Abstract
Naïve T helper cells differentiate into two subsets, T helper 1 and 2, which either transcribe the Ifng gene and silence the Il4 gene or transcribe the Il4 gene and silence the Ifng gene, respectively. This process is an essential feature of the adaptive immune response to a pathogen and the development of long-lasting immunity. The ‘histone code’ hypothesis proposes that formation of stable epigenetic histone marks at a gene locus that activate or repress transcription is essential for cell fate determinations, such as T helper 1/T helper 2 cell fate decisions. Activation and silencing of the Ifng gene are achieved through the creation of stable epigenetic histone marks spanning a region of genomic DNA over 20 times greater than the gene itself. Key transcription factors that drive the T helper 1 lineage decision, signal transducer and activator 4 (STAT4) and T-box expressed in T cells (T-bet), play direct roles in the formation of activating histone marks at the Ifng locus. Conversely, STAT6 and GATA binding protein 3, transcription factors essential for the T helper 2 cell lineage decision, establish repressive histone marks at the Ifng locus. Functional studies demonstrate that multiple genomic elements up to 50 kilobases from Ifng play critical roles in its proper transcriptional regulation. Studies of three-dimensional chromatin conformation indicate that these distal regulatory elements may loop towards Ifng to regulate its transcription. We speculate that these complex mechanisms have evolved to tightly control levels of interferon-γ production, given that too little or too much production would be very deleterious to the host.
Keywords: differentiation, epigenetics, interferon-γ, T helper 1, transcription
Introduction
In the periphery, naïve CD4 T cells have the potential to differentiate into distinct lineages of effector cells that are primarily defined by the cytokines they produce.1–3 These effector lineages orchestrate the adaptive immune response, playing key roles in directing the elimination of both intracellular and extracellular pathogens. For example, effector T helper 1 (Th1) cells produce high levels of interferon-γ (IFN-γ) and very low levels of interleukin-4 (IL-4) in response to antigenic stimulation and effector Th2 cells produce high levels of IL-4 and very low levels of IFN-γ in response to identical antigenic stimulation. In contrast, naïve CD4 T cells produce only extremely low levels of IFN-γ and IL-4 after stimulation by antigen. For the most part, this differentiation process is driven by the combination of T-cell receptor signalling through antigenic stimulation and by cytokines produced by the innate immune system. Interleukin-12 and IL-4 are among the more effective cytokines at driving Th1 and Th2 differentiation, respectively.2,4 This developmental process imparts a permanent change upon the cell, making it possible for the differentiated cell to produce a markedly different cytokine expression pattern following identical T-cell receptor stimulation.
Expression of messenger RNA (mRNA) transcripts encoding IFN-γ and IL-4 proteins in naïve CD4 T cells and effector Th1 and Th2 cells mirrors protein production, arguing that these differentiation paths are controlled at the level of transcription. This has led to a search for transcriptional regulators of Th1 and Th2 differentiation. The transcription factor signal transducer and activator 4 (STAT4) plays an essential role in Th1 differentiation driven by IL-12 and STAT6 plays an essential role in Th2 differentiation driven by IL-4.4,5 In addition, T-box expressed in T cells (T-bet) and GATA-binding protein 3 (GATA-3) are key regulators of Th1 and Th2 differentiation, respectively.6,7 Additional transcription factors contribute to these processes and to cytokine gene expression patterns. These have been covered in several recent excellent reviews and will not be covered in further detail herein.3,8,9
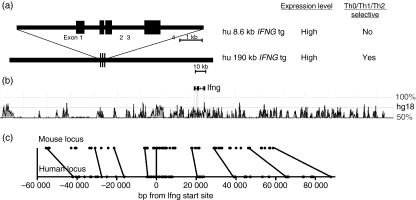
To more completely understand the mechanisms underlying the Th1/Th2 cell fate decision, it is also necessary to identify the functional DNA elements that confer proper transcriptional regulation upon the Ifng gene; high expression in Th1 cells, silenced expression in Th2 cells, and inefficient expression in naïve T cells. A general approach to the identification and analysis of DNA elements necessary for transcriptional expression and silencing of genes is the use of reporter genes linked to suspected promoters and enhancers, typically located proximal to the transcriptional initiation start site. Our goal was to use this approach to identify promoter and enhancer elements capable of conferring Th1-selective expression upon a reporter gene. We used a transgenic approach because cell lines do not recapitulate Th1 differentiation. Although initial studies showed promise, it was not possible to recapitulate both high-level and Th1/Th2-selective expression using this approach.10–12 Therefore, we turned to a different system using transgenic constructs containing the complete human IFNG gene.13 A transgene with the human IFNG gene and ∼1500 base pairs (bp) of both upstream and downstream sequences is sufficient to confer high-level expression of human IFNG mRNA and human IFN-γ but does not confer Th1/Th2-selective expression of human IFNG. In contrast, a transgenic construct derived from a human bacterial artificial chromosome (BAC) of about 190 kilobases (kb) containing the human IFNG gene in the middle of the construct and approximately 90 kb of both upstream and downstream sequence is able to confer both high levels of expression of human IFNG mRNA and human IFN-γ protein as well as Th1/Th2 selectivity (Fig. 1a). The major difference in the activity of the two transgenes was not the level of expression but the ability to achieve Th1/Th2-selective expression. We therefore concluded from these experiments that Th1/Th2-selective expression of IFN-γ must be achieved by the complex interplay between transcriptional activators and repressors acting over very large distances.
Figure 1.
The Ifng locus. (a) Schematic and functional properties of two human IFNG transgenes. The upper transgene is an ∼8·6 kilobase (kb) fragment containing the human IFNG gene and 2·0–2·5 of upstream and downstream sequence. The lower transgene is an ∼190 kb bacterial artificial chromosome with the human IFNG gene and flanking sequence. (b) Positions of evolutionarily conserved non-coding DNA elements relative to the Ifng gene from the dcode website (http://www.dcode.org).38 (c) Positions of conserved non-coding sequences (CNS), filled circles, relative to the mouse (upper) and human (lower) Ifng/IFNG genes. Lines connect representative individual CNS within the mouse and human loci.
The 190-kb genomic region making up the BAC transgene is void of other known genes but is enriched in sequences exhibiting evolutionary conservation between mouse and human genomes, also referred to as conserved non-coding sequences (CNS) (Fig. 1b). Although no strict definition exists, CNS typically exhibit > 70% sequence conservation between two distant species, such as humans and rodents, and span > 100 bp of genomic DNA.14,15 The presence of evolutionary conservation in CNS is also a proven method to identify transcriptional promoters, enhancers, insulators and repressors. Many distant sequences within this region of genomic DNA exhibit evolutionary sequence conservation similar in both magnitude and size to the Ifng promoter. Using this logic, distant CNS may also possess activities capable of regulating Ifng transcription. A further characteristic of the multiple CNS across the Ifng locus is that the order of each CNS relative to the Ifng transcriptional start site, and every other CNS, exhibits absolute conservation between humans and rodents (Fig. 1c). The 5′–3′ orientation of each CNS is also absolutely conserved. In contrast, the absolute distance in base pairs of each CNS from the transcriptional start site can vary by up to 20 000 bp. Although not experimentally proven, this seems to argue that proper Ifng transcription absolutely requires a specific order of CNS and orientation of CNS across a region spanning > 150 000 bp. This is in marked contrast to the general rules of enhancers proximal to a promoter where order and orientation are not critical for enhancer activity. Therefore, genes with these features, such as the Ifng gene, may give rise to unique forms of transcriptional regulation not required by genes that do not exhibit cell-type or developmental regulation.
Th1 differentiation and the histone code
Th1/Th2 differentiation, as with other developmental processes, represents a heritable change to the cell passed on to daughter generations. Since these changes do not represent genetic events or changes in the DNA sequence, the term epigenetics arose to define developmental changes that produce heritable changes in gene expression in the absence of changes in DNA sequence.16 Epigenetic changes playing key roles in development include the chemical modification of histones and the methylation of DNA at CpG dinucleotides.17 Amino terminal ‘tails’ of the core histones, H2A, H2B, H3 and H4, undergo enzymatically catalysed post-translational modifications including methylation, acetylation, phosphorylation, sumoylation and ubiquitination.18–23 The recognition that specific post-translational modifications of histones are associated with the activation and silencing of specific genes has given rise to the histone code hypothesis, which proposes that histone modifications at specific gene loci are key elements in the acquisition and maintenance of developmental and cell-type specific expression and silencing of genes. Besides the enzymes that catalyse the formation of these histone marks, there are also enzymes that catalyse the removal of most histone marks.24,25 Consequently, where a previous view was that formation of histone marks at gene loci was a relatively irreversible event, the discovery of enzymes that remove histone marks has dramatically changed the chromatin landscape and suggests that epigenetics may represent a highly dynamic process.
In most instances, these histone modifications are localized to promoters and transcribed regions of genes.26–28 Enzymes that catalyse the formation and removal of histone marks do not directly bind DNA but are tethered to DNA by their ability to bind to an array of DNA-binding proteins such as transcription factors, and so are able to catalyse the formation of histone marks at adjacent nucleosomes in chromatin.29 These enzymes also possess domains that allow them to be recruited to their modified histone products, so permitting the spread of histone marks across chromatin.30,31 For example, histone acetyltransferases have bromodomains that recognize acetylated histone H4. Similarly, histone methyltransferases possess domains such as chromodomains that recognize methylated histones. Although less well understood, it is likely that additional mechanisms exist, either at genetic or epigenetic levels, which stop the spread of modified histone domains.
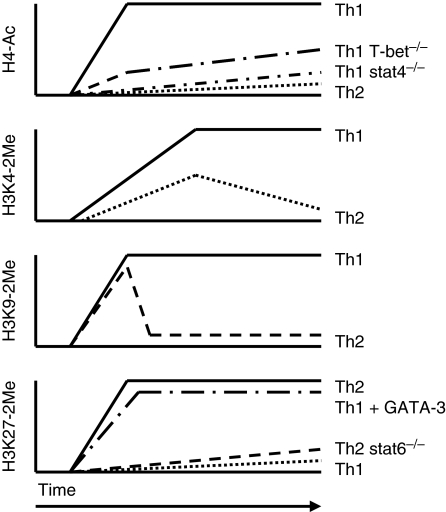
Promoters of both the Ifng and Il4 genes are selectively H4 acetylated in effector Th1 and Th2 cells, respectively.32 Therefore, we reasoned that if distal CNS elements across the Ifng locus contribute to the Th1 differentiation programme, they should also undergo H4 acetylation as part of this cell fate decision. This is exactly what occurs. The Th1 differentiation programme drives H4 acetylation of individual CNS spanning distances of > 50 kb in both upstream and downstream directions of the Ifng gene.33 The Th1 differentiation programme also stimulates H3K4 methylation, another histone mark associated with actively transcribed genes, at these CNS across a similar span of genomic DNA.34,35 Careful analysis of H3K4-methylation marks at these CNS demonstrates another property of the histone-methylation pattern across the locus. The highest levels of H3K4-methylation are centred at individual CNS elements across the locus and decline in magnitude in both the 5′ and 3′ directions moving away from each individual CNS. This creates an image of peaks and valleys of H3K4-methylation and presumably H4 acetylation across the entire locus. Formation of H4 acetylation marks across the entire locus is dependent upon the transcription factor STAT4, in a Th1 differentiation system that is driven by IL-12.36 While H4 acetylation across the entire locus is STAT4-dependent, only H4 acetylation at CNS 3′ of Ifng is dependent upon T-bet. The functional significance of or mechanisms underlying the differential STAT4 and T-bet requirements for H4 acetylation across the Ifng locus are not clear. The Ifng promoter and multiple CNS across the locus bind either STAT4 or T-bet or both.36,37 Other transcription factors also associate with distal CNS across the Ifng locus and certain CNS have been studied in greater detail than others (below). Acquisition of histone marks favourable to transcription across the entire Ifng locus is therefore an essential feature of Th1 differentiation driven by transcription factors necessary to complete the Th1 developmental programme.
In contrast to T cells, natural killer (NK)38 cells do not have to endure additional differentiation programmes in the periphery before they are able to produce high levels of IFN-γ after appropriate stimulation. Also in contrast to T cells, histone acetylation marks are already established across the Ifng locus in mature NK cells.36 This pattern of marks, with a few exceptions, is similar in distribution and magnitude to that of effector Th1 cells. Developmental programmes that give rise to mature peripheral NK cells include programmes to establish histone acetylation marks across the Ifng locus.
Differentiation along the Th2 pathway also establishes histone marks across the Ifng locus.34,39 These marks are H3K27-dimethylation and -trimethylation marks, which are repressive marks. Consequently, silencing of Ifng in effector Th2 cells, like activation of Ifng in effector Th1 cells, is an active process. Establishing H3K27-methylation marks across the Ifng locus involves both STAT6 and GATA-3, two major transcription factors driving the Th2 lineage choice. STAT6 binds to the Ifng promoter and is necessary to establish H3K27-methylation marks across the entire locus. GATA-3 also binds to the Ifng promoter and a distal site 53 kb 5′ of the promoter. GATA-3 induces H3K27-methylation across the entire locus in Th1 cells. Furthermore, GATA-3 directly recruits EZH2, the enzyme that catalyses formation of H3K27-methylation marks, to the Ifng promoter. This expands the function of GATA-3 to not only that of a transcriptional activator (Il4) but also of a transcriptional repressor (Ifng). It is clear that a key element of the Th2 differentiation process is both activation of the Il4 locus and silencing of the Ifng locus and the histone code plays key roles in both these processes. The question of how the enzymes that catalyse this series of histone covalent marks know which locus to go to is not well understood. For example, STAT6 and GATA-3 are recruited to both the Il4 and Ifng loci in effector Th2 cells. However, STAT6 and GATA-3 recruit enzymes to the Il4 locus that produce histone marks favouring transcription and recruit enzymes to the Ifng locus that produce histone marks silencing transcription. One difference may be the density of binding sites. Multiple binding sites for T-bet and STAT4 exist across the Ifng locus but very few GATA-3- and STAT6-binding sites across the Ifng locus have been identified. The converse may be true for the Il4 locus and many more binding sites for GATA-3 and STAT6 may exist across the Il4 locus than T-bet- and STAT4-binding sites. Whether differences in total binding sites explain the ability of these transcription factors to recruit different histone-modifying enzymes to the Ifng and Il4 loci is not known.
The Ifng locus also acquires H3K9-dimethylation marks under both Th1 and Th2 differentiation conditions.39 These marks are sustained in differentiating effector Th1 cells but are rapidly extinguished in effector Th2 cells. In contrast to H4-acetylation marks or H3K4-methylation marks, key elements of the Th1 differentiation programme, formation of H3K9-methylation marks does not appear to depend upon Th1 differentiation signals. The interesting thing about H3K9-dimethylation marks is that they are typically associated with transcriptionally repressed rather than transcriptionally active genes. The functional significance of the presence of H3K9-dimethylation marks at the Ifng locus in Th1 cells is unclear. The presence of H3K9 trimethylation at transcribed regions of genes, but not their promoters, undergoing active transcription has been described.28 Alternatively, H3K9 methylation marks across the locus of an actively transcribed gene may actually serve a suppressive function. In the case of Ifng, it is easy to see how production of too much or too little IFN-γ in an inflammatory setting may be deleterious to the host. Production of too little IFN-γ may compromise the functions of the adaptive immune system leading to failure to curb an infection. Production of too much IFN-γ could contribute to inflammatory disease or even cytokine sickness that, if unchecked, would produce extreme morbidity or even mortality.
Establishing the histone code across the Ifng locus in differentiating effector Th1 and Th2 cells is a complex, multi-stage, dynamic process. These results also suggest that the regulatory region of the Ifng gene spans > 100 kb. Figure 2 provides a crude schematic to illustrate epigenetic dynamics at the Ifng locus during Th1 and Th2 cell fate decisions.
Figure 2.
Kinetics of histone modifications at the Ifng locus during T helper 1 (Th1)/Th2 differentiation. Initiation of Th1 and Th2 differentiation programmes induces formation of markedly different histone ‘marks’ at the Ifng locus. Both signal transducer and activator 4 (STAT4) and T-box expressed in T cells (T-bet) play critical roles in directing the formation of histone 4 (H4)-acetylation marks in developing Th1 cells and STAT6 and GATA-binding protein 3 (GATA-3) play critical roles in directing the formation of H3K27-methylation marks in developing Th2 cells. Changes in H3K9-methylation marks during differentiation illustrate the dynamic nature of the histone code in developing effector T cells.
Functional properties of distal CNS across the Ifng locus
Several CNS across the Ifng locus that undergo histone modifications in developing Th1 and Th2 cells exhibit defined functional activity in different assay systems. The CNS 6 kb upstream of the Ifng start site, CNS-6 (also called CNS1), possesses a DNAse hypersensitivity site in naive T cells, suggesting that it is a site critical for early events in Ifng remodelling.40–42 The transcription factors T-bet, STAT5 and nuclear factor of activated T cells 1 (NFAT1) bind to the CNS-6 site. Early, within 24–72 hr of Th1-cell development, a Jak3-dependent cytokine signal, probably IL-2, stimulates recruitment of STAT5 to the CNS-6 site. This STAT5 signal is thought to promote T-bet binding to the promoter and H3 and H4 acetylation. By 72 hr, the chromatin containing CNS-6 has looped into the promoter, possibly explaining why T-bet is known to bind at each CNS.43 How CNS-6 functions in a differentiated Th1 cell is unclear. CNS+18–20 (located 18–20 kb upstream of the Ifng start site, also called CNS2) does not develop DNAse hypersensitivity until Th1 or Th2 polarization, suggesting a role after the initial differentiation signal.44 CNS+18–20 does not have enhancer activity in various reporter assays. Rather, CNS+18–20 augments the enhancer activity of CNS-6. Along these lines, the chromatin containing CNS+18–20 loops into the Ifng promoter upon Th1 differentiation.45 From these data it appears that CNS+18–20 functions by enhancing the activity of CNS-6 after the Th1 differentiation signal.
Analyses of CNS-22 (located 22 kb upstream of the Ifng transcriptional start site) in reporter models suggest that this CNS is necessary for IFN-γ production by Th1 cells.37 CNS-22 lacks a DNAse hypersensitivity site in naive CD4 cells, but acquires a very strong hypersensitivity site upon Th1 or Th2 differentiation. So, this CNS has a regulatory role specific for events after the initial differentiation signal. CNS-22 functions as a strong T-bet-dependent enhancer in vitro. In vivo, T-bet binds to CNS-22 in both resting and stimulated Th1 cells. Because the site contains a permissive chromatin environment in both Th1 and Th2 cells, and strongly reacts to T-bet in an activation-independent manner, it is theorized that CNS-22 functions to create an environment that is favourable to transcription in Th1 cells. The promoter, CNS-34, and CNS-54 also possess T-bet-binding sites and have T-bet-dependent enhancer functions in reporter assays. Numerous CNS exist across large distances of genomic DNA spanning the Ifng locus capable of stimulating transcription by a T-bet-dependent mechanism. Experimental analyses also suggest that silencers play a critical role in preventing Ifng transcription in proliferating T cells that have not received a polarizing differentiation signal and in effector Th2 cells that have received a polarizing differentiation signal. These CNS have not been identified or characterized.
Looping of chromatin domains
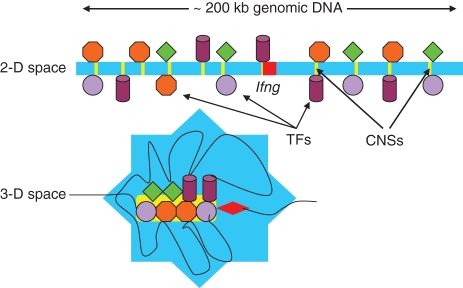
A significant question raised by the above experimental analyses is the mechanism by which these distal elements communicate with each other and with Ifng. Evidence has emerged from a variety of investigations to demonstrate that cellular signalling events result in changes in the three-dimensional conformation of chromatin.43,45–49 In this regard, the Ifng genomic locus associates with the Th2 cytokine genomic locus in naïve T cells and exists in an open conformation.43,45 During Th1 differentiation, the Ifng locus is freed from the Th2 cytokine locus, adopts a looped conformation, and is repositioned to different nuclear compartments via nuclear attachment DNA elements.43,50 T-cell activation, in the absence of polarizing signals, induces chromatin looping at the Ifng locus. Differences exist in conformation of the Ifng locus chromatin between Th1 and Th2 cells and these differences may become more pronounced as larger chromatin domains are examined. A looping model such as this provides a mechanism by which transcriptional enhancers and repressors many kilobases from a gene can affect its expression (Fig. 3). Changes in chromosome conformation also provide a mechanism by which genomic elements on different chromosomes can regulate gene expression. An example is the association of Ifng and Il4 loci in naïve T cells. Much like proteins, the three-dimensional structure may be critical to the regulation of gene transcription, which is the essence of cell lineage decisions.
Figure 3.
Changes in the three-dimensional (3-D) conformation of the Ifng locus may recruit distal conserved non-coding sequences (CNS) to the gene to regulate transcription. Distal evolutionarily conserved DNA elements are occupied by transcription factors (TF) after initiation of T helper type 1 (Th1)/Th2 differentiation programmes. These transcription factors can tether enzymes that catalyse histone modifications, chromatin remodelling and other functions to these DNA elements. Changes in three-dimensional conformation of the locus may serve to localize these DNA elements and their associated proteins to the Ifng gene. CNS, conserved non-coding sequences.
Positioning of chromatin regions within the nucleus also plays a critical role in transcriptional regulation and specific DNA elements are required for nuclear positioning.51 A new concept is that ‘transcription factories’ exist within the nucleus as independent subnuclear structures, which are characterized by high concentrations of hyperphosphorylated RNA polymerase II.52–54 Actively transcribed genes are localized to these transcription factories. Therefore, an essential feature of transcription may be to recruit genomic loci to a transcription factory, for example the Ifng locus in effector Th1 cells, or repel a genomic locus from a transcription factory, for example the Ifng locus in effector Th2 cells. The series of covalent histone marks at a locus may also contribute to the nuclear positioning of the locus into or away from the transcriptional factory.
Perspectives
Interferon-γ is critically important in controlling both the adaptive and innate arms of the immune system. Underexpression of Ifng would reduce the ability of the immune system to control pathogen replication and invasion, leading to bacteraemia, viraemia or even death. Overexpression of Ifng could lead to cytokine-mediated inflammation, high morbidity and even mortality. It is easy to see how expression of Ifng needs to be tightly controlled and the immune system employs a series of regulatory elements separated by significant genomic distances and complex epigenetic mechanisms to achieve proper transcriptional control. It is relevant to consider that many cytokines and chemokines are equally potent effector proteins, so it is easy to see how their expression also needs to be under very strict control. Failures in the long-range epigenetic regulation of transcription of Ifng and other cytokine genes could also contribute to autoimmunity by producing too much or too little of the key cytokines or chemokines in peripheral tissues. Consequently, complex long-range epigenetic transcriptional regulation may be a common occurrence in haematopoietic cells to achieve proper control of key genes that regulate the function of the immune system.
Acknowledgments
Work from the authors’ laboratory was supported by grants RO1 AI 044924 and T32 HL069765 from the National Institutes of Health, Bethesda, MD.
Glossary
Abbreviations
- BAC
bacterial artificial chromosome
- CNS
conserved non-coding sequences
- EZH2
Enhancer of Zeste homologue 2
- GATA3
GATA-binding protein 3
- h
helper
- H
histone
- NK
natural killer
- STAT
signal transducer and activator
- T-bet
T-box expressed in T cells
References
- 1.Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–73. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 2.Seder RA, Paul WE. Acquisition of lymphokine-producing phenotype by CD4+ T cells. Annu Rev Immunol. 1994;12:635–73. doi: 10.1146/annurev.iy.12.040194.003223. [DOI] [PubMed] [Google Scholar]
- 3.Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–44. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 4.Thierfelder WE, van Deursen JM, Yamamoto K, et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382:171–4. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 5.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for the development of Th2 cells. Immunity. 1996;4:313–9. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 6.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–96. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 7.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–69. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 8.Ho IC, Glimcher LH. Transcription: tantalizing times for T cells. Cell. 2002;109:109–20. doi: 10.1016/s0092-8674(02)00705-5. [DOI] [PubMed] [Google Scholar]
- 9.Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Annu Rev Immunol. 2003;21:713–58. doi: 10.1146/annurev.immunol.21.120601.140942. [DOI] [PubMed] [Google Scholar]
- 10.Aune TM, Penix LA, Rincon MR, Flavell RA. Differential transcription directed by discrete gamma interferon promoter elements in naive and memory (effector) CD4 T cells and CD8 T cells. Mol Cell Biol. 1997;17:199–208. doi: 10.1128/mcb.17.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soutto M, Zhang F, Enerson B, Tong Y, Boothby M, Aune TM. A minimal IFN-gamma promoter confers Th1 selective expression. J Immunol. 2002;169:4205–12. doi: 10.4049/jimmunol.169.8.4205. [DOI] [PubMed] [Google Scholar]
- 12.Zhu H, Yang J, Murphy TL, Ouyang W, Wagner F, Saparov A, Weaver CT, Murphy KM. Unexpected characteristics of the IFN-gamma reporters in nontransformed T cells. J Immunol. 2001;167:855–65. doi: 10.4049/jimmunol.167.2.855. [DOI] [PubMed] [Google Scholar]
- 13.Soutto M, Zhou W, Aune TM. Cutting edge: distal regulatory elements are required to achieve selective expression of IFN-gamma in Th1/Tc1 effector cells. J Immunol. 2002;169:6664–7. doi: 10.4049/jimmunol.169.12.6664. [DOI] [PubMed] [Google Scholar]
- 14.Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dermitzakis ET, Reymond A, Antonarakis SE. Conserved non-genic sequences – an unexpected feature of mammalian genomes. Nat Rev Genet. 2005;6:151–7. doi: 10.1038/nrg1527. [DOI] [PubMed] [Google Scholar]
- 16.Van Speybroeck L. From epigenesis to epigenetics: the case of C. H. Waddington. Ann N Y Acad Sci. 2002;981:61–81. [PubMed] [Google Scholar]
- 17.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–32. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 18.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–40. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 21.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 22.Calestagne-Morelli A, Ausio J. Long-range histone acetylation: biological significance, structural implications, and mechanisms. Biochem Cell Biol. 2006;84:518–27. doi: 10.1139/o06-067. [DOI] [PubMed] [Google Scholar]
- 23.van Holde KE. Chromatin. New York: Springer Verlag; 1988. [Google Scholar]
- 24.Trojer P, Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell. 2006;125:213–7. doi: 10.1016/j.cell.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–9. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 26.Bernstein BE, Kamal M, Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–81. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–91. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Bulger M. Hyperacetylated chromatin domains: lessons from heterochromatin. J Biol Chem. 2005;280:21689–92. doi: 10.1074/jbc.R500004200. [DOI] [PubMed] [Google Scholar]
- 30.Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–76. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Bottomley MJ. Structures of protein domains that create or recognize histone modifications. EMBO Rep. 2004;5:464–9. doi: 10.1038/sj.embor.7400146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol. 2002;169:647–50. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 33.Zhou W, Chang S, Aune TM. Long-range histone acetylation of the Ifng gene is an essential feature of T cell differentiation. Proc Natl Acad Sci USA. 2004;101:2440–5. doi: 10.1073/pnas.0306002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–42. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamalainen-Laanaya HK, Kobie JJ, Chang C, Zeng WP. Temporal and spatial changes of histone 3 K4 dimethylation at the IFN-gamma gene during Th1 and Th2 cell differentiation. J Immunol. 2007;179:6410–5. doi: 10.4049/jimmunol.179.10.6410. [DOI] [PubMed] [Google Scholar]
- 36.Chang S, Aune TM. Histone hyperacetylated domains across the Ifng gene region in natural killer cells and T cells. Proc Natl Acad Sci USA. 2005;102:17095–100. doi: 10.1073/pnas.0502129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatton RD, Harrington LE, Luther RJ, et al. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25:717–29. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W253–W6. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang S, Aune TM. Dynamic changes in histone-methylation ‘marks’ across the locus encoding interferon-gamma during the differentiation of T helper type 2 cells. Nat Immunol. 2007;8:723–31. doi: 10.1038/ni1473. [DOI] [PubMed] [Google Scholar]
- 40.Bream JH, Hodge DL, Gonsky R, et al. A distal region in the interferon-gamma gene is a site of epigenetic remodeling and transcriptional regulation by interleukin-2. J Biol Chem. 2004;279:41249–57. doi: 10.1074/jbc.M401168200. [DOI] [PubMed] [Google Scholar]
- 41.Lee DU, Avni O, Chen L, Rao A. A distal enhancer in the interferon-gamma (IFN-gamma) locus revealed by genome sequence comparison. J Biol Chem. 2004;279:4802–10. doi: 10.1074/jbc.M307904200. [DOI] [PubMed] [Google Scholar]
- 42.Shi M, Lin TH, Appell KC, Berg LJ. Janus-kinase-3-dependent signals induce chromatin remodeling at the Ifng locus during T helper 1 cell differentiation. Immunity. 2008;28:763–73. doi: 10.1016/j.immuni.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eivazova ER, Aune TM. Dynamic alterations in the conformation of the Ifng gene region during T helper cell differentiation. Proc Natl Acad Sci USA. 2004;101:251–6. doi: 10.1073/pnas.0303919101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shnyreva M, Weaver WM, Blanchette M, Taylor SL, Tompa M, Fitzpatrick DR, Wilson CB. Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proc Natl Acad Sci USA. 2004;101:12622–7. doi: 10.1073/pnas.0400849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–45. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 46.de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–59. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 47.Lomvardes S, Barnes G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–13. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 48.Oestreich KJ, Cobb RM, Pierce S, Chen J, Ferrier P, Oltz EM. Regulation of TCRbeta gene assembly by a promoter/enhancer holocomplex. Immunity. 2006;24:381–91. doi: 10.1016/j.immuni.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 49.Jhunjhunwala S, van Zelm MC, Peak MM, et al. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–79. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eivazova ER, Vassetzky YS, Aune TM. Selective matrix attachment regions in T helper cell subsets support loop conformation in the Ifng gene. Genes Immun. 2006;8:35–43. doi: 10.1038/sj.gene.6364349. [DOI] [PubMed] [Google Scholar]
- 51.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–7. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 52.Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitchell JA, Fraser P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2008;22:20–5. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–57. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]