SUMMARY
Mapping DNase I hypersensitive (HS) sites is an accurate method of identifying the location of genetic regulatory elements, including promoters, enhancers, silencers, insulators, and locus control regions. We employed high-throughput sequencing and whole-genome tiled array strategies to identify DNase I HS sites within human primary CD4+ T cells. Combining these two technologies, we have created a comprehensive and accurate genome-wide open chromatin map. Surprisingly, only 16%–21% of the identified 94,925 DNase I HS sites are found in promoters or first exons of known genes, but nearly half of the most open sites are in these regions. In conjunction with expression, motif, and chromatin immunoprecipitation data, we find evidence of cell-type-specific characteristics, including the ability to identify transcription start sites and locations of different chromatin marks utilized in these cells. In addition, and unexpectedly, our analyses have uncovered detailed features of nucleosome structure.
INTRODUCTION
The discovery and characterization of noncoding functional elements in genomes is paramount to understanding the complexities of gene expression in different biological systems. One established and robust method to locate active functional elements is through the identification of regions of the genome that are hypersensitive to DNase I cleavage (Keene et al., 1981; McGhee et al., 1981). In the nucleus, the vast majority of genomic DNA is wrapped around regularly spaced protein complexes called nucleosomes that serve to package DNA and also affect important biological processes such as gene transcription (Felsenfeld and Groudine, 2003). Regions where local modifications to this chromatin structure displace these nucleosomes (such as for the activation of promoters) allow for easier digestion by DNase I. DNase I hyper-sensitive (HS) sites have been shown over the last 28 years to be markers for many different types of genetic regulatory elements, including promoters, enhancers, silencers, insulators, and locus control regions (Felsenfeld and Groudine, 2003; Gross and Garrard, 1988; Stalder et al., 1980). More recently, the ENCODE consortium has shown that DNase I HS sites identified in 1% of the human genome were robust markers for histone modifications, regions of early replication, transcription start sites, and transcription factor binding sites (ENCODE, 2007). Accurately identifying the locations of DNase I HS sites across the entire genome will help us to understand the biological basis for gene regulation expression patterns in different cell types within and across species, in response to external stimuli, and in diseased tissues.
Until recently, individual DNase I HS sites were identified using traditional Southern blot assays (Wu, 1980). While this method has provided key insights about gene regulation, the low throughput nature of this strategy limits analysis to small regions of the genome. More recently, we developed two high-throughput strategies to simultaneously identify thousands of DNase I HS sites in an undirected and nonbiased manner (Crawford et al., 2006a, 2006b). Both methods start with chromatin that is digested with a small amount of DNase I that preferentially cuts at a DNase I HS site, followed by the attachment of a biotinylated linker to the DNase I-digested ends. The linker is used to extract short adjacent DNA fragments that can be identified by either next generation sequencing (DNase-seq) or labeling and hybridization to tiled microarrays (DNase-chip). Initial experiments using these and similar strategies, though an advance over the Southern blot, were not comprehensive (Crawford et al., 2006a, 2004, 2006b; Sabo et al., 2004a, 2004b, 2006). For example, our DNase-seq experiments produced approximately 230,000 sequence tags and identified an estimated 20% of sites (Crawford et al., 2006b) while our DNase-chip strategy covered only 1% of the genome (Crawford et al., 2006a).
The development of higher throughput sequencing platforms by Illumina (formerly Solexa) and Roche (454 Life Sciences), as well as the availability of genome-wide tiled genomic microarrays by NimbleGen, has now allowed us to create the first genome-wide map of DNase I HS sites in human cells. The different readout platforms for DNase-seq and DNase-chip provide for independent and complementary whole-genome validation. A unique property of DNase-seq is that it generates basepair resolution of DNase I digestion. While DNase-chip has slightly lower resolution (limited to size of sheared fragments that range from 200–500 bases), this method has also been shown to be highly sensitive and specific at identifying valid DNase I HS sites (Crawford et al., 2006a). By combining whole-genome data from both DNase-seq and DNase-chip, we provide an unprecedented, high-quality view of human chromatin.
RESULTS
Comprehensive Genome-Wide Identification of DNase I HS Sites
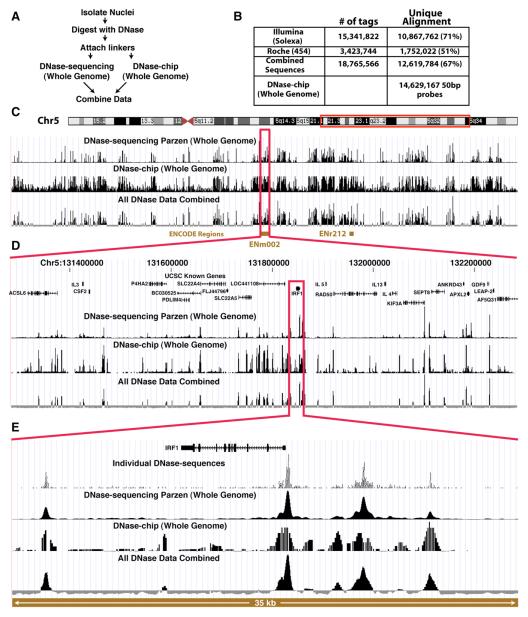
To perform DNase-seq, we generated a single DNase I library from primary human CD4+ T cells and sequenced the same sample using both the Illumina and 454 platforms (Figure 1A). Over 18 million unique sequence tags were generated and over 12 million (∼70%) were uniquely mapped to the NCBI Build 35 genome sequence assembly (Figure 1B). We employed a kernel density estimation function called Parzen windowing (Parzen, 1962) to identify DNase I HS sites from uniquely mapping tags. This technique allows for each base position to be scored based on the number of sequences in the immediate area with those a short distance away having a greater weight than those more distant. The resulting annotation, shown in the UCSC Genome Browser (Kent et al., 2002), more accurately reflects the boundaries of DNase I HS sites (Figure 1).
Figure 1. DNase-Chip and DNase-Seq Identify DNase I Hypersensitive Sites on a Whole-Genome Scale.
(A) Each method begins with the digestion of intact nuclei with DNase I followed by the attachment of linkers. Each technology is then used to independently identify DNase I HS sites. Finally, the data are combined into a comprehensive, high-resolution and low-noise map of HS sites on a genome-wide scale.
(B) Number of sequence tags generated using Illumina and 454 technologies, as well as probes for whole-genome DNase-chip studies.
(C) UCSC genome browser view of the q arm of chromosome 5 showing a large-scale view of each technology along with the combined set.
(D) Browser view of ENCODE region ENm002.
(E) Browser view of the DNase I HS sites around the IRF1 gene. Each of these views shows the high correlation between the peak size and location for both the sequencing and chip technologies.
To perform whole-genome DNase-chip, a DNase I library was generated from the identical cells used for sequencing, as well as from a second biological replicate (Figure 1A). DNase I-treated DNA was hybridized to two 38-array sets (NimbleGen) consisting of 100-bp-spaced oligonucleotides that cover the entire nonrepetitive fraction of the genome. The resulting data were averaged across the two biological replicates and processed as previously described (Scacheri et al., 2006).
Based on previous work, we predict that signal strength in both methods reflects the degree of openness at that site. Accordingly, we find that raw ratio DNase-chip intensity values and corresponding DNase-seq Parzen window scores are correlated (Pearson’s R = 0.45, Figures 1C–1E, and see Figure S1 available online). We also compared DNase-seq and DNase-chip data to a third independent readout platform that validates DNase I HS sites using quantitative PCR (qPCR). QPCR has been shown to be an acceptable proxy for Southern blotting and allows for semi-high throughput validation (McArthur et al., 2001). Previously, we employed qPCR to validate 287 DNase I HS sites and 321 DNase I-resistant sites in CD4+ T cells (Crawford et al., 2006a, 2006b). We find that qPCR signal intensity is highly correlated to both DNase-seq and DNase-chip (Figures 2A and 2B).
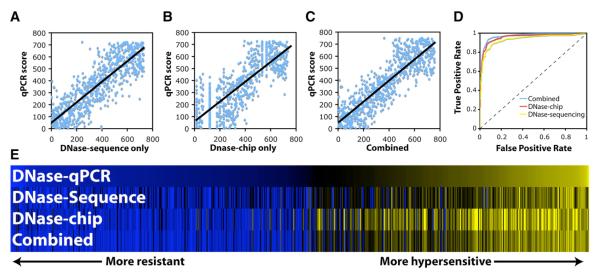
Figure 2. Each Individual Technology Is Highly Correlated with the Entire Spectrum of qPCR Values.
(A) DNase-seq data are correlated with qPCR with Spearman’s ρ = 0.744.
(B) DNase-Chip is correlated with qPCR with ρ = 0.812.
(C) The combined DNase-seq/DNase-chip dataset is even more correlated with qPCR with ρ = 0.874.
(D) Receiver Operating Characteristic (ROC) curves showing sensitivity and specificity. DNase-seq has an area under the curve (AUC) of 0.937, DNase-Chip has an AUC of 0.956, and the combined dataset has an AUC of 0.971. A perfect discriminator would have an AUC of 1, while a random test would have an AUC of 0.5 (dashed line).
(E) Heat map showing the range of qPCR experiments (n = 608) with yellow showing true DNase I HS and blue showing DNase resistant sites. Note that in some cases both DNase-Chip and DNase-seq agree on a qPCR negative call, indicating that this site may in fact be hypersensitive.
These correlations support the view that DNase I hypersensitivity is not a binary property but rather reflects a continuous range of chromatin accessibility or “openness.” The significance of differential hypersensitivity is unknown, but may represent true biological phenomena such as protein occupancy. This makes validation by independent methods especially critical for identifying a weak but significant signal.
For many analyses, such as determining sensitivity and specificity, it is necessary to define discrete DNase I HS site regions. This primarily requires determining appropriate thresholds to distinguish positives from negatives. To do this, we used our previously published qPCR data (Crawford et al., 2006a). Figure 2D shows a Receiver Operating Characteristic (ROC) curve showing how sensitivity and specificity changes at different thresholds for DNase-seq and DNase-chip annotations. The large areas under the ROC curves for both emphasize the high sensitivity and specificity of both methods.
Given the high quality of both data sets, we combined them to produce a single global DNase I HS map. To do this, we independently mapped the scores from each set to a similar distribution, normalized each based on sensitivity and specificity, and summed the resulting scores. The new combined set was more highly correlated with qPCR values (Spearman’s ρ = 0.87, Figure 2C), indicating that it is accurate at detecting the degree of hypersensitivity. We also found the combined set to be more accurate than either of the individual datasets by ROC curve analysis (Figure 2D), and overall generated a sensitivity of 92% with a specificity of 94%. Furthermore, assuming that the top and bottom 20% of the qPCR data represent the most accurate positive and negative annotations, we find a sensitivity of > 99.9% and a specificity of 98%. The correlation among these data can be visualized as a heat map showing the labeling of individual samples (Figure 2E). These combined data also accurately identify many previously mapped DNase I HS sites (using Southern blots and qPCR) from T cells such as in the interferon gamma, Il-3, Il-2, and Il-4 loci (Agarwal and Rao, 1998; Cockerill et al., 1993; Lee et al., 2004; Schoenborn et al., 2007; Siebenlist et al., 1986, and Figure S2). Together, these show that combining data from multiple, independent high-throughput platforms can increase both sensitivity and specificity, as well as provide for a more accurate measure of degree of DNase I hypersensitivity. Our DNase I HS map can be viewed and/or downloaded from the Duke DN-ase I track on the UCSC genome browser (Kent et al., 2002,http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17).
Functional Annotation of DNase I HS Sites
In agreement with our previous estimates (Crawford et al., 2004, 2006b), our combined DNase I HS annotation identifies 94,925 DNase I HS sites covering 60 million bases (2.1%) across the genome. We find that only 43% of all DNase I HS sites overlap evolutionarily constrained regions of the genome, which is similar to that found recently in 1% of the genome (ENCODE, 2007). In addition, 75% of DNase I HS sites within 2 kb of a transcription start site are constrained (Figure S3). This indicates that the majority of DNase I HS sites have not evolved under selection.
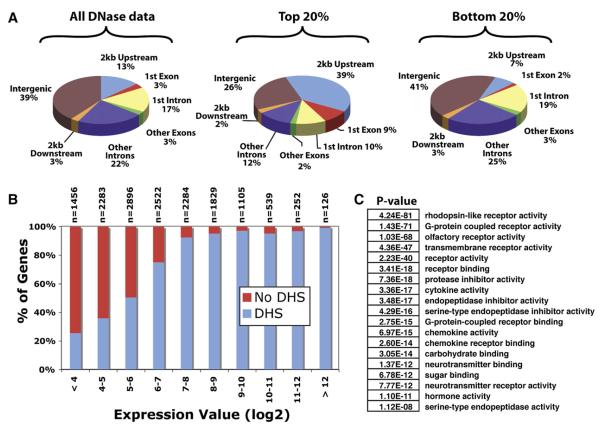
We have analyzed the distribution of DNase I HS sites across the genome with respect to genes based on the Known Genes annotation in the UCSC Genome Browser (Figure 3A). Only 16% of all DNase I HS sites map to a first exon or within 2 kb upstream (promoter region) of a gene, while 17% map to a first intron and 25% map within other exonic or intronic regions. However, the promoter and first exon HS sites are more than twice the size and twice as hypersensitive on average than in other regions (Table S1). In fact, looking specifically at the strongest 20% of all DNase I HS signals, we find that almost half of these map to a first exon or within 2 kb upstream of a gene (Figure 3A and Table S1). This indicates that promoter regions for protein-coding genes are extremely hypersensitive to DNase I digestion, while presumptive regulatory elements unrelated to known promoters are less susceptible to digestion (although still significantly more susceptible than the genome average). At the opposite extreme, the categorical distribution of the weakest DNase I HS sites does not differ substantially from what is seen for all sites (Figure 3A). The fact that these are not randomly distributed but rather reflect what is seen in stronger sites supports the validity of these smaller, less-sensitive regions.
Figure 3. Location of DNase I Hypersensitive Sites Relative to Annotated Genes.
(A) The locations of DNase I hypersensitive (DHS) sites relative to gene annotations. Shown are the locations of all DNase I HS sites, the strongest scoring DNase I HS sites (top 20%), and the weakest scoring DNase I HS sites (bottom 20%).
(B) Genes that have high expression (>9) are likely to have a DNase I HS site at the 5′ end, while genes lacking a 5′ DNase I HS site are more likely to have low expression.
(C) GO categories and probabilities related to genes that are lacking 5′ DNase I HS sites.
The Known Genes annotation in the UCSC Genome Browser is not complete and therefore some DNase I HS sites may not be correctly categorized in the above analysis. To help correct for this, we compared the DNase I HS sites to recently generated genome-wide data for RNA polymerase II (Pol II) binding in CD4+ T cells (Barski et al., 2007). We find that an additional 5.5% of all DNase I HS sites that map within the intergenic category (Figure 3A) are within 2 kb of a strong Pol II signal indicating these may be in the promoter, first exon, or first intron of an unannotated gene. Additionally, if we consider all mRNA and EST sequences as evidence of transcribed regions, only 15.5% of the DNase I HS sites would be classified as being intergenic (data not shown). This shows that a more complete gene annotation is necessary to fully understand the distribution of HS sites within and around genes.
We explored the relationship between DNase I HS sites and levels of gene expression using CD4+ T cell Affymetrix expression array data previously generated by us from CD4+ T cells. Using these data, we assigned expression levels to 15,293 Known Genes that had Protein Databank, RefSeq, and/or Swiss-Prot supporting evidence. For each gene, we determined if there existed a DNase I HS site overlapping a 200 bp window centered on any annotated transcription start site (TSS) in the Known Genes annotation. Nearly all highly expressed genes were found to have an associated HS site (Figure 3B). We investigated all 67 of the 2000 highest expressed genes (Figure 3B, expression value > 9) that appeared to lack a DNase I HS site. In 41 instances (61%), there is ample EST and mRNA evidence for an unannotated TSS for which we do see a DNase I HS site. Another eight have a nearby DNase I HS site that may indicate an unannotated TSS but without other transcript support, eight are contained in recent segmental duplications where we lack the ability to uniquely map sequences, and one is a pseudogene. The explanation for the remaining nine highly expressed genes is not clear but may be due to unknown annotation errors, biological variations in the expression programs in the separate T cell populations, or possibly a low false-negative rate of our assays.
We conclude that the TSS of essentially all highly expressed protein-coding genes, and possibly all expressed genes, is marked by a DNase I HS site, nearly all of which can be identified by DNase-seq and/or DNase-chip. This implies that these results can be used to confirm currently annotated TSSs, identify novel TSSs, or help determine which alternative TSSs are being used in a particular cell type (Carninci et al., 2005). However, though DNase I HS sites might be necessary, they are clearly not sufficient for gene expression (Figure 3B). Those genes that have a DNase I HS site but are not expressed may be in a transcriptionally poised state (Gross and Garrard, 1988).
One might expect that the degree of hypersensitivity would be related to the amount of transcriptional activity, but that relationship is not clear. When looking simply at expression levels versus degree of hypersensitivity, we find little correlation (Pearson’s R = 0.09, data not shown). Interestingly, the average level of hypersensitivity is significantly less at the TSSs of low or non-expressed genes (expression < 5) than moderately to highly expressed genes (Student’s t test, p < 2e-16). This is supportive evidence that actively transcribed genes are more hypersensitive due to the presence of the full complement of transcriptional machinery. It is possible that genes that are poised lack some or all of these elements leading to reduced levels of hypersensitivity.
Histone Modifications and Transcription Factor Binding Sites
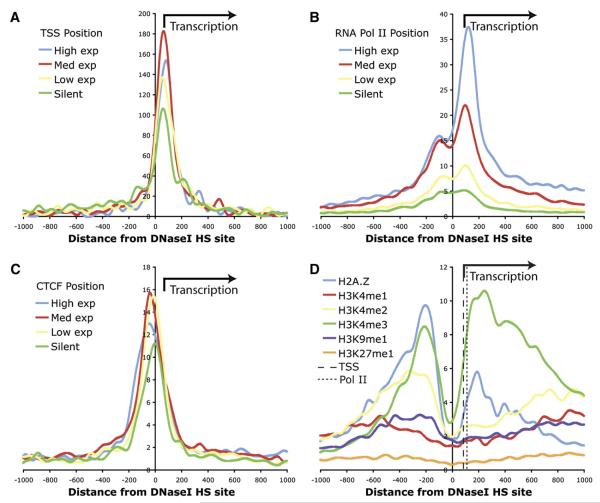
A recent study used high-throughput sequencing to find genomic regions enriched for several different histone methylations, RNA Pol II, CTCF, and the histone variant H2AZ in CD4+ T cells (Barski et al., 2007). Their analysis focused on the relationship of these data as compared to TSS. We performed similar analyses but instead centered these chromatin marks on the strongest hypersensitivity signal within our proximal DNase I HS sites, defined as within 2 kb of an annotated TSS or RNA Pol II signal (Figure S4). We find that the peak DNase I signal within each DN-ase I HS site is shifted 5′ from annotated TSS (Figure 4A), RNA Pol II (Figure 4B), and RIKEN CAGE tags (Kawaji et al., 2006 and Figure S5), indicating that on average DNase I HS sites are directional relative to transcription. This is consistent regardless of the level of transcription in the associated gene. In contrast, CTCF is on average located preferentially on the opposite side of the DNase I HS site with respect to the transcription start site (Figure 4C). Other groups have noticed dips in active histone marks near the transcription start sites, indicating nucleosome depleted regions (Barski et al., 2007; Heintzman et al., 2007). We find that these modified histone troughs do not directly overlap transcription start sites or RNA Pol II, but are instead directly overlapping the strongest portion of each DNase I HS site (Figure 4D), providing supportive evidence that these regions are nucleosome depleted.
Figure 4. TSS and ChIP-Seq Data Related to the Strongest Portion of Each DNase I HS Site.
(A) Transcription start site for annotated genes in UCSC Genome Browser Known Genes track are on average 85 bp downstream from the DNase I HS sites.
(B) RNA Polymerase II ChIP-seq data are enriched on average 123 bp downstream from DNase I HS sites.
(C) CTCF ChIP-seq data are enriched for a peak slightly upstream of the DNase I HS sites. TSS, Pol II, and CTCF datasets are divided into four groups based on expression level (high, medium, low, and silent).
(D) Histone modifications and H2A.Z are enriched for the DNase I HS sites that are near highly expressed genes. A trough for each histone modification and H2A.Z directly overlaps the strongest portion of each DNase I HS site, and not TSS (thick dotted line) or Pol II (thin dotted line). Note that for ease of comparison, H3K4me3 normalized counts were halved.
To compare to specific histone marks, we divided DNase I HS sites into three groups: (1) those proximal to TSS, (2) those that overlap transcribed regions (mRNA and/or EST evidence), and (3) distal sites that make up the remainder. As expected, we find that proximal HS sites are associated with activating histone marks found at promoters (Figure S6). Many of these marks associated with active promoters were found at different levels around DNase I HS sites that overlap transcribed regions compared to distal sites. Those overlapping transcribed regions showed an enrichment for H3K36me3 in agreement with a recent study (Mikkelsen et al., 2007), but this modification was not found in distal HS sites. Distal sites were more enriched in H3K27me3, H3K9me2, and H3K9me3 marks, while those found in transcribed regions were more enriched in H3K27me1 and H3K9me1 modifications (Figure S6). This shows that nonpromoter DNase I HS sites can be broadly classified into at least two distinct categories based on histone methylations and their relationship to transcription.
As mentioned earlier, many genes with essentially little or no detectable expression in microarray experiments (exp < 4.5) have DNase I HS sites in their promoter regions, while others do not. We found that within these transcriptionally silent genes, the presence of a DNase I HS sites was accompanied by activating histone marks and binding of RNA Pol II (Figure S7), but these signals were not as strong as those around HS sites near more highly expressed genes (Figure S4). In contrast, promoter regions near silenced genes with no HS site showed no evidence of these marks (Figure S7). This suggests that these genes that are silent but are marked with a DNase I HS sites are either poised for expression, or that they are expressed at a level too low to be reliably detected by current microarray technology.
Cell-Type-Specific Gene Ontology and Motif Analysis
To characterize genes and gene families with respect to presence or absence of a DNase I HS site, we performed Gene Ontology (GO) analyses using GoStat (Beissbarth and Speed, 2004). Genes that lack a DNase I HS site at the TSS are significantly enriched for non-CD4+ T cell related GO categories, such as visual, olfactory, digestive, and neurotransmitter activity (Figure 3C). In contrast, genes that are both highly expressed and have an extremely hypersensitive promoter region are enriched for housekeeping-like GO categories, such as mRNA processing and splicing and general metabolic processes (data not shown).
To determine if we could detect any cell-type specific function, we searched for enriched motifs in the 75,954 DNase I HS sites that are more than 2 kb away from a TSS (TSS were removed to avoid simply identifying core promoter elements). Using the Clover algorithm (Frith et al., 2004) and all motifs in the TRANS-FAC database (Wingender et al., 1996), we detected enrichment for several families of transcription factors known to be involved in the immune system: AML, PU.1, ETS, C/EBP, STAT, IRF, and TAL1 (Meraro et al., 1999; Smith et al., 2006; Wadman et al., 1997; Yao et al., 2006). We also detected enrichment for a motif corresponding to CTCF, a known insulator protein that is not currently in the TRANSFAC database (Bell et al., 1999; Kim et al., 2007). An example of the Clover scores for AML in DNase I HS sites and background sequences are plotted across all chromosomes in Figure S8. To verify that the motif discovery and enrichment represented in vivo biology, we compared our enriched motifs to recently published ChIP-seq data. We confirmed that a significant fraction (p value < 0.0001) of DNase I HS sites that have the STAT motif overlap with STAT1 ChIP-seq data from HeLa S3 cells (Robertson et al., 2007) and the CTCF motif corresponding to CTCF ChIP-seq from CD4+ T cells (Barski et al., 2007) (Figure S9). Thus, we conclude that motifs that likely regulate global CD4+ T cell gene expression are enriched in DNase I HS sites identified within CD4+ T cells.
Genes that are specifically expressed in CD4+ T cells have on average more DNase I HS sites. While all genes that have a DNase I HS site at the TSS have on average of 4.71 (±6.86 SD) sites, 158 genes that are uniquely expressed in CD4+ T cells were found to have significantly more DNase I HS sites (11.03 ± 15.90 SD) within their promoter (2 kb upstream) and transcribed regions. The higher number of DNase I HS sites in CD4+ specific genes suggests that cell type gene expression has more complex combinatorial control.
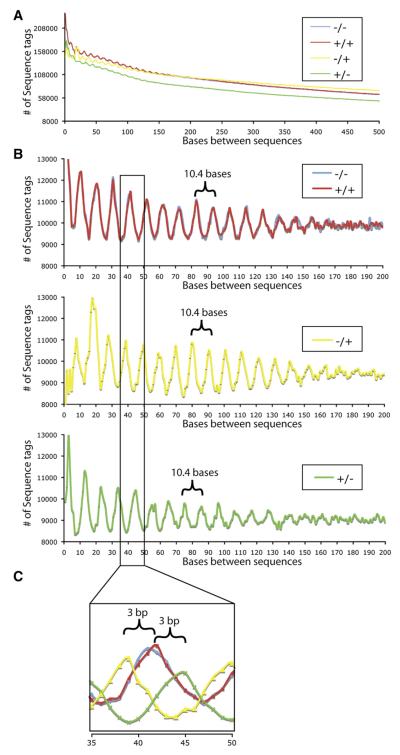
Identification of Fine-Scale Nucleosome Structure
Approximately 30% of all DNase I sequences map within a DNase I HS site. The remaining sequences are likely a result of background DNase I nicking. To determine whether background DNase-seq data displayed patterns that might result from positioned nucleosomes, we tallied distances between digestion sites, taking into account the sequence strand. We noticed a striking, repeating, oscillating pattern that has an average frequency of 10.4 bases (Figure 5A) that is highly enriched for regions outside of DNase I HS sites (Figure 5B). We note that this oscillation frequency is the exact distance in bases of a single turn of the double helix. This pattern was first shown over 30 years ago by gel electrophoresis believed to result from DNase I cleaving the exposed minor groove of the double helix wrapped around nucleosomes (Noll, 1974). Interestingly, this oscillation pattern occurs for approximately 14–15 periods, matching the amount of DNA that is directly associated with a single nucleosome (Felsenfeld and Groudine, 2003).
Figure 5. Ultra-High-Resolution View of Chromatin Structure.
(A) A clear oscillation pattern is visible for sites < 150 bp apart.
(B) A 10.4 base pair oscillation frequency is observed only in DNase sequences that do not map within DNase I HS sites. This pattern exists between sequences that are on the same strand (+/+ and -/-) and on opposite strands (-/+ and +/-).
(C) By overlaying the data from the different strand sets, we find that the two opposite stranded sets each have an approximately three base offset from the same stranded set.
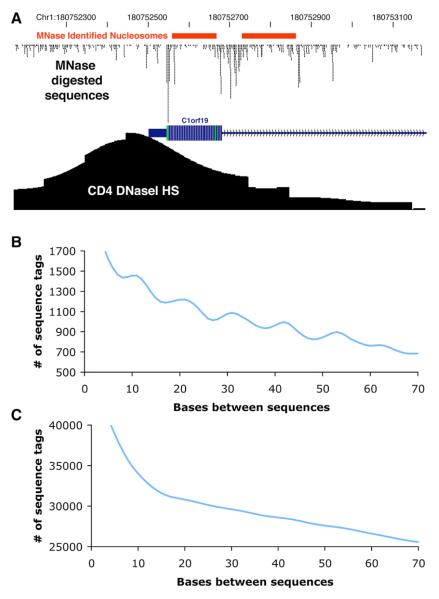
Although we detect no oscillation pattern within DNase I HS sites as a whole (Figure S10), we hypothesized that there may be well-positioned nucleosomes on the boundaries of DNase I HS sites that should also be enriched for minor groove cutting. We mapped putative nucleosome positions using data from the histone methylation ChIP-seq experiments that initially digested CD4+ T cell chromatin with MNase to isolate mononucleosomes (Barski et al., 2007). By pooling data from all experiments, we were able to identify dense clusters of sequences aligned in the same orientation indicating the boundaries of nucleosomes (Figure 6A). We then mapped ∼3,000 putative nucleosomes of length 100–160 bps that were bracketed on both sides by clusters of at least 45 sequences where each sequence is separated by no more than one basepair. The majority of these nucleosomes were contained within of the boundaries of DNase I HS sites, but were offset from the strongest DNase I signal and generally located near the edges. We were able to detect the oscillation pattern (previously discernable only outside of DNase I HS sites) within portions of DNase I HS sites that display nucleosome positioning (Figure 6B) but not within the remainder of the DNase I HS site (Figure 6C). The ability to detect this pattern within a population of cells indicates that a significant percentage of nucleosomes are precisely positioned and implies that there is a sequence or chromatin signal that regulates this process.
Figure 6. MNase-Derived Sequence Tags Define Nucleosomes Near the Boundaries of DNase I Hypersensitive Sites.
(A) Representative example of MNase sequence tags and MNase identified positioned nucleosomes (red bars).
(B) Regions of DNase I HS sites that overlap positioned nucleosomes are enriched for the 10.4 bp periodicity also detected in the whole-genome data.
(C) Regions of DNase I HS sites not associated with a positioned nucleosome do not display the 10.4 bp oscillation pattern.
Another unique property of DNase I is that in the presence of Mg2+, this enzyme nicks one strand of DNA at a time, often leaving a 2–4 base pair overhang when digesting intact chromatin (Cousins et al., 2004; Sollner-Webb et al., 1978). As DNase-seq allows us to identify the DNA strand that was cut, we have detected a 3 bp average overhang created by DNase I in the non-HS regions (Figure 5C, detailed explanation in Figure S11).
While the depth of sequencing that we have performed does not allow robust characterization of DNase I cutting within individual regions of the genome (that is, we can only detect oscillating patterns as a composite of all sequences), this might be possible with extremely deep sequencing. Therefore, we believe DNase-seq, in conjunction with ChIP-seq for MNase experiments, has the potential to identify the exact location of each basepair with respect to its precise positioning around a stably positioned nucleosome.
DISCUSSION
The accurate and comprehensive DNase I HS map presented here offers an unprecedented view of open chromatin structure at extremely high resolution. We have shown that both tiled microarrays and high-throughput sequencing are very accurate at identifying DNase I HS sites across the genome and combining these platforms improves the sensitivity, specificity, and the ability to determine the degree of hypersensitivity. We believe this is especially important for correctly calling DNase I HS sites that are more moderately hypersensitive. In future studies, most would find it undesirable and likely cost prohibitive to employ both technologies. While both methods are very accurate at identifying DNase I HS sites, each method has unique benefits and limitations. For example, we have demonstrated that DNase-seq can also be used to detect sub-nucleosome structure, something not possible with current tiling arrays. However, DNase-seq analysis on aneuploid cell lines will be difficult without performing extensive sequencing of the input DNA from each cell type. In contrast, tiled arrays readily normalize for DNA content and thus are suitable for cells with abnormal karyotypes. In addition, while DNase-seq can currently only be used to study the whole genome, tiling arrays can be used for inexpensive validation and for studying smaller targeted regions of the genome. Neither platform is well suited for duplicated sequences such as those found in recent segmental duplications, or for other highly repetitive sequences. Therefore, it is difficult to compare costs of the two technologies, since it depends on the particular application.
DNase I HS maps provide a scaffold on which to combine and analyze data from ChIP-chip/ChIP-Seq and gene expression experiments to better understand complex gene regulation. In our limited study in a single cell type, we are able to show previously undescribed positional relationships between DNase I HS peaks, transcription start sites, and sites of RNA PolII binding. We also describe differences in histone modifications around different categories of HS sites based on their degree of hypersensitivity, their positional relationship to transcription start sites, and the expression level of associated genes.
As similar types of data continue to be produced from different cell types, we anticipate the development of regulatory maps consisting of DNase I HS sites that are characterized by the presence or absence of features such as histone modifications, DNA binding proteins, DNA methylation, nucleosome positions, SNPs, insertions and deletions that collectively explain the transcriptional status of associated genes in particular cell types under particular conditions. In addition, DNase I HS maps can be used to focus computational motif discovery and analyses on those regions of the genome most likely to contain functional binding, a role that evolutionary conservation has not satisfactorily filled (ENCODE, 2007).
The data resource presented here should be of particular interest to those studying the biology of CD4+ T cells, the regulation of genes that are expressed in many cell types, and those studying comparative genomics. We have shown that we efficiently identify previously characterized HS sites in these cells, and our data should therefore benefit future research. The further generation of genome-wide DNase I HS maps from a diverse set of normal and diseased human cell types, as well as from those from other species, will continue to reveal how chromatin structure, and underlying primary sequence differences, contribute to cell-type specific gene expression and cell fate decisions.
EXPERIMENTAL PROCEDURES
Preparation of DNase I-Treated DNA
Intact nuclei from primary human CD4+ T cells were digested with DNase I, and prepared for DNase-seq and DNase-chip. For DNase-seq using Solexa, biotinylated linker I (5′ Bio-ACAGGTTCAGAGTTCTACAGTCCGAC and 5′ P- GTCGGACTGTAGAACTCTGAAC) that has a MmeI site at the 3′ end was ligated to the DNase-digested ends. After MmeI digestion, DNase ends were enriched on streptavidin beads (Invitrogen) and ligated to linker II (5′ P-TCGTATGCCGTCTTCTGCTTG and 5′ CAAGCAGAAGACGGCATACGANN). The DNase material was amplified by PCR using linker-specific primers (5′ CAAGCAGAAGACGGCATACGA and 5’ AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGA), purified by PAGE, and sequenced using a primer specific to linker I (5′ CCACCGACAGGTTCAGAGTTCTACAGTCCGAC). DNase-chip was performed exactly as was previously described (Crawford et al., 2006a). Briefly, DNase-digested ends were ligated to biotinylated linkers, followed by sonication. DNase-digested ends were enriched on a streptavidin column, and a second set of linkers was ligated to the sheared ends. DNase-enriched DNA was amplified by PCR, labeled, and hybridized to NimbleGen arrays.
Hybridization to Tiled Microarrays and Data Analysis
DNase-chip material from 2 biological replicates (as well as randomly sheared DNA used as a reference control) was hybridized to whole-genome NimbleGen 38 array sets that contain ∼14,629,167 50 bp probes spaced approximately every 100 bp of unique sequence. Raw log2 ratio-transformed data from each array were centered to have median of 0 and scaled to have median absolute deviation of 1 and then placed into chromosome order for further analysis. Data from the 2 biological replicates were averaged and analyzed using ACME as described in previous work (Crawford et al., 2006a; Scacheri et al., 2006). ACME produces a p-value for each region covered by chip across the genome. As this is a continuous value, we must set a threshold to obtain discrete regions for analysis. We use this to generate the ROC curve by increasing the threshold and delineating sites where the values drop below our threshold. Raw data can be downloaded from the Gene Expression Omnibus (GEO) accession number GSE8486.
High-Throughput Sequencing and Data Analysis
The 15,341,822 × 20 bp Illumina sequence tags were aligned to the National Center for Biotechnology Information (NCBI) human genome build 35 using Mummer (Kurtz et al., 2004). In addition, 3,423,744 ∼108 bp tags were generated by 454 Life Sciences. 454 sequences were trimmed of poor quality sequence and were required to have at least 50 bp of continuous high-quality sequence beginning at the 5′ end. These sequences were then aligned to the NCBI human genome build using BLAT (Kent, 2002), requiring an alignment percentage > 85%, a coverage percentage > 70%, and allowing a gap size of 5 bp. For both Illumina and 454 data, only sequences with unique genomic positions were used for analysis. Within each set, any 5′ end shared by more than one sequence on the same strand was only counted once to remove duplications caused by PCR during the library preparation. To identify DNase I HS sites, the combined set of 12,619,784 uniquely aligned sequences was used generate a kernel-density estimation (Parzen window method [Parzen, 1962]) with a variance of .5 and a bandwidth of 200. This process produces a continuous value of hypersensitivity across the genome. To determine discrete regions for analysis (such as for the ROC curve) a threshold is set and each region above the threshold is considered a single DNase I HS site. In the case of the ROC curve, this is recomputed for many thresholds to give a range of sensitivity and specificity. Raw data can be downloaded from the Gene Expression Omnibus (GEO) accession number GSE8486.
Combination of Data
Our ultimate goal was to use information provided by DNase-seq and DNase-chip to complement each other in the identification of DNase I HS sites. To combine the data, we first rescaled the sequencing and chip data into the same range. Because the DNase-chip data was generated from probes approximately 50 bp long and with a 50 bp gap between probes, we filled in the spaces between near probes (<130 bp apart) by “virtually” expanding the probes on each side of the gap. We then combined the scores by converting each base pair score into a Z score using the population mean and standard deviation from each set. We used the information from our known qPCR positives and negatives to equate the two sets by determining the point where sensitivity was equal to specificity (maximal sensitivity/specificity). This value was subtracted from our Z score values to give a distribution of scores that we would consider anything above zero to be a positive call. These two sets were then summed to generate a combined DNase-seq/DNase-chip score. For any regions in which there was sequence but no tiled probe coverage, the sequence score was doubled to provide an equivalent range of scores in those regions. The final combined score set was found to have a sensitivity and specificity intersect at a value slightly greater than zero so this difference was subtracted from the final set leaving any positive call to have a value greater than zero.
Comparison to ChIP-Seq Data
Chromatin immunoprecipitation data for RNA Polymerase II, CTCF, H2A.Z, and histone modifications for CD4 cells was obtained from http://dir.nhlbi.nih.gov/papers/lmi/epigenomes/hgtcell.html. Only hypersensitive sites that were considered unidirectional (within 5 kb of the 5′ end of only one gene) and not near other strong hypersensitive sites were used for this analysis. Proximal hypersensitive sites (within 2 kb of the TSS) were classified as being near high, medium, low, or silent based on comparison of the related gene’s expression to the full distribution of expression levels. Exact intervals were determined to obtain a relatively high and equal number of genes in each bin but with the goal of keeping the bins relatively separate. The High class consists of the 570 genes with an expression level > 10, the Medium class consists of 791 expressed genes with an expression level between 8.2 and 8.9, the Low class consists of 727 genes with an expression level between 5.5 and 5.8, and the Silent class consists of 698 genes with an expression level < 4.5. Expressed sites were also classified as those within a transcribed region but outside of the promoter and distal sites were those not classified as being either proximal or expressed. Plots were generated centered on the highest value of the HS site and sequence tags were counted and normalized from each data type.
Motif Analysis
To identify the putative transcription factors that bind within DNase I HS sites, we searched the 75,954 DNase I HS sites that were more than 2 kb away from a TSS for enriched motifs. We grouped these distal HS sites by chromosome and scanned each set of sequences using the Clover algorithm (Frith et al., 2004) to identify enriched motifs from the TRANSFAC database (Wingender et al., 1996). The enrichment was measured by comparison to two background sets of sequences: the union set of all ChIP-chip hits generated by the ENCODE Transcription regulation group at the 5% false discovery rate cut-off (ENCODE, 2007), and random dinucleotide shuffling of the input sequence set (DNase I HS sites in a chromosome). We generated 1000 sets of random sequences for each input sequences set and any motif with P value < 0.01 was deemed significantly enriched. All motifs that are enriched in the DNase I HS sites in more than half of the chromosomes by comparison to both background sets are listed in Table S2.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shujun Luo, Daixing Zhou, Gary Schroth, and David Bentley from Illumina, and we thank Brian Godwin, Jan Simons, Jennifer Tsai, Lei Du, and Michael Egholm from 454 Life Sciences for generation of DNase I sequence tags. We thank Pablo Alvarez and Chad Nusbaum from the Broad Institute for help with analysis of 454 sequences, and Kyle Munn and Roland Green from NimbleGen for whole-genome DNase-chip. Finally, we would like to thank Francis Collins for helpful comments and advice. This project was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (E.H.M.), a National Science Foundation Graduate Research Fellowship (A.P.B.), and by NIH grant HG003169 (G.E.C.).
REFERENCES
- Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the Mamm. Genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Cockerill PN, Shannon MF, Bert AG, Ryan GR, Vadas MA. The granulocyte-macrophage colony-stimulating factor/interleukin 3 locus is regulated by an inducible cyclosporin A-sensitive enhancer. Proc. Natl. Acad. Sci. USA. 1993;90:2466–2470. doi: 10.1073/pnas.90.6.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins DJ, Islam SA, Sanderson MR, Proykova YG, Crane-Robinson C, Staynov DZ. Redefinition of the cleavage sites of DNase I on the nucleosome core particle. J. Mol. Biol. 2004;335:1199–1211. doi: 10.1016/j.jmb.2003.11.052. [DOI] [PubMed] [Google Scholar]
- Crawford GE, Davis S, Scacheri PC, Renaud G, Halawi MJ, Erdos MR, Green R, Meltzer PS, Wolfsberg TG, Collins FS. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat. Methods. 2006a;3:503–509. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford GE, Holt IE, Mullikin JC, Tai D, Blakesley R, Bouffard G, Young A, Masiello C, Green ED, Wolfsberg TG, et al. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc. Natl. Acad. Sci. USA. 2004;101:992–997. doi: 10.1073/pnas.0307540100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford GE, Holt IE, Whittle J, Webb BD, Tai D, Davis S, Margulies EH, Chen Y, Bernat JA, Ginsburg D, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006b;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Kawaji H, Kasukawa T, Fukuda S, Katayama S, Kai C, Kawai J, Carninci P, Hayashizaki Y. CAGE Basic/Analysis Databases: the CAGE resource for comprehensive promoter analysis. Nucleic Acids Res. 2006;34:D632–D636. doi: 10.1093/nar/gkj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene MA, Corces V, Lowenhaupt K, Elgin SC. DNase I hypersensitive sites in Drosophila chromatin occur at the 5′ ends of regions of transcription. Proc. Natl. Acad. Sci. USA. 1981;78:143–146. doi: 10.1073/pnas.78.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DU, Avni O, Chen L, Rao A. A distal enhancer in the interferon-gamma (IFN-gamma) locus revealed by genome sequence comparison. J. Biol. Chem. 2004;279:4802–4810. doi: 10.1074/jbc.M307904200. [DOI] [PubMed] [Google Scholar]
- McArthur M, Gerum S, Stamatoyannopoulos G. Quantification of DNaseI-sensitivity by real-time PCR: quantitative analysis of DNaseI-hyper-sensitivity of the mouse beta-globin LCR. J. Mol. Biol. 2001;313:27–34. doi: 10.1006/jmbi.2001.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, Wood WI, Dolan M, Engel JD, Felsenfeld G. A 200 base pair region at the 5′ end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981;27:45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- Meraro D, Hashmueli S, Koren B, Azriel A, Oumard A, Kirchhoff S, Hauser H, Nagulapalli S, Atchison ML, Levi BZ. Protein-protein and DNA-protein interactions affect the activity of lymphoid-specific IFN regulatory factors. J. Immunol. 1999;163:6468–6478. [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974;1:1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parzen E. On estimation of a probability density function and mode. Ann. Math. Stat. 1962;33:1065–1076. [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Sabo PJ, Hawrylycz M, Wallace JC, Humbert R, Yu M, Shafer A, Kawamoto J, Hall R, Mack J, Dorschner MO, et al. Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc. Natl. Acad. Sci. USA. 2004a;101:16837–16842. doi: 10.1073/pnas.0407387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo PJ, Humbert R, Hawrylycz M, Wallace JC, Dorschner MO, McArthur M, Stamatoyannopoulos JA. Genome-wide identification of DNaseI hypersensitive sites using active chromatin sequence libraries. Proc. Natl. Acad. Sci. USA. 2004b;101:4537–4542. doi: 10.1073/pnas.0400678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- Scacheri PC, Crawford GE, Davis S. Statistics for ChIP-chip and DNase hypersensitivity experiments on NimbleGen arrays. Methods Enzymol. 2006;411:270–282. doi: 10.1016/S0076-6879(06)11014-9. [DOI] [PubMed] [Google Scholar]
- Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat. Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U, Durand DB, Bressler P, Holbrook NJ, Norris CA, Kamoun M, Kant JA, Crabtree GR. Promoter region of interleukin-2 gene undergoes chromatin structure changes and confers inducibility on chloramphenicol acetyltransferase gene during activation of T cells. Mol. Cell. Biol. 1986;6:3042–3049. doi: 10.1128/mcb.6.9.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD, Sumazin P, Xuan Z, Zhang MQ. DNA motifs in human and mouse proximal promoters predict tissue-specific expression. Proc. Natl. Acad. Sci. USA. 2006;103:6275–6280. doi: 10.1073/pnas.0508169103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B, Melchior W, Jr., Felsenfeld G. DNAase I, DNAase II and staphylococcal nuclease cut at different, yet symmetrically located, sites in the nucleosome core. Cell. 1978;14:611–627. doi: 10.1016/0092-8674(78)90246-5. [DOI] [PubMed] [Google Scholar]
- Stalder J, Larsen A, Engel JD, Dolan M, Groudine M, Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introxsduced by DNAase I. Cell. 1980;20:451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, Li D, Durum SK, Jiang Q, Bhandoola A, et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc. Natl. Acad. Sci. USA. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.