Abstract
RNA interference (RNAi) is an effective tool for studying gene function in oocytes, but no studies have targeted somatic cells of primary cultured cumulus cell-oocyte complexes (COCs). This is probably due to difficulty in introducing RNAi-inducing molecules, such as a short-hairpin RNA (shRNA) gene, into COCs by commonly used transfection reagents. We therefore tested whether a developmental process of intact COCs could be suppressed by adenovirus-mediated shRNA expression. Has2, encoding hyaluronan synthase 2, was selected as the target transcript, because the process of cumulus expansion depends upon expression of Has2 mRNA and this process is easily evaluated in vitro. Intact COCs were infected with replication-incompetent adenoviruses containing an expression sequence of shRNA targeting either Has2 (Has2 shRNA) or a control transcript not expressed in cumulus cells, and the effects on epidermal growth factor (EGF)-stimulated cumulus expansion were determined. Has2 shRNA expression suppressed Has2 mRNA levels in COCs by more than 70%, without affecting expression levels of Ptgs2, Ptx3, Tnfaip6 mRNAs, which are also required for cumulus expansion, or other transcripts not related to expansion. Interestingly, levels of Areg and Ereg mRNAs were decreased in COCs expressing Has2 shRNA when compared with those in controls, while Btc mRNA levels remained unaffected. Furthermore, the degree of cumulus expansion by Has2 shRNA-expressing COCs was significantly less than that of controls. Thus adenovirus-mediated introduction of shRNA produces specific gene silencing and a phenotype in intact COCs, providing proof of principle that this method will be a helpful tool for understanding mechanisms of COC development.
Keywords: RNA interference, cumulus expansion, hyaluronan synthase 2, shRNA
INTRODUCTION
RNA interference (RNAi), initially discovered in Caenorhabditis elegans (Fire et al. 1998), is a conserved mechanism by which small interfering RNAs (siRNAs) derived from double-stranded RNA bring about sequence-specific gene silencing via transcript degradation. Several studies have used the RNAi technology to examine gene function in primary cultured granulosa cells of pig (Hirano et al. 2004), rat (Jo and Curry 2006; Kwintkiewicz et al. 2007; Parakh et al. 2006; Tamura et al. 2007), cattle (Kobayashi et al. 2007) and mouse (Shimada et al. 2007). Since these studies were conducted using granulosa cells cultured as mono-layers, the protocols used are not necessarily applicable to studying cumulus cell processes that require communication between oocytes and companion cumulus cells in a three dimensional cumulus cell-oocyte complex (COC). One of the reasons for the absence of studies using intact COCs has been the difficulty of commonly used transfection reagents in introducing RNAi producing molecules, such as a vector containing an expression sequence of short hairpin RNA (shRNA), into primary cultured COCs.
shRNA is an artificially designed RNA molecule that contains an intramolecular stem-loop structure. Once expressed in cells, specific shRNA is processed into siRNA by the endogenous DICER1 enzyme, and triggers subsequent cleavage and degradation of target transcripts, thus “silencing” specific gene expression (Brummelkamp et al. 2002; Paddison et al. 2002; Paul et al. 2002; Sui et al. 2002; Yu et al. 2002). shRNA can be introduced into cells by viral vectors, such as adenoviruses. The relative ease in preparing high-titer viral stocks and the high efficiency in delivering shRNA into both actively dividing and non-dividing cells are major advantages of using adenoviral vectors (Hommel et al. 2003; Xia et al. 2002).
Oocytes and companion cumulus cells communicate via paracrine regulatory factors and gap junctions (Eppig 2001). Removal of oocytes from COCs impairs some cumulus cell functions such as cumulus expansion requiring oocyte-derived paracrine factors (Buccione et al. 1990; Eppig et al. 1993; Vanderhyden et al. 1990). The production of hyaluronan, a non-sulfated glycosaminoglycan, is necessary for cumulus expansion (Richards 2005). Hyaluronan synthase 2 (HAS2) is one of the enzymes required for hyaluronan synthesis (Weigel et al. 1997). Since Has2 mRNA expression in cumulus cells is well correlated with the cumulus expansion process (Fulop et al. 1997), it is generally accepted that HAS2 is one of the key enzymes required for this process (Richards 2005).
The objective of this study was to determine whether a maturational process of intact oocyte-cumulus cell complexes, cumulus expansion, could be suppressed by adenoviral vector-mediated expression of shRNA, without disrupting the three dimensional COC structure, thus with maintaining the cumulus cell-oocyte communication. Has2 was therefore selected as the specific shRNA target transcript because the functional consequence of Has2 silencing could be easily evaluated by assessing the degree of cumulus expansion. Recombinant adenoviruses containing an expression sequence of shRNA targeting Has2 (Has2 shRNA) or human lamin A/C (LMNA shRNA, as control) were produced. Then, the effects of infecting intact COCs with these viruses on the mRNA levels of Has2 and other cumulus expansion-related transcripts, Ptgs2, Ptx3 and Tnfaip6, encoding prostaglandin synthase 2, pentraxin 3 and tumor necrosis factor alpha induced protein 6, respectively (Fulop et al. 2003; Ochsner et al. 2003a; Ochsner et al. 2003b; Varani et al. 2002), as well as the process of cumulus expansion itself, were examined.
RESULTS
Effect of Has2 shRNA expression on steady-state Has2 mRNA levels during cumulus expansion process
As a preliminary experiment, three shRNA sequences targeting Has2 mRNA were designed and tested for the efficiency of gene silencing. The Has2 shRNA sequence (see Materials and Methods) that exhibited the greatest efficiency in targeting Has2 mRNA expression was selected for use in the present study (data not shown).
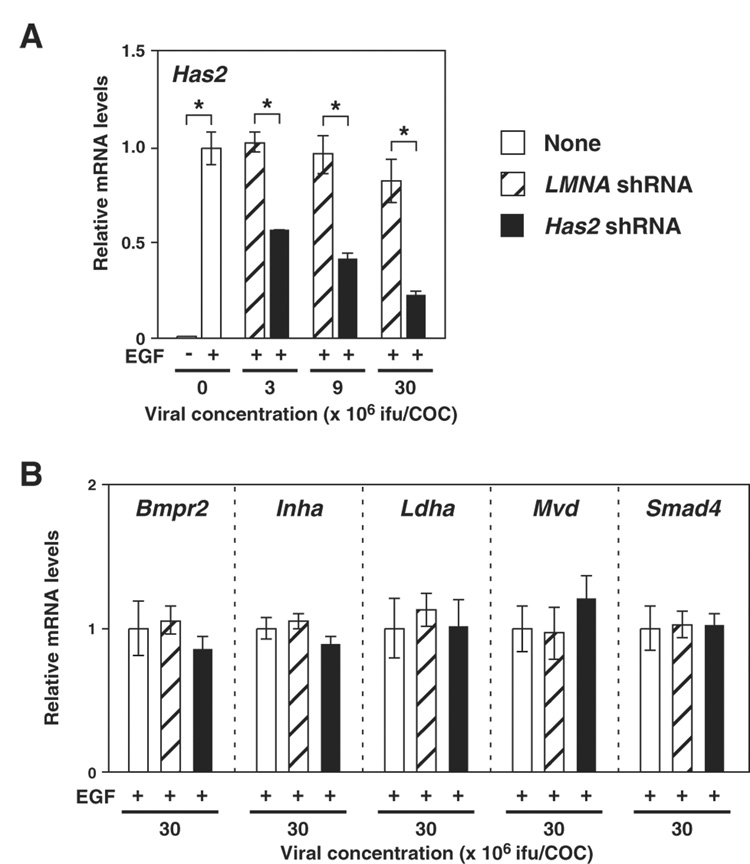
Effects of Has2 shRNA expression on Has2 mRNA levels in COCs were determined. As shown in Fig. 1A, infecting with adenovirus containing expression sequence of Has2 shRNA decreased Has2 mRNA levels in COCs in a dose dependent manner, whereas control virus infection had no effect on Has2 mRNA levels. The degree of Has2 mRNA knockdown was more than 70% when COCs were infected at a viral concentration of 30 × 106 ifu/COC. Infecting at a higher viral concentration (90 × 106 ifu/COC) did not improve knockdown efficiency (data not shown).
Figure 1. Effect of Has2 shRNA expression on levels of transcripts encoding HAS2 or proteins not involved in the cumulus expansion process.
COCs infected with adenoviruses containing expression sequences of either Has2 shRNA (solid bar) or LMNA shRNA (hatched bar) or without any viruses (open bar) were sampled after 6 hr of EGF treatment, and expression levels of (A) Has2 mRNA and (B) mRNAs encoding proteins not involved in the cumulus expansion process, but are known to play important roles in functions of cumulus cells, were examined with real-time PCR. Data are presented as the relative levels to the control non-virus-infected (+EGF) groups (mean ± SEM). An asterisk denotes a significant difference between the two indicated groups (p<0.05).
To examine specificity of Has2 mRNA silencing, levels of other transcripts encoding proteins known to play important roles in cumulus cell functions were examined. The transcripts examined were: Bmpr2, encoding BMP type 2 receptor (Shimasaki et al. 2004); Inha, encoding inhibin alpha subunit (Matzuk et al. 1992); Ldha, encoding a glycolytic enzyme (Sugiura et al. 2005); Mvd, encoding an enzyme involved in cholesterol biosynthesis pathway (Su et al. 2008); and Smad4, encoding MAD homolog 4 (Diaz et al. 2007). As shown in Fig. 1B, infecting with either the Has2 targeting virus or the control virus had no effect on the mRNA levels of these transcripts, suggesting that the silencing induced by Has2 shRNA expression was specific to Has2 mRNA.
Effect of Has2 shRNA expression on cumulus expansion
To assess functional significance of silencing Has2 expression, the effect of Has2 shRNA expression on the cumulus expansion was determined. As shown in Fig. 2 and Fig. 3, Has2 shRNA expression significantly reduced the cumulus expansion index (about 70%) when compared with the LMNA shRNA expression or the not infected control. Control shRNA-expressing COCs exhibited comparable levels of cumulus expansion to COCs not infected with any viruses (Fig. 2 and Fig. 3).
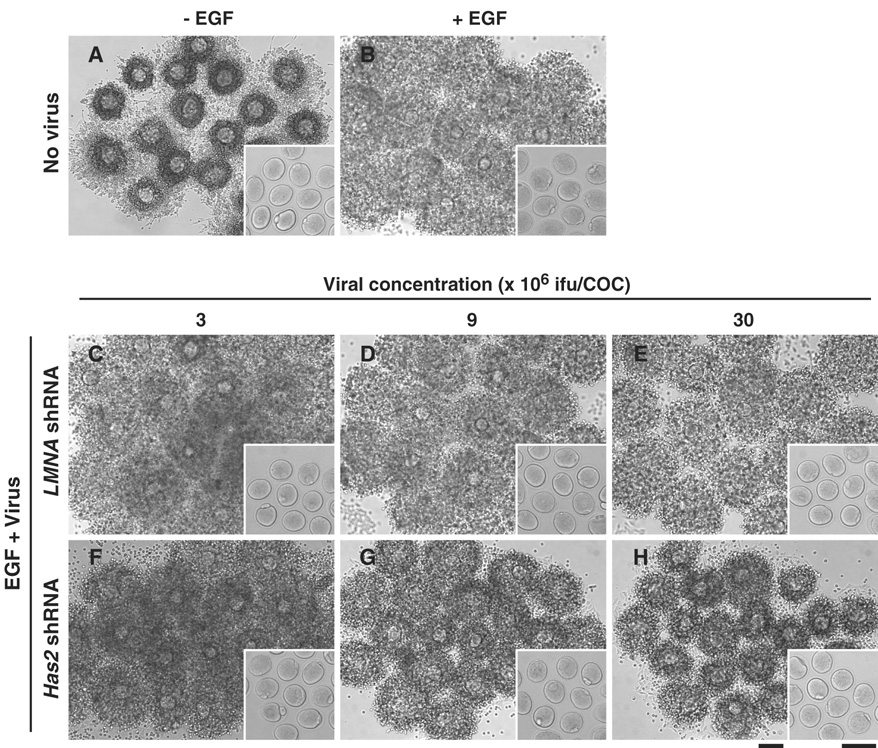
Figure 2. Representative photographs of virus-infected COCs after induction of cumulus expansion.
COCs infected with adenoviruses containing expression sequences of either Has2 shRNA (F–H) or LMNA shRNA (C–E) or without any viruses (A, B) were treated with (B–H) or without (A) EGF for 16 hr. After 16 hr, oocytes were denuded of cumulus cells and assessed for GVB and the first polarbody emission (insert). Bars, 100 µm.
Figure 3. Effect of Has2 shRNA expression on cumulus expansion.
COCs infected with adenoviruses containing expression sequences of either Has2 shRNA (solid bar) or LMNA shRNA (hatched bar) or without any viruses (open bar) were treated with EGF, and cumulus expansion index was assessed after 16 hr. Mean ± SEM. An asterisk denotes a significant difference between the two indicated groups (p <0.05).
To test whether viral infection caused any deleterious effects on the viability of oocytes, oocytes in the COCs not infected or infected with viruses were fertilized in vitro and assessed for their ability to develop to blastocyst stage (Fig. 4). As shown in Fig. 4, the percentages of the oocytes resumed meiosis (GVB), emitted the first polarbody (Pb1), developed to 2-cell (2-cell) and blastocyst (Blast) embryos were comparable between viral infected and non-infected COCs. This suggests that viral infection has no obvious effect on viability and developmental ability of the oocytes, therefore, the reduction of the degree of the cumulus expansion was not attributed to the lower viability of the oocytes. These results strongly suggest that Has2 mRNA silencing with Has2 shRNA expression was functionally significant.
Figure 4. Effect of exposure to virus on the developmental competence of the oocytes.
Oocytes in COCs infected with adenoviruses containing expression sequences of either Has2 shRNA (solid bar) or LMNA shRNA (hatched bar) or without any viruses (open bar) were matured and fertilized in vitro, and the preimplantation embryonic development were assessed. GVB, oocytes underwent germinal vesicle breakdown. Pb1, oocytes emitted the first polarbody. 2-cell, 2-cell embryo. Blast, blastocyst embryo.
Effect of Has2 shRNA expression on levels of cumulus expansion-related transcripts
Expression levels of Ptgs2, Ptx3 and Tnfaip6 transcripts were not significantly different between COCs expressing Has2 shRNA and LMNA shRNA; however, these mRNA levels increased when compared with those in COCs not infected with viruses (Fig. 5). The levels of Tnfaip6 mRNA in COCs, infected with either viruses at concentrations of 9 × 106 and 30 × 106 ifu/COC, were significantly higher than those in COCs not exposed to the viruses.
Figure 5. Effect of Has2 shRNA expression on mRNA levels of Ptgs2, Ptx3 and Tnfaip6 during cumulus expansion.
COCs infected with adenoviruses containing expression sequences of either Has2 shRNA (solid bar) or LMNA shRNA (hatched bar) or without any viruses (open bar) were sampled after 6 hr of EGF treatment, and expression levels of Ptgs2, Ptx3 and Tnfaip6 mRNA were examined with real-time PCR. Data are presented as the relative levels to the control non-virus-infected (+EGF) groups (mean ± SEM). The letter “a” denotes a significant difference from the control groups (p<0.05). An asterisk denotes a significant difference between the two indicated groups (p<0.05).
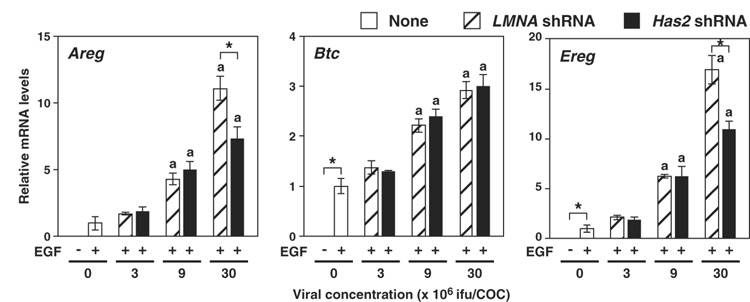
To assess whether the increases in mRNA levels were restricted to Ptgs2, Ptx3 and Tnfaip6 transcripts, mRNA levels of Areg, Btc and Ereg, encoding EGF-like growth factors, amphiregulin, betacellulin and epiregulin, involved in the cumulus expansion process (Park et al. 2004) were examined. As was the case in other cumulus expansion-related transcripts, the mRNA levels of Areg, Btc and Ereg increased as viral concentration increased (Fig. 6). Therefore, it appears that viral infection causes up-regulation of many transcripts involved in cumulus expansion process, except for Has2, in COCs during cumulus expansion.
Figure 6. Effect of Has2 shRNA expression on levels of transcripts encoding EGF-like growth factors during cumulus expansion.
COCs infected with adenoviruses containing expression sequences of either Has2 shRNA (solid bar) or LMNA shRNA (hatched bar) or without any viruses (open bar) were sampled after 6 hr of EGF treatment, and expression levels of Areg, Btc and Ereg mRNA were examined with real-time PCR. Data are presented as the relative levels to the control non-virus-infected (+EGF) groups (mean ± SEM). The letter “a” denotes a significant difference from the control groups (p<0.05). An asterisk denotes a significant difference between the two indicated groups (p<0.05).
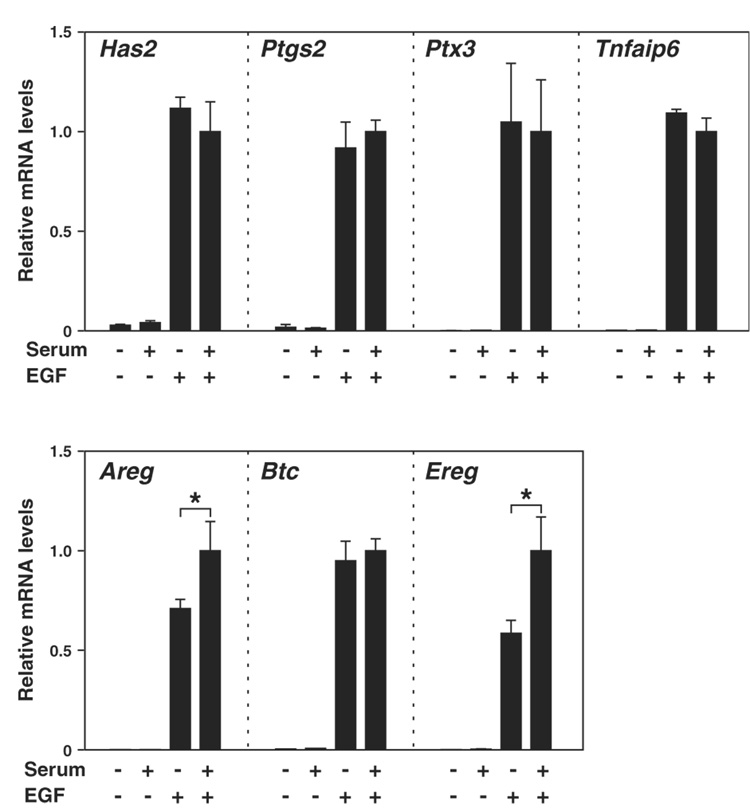
Levels of Areg and Ereg transcripts in COCs expressing Has2 shRNA were significantly lower than those in control shRNA-expressing cells (Fig. 6). This suggests the possibility that typical formation of extracellular matrix may be required to allow the normal levels of Areg and Ereg transcripts during cumulus expansion process. To assess this hypothesis, effect of serum removal from culture medium on expression levels of cumulus expansion-related transcripts were examined (Fig. 7). Since serum derived inter alpha trypsin inhibitor is required for the formation of extracellular matrix during cumulus expansion, normal expansion was inhibited when serum was removed from medium (Chen et al. 1992). As shown in Fig. 7, removing serum from medium significantly decreased transcript levels of Areg and Ereg, but not Btc nor the other transcripts related to cumulus expansion. Taking together, these results suggest that normal formation of extracellular matrix may be required for greater expression levels of Areg and Ereg mRNA during cumulus expansion process. Furthermore, these results support the hypothesis that steady-state levels of Areg and Ereg mRNAs decreased when Has2 mRNA levels were silenced as a consequence of silencing this transcript on matrix formation.
Figure 7. Effect of serum removal from culture medium on levels of the cumulus expansion-related transcripts.
Freshly isolated COCs were cultured with or without EGF or serum and sampled after 6 hr of EGF treatment, and expression levels of the cumulus expansion-related transcripts were examined with real-time PCR. An asterisk denotes a significant difference between the two indicated groups (p<0.05).
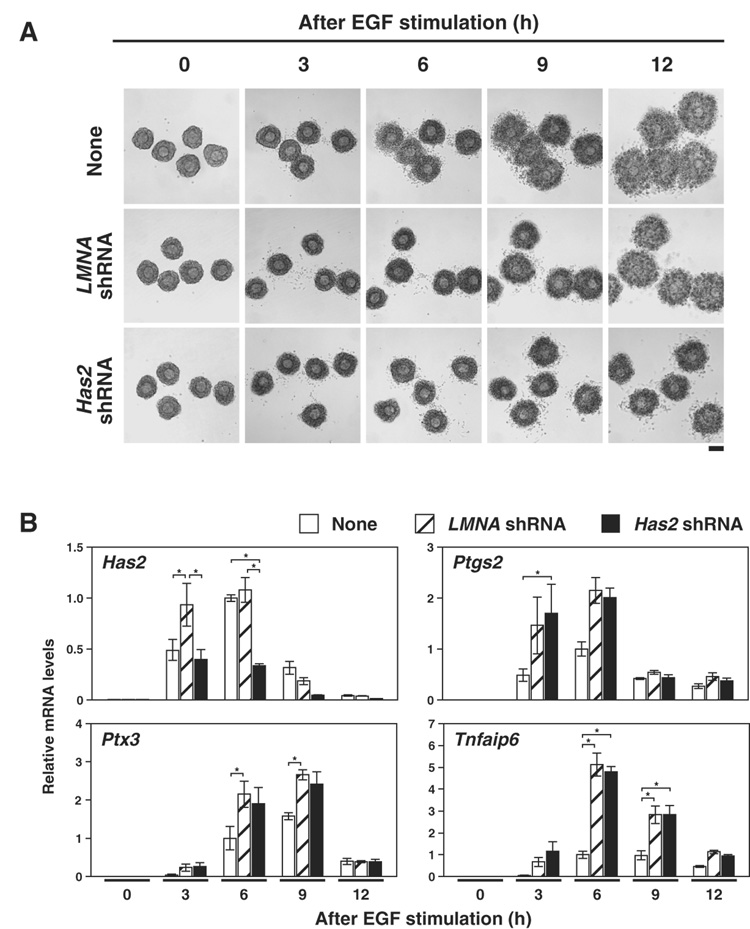
Effect of Has2 shRNA expression on the kinetics of cumulus expansion and mRNA levels
Since the levels of Has2, Ptgs2, Ptx3 and Tnfaip6 mRNA are dynamically regulated during cumulus expansion, it was possible that the decreased level of Has2 mRNA caused by Has2 shRNA expression observed at the 6 hr time point could be explained by differences in the kinetics of expression rather than RNAi-mediated silencing. Therefore, to examine the kinetics of mRNA expression in COCs expressing Has2 shRNA in more detail, mRNA levels of Has2, Ptgs2, Ptx3 and Tnfaip6 at various time points throughout the cumulus expansion process were determined (Fig. 8). As shown in Fig. 8B, expression levels of these transcripts were low before EGF-treatment (0 hr), and EGF-treatment induced dramatic increase of these transcripts in all groups, as expected. Notably, Has2 shRNA expression prevented Has2 mRNA levels from ever reaching levels reached in the LMNA shRNA expressing group. Moreover, expression levels of Ptgs2, Ptx3 and Tnfaip6 after EGF treatment were not significantly different between Has2 shRNA and LMNA shRNA expressing COCs.
Figure 8. The kinetics of Has2, Ptgs2, Ptx3 and Tnfaip6 mRNA expression in Has2 shRNA expressing COCs during cumulus expansion.
(A) Representative photographs of COCs at different time points were shown. Bar, 100 µm. (B) COCs infected with adenoviruses containing expression sequences of either Has2 shRNA (solid bar) or LMNA shRNA (hatched bar) or without any viruses (open bar) were sampled at the time points indicated, and expression levels of Has2, Ptgs2, Ptx3 and Tnfaip6 mRNAs were examined with real-time PCR. Data are presented as the relative levels to the control non-virus-infected groups at 6 hr (mean ± SEM). *, p<0.05.
DISCUSSION
The effects of infecting with adenoviruses containing expression sequence of Has2 shRNA on expression levels of cumulus expansion-related and non-related transcripts as well as cumulus expansion by intact COCs were examined. Has2 shRNA expression suppressed Has2 mRNA expression in intact COCs, without affecting expression levels of Ptgs2, Ptx3, Tnfaip6 and other transcripts not related to cumulus expansion process. Interestingly, levels of Areg and Ereg mRNAs were decreased in Has2 mRNA-targeted COCs when compared with those in control shRNA-expressing COCs, while Btc mRNA levels remained unaffected. Furthermore, the degree of cumulus expansion by Has2 shRNA-expressing COCs was lower than those by control shRNA-expressing COCs, indicating that Has2 transcript silencing was functionally significant. The reduction of Has2 mRNA levels and degree of cumulus expansion were not attributable to potential deleterious effects of viral infection on the COC viability; since (1) the control virus did not cause significant effects on the Has2 mRNA or cumulus expansion; (2) Has2 shRNA-expressing COCs responded to EGF by elevating mRNA levels of the cumulus expansion-related transcripts; (3) expression levels of the transcripts not involved in cumulus expansion process were not affected by the viral infection; and (4) comparable percentages of the oocytes in COCs infected with the viruses were capable in completing meiotic maturation and developing to blastocyst stage after in vitro fertilization compared with oocytes in COCs not infected with viruses.
Compared with other studies using adenoviral vectors, relatively higher viral concentration was required to achieve the effect observed in this study. This is probably because of the difference in the condition of culture (e.g. two-dimensional mono-layers vs. three-dimensional COCs). Since cells cultured as mono-layer are likely to have more opportunity for contact with viral particles than those cultured as COCs, the efficiency of viral infection into cells cultured as intact COCs appears to be significantly lower than that into mono-layer cells. In addition, a recent study has shown that a rodent specific gene, Clsp (also known as Gm1123) encoding Car-like soluble protein that might play a role in the anti-viral defense, is expressed in mouse brain and ovary (Kawabata et al. 2007). Although the identity of the cell types expressing CLSP in the ovary is currently not determined, it is possible that mouse granulosa cells are more protected from viral infection by this protein.
Levels of cumulus expansion-related transcripts in COCs infected with viruses tended to be up-regulated compared with those in non-infected COCs. Interestingly, the PTGS2, PTX3, and TNFAIP6, which are necessary for cumulus expansion, also participate in immune/inflammatory response (Richards et al. 2002), and several studies have shown that infecting with recombinant virus itself induced immune-like responses (Culver and Laster 2007; Russell 2000). However, it is important to note that levels of Ptgs2, Ptx3, Tnfaip6 mRNAs in adenovirus infected COCs were considerably lower before EGF treatment, and the robust increase of these transcripts occurred only after EGF treatment. Therefore, expression of these transcripts after viral infection still requires EGF stimulation. Clearly, further studies are required for assessing the reasons for the elevated transcript levels. However, one explanation could be that viral infection causes greater sensitivity of cumulus cells to EGF stimulation resulting in higher levels of expression of these immune/inflammatory-related and cumulus expansion-related transcripts. Importantly, levels of the other transcripts not related to cumulus expansion that were assessed in this study were not stimulated by viral infection.
The mRNA levels of Areg and Ereg, but not Btc in Has2 shRNA-expressing COCs were significantly lower than those in COCs expressing control shRNA. Although the mechanisms underlying gene regulation during cumulus expansion are not fully understood, a reasonable model was proposed by Shimada et al. (2006). According to their model, expression of the EGF-like growth factors in cumulus cells is promoted by prostaglandin E2 secreted from cumulus and mural granulosa cells by autocrine and paracrine mechanisms. Supporting this model, they showed that mRNA levels of Areg and Ereg, but not Btc, in COCs in Ptgs2 null mice were decreased compared with those in wild-type mice. Coincidentally, in the present study, mRNA levels of Areg and Ereg, but not Btc, were decreased by specific suppression of Has2 expression, whereas Ptgs2 mRNA levels were not affected. One possibility is that proper formation of extracellular matrix during cumulus expansion is required for retaining prostaglandin E2 produced by cumulus cells within the cumulus oophorus; thus, without sufficient production of hyaluronan, prostaglandin E2 diffuses into medium, impairing efficient transduction of its signal cascade. Supporting this hypothesis, Areg and Ereg mRNA levels, but not Btc mRNA, were significantly reduced when absence serum from medium did not allow proper formation of extracellular matrix. Alternatively, it is possible that hyaluronan/hyaluronan receptor signaling plays a critical role in Areg and Ereg expression, since several studies have shown that mRNA encoding receptors for hyaluronan, Cd44 and Hmmr, are expressed in porcine and bovine cumulus cells, and these transcripts were also detectable in mouse COCs (Hernandez-Gonzalez et al. 2006; Kimura et al. 2002; Schoenfelder and Einspanier 2003).
Although it is generally accepted that hyaluronan synthase is a critical enzyme required for cumulus expansion, no studies have demonstrated participation of the HAS2 isoform in the cumulus expansion process. Homozygous mutation of Has2 gene results in embryonic lethality during midgestation (Camenisch et al. 2000), thus HAS2 requirement in cumulus expansion could not be assessed. Historically, based on the observations that the expanded cumulus oophorus contains hyaluronan and is readily lysed to single cells by specific hyaluronidase treatment (Ball et al. 1982; Eppig 1979; Leonard and Kurzrok 1945; McClean and Rowlands 1942; Talbot 1984), and a potent inhibitor of glucosamine synthesis completely inhibits cumulus expansion in vitro (Buchanan 1973; Chen et al. 1993), it has been accepted that activity of hyaluronan synthase is required for cumulus expansion. Subsequently, in 1997, Has2 was cloned as a novel isoform of hyaluronan synthase family, of which expression increases during cumulus expansion process (Fulop et al. 1997). Therefore, it is likely that HAS2 is one of the key enzymes required for hyaluronan synthesis during cumulus expansion. However, recent studies have shown that other hyaluronan synthase isoforms are also expressed in bovine COCs and porcine oocytes (Kimura et al. 2002; Schoenfelder and Einspanier 2003). It was possible that these hyaluronan synthase isoforms and/or other unidentified homologues of the hyaluronan synthase related proteins could compensate for the reduction of HAS2 activity, and specific inhibition of the HAS2 isoform could have little effect on the cumulus expansion. The shRNA sequence targeting Has2 mRNA used in the present study has no homology to other known hyaluronan synthase genes and therefore do not target them. That specific silencing of Has2 mRNA levels significantly attenuated the cumulus expansion, strongly suggests that HAS2 is the primary hyaluronan synthase isoform required for cumulus expansion and no other hyaluronan synthase isoforms can compensate for the absence of HAS2 activity.
In summary, we have demonstrated that expression of a transcript, Has2, in cumulus cells cultured as intact COCs can be specifically suppressed by adenovirus-mediated shRNA expression. The efficiency of the Has2 transcript knockdown in the present study was more than 70% and this reduction was functionally significant. This study shows that adenovirus-mediated introduction of shRNA produces specific gene silencing in intact oocyte-cumulus cell complexes, and provides proof of principle that this method will be a helpful tool for understanding mechanisms and processes involved in development of the oocyte-granulosa cell complex. However, when implementing this approach, especially when studying protein functions during cumulus expansion/ovulation process, the response of cumulus cells to viral infection, independently of the function of delivered silencing constructs, must be properly controlled.
MATERIALS AND METHODS
Adenovirus preparation
Replication incompetent recombinant adenoviruses expressing shRNAs were constructed using BLOCK-iT™ Adenoviral RNAi Expression System (Invitrogen, Carlsbad CA) that consists of BLOCK-iT™ U6 RNAi Entry Vector Kit, Gateway® LR Clonase™ II Enzyme Mix, 293A cell line and pAd/BLOCK-iT™-DEST Gateway® Vector Kit, according to the manufacturer’s instructions. The adenoviral vectors produced with this system are based on the second-generation vectors developed by Bett et al. (1994) and expression of the shRNA is controlled by the human U6 promoter (Paddison et al. 2002; Paul et al. 2002).
Sequences of shRNA targeting mouse Has2 (accession number NM_008216) were designed using Invitrogen’s RNAi Designer (www.invitrogen.com/rnai). Two single-stranded DNA oligonucleotides, one encoding a shRNA sequence targeting Has2 and an additional sequence CACC at 5’ end (5’-CACCGCATTGTGAGAGGTTTCTATGCGAACATAGAAACCTCTCACAATGC-3’) and the other encoding complementary sequences and an additional sequence AAAA at 5’ end (5’-AAAAGCATTGTGAGAGGTTTCTATGTTCGCATAGAAACCTCTCACAATGC-3’) were purchased (Integrated DNA Technologies, Inc, Coralville, IA). The CACC and AAAA sequences are complementary to the overhang sequences in the pENTR™/U6 vector. These two single-stranded DNA oligonucleotides were annealed and then the generated double-stranded DNA oligonucleotide was cloned into the pENTR™/U6 vector to produce pENTR™/U6-Has2 shRNA vector using BLOCK-iT™ U6 RNAi Entry Vector Kit (Invitrogen). The ligation reaction product was introduced into competent E. coli, and individual clones were isolated and sequenced to confirm the shRNA coding sequence.
The U6 RNAi cassette in the pENTR™/U6-Has2 shRNA vector (consisting of the human U6 promoter, the sequence encoding Has2 shRNA and the polymerase III terminator (Bogenhagen and Brown 1981)) was transferred to the adenoviral expression plasmid by LR recombination reaction using Gateway® LR Clonase™ II Enzyme Mix and pAd/BLOCK-iT™-DEST Gateway® Vector Kit (Invitrogen). The LR recombination reaction product was introduced into competent E. coli and individual clones were isolated. Then the isolated adenoviral expression and pAd-GW/U6-laminshRNA plasmids, that was included in the pAd/BLOCK-iT™-DEST Gateway® Vector Kit, were digested with the restriction enzyme, Pac I (New England Biolabs, Inc., Ipswich, MA). Pac I-digested plasmids were transfected into 293A cells using Lipofectamine™ 2000 (Invitrogen) to produce crude adenoviral stocks. The crude adenoviral stocks were used to infect 293A cells to amplify the adenoviral stocks. These stocks were purified using the Adeno-X™ Virus Purification Kit (Clontech Laboratories, Inc., Palo Alto, CA) and stored at −80°C. The titers of the adenoviral stocks were determined using Adeno-X™ Rapid Titer Kit (Clontech Laboratories) with 293A cells, and adjusted to 3× 1011 infectious units (ifu)/ml using the Elution buffer provided in the Adeno-X™ Virus Purification Kit.
COC isolation and viral infection
B6SJLF1 mice were bred and raised in the research colony of the authors at The Jackson Laboratory and used for all experiments. Animals were maintained according to the Guide for the Care and Use of Laboratory Animals (Institute for Learning and Animal Research). COCs were isolated from antral follicles of 22 to 24-day-old mice that had been primed with 5 IU equine chorionic gonadotropin 44 to 48 hr earlier as reported previously (Sugiura et al. 2005). The basic culture medium used was bicarbonate-buffered MEMα (Life Technologies, Inc., Grand Island, NY) with Earle’s salts, supplemented with 75 µg/ml penicillin G, 50 µg/ml streptomycin sulfate, 0.23 mM pyruvate, and 3 mg/ml bovine serum albumin (Sigma-Aldrich Co., St. Louis, MO).
Twenty COCs were infected with the viruses for 24 hr in one well of 4-well (Nalge Nunc International, Rochester, NY) or 24-well plates (Corning Inc., Lowell, MA) in 250 µl volume of the basic culture medium supplemented with the phosphodiesterase inhibitor, milrinone (10 µM) (Sigma) (to maintain the oocytes at germinal vesicle stage) and 10−8 M 17β-estradiol (Sigma) with the viral concentrations indicated in the text. After 24 hr of infection, the cells were washed thoroughly and cultured for additional 24 hr in fresh medium. All the cultures were maintained at 37°C in a modular incubation chamber (Billups Rothenberg, Del Mar, CA) infused with 5% O2, 5% CO2 and 90% N2.
Induction and evaluation of cumulus expansion
Twenty four hr after the viral infection period, the COCs were washed thoroughly in the basic culture medium, without milrinone (allowing oocyte maturation to occur) or 17β-estradiol but supplemented with 5% fetal bovine serum (FBS) and then cultured in 50 µl of culture medium supplemented with 5% FBS and with or without 10 ng/ml epidermal growth factor (EGF, 10 ng/ml, BD Biosciences, San Jose, CA) under washed mineral oil in 35-mm Petri dishes. In some experiments, freshly isolated COCs were cultured in the basic culture medium supplemented with or without 5% FBS or 10 ng/ml EGF. Samples of COCs were taken before (0 hr) and after (3, 6, 9 and 12 hr) induction of cumulus expansion to assess levels of mRNAs with real-time PCR. The cumulus expansion index was evaluated after 16 hr of culture as reported previously (Eppig et al. 1993).
Fertilization and preimplantation embryonic development in vitro
For in vitro fertilization of oocytes, fetuin (1 mg/ml) was added to the culture medium during viral infection- and subsequent 24 h-culture periods to prevent zona pellucida hardening (Eppig et al. 1992). In vitro maturation and fertilization of oocytes and preimplantation embryonic development were conducted as described previously (Eppig and O'Brien 1996). After maturation culture, prior to in vitro fertilization, oocytes were denuded of cumulus cells by hyaluronidase treatment (Sigma) and pipetting to assess germinal vesicle breakdown (GVB, a marker of resumption of meiosis) and emission of the first polarbody (Pb1) as described previously (Eppig and O'Brien 1996).
Reverse-transcription (RT) real-time polymerase chain reaction (PCR)
RT-real-time PCR analysis was conducted as previously reported (Sugiura et al. 2007). The PCR primers used are presented in Table 1. The other PCR primers used are reported previously (Has2, Ptgs2, Ptx3, Tnfaip6 (Diaz et al. 2006), Rpl19, Mvd (Su et al. 2008), and Ldha (Sugiura et al. 2007)). The results are presented as the relative expression levels to the transcript amount of a standard sample, as indicated in the text, after normalization to the expression levels of a housekeeping gene, Rpl19, by the 2−ΔΔCt method (Livak and Schmittgen 2001). To avoid false positive signals, dissociation-curve analyses were performed at the end of analyses, and the PCR products were applied to agarose gel electrophoresis to confirm the sizes. Moreover, the PCR products were purified and were sequenced to verify sequence identity as preliminary experiments. The reactions were conducted at least in duplicate.
Table 1.
Sequence of PCR primers used for real-time PCR
| Gene | Forward | Reverse |
|---|---|---|
| Areg | TCCAAGATTGCAGTAGTAGCTGTCA | TATCGTTTCCAAAGGTGCACTGT |
| Btc | AACTGCACAGGTACCACCCCTAGA | ACAGATGCAGGAGGGAGTTTGC |
| Ereg | CCATCATGCATCCCAGGAGAA | TAGCCGTCCATGTCAGAACTACACT |
| Bmpr2 | CTTTCCACCCCCTGACACAA | GCGACTATCAAAACAGCTAACACAGA |
| Inha | GAAGATGTCTCCCAGGCTATCCTT | TGGCCGGAATACATAAGTGAAGA |
| Smad4 | GCCAGCTTCTCTGTCCAGGTAGTA | TGTCCACAGGACAGAAGCGATT |
Statistical analysis
All experiments were repeated at least 3 times and statistical analyses were conducted using computer software JMP (SAS Institute, Inc., Cary, NC). The Tukey-Kramer HSD test was used to compare multiple groups, and a student’s t-test was used for paired comparison. A p value <0.05 was considered statistically significant.
ACKNOWLEDGMENT
We thank Mary Ann Handel, Marilyn J. O'Brien and Karen Wigglesworth for their help in this study. This research was supported by grant HD21970 from the NICHD to J.J.E.
REFERENCES
- Ball GD, Bellin ME, Ax RL, First NL. Glycosaminoglycans in bovine cumulus-oocyte complexes: morphology and chemistry. Mol Cell Endocrinol. 1982;28(1):113–122. doi: 10.1016/0303-7207(82)90045-4. [DOI] [PubMed] [Google Scholar]
- Bett AJ, Haddara W, Prevec L, Graham FL. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc Natl Acad Sci U S A. 1994;91(19):8802–8806. doi: 10.1073/pnas.91.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen DF, Brown DD. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296(5567):550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Buccione R, Vanderhyden BC, Caron PJ, Eppig JJ. FSH-induced expansion of the mouse cumulus oophorus in vitro is dependent upon a specific factor(s) secreted by the oocyte. Dev Biol. 1990;138:16–25. doi: 10.1016/0012-1606(90)90172-f. [DOI] [PubMed] [Google Scholar]
- Buchanan JM. The amidotransferases. Adv Enzymol Relat Areas Mol Biol. 1973;39:91–183. doi: 10.1002/9780470122846.ch2. [DOI] [PubMed] [Google Scholar]
- Camenisch TD, Spicer AP, Brehm-Gibson T, Biesterfeldt J, Augustine ML, Calabro A, Jr, Kubalak S, Klewer SE, McDonald JA. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest. 2000;106(3):349–360. doi: 10.1172/JCI10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Mao SJ, Larsen WJ. Identification of a factor in fetal bovine serum that stabilizes the cumulus extracellular matrix. A role for a member of the inter-alpha-trypsin inhibitor family. J Biol Chem. 1992;267(17):12380–12386. [PubMed] [Google Scholar]
- Chen L, Russell PT, Larsen WJ. Functional significance of cumulus expansion in the mouse: roles for the preovulatory synthesis of hyaluronic acid within the cumulus mass. Molecular Reproduction and Development. 1993;34(1):87–93. doi: 10.1002/mrd.1080340114. [DOI] [PubMed] [Google Scholar]
- Culver CA, Laster SM. Adenovirus type 5 exerts multiple effects on the expression and activity of cytosolic phospholipase A2, cyclooxygenase-2, and prostaglandin synthesis. J Immunol. 2007;179(6):4170–4179. doi: 10.4049/jimmunol.179.6.4170. [DOI] [PubMed] [Google Scholar]
- Diaz FJ, O'Brien MJ, Wigglesworth K, Eppig JJ. The preantral granulosa cell to cumulus cell transition in the mouse ovary: development of competence to undergo expansion. Dev Biol. 2006;299(1):91–104. doi: 10.1016/j.ydbio.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Diaz FJ, Wigglesworth K, Eppig JJ. Oocytes determine cumulus cell lineage in mouse ovarian follicles. J Cell Sci. 2007;120(Pt 8):1330–1340. doi: 10.1242/jcs.000968. [DOI] [PubMed] [Google Scholar]
- Eppig JJ. FSH stimulates hyaluronic acid synthesis by oocyte-cumulus cell complexes from mouse preovulatory follicles. Nature. 1979;281(5731):483–484. doi: 10.1038/281483a0. [DOI] [PubMed] [Google Scholar]
- Eppig JJ. Oocyte control of ovarian follicular development and function in mammals. Reproduction. 2001;122(6):829–838. doi: 10.1530/rep.0.1220829. [DOI] [PubMed] [Google Scholar]
- Eppig JJ, O'Brien MJ. Development in vitro of mouse oocytes from primordial follicles. Biol Reprod. 1996;54(1):197–207. doi: 10.1095/biolreprod54.1.197. [DOI] [PubMed] [Google Scholar]
- Eppig JJ, Wigglesowrth K, O'Brien MJ. Comparison of embryonic developmental competence of mouse oocytes grown with and without serum. Mol Reprod Dev. 1992;32(1):33–40. doi: 10.1002/mrd.1080320107. [DOI] [PubMed] [Google Scholar]
- Eppig JJ, Wigglesworth K, Chesnel F. Secretion of cumulus expansion enabling factor by mouse oocytes: relationship to oocyte growth and competence to resume meiosis. Dev Biol. 1993;158(2):400–409. doi: 10.1006/dbio.1993.1198. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fulop C, Salustri A, Hascall VC. Coding sequence of a hyaluronan synthase homologue expressed during expansion of the mouse cumulus-oocyte complex. Arch Biochem Biophys. 1997;337(2):261–266. doi: 10.1006/abbi.1996.9793. [DOI] [PubMed] [Google Scholar]
- Fulop C, Szanto S, Mukhopadhyay D, Bardos T, Kamath RV, Rugg MS, Day AJ, Salustri A, Hascall VC, Glant TT, Mikecz K. Impaired cumulus mucification and female sterility in tumor necrosis factor-induced protein-6 deficient mice. Development. 2003;130(10):2253–2261. doi: 10.1242/dev.00422. [DOI] [PubMed] [Google Scholar]
- Hernandez-Gonzalez I, Gonzalez-Robayna I, Shimada M, Wayne CM, Ochsner SA, White L, Richards JS. Gene expression profiles of cumulus cell oocyte complexes during ovulation reveal cumulus cells express neuronal and immune-related genes: does this expand their role in the ovulation process? Mol Endocrinol. 2006;20(6):1300–1321. doi: 10.1210/me.2005-0420. [DOI] [PubMed] [Google Scholar]
- Hirano T, Yamauchi N, Sato F, Soh T, Hattori MA. Evaluation of RNA interference in developing porcine granulosa cells using fluorescence reporter genes. J Reprod Dev. 2004;50(5):599–603. doi: 10.1262/jrd.50.599. [DOI] [PubMed] [Google Scholar]
- Hommel JD, Sears RM, Georgescu D, Simmons DL, DiLeone RJ. Local gene knockdown in the brain using viral-mediated RNA interference. Nat Med. 2003;9(12):1539–1544. doi: 10.1038/nm964. [DOI] [PubMed] [Google Scholar]
- Jo M, Curry TE., Jr Luteinizing hormone-induced RUNX1 regulates the expression of genes in granulosa cells of rat periovulatory follicles. Mol Endocrinol. 2006;20(9):2156–2172. doi: 10.1210/me.2005-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata K, Tashiro K, Sakurai F, Osada N, Kusuda J, Hayakawa T, Yamanishi K, Mizuguchi H. Positive and negative regulation of adenovirus infection by CAR-like soluble protein, CLSP. Gene Ther. 2007;14(16):1199–1207. doi: 10.1038/sj.gt.3302975. [DOI] [PubMed] [Google Scholar]
- Kimura N, Konno Y, Miyoshi K, Matsumoto H, Sato E. Expression of hyaluronan synthases and CD44 messenger RNAs in porcine cumulus-oocyte complexes during in vitro maturation. Biol Reprod. 2002;66(3):707–717. doi: 10.1095/biolreprod66.3.707. [DOI] [PubMed] [Google Scholar]
- Kobayashi SI, Sakatani M, Kobayashi S, Okuda K, Takahashi M. Gene Silencing of Cyclooxygenase-2 mRNA by RNA Interference in Bovine Cumulus-Granulosa cells. J Reprod Dev. 2007 doi: 10.1262/jrd.19050. [DOI] [PubMed] [Google Scholar]
- Kwintkiewicz J, Cai Z, Stocco C. Follicle-stimulating hormone-induced activation of Gata4 contributes in the up-regulation of Cyp19 expression in rat granulosa cells. Mol Endocrinol. 2007;21(4):933–947. doi: 10.1210/me.2006-0446. [DOI] [PubMed] [Google Scholar]
- Leonard SL, Kurzrok R. A study of the hyaluronidase effect in the collicle cells of ovulated rat ova. Endocrinology. 1945;37:171–176. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (Duluth) 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Finegold MJ, Su JG, Hsueh AJ, Bradley A. Alpha-inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature. 1992;360(6402):313–319. doi: 10.1038/360313a0. [DOI] [PubMed] [Google Scholar]
- McClean D, Rowlands IW. Role of hyaluronidase in fertilization. Nature. 1942;150:627–628. [Google Scholar]
- Ochsner SA, Day AJ, Rugg MS, Breyer RM, Gomer RH, Richards JS. Disrupted function of tumor necrosis factor-alpha-stimulated gene 6 blocks cumulus cell-oocyte complex expansion. Endocrinology. 2003a;144(10):4376–4384. doi: 10.1210/en.2003-0487. [DOI] [PubMed] [Google Scholar]
- Ochsner SA, Russell DL, Day AJ, Breyer RM, Richards JS. Decreased expression of tumor necrosis factor-alpha-stimulated gene 6 in cumulus cells of the cyclooxygenase-2 and EP2 null mice. Endocrinology. 2003b;144(3):1008–1019. doi: 10.1210/en.2002-220435. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16(8):948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parakh TN, Hernandez JA, Grammer JC, Weck J, Hunzicker-Dunn M, Zeleznik AJ, Nilson JH. Follicle-stimulating hormone/cAMP regulation of aromatase gene expression requires beta-catenin. Proc Natl Acad Sci U S A. 2006;103(33):12435–12440. doi: 10.1073/pnas.0603006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Su YQ, Ariga M, Law E, Jin SL, Conti M. EGF-like growth factors as mediators of LH action in the ovulatory follicle. Science. 2004;303(5658):682–684. doi: 10.1126/science.1092463. [DOI] [PubMed] [Google Scholar]
- Paul CP, Good PD, Winer I, Engelke DR. Effective expression of small interfering RNA in human cells. Nat Biotechnol. 2002;20(5):505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- Richards JS. Ovulation: new factors that prepare the oocyte for fertilization. Mol Cell Endocrinol. 2005;234(1–2):75–79. doi: 10.1016/j.mce.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Richards JS, Russell DL, Ochsner S, Espey LL. Ovulation: new dimensions and new regulators of the inflammatory-like response. Annu Rev Physiol. 2002;64:69–92. doi: 10.1146/annurev.physiol.64.081501.131029. [DOI] [PubMed] [Google Scholar]
- Russell WC. Update on adenovirus and its vectors. J Gen Virol. 2000;81(Pt 11):2573–2604. doi: 10.1099/0022-1317-81-11-2573. [DOI] [PubMed] [Google Scholar]
- Schoenfelder M, Einspanier R. Expression of hyaluronan synthases and corresponding hyaluronan receptors is differentially regulated during oocyte maturation in cattle. Biol Reprod. 2003;69(1):269–277. doi: 10.1095/biolreprod.102.011577. [DOI] [PubMed] [Google Scholar]
- Shimada M, Hernandez-Gonzalez I, Gonzalez-Robayna I, Richards JS. Paracrine and autocrine regulation of epidermal growth factor-like factors in cumulus oocyte complexes and granulosa cells: key roles for prostaglandin synthase 2 and progesterone receptor. Mol Endocrinol. 2006;20(6):1352–1365. doi: 10.1210/me.2005-0504. [DOI] [PubMed] [Google Scholar]
- Shimada M, Yanai Y, Okazaki T, Yamashita Y, Sriraman V, Wilson MC, Richards JS. Synaptosomal-associated protein 25 gene expression is hormonally regulated during ovulation and is involved in cytokine/chemokine exocytosis from granulosa cells. Mol Endocrinol. 2007;21(10):2487–2502. doi: 10.1210/me.2007-0042. [DOI] [PubMed] [Google Scholar]
- Shimasaki S, Moore RK, Otsuka F, Erickson GF. The bone morphogenetic protein system in mammalian reproduction. Endocr Rev. 2004;25(1):72–101. doi: 10.1210/er.2003-0007. [DOI] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Wigglesworth K, O'Brien MJ, Affourtit JP, Pangas SA, Matzuk MM, Eppig JJ. Oocyte regulation of metabolic cooperativity between mouse cumulus cells and oocytes: BMP15 and GDF9 control cholesterol biosynthesis in cumulus cells. Development. 2008;135(1):111–121. doi: 10.1242/dev.009068. [DOI] [PubMed] [Google Scholar]
- Sugiura K, Pendola FL, Eppig JJ. Oocyte control of metabolic cooperativity between oocytes and companion granulosa cells: energy metabolism. Dev Biol. 2005;279(1):20–30. doi: 10.1016/j.ydbio.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Sugiura K, Su YQ, Diaz FJ, Pangas SA, Sharma S, Wigglesworth K, O'Brien MJ, Matzuk MM, Shimasaki S, Eppig JJ. Oocyte-derived BMP15 and FGFs cooperate to promote glycolysis in cumulus cells. Development. 2007;134(14):2593–2603. doi: 10.1242/dev.006882. [DOI] [PubMed] [Google Scholar]
- Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC, Shi Y. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci U S A. 2002;99(8):5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot P. Hyaluronidase dissolves a component in the hamster zona pellucida. J Exp Zool. 1984;229(2):309–316. doi: 10.1002/jez.1402290216. [DOI] [PubMed] [Google Scholar]
- Tamura K, Matsushita M, Endo A, Kutsukake M, Kogo H. Effect of insulin-like growth factor-binding protein 7 on steroidogenesis in granulosa cells derived from equine chorionic gonadotropin-primed immature rat ovaries. Biol Reprod. 2007;77(3):485–491. doi: 10.1095/biolreprod.106.058867. [DOI] [PubMed] [Google Scholar]
- Vanderhyden BC, Caron PJ, Buccione R, Eppig JJ. Developmental pattern of the secretion of cumulus-expansion enabling factor by mouse oocytes and the role of oocytes in promoting granulosa cell differentiation. Dev Biol. 1990;140:307–317. doi: 10.1016/0012-1606(90)90081-s. [DOI] [PubMed] [Google Scholar]
- Varani S, Elvin JA, Yan C, DeMayo J, DeMayo FJ, Horton HF, Byrne MC, Matzuk MM. Knockout of pentraxin 3, a downstream target of growth differentiation factor-9, causes female subfertility. Mol Endocrinol. 2002;16(6):1154–1167. doi: 10.1210/mend.16.6.0859. [DOI] [PubMed] [Google Scholar]
- Weigel PH, Hascall VC, Tammi M. Hyaluronan synthases. J Biol Chem. 1997;272(22):13997–14000. doi: 10.1074/jbc.272.22.13997. [DOI] [PubMed] [Google Scholar]
- Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20(10):1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci U S A. 2002;99(9):6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]