingner J, Cech TR, Hughes TR, Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. U S A. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beernink HT, Miller K, Deshpande A, Bucher P, Cooper JP. Telomere maintenance in fission yeast requires an Est1 ortholog. Curr Biol. 2003;13:575–580. doi: 10.1016/s0960-9822(03)00169-6. [DOI] [PubMed] [Google Scholar]
- 10.Leonardi J, Box JA, Bunch JT, Baumann P. TER1, the RNA subunit of fission yeast telomerase. Nat Struct Mol Biol. 2008;15:26–33. doi: 10.1038/nsmb1343. [DOI] [PubMed] [Google Scholar]
- 11.Livengood AJ, Zaug AJ, Cech TR. Essential regions of Saccharomyces cerevisiae telomerase RNA: separate elements for Est1p and Est2p interaction. Mol Cell Biol. 2002;22:2366–2374. doi: 10.1128/MCB.22.7.2366-2374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 2002;16:2800–2812. doi: 10.1101/gad.1029302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webb CJ, Zakian VA. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat Struct Mol Biol. 2008;15:34–42. doi: 10.1038/nsmb1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 15.Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 16.Bianchi A, Negrini S, Shore D. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol Cell. 2004;16:139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Bianchi A, Shore D. Increased association of telomerase with short telomeres in yeast. Genes Dev. 2007;21:1726–1730. doi: 10.1101/gad.438907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 19.Taggart AK, Teng SC, Zakian VA. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- 20.Friedman KL, Heit JJ, Long DM, Cech TR. N-terminal domain of yeast telomerase reverse transcriptase: recruitment of Est3p to the telomerase complex. Mol Biol Cell. 2003;14:1–13. doi: 10.1091/mbc.E02-06-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu M, Yu EY, Singh SM, Lue NF. Mutual dependence of Candida albicans Est1p and Est3p in telomerase assembly and activation. Eukaryot Cell. 2007;6:1330–1338. doi: 10.1128/EC.00069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osterhage JL, Talley JM, Friedman KL. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat Struct Mol Biol. 2006;13:720–728. doi: 10.1038/nsmb1125. [DOI] [PubMed] [Google Scholar]
- 23.Fang GW, Cech TR. Molecular cloning of telomere-binding protein genes from Stylonychia mytilis. Nucleic Acids Research. 1991;19:5515–5518. doi: 10.1093/nar/19.20.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray JT, Celander DW, Price CM, Cech TR. Cloning and expression of genes for the Oxytricha telomere-binding protein: specific subunit interactions in the telomeric complex. Cell. 1991;67:807–814. doi: 10.1016/0092-8674(91)90075-a. [DOI] [PubMed] [Google Scholar]
- 25.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 26.Ye JZ, et al. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu D, et al. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 28.Houghtaling BR, Cuttonaro L, Chang W, Smith S. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol. 2004;14:1621–1631. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 29.Churikov D, Wei C, Price CM. Vertebrate POT1 restricts G-overhang length and prevents activation of a telomeric DNA damage checkpoint but is dispensable for overhang protection. Mol Cell Biol. 2006;26:6971–6982. doi: 10.1128/MCB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacob NK, Lescasse R, Linger BR, Price CM. Tetrahymena POT1a regulates telomere length and prevents activation of a cell cycle checkpoint. Mol Cell Biol. 2007;27:1592–1601. doi: 10.1128/MCB.01975-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hockemeyer D, et al. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat Struct Mol Biol. 2007 doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- 32.Guo X, et al. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. Embo J. 2007;26:4709–4719. doi: 10.1038/sj.emboj.7601893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Else T, et al. Tpp1/Acd maintains genomic stability through a complex role in telomere protection. Chromosome Res. 2007;15:1001–1013. doi: 10.1007/s10577-007-1175-5. [DOI] [PubMed] [Google Scholar]
- 34.Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- 35.Morris DK, Lundblad V. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr Biol. 1997;7:969–976. doi: 10.1016/s0960-9822(06)00416-7. [DOI] [PubMed] [Google Scholar]
- 36.Singh SM, Steinberg-Neifach O, Mian IS, Lue NF. Analysis of telomerase in Candida albicans: potential role in telomere end protection. Eukaryot Cell. 2002;1:967–977. doi: 10.1128/EC.1.6.967-977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang F, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 39.Karplus K, et al. SAM-T04: what is new in protein-structure prediction for CASP6. Proteins. 2005;61(Suppl 7):135–142. doi: 10.1002/prot.20730. [DOI] [PubMed] [Google Scholar]
- 40.Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 41.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans SK, Lundblad V. The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics. 2002;162:1101–1115. doi: 10.1093/genetics/162.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xin H, et al. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 45.Singh SM, Lue NF. Ever shorter telomere 1 (EST1)-dependent reverse transcription by Candida telomerase in vitro: evidence in support of an activating function. Proc Natl Acad Sci U S A. 2003;100:5718–5723. doi: 10.1073/pnas.1036868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Armougom F, et al. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34:W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pei J, Kim BH, Tang M, Grishin NV. PROMALS web server for accurate multiple protein sequence alignments. Nucleic Acids Res. 2007;35:W649–W652. doi: 10.1093/nar/gkm227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moretti S, et al. The M-Coffee web server: a meta-method for computing multiple sequence alignments by combining alternative alignment methods. Nucleic Acids Res. 2007;35:W645–W648. doi: 10.1093/nar/gkm333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 50.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.