Abstract
Statement of Translational Relevance
This work has demonstrated that a microRNA expression ratio can distinguish between non-diseased tissue and tumor tissue with great accuracy in the context of head and neck squamous cell carcinoma (HNSCC), an important public health concern worldwide. The ratio of miR-221:miR-375 showed high discriminatory potential, with a sensitivity of 92% and specificity of 93% in distinguishing tumor from normal tissue, and suggests that this simple molecular marker may hold significant clinical potential as a diagnostic tool.
Purpose
The involvement of microRNAs (miRNAs) in cancer and their potential as biomarkers of diagnosis and prognosis are becoming increasingly appreciated. We sought to identify miRNAs altered in head and neck squamous cell carcinoma (HNSCC) and to determine whether miRNA expression is predictive of disease.
Experimental Design
RNA isolated from fresh frozen primary tumors, fresh frozen non-diseased head and neck epithelial tissues, and HNSCC cell lines was profiled for the expression of 662 miRNAs by microarray. The miRNAs that were both differentially expressed on the array and by qRT-PCR were subsequently validated by qRT-PCR using a total of 99 HNSCC samples and 14 normal epithelia.
Results
A marked difference in miRNA expression pattern was observed between tumors and cell lines. Eighteen miRNAs were significantly altered in their expression between normal tissues and tumors. Four of these miRNAs were validated in the larger sample series, and each showed significant differential expression (P < 0.0001). Further, an expression ratio of miR-221:miR-375 demonstrated a high sensitivity (0.92) and specificity (0.93) for disease prediction.
Conclusions
These data suggest that cultured tumor cell lines are inappropriate for miRNA biomarker identification, and that the pattern of miRNA expression in primary head and neck tissues is reflective of disease status, with certain miRNAs exhibiting strong predictive potential. These results indicate that miR-221 and miR-375 should be evaluated further as diagnostic biomarkers, as they may hold utility in defining broadly responsive prevention and treatment strategies for HNSCC.
Keywords: Head and Neck Cancer, microRNA, microarray, carcinogenesis
Introduction
Head and neck squamous cell carcinoma (HNSCC) includes carcinomas arising from the epithelium of the oral cavity, pharynx, and larynx, and is the sixth most common malignancy worldwide (1). The major risk factors for the disease are tobacco and alcohol use, and human papillomavirus (HPV) infection (2-4). Despite advances in detection, as well as surgical and chemotherapeutic treatments over recent decades, the five year survival rate for HNSCC has remained around 50%, one of the lowest of the major cancers (5). Frequent late stage diagnosis, formation of additional primary tumors and regional and distant metastases all contribute to this poor survival rate (2).
A better understanding of the molecular pathways that give rise to HNSCC is essential in the identification of novel molecular biomarkers that have clinical utility in predicting prognosis and therapeutic efficacy, as well as in designing targeted therapy for this disease. In recent years, gene expression profiling technologies have become increasingly sophisticated, allowing investigators to explore their diagnostic and therapeutic potential as biomarkers in HNSCC and other cancers (6-8). These biomarkers, however, have had limited success in the clinical setting and, to date, limited utility in further elucidating mechanisms of HNSCC carcinogenesis.
The discovery of microRNAs (miRNAs), ~22 nucleotide long, non-coding RNA molecules, has revolutionized our understanding of the modulation of gene expression. Nearly 700 miRNAs have been identified in humans (miRBase: http://microrna.sanger.ac.uk/), a number that is rapidly growing and expected to reach 1,000 or higher (9). Highly ubiquitous and largely conserved across species, miRNAs regulate gene expression post-transcriptionally by base-pairing, usually imperfectly, to the 3'-untranslated region (10) of a cognate messenger RNA (11). The interaction of a miRNA with a target mRNA transcript results either in translational repression of the mRNA or in its direct degradation (11). Due to the partial complementarity between miRNAs and their target transcripts, a single miRNA is capable of simultaneously regulating up to hundreds of genes, giving rise to an enormous modulatory potential (12).
Through their targets, miRNAs are known to play important roles in cell differentiation, proliferation, and apoptosis (13). As these processes are known to be deregulated in cancer, it is not surprising that many studies have now identified a role for miRNAs in carcinogenesis (14-16), including head and neck carcinogenesis (17, 18). Indeed, recent work on different cancer types has illustrated the existence of distinct miRNA expression profiles between tumor tissues and their corresponding normal tissue (19-21). Moreover, some studies have identified miRNA expression profiles that can distinguish different tumor subtypes or developmental lineages, findings that may have clinical applications in diagnostics and tumor staging (16, 22).
Here we have compared the miRNA expression of normal head and neck epithelia with primary HNSCC tumors and cultured HNSCC cell lines in order to define those miRNA most capable of differentiating disease, and thus those with the greatest potential as biomarkers. Upon identifying miRNAs specifically altered in head and neck cancer, we sought to validate a subset of these in a larger population of tumors with the aim of identifying a clinically-applicable diagnostic tool.
Materials and Methods
HNSCC samples and cell lines
Non-diseased head and neck epithelial tissue were obtained from the National Disease Research Interchange (NDRI) and consisted of fresh-frozen tongue, larynx and uvula samples. All fresh-frozen HNSCC samples were obtained, with informed consent after Institutional Review Board approval at participating hospitals, as part of a population-based case-control study of HNSCC spanning December 1999 to December 2003 in the Greater Boston Metropolitan area. The fresh-frozen tumors originated from uvula, larynx, floor of mouth, and tongue resections. Details of this study have been described previously (23). Study pathologists confirmed >75% tumor in all HNSCC samples tested. FaDu and Cal27, HNSCC cell lines, were obtained from American Type Culture Collection (ATCC) and maintained in Eagle's Minimum Essential Medium and Dulbecco's Modified Eagle's Medium, respectively, both supplemented with fetal bovine serum to a final concentration of 10%.
RNA isolation and microarray profiling
Total RNA was isolated from normal tissues, tumors, and cell lines using the mirVANA RNA Isolation Kit (Ambion, Inc., Austin, TX) according to the manufacturer's protocol. RNA was quantified using the Nanodrop ND-1000 spectrophotometer (Nanodrop, Wilmington, DE), aliquoted, and stored at -80°C briefly until needed. 5 μg of total RNA was sent for miRNA profiling studies at Asuragen Services using the mirVANA miRNA Bioarrays platform v2 (Ambion, Inc) as single channel format according to the standard operating procedures of the company, including pre-array qualitative Bioanalyzer (Agilent, Santa Clara, CA) RNA analysis, as previously described (24). The Bioarrays platform v2 contains probes specific to miRNA identified in human, mouse, and rat, as well as a number of proprietary miRNAs identified through cloning at Ambion, Inc. The Cy5 fluorescence on the arrays was scanned at an excitation wavelength of 635 nm using a GenePix 4200AL scanner (Molecular Devices, Union City, CA). The fluorescent signal associated with the probes and local background was extracted using GenePix Pro (version 6.0, Molecular Devices). Raw signal data were normalized by first log2 transformation of signal intensity followed by global Variance Stabilization Normalization (25) of all the arrays within the project. Normalized data were submitted to the GEO archive (accession #GSE11163).
Quantitative reverse transcription-PCR
TaqMan miRNA Assays (Applied Biosystems) were used to quantify mature miRNA. cDNA was synthesized by priming with a pool of gene-specific looped primers including the primers of the miRNAs of interest and RNU48, as a universally-expressed endogenous control (Applied Biosystems). 10 μl of total RNA diluted to a final concentration of 5 ng/μl was used for each reverse transcription (RT) reaction along with other RT components, per manufacturer's specifications. 40μl reactions were incubated in an Applied Biosystems GeneAmp PCR system 9700 for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C, and held at 4°C. qRT-PCR was performed as previously described (26) with the following exception: all reactions, excluding no-template controls and non-reverse-transcribed controls, were run in triplicate on an ABI 7500 Fast Real Time PCR Detection System. All real-time PCR data were analyzed using the comparative CT method, normalizing against expression of RNU48.
Statistical analysis
Differential expression of miRNAs by microarray was determined with Significance Analysis of Microarray (SAM) software (Stanford University Labs) using 1000 permutations of the data and with delta adjusted to minimize false discovery rate. All hierarchical clustering analyses were carried out using Cluster 3.0 (Stanford University) with Euclidian distance as the distance metric and centroid linkage between clusters. A two-tailed Student's t test was used to compare miRNA expression levels determined by real-time PCR. miRNA expression ratios were calculated following the methods of Gordon et al (27), and receiver operating curve (ROC) analyses were used to assess the predictive power of the miRNA quotients.
Results
miRNA expression patterns differentiate HNSCC cell lines from primary tissues
A microarray platform was used to determine miRNA expression of 662 miRNAs in 16 fresh frozen HNSCC tumors, 5 non-diseased head and neck epithelial tissues, and 2 individual HNSCC cell lines. The normalized data has been deposited in the GEO archive (accession #GSE11163).
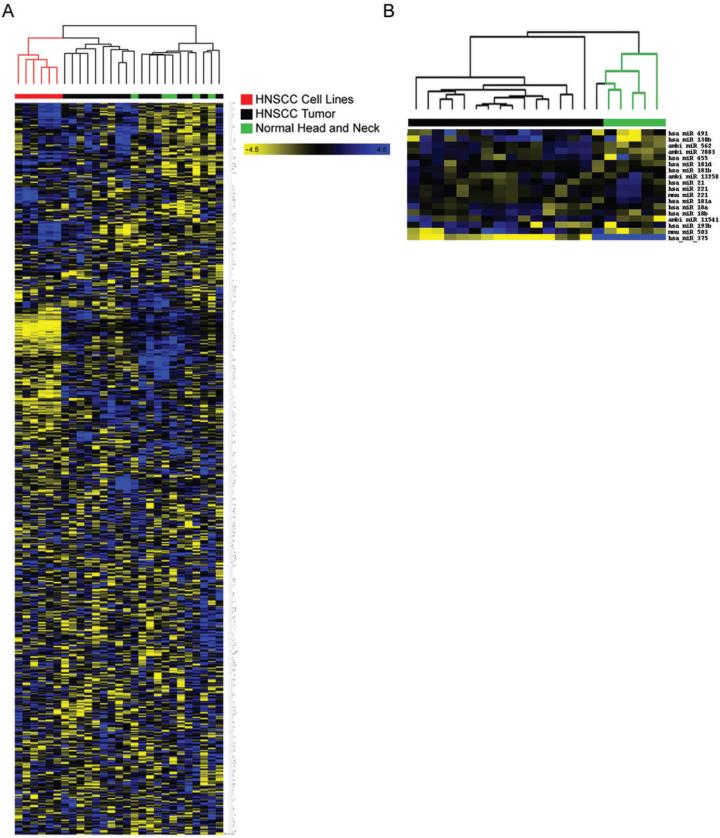
Unsupervised hierarchical clustering based on all the miRNAs spotted on the chip revealed a marked, very distinct separation of the cell line miRNA profiles compared to those of primary tissues (Figure 1A). SAM identified 67 significantly differentially expressed miRNAs (Q < 0.0001) between cell lines and primary tissues, consistently showing lower expression of miRNAs in cell lines compared to tumors (Supplementary Table 1).
Figure 1.
Heat maps created by unsupervised hierarchical clustering analysis between groups of samples. Blue and yellow represent higher and lower relative expression, respectively (A) Clustering analysis of HNSCC cell lines, HNSCC tumors and normal head and neck epithelia across 662 individual miRNAs shows separation of cell lines and primary tissues. (B) Separation of HNSCC tumors and normal tissues based on differential expression of 18 significantly altered miRNAs.
18 miRNAs are differentially expressed in HNSCC tumor tissue compared to normal head and neck epithelia
SAM analysis identified 18 miRNAs to be significantly altered in their expression between non-diseased tissues and primary HNSCC tumors, with 17 being up-modulated and 1 down-modulated in tumors (Q < 0.0001) (Supplementary Table 2). Of the 18 differentially expressed miRNAs, 12 were human miRNAs, four were proprietary miRNAs of Ambion Inc. and two were mouse orthologues, both of which have known human counterparts with identical mature sequences. Hierarchical clustering based on this limited set of miRNAs showed clear separation of tumors from normal tissues (Figure 1B).
All human miRNAs identified by SAM method showed greater than two-fold difference between normal and tumor tissue (Table 1). Of the human miRNAs, six (miR-21, miR-181d, miR-181b, miR-18a, miR-221, and miR-375) were chosen for confirmation of the microarray results using stem-loop based RT-PCR followed by conventional Taqman real-time PCR using miRNA-specific probes, as validated assays were available for these six miRNA and they constituted one primer pool, allowing for examination using a single reverse transcription step. Confirmation assays utilized only the samples screened on the array. Of the six miRNAs tested, four were reliably confirmed (miR-21, miR-18a, miR-221, and miR-375), showing significant differential expression between tumor and normal (P < 0.01, Supplementary Figure 1).
Table 1.
Annotated human miRNAs differentially expressed in Tumor vs Normal by SAM (Q < 0.001)
| miRNAs | Fold change |
|---|---|
| miR-21 | 3.67 |
| miR-181d | 2.86 |
| miR-181b | 2.97 |
| miR-491 | 5.08 |
| miR-455 | 2.58 |
| miR-18a | 2.78 |
| miR-130b | 4.17 |
| miR-221 | 2.26 |
| miR-193b | 3.44 |
| miR-181a | 2.11 |
| miR-18b | 2.5 |
| miR-375 | -21.88 |
miRNA expression ratio demonstrates high specificity and sensitivity in predicting disease
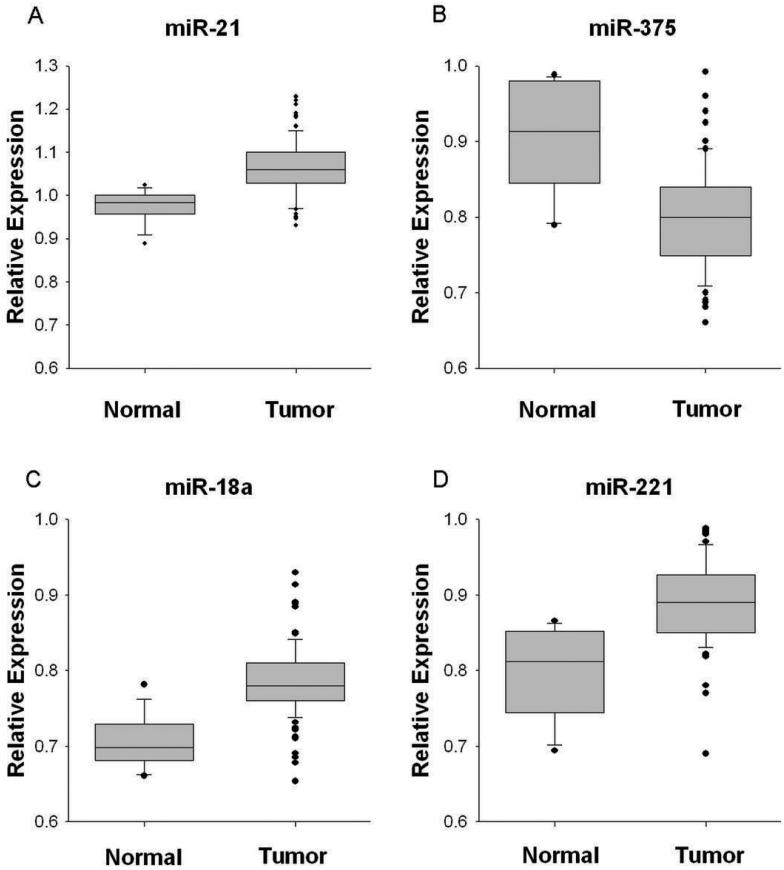
Validation of the four confirmed miRNAs was performed in nine additional normal tissue samples (total n = 14), and 83 additional tumor samples (total n = 99). Quantification of miRNA expression in this large set of samples showed strong validation of the results seen in the smaller population. miR-21, miR-18a, and miR-221 showed significant upregulation in tumors (P < .0001; Figure 2a, 2c, and 2d, zrespectively), whereas miR-375 was significantly downregulated in tumors (P < .0001; Figure 2b).
Figure 2.
Validation of differential expression of four microRNAs by quantitative real-time PCR in tumor (n=99) and normal (n=14) samples. For each miRNA, boxes denote distribution of expression values from the 25th to 75th percentile. Horizontal lines in boxes represent median values, whiskers represent 5th and 95th percentiles and outliers are denoted by dots. P < 0.0001 in all comparisons.
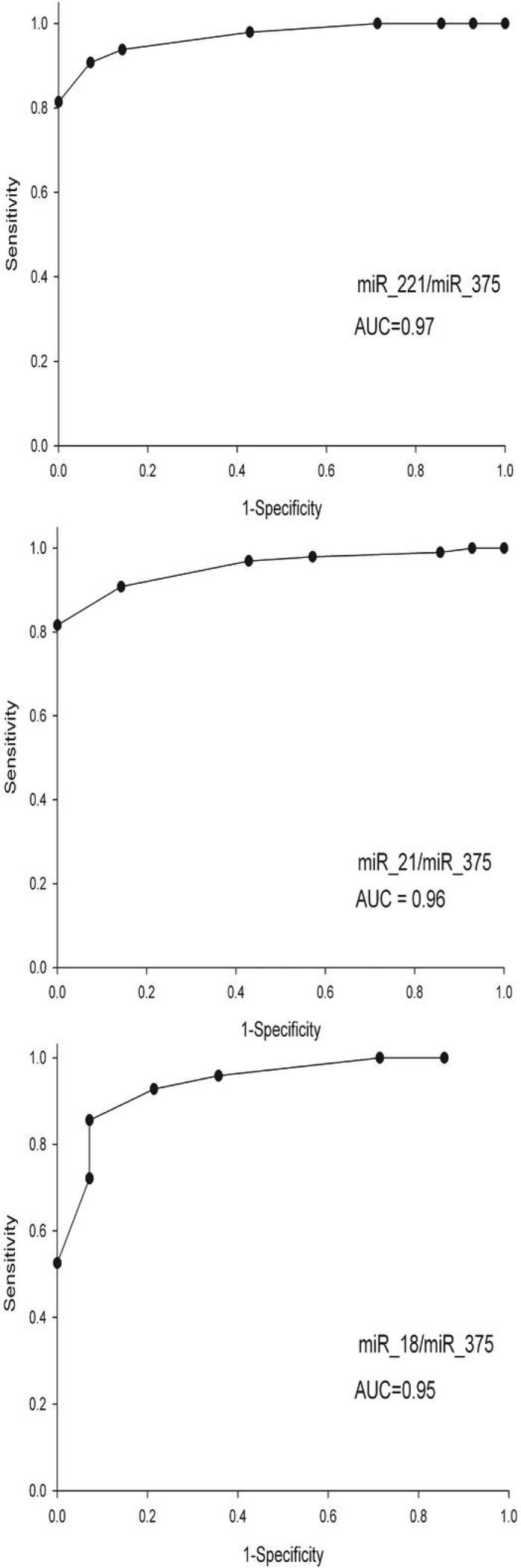
As miR-21, miR-18a, and miR-221 demonstrated consistent upregulation and miR-375 consistent downregulation, we next sought to determine if expression ratios constructed between these miRNA could improve their predictive potential for differentiating HNSCC tumor from non-diseased epithelia. Following the methods of Gordon et. al (27), miRNA. expression ratios were calculated by dividing the relative expression value of each of the three miRNAs showing upregulation in tumors by the expression value of the only downregulated miRNA, miR-375. ROC analysis was performed to determine which of these ratios demonstrated the greatest predictive power (Figure 3). Table 2 lists the representative sensitivity and specificity of ratios using the cutoff value of 1 for each of the upregulated miRNA to the downregulated miR-375 in differentiating between non-diseased tissue and HNSCC, using the validation series. miR-21:miR-375 ratios above 1.0 exhibited high specificity (0.99) but low sensitivity (0.14) whereas the relationship of miR-18a:miR-375 showed high sensitivity (1.00) but low specificity (0.52) (Table 2). However the ratio of miR-221:miR-375 exhibited the strongest predictive ability with both high sensitivity and specificity (0.92 and 0.93 respectively; Table 2).
Figure 3.
ROC analysis of expression ratios of miR-221, miR-21 and miR-18a to miR-375 for differentiation of HNSCC tumors and normal tissues. Curves were constructed using ratio value cut-offs ranging from 0.75 to 1.25 for each ratio. AUC value is indicated in each panel.
Table 2.
Examination of sensitivity and specificity of disease prediction using a miRNA expression ratio greater than 1.0
| Ratio | Sensitivity | Specificity |
|---|---|---|
| miR-21/miR-375 | 0.99 | 0.14 |
| miR-18a/miR-375 | 0.52 | 1 |
| miR-221/miR-375 | 0.92 | 0.93 |
Discussion
The present study revealed a number of miRNAs to be aberrantly expressed in HNSCC tumors, including an extensively validated subset that may hold utility as clinical biomarkers of disease. Microarray profiling of over 600 miRNAs identified 18 miRNAs that were significantly differentially expressed in tumor tissues compared to analogous non-diseased head and neck epithelia. Of the 12 human miRNAs in this group, 4 miRNA which were validated by qRT-PCR were used for in-depth examination of a larger population of fresh frozen HNSCC tumors and normal head and neck tissue.
The permutation-based software SAM was used to identify differentially expressed genes by pair-wise comparisons between groups of interest. It should be noted that SAM was designed (and may be better suited) for identification of important genes from high density microarrays, as it allows the user to control the number of findings based on a desired false discovery rate (FDR) while avoiding parametric assumptions about the data, inherent in tests such as the Analysis of Variance (ANOVA) (28).
The microarray data revealed that HNSCC cell lines show a distinct pattern of miRNA expression compared to primary tumors. This is consistent with past reports demonstrating a clear segregation of cell lines away from primary tumors upon high-throughput analysis of their miRNA expression (22, 29), suggesting that cell lines have limited utility as a model system for the identification of clinically relevant miRNA biomarkers. There remains, though, significant utility in utilizing in-vitro approaches for defining the biological mechanisms of these miRNAs, with appropriate understanding that the pattern of their expression is markedly different from that observed in the parent primary tissue.
Some of the miRNAs identified as differentially expressed in HNSCC compared to normal tissues have been characterized in past reports, particularly in relation to cancer. One of these, miR-21, was shown by quantitative real-time PCR analysis to be significantly upregulated in HNSCC tumors (17). miRNA profiling of breast, cervical, and ovarian tumors, glioblastomas, and head and neck primary tumors and cell lines, amongst others, has shown that miR-21 is commonly upregulated in cancer (17, 18, 21, 30). In fact, the relative levels of miR-21 and other miRNAs were reported to have prognostic relevance for predicting patient survival in lung cancer (31). Some known targets of miR-21 include the tumor suppressor genes tropomyosin 1 (TPM1) and programmed cell death 4 (PDCD4) (32, 33). Additionally, miR-221 and miR-18a, both shown to be upregulated in HNSCC tumors, have previously been implicated in hepatocellular, prostate, and other cancers (34-37).
miR-375 was the only downregulated miRNA found when comparing tumors and normal tissues, showing a ~22 fold decrease in tumors. Validation experiments also showed it to be sharply and significantly downregulated in tumors relative to normal tissues. miR-375 has been found to regulate insulin secretion in mice and its downregulation has been implicated in aberrant morphology of pancreatic islet cells in zebrafish (38, 39). Recently, miR-375 downregulation has also been associated with β-catenin mutation in hepatocellular adenoma (40). The downregulation of miR-375 in HNSCC tumors, as shown in this work, is consistent with a possible role for miR-375 in the transcriptional repression of oncogenes, analogous to the regulation of oncogenic KRAS by the let-7 miRNA (41).
In order to improve the predictive potential for individual miRNA alterations, we used expression ratios as described by Gordon et al (27). The ratio of miR-221:miR-375 demonstrated high discriminatory potential, with a sensitivity of 92% and specificity of 93% in distinguishing tumor from normal tissue. These data suggest that the ratio of these miRNAs may hold significant clinical potential, but further validation is necessary in an independent series of HNSCC tumors.
The identification of an HNSCC-specific miRNA signature indicates a plausible role for miRNAs in the development or progression of this disease. Finding abnormally expressed miRNAs may prove to be an important step in identifying the specific mechanisms of HNSCC carcinogenesis, as these aberrations may constitute early events in initiation or progression of the disease (42). As such, the utilization of a miRNA expression ratio that distinguishes disease tissue from non-diseased tissue holds potential as a simple, early diagnostic for HNSCC. An examination of these miRNA expression ratios in preneoplastic lesions, early stage tumors, and samples obtained for screening such as saliva and mouthwash would clarify the true clinical applicability of these findings in the realm of early diagnostics. Future studies should also examine the potential for associations between miRNA expression and clinical criteria such as tumor stage, metastasis, and prognosis in a larger series of tumors than what has been examined here. Finally, a validation of the miRNAs identified in this study in an independent test population of HNSCCs could be instrumental both in demonstrating that the individual miRNA aberrations are globally specific to HNSCC, and in resolving the true diagnostic potential of the miR-221:miR-375 ratio.
Supplementary Material
Acknowledgments
The authors thank the individuals involved in the head and neck cancer study, including hospital site investigators Marshall Posner (Dana-Farber Cancer Institute), John Clark (Massachusetts General Hospital), Gregory Grillone (Boston Medical Center), Richard Wein (Tufts Medical Center), and John Gooey (Veterans Administration Boston Healthcare System) as well as the NDRI for procurement of tissue samples.
Grant support: NIH/NCI R01CA078609, R01CA100679, NIH/NIEHS T32ES007272 (M.A.), the Flight Attendants Medical Research Institute (C.J.M), the Friends of the Dana-Farber Cancer Institute, and the Mesothelioma Applied Research Foundation.
References
- 1.Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol. 2001;2:533–43. doi: 10.1016/S1470-2045(01)00486-7. [DOI] [PubMed] [Google Scholar]
- 2.Ragin CC, Modugno F, Gollin SM. The epidemiology and risk factors of head and neck cancer: a focus on human papillomavirus. J Dent Res. 2007;86:104–14. doi: 10.1177/154405910708600202. [DOI] [PubMed] [Google Scholar]
- 3.Loning T, Ikenberg H, Becker J, Gissmann L, Hoepfer I, zur Hausen H. Analysis of oral papillomas, leukoplakias, and invasive carcinomas for human papillomavirus type related DNA. J Invest Dermatol. 1985;84:417–20. doi: 10.1111/1523-1747.ep12265517. [DOI] [PubMed] [Google Scholar]
- 4.Furniss CS, McClean MD, Smith JF, et al. Human papillomavirus 16 and head and neck squamous cell carcinoma. Int J Cancer. 2007;120:2386–92. doi: 10.1002/ijc.22633. [DOI] [PubMed] [Google Scholar]
- 5.Hardisson D. Molecular pathogenesis of head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2003;260:502–8. doi: 10.1007/s00405-003-0581-3. [DOI] [PubMed] [Google Scholar]
- 6.Al Moustafa AE, Alaoui-Jamali MA, Batist G, et al. Identification of genes associated with head and neck carcinogenesis by cDNA microarray comparison between matched primary normal epithelial and squamous carcinoma cells. Oncogene. 2002;21:2634–40. doi: 10.1038/sj.onc.1205351. [DOI] [PubMed] [Google Scholar]
- 7.Schlecht NF, Burk RD, Adrien L, et al. Gene expression profiles in HPV-infected head and neck cancer. J Pathol. 2007;213:283–93. doi: 10.1002/path.2227. [DOI] [PubMed] [Google Scholar]
- 8.Pyeon D, Newton MA, Lambert PF, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67:4605–19. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blenkiron C, Miska EA. miRNAs in cancer: approaches, aetiology, diagnostics and therapy. Hum Mol Genet. 2007;16 Spec No 1:R106–13. doi: 10.1093/hmg/ddm056. [DOI] [PubMed] [Google Scholar]
- 10.Bunz F, Dutriaux A, Lengauer C, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 11.Cowland JB, Hother C, Gronbaek K. MicroRNAs and cancer. Apmis. 2007;115:1090–106. doi: 10.1111/j.1600-0463.2007.apm_775.xml.x. [DOI] [PubMed] [Google Scholar]
- 12.Du T, Zamore PD. Beginning to understand microRNA function. Cell Res. 2007;17:661–3. doi: 10.1038/cr.2007.67. [DOI] [PubMed] [Google Scholar]
- 13.Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15:563–8. doi: 10.1016/j.gde.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 14.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075–80. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–45. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 16.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 17.Chang SS, Jiang WW, Smith I, et al. MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer. 2008;123:2791–7. doi: 10.1002/ijc.23831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran N, McLean T, Zhang X, et al. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun. 2007;358:12–7. doi: 10.1016/j.bbrc.2007.03.201. [DOI] [PubMed] [Google Scholar]
- 19.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Y, Chen Z, Zhang L, et al. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26–33. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 21.Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 22.Blenkiron C, Goldstein LD, Thorne NP, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters ES, McClean MD, Liu M, Eisen EA, Mueller N, Kelsey KT. The ADH1C polymorphism modifies the risk of squamous cell carcinoma of the head and neck associated with alcohol and tobacco use. Cancer Epidemiol Biomarkers Prev. 2005;14:476–82. doi: 10.1158/1055-9965.EPI-04-0431. [DOI] [PubMed] [Google Scholar]
- 24.Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–8. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- 25.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 26.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon GJ, Jensen RV, Hsiao LL, et al. Translation of microarray data into clinically relevant cancer diagnostic tests using gene expression ratios in lung cancer and mesothelioma. Cancer Res. 2002;62:4963–7. [PubMed] [Google Scholar]
- 28.Pan W. A comparative review of statistical methods for discovering differentially expressed genes in replicated microarray experiments. Bioinformatics. 2002;18:546–54. doi: 10.1093/bioinformatics/18.4.546. [DOI] [PubMed] [Google Scholar]
- 29.Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–52. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 30.Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 32.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–33. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 33.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–36. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 34.Fornari F, Gramantieri L, Ferracin M, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–61. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 35.Galardi S, Mercatelli N, Giorda E, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–24. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 36.Nam EJ, Yoon H, Kim SW, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–5. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 37.Takakura S, Mitsutake N, Nakashima M, et al. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008;99:1147–54. doi: 10.1111/j.1349-7006.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–30. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 39.Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ladeiro Y, Couchy G, Balabaud C, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–63. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 41.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 42.Gomes CC, Gomez RS. MicroRNA and oral cancer: future perspectives. Oral Oncol. 2008;44:910–4. doi: 10.1016/j.oraloncology.2008.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.