Abstract
Initially isolated as the dominant suppressor of the mutant epidermal growth factor receptor (ellipse), the Dachshund gene plays a key role in metazoan development regulating the Retinal Determination Gene Network (RDGN). Herein the DACH1 gene was expressed in normal prostate epithelial cells with reduced expression in human prostate cancer. DACH1 inhibited prostate cancer cellular DNA synthesis, growth in colony forming assays, and blocked contact-independent growth in soft agar assays. DACH1 inhibited Androgen Receptor (AR) activity, requiring a conserved DS Domain (Dachshund domain conserved with Ski/Sno) that bound NCoR/HDAC and was recruited to an androgen-responsive gene promoter. DACH1 inhibited ligand-dependent activity of AR mutations identified in patients with androgen insensitive prostate cancer. The DS domain was sufficient for repression of the AR wt but failed to repress an AR acetylation site point mutant. These studies demonstrate a role for the RDGN in regulating cellular growth and signaling in prostate cancer.
Keywords: DACH1, AR, Prostate Cancer, Proliferation
INTRODUCTION
Prostate cancer is the most frequently diagnosed cancer of men in the United States and the second leading cause of male cancer deaths (1–2). Hyperactivation of the Androgen Receptor (AR) is a fundamental driver of prostate cancer progression (3) and AR antagonists are the frontline of therapeutic intervention (4). Increased expression and/or activity of the AR is a common feature during prostate cancer onset and progression. Androgen ablation therapy results in a 60–80% initial response rate. Understanding the molecular mechanisms governing the growth property of the AR is essential to devise targeted therapies for prostate cancer, including androgen insensitive prostate cancer.
The AR is a ligand activated transcription factor that belongs to the superfamily of steroid receptors (5). These receptors have similar protein structures and are composed of an N-terminal domain that contains an activation function (AF-1), a DNA-binding domain (DBD), a hinge region and a ligand binding domain (LBD) that contains a second activation function domain (AF-2). Upon binding of ligands, such as dihydrotestosterone (DHT), the AR translocates to the nucleus and undergoes a conformational change that results in formation of a homodimer and the recruitment of multiple transcription factors that activate the expression of androgen-dependent genes (5). Activity of the AR is mediated via a balance between the function of co-activators (CBP/p300, Tip-60 and AR activator proteins (ARA)) (6, 7) and co-repressor proteins (including NCoR, Smads and histone deacetylases (HDACs, SIRT1)(8–10). In contrast with the wild type AR, mutant AR, that arise in patients with prostate cancer are promiscuously activated by a variety of hormones (estrogen, progesterone) that would normally have no effect on the wild type AR (11). These AR ligand binding mutations cause an androgen antagonist such as flutamide to behave as androgen agonist (12).
Upon ligand binding to the AR, post-translational modifications are induced including phosphorylation, acetylation, and ubiquitination, which in turn determine transcriptional and growth regulatory functions (13). The AR, like several other nuclear receptors, is directly acetylated at a motif that is widely conserved among various species (14–17). AR acetylation, which is induced by agonists including DHT and bombesin (18), governs prostate cancer cellular growth (15). Thus, gain of function mutants of the AR-acetylation site promote growth of human prostate cancer cells in tissue culture and when implanted in nude mice (15). The enhanced cellular growth is mediated by reduced MEKK1-induced apoptosis (16) and enhanced cellular proliferation through induction of cell-cycle regulatory genes (16).
Conversely, ligand-dependent AR activity is repressed by histone deacetylases of both the TSA-dependent type (Class I and Class II) and by the Class III HDAC. Co-repressors of the AR, including HDAC1, HDAC3 and NCoR can bind the AR in the context of local chromatin and disengage in the presence of ligand (19–21). The Class III HDACS are homologous to the transcriptional repressor Sir2P. It is known that Class I and Class II histone deacetylases are sensitive to the inhibitor Trichostatin A (TSA). Class III HDAC activity is nicotinamide-adenine dinucleotide-dependent, and cannot be inhibited by TSA. SIRT1 represses ligand-dependent AR activity and SIRT1 deacetylates the AR with similar kinetics and Km to that described for p53 (10). Inhibition of Class III histone deacetylates activity in prostate cancer cells enhances liganded AR activity and AR expression (10) suggesting an important biological role for endogenous Class III histone deacetylases in AR signaling (22). SIRT1 inhibits prostate cancer cellular growth in the presence of the AR (10).
Although AR co-repressors inhibit AR function, studies to date have not revealed significant alteration in the abundance of AR co-repressor proteins during prostate cancer onset and progression. Recent studies have identified a protein resembling the NCoR/Ski family that physically associates with HDAC1 and HDAC3, known as Dachshund1 (23). DACH1 repression function has been linked to its association with HDAC1, HDAC3, Sin3 and NCoR which colocalized with DACH1 at the promoters of target genes repressed by DACH1 (23, 24). The Dachshund1(DACH1) gene was initially identified in Drosophila as a dominant suppressor of Ellipse, a constitutively active epidermal growth factor receptor (25). The Dac gene is known to play a key role in metazoan development regulating ocular, limb and gonadal development. In the Drosophila eye, Dac encodes a key component of the Retinal Determination Gene Network (RDGN). Loss of function mutations of the gene result in defective eye development, whereas ectopic Dac expression is sufficient for the induction of eye development implicating Dac in organizing cellular fate (25).
The DACH1 gene encodes a winged helix-Forkhead subgroup structure of the helix-turn-helix family (26). Although no specific DNA binding characteristics have been identified, DACH1 has been identified in the context of local chromatin by ChIP assays within distinct DNA binding sites for the transcription factors Smad, c-Jun, CREB, and Six1 (23, 24, 27). Thus, the DACH1 gene product uses other DNA-binding transcription factors to regulate gene expression. A conserved domain (Dachshund domain 1) has significant identity with the Ski/Sno (Dachshund DS domain), which is conserved from Drosophila to human. DACH1 gene expression is lost in metastatic human breast cancer (23). The loss of DACH1 expression correlates with poor prognosis. DACH1 inhibited cellular migration and invasion of oncogene (Ras, Myc, ErbB2)-transformed human mammary epithelial cells. IL-8 is a critical target of DACH1 mediated breast cancer cellular migration in vitro and metastasis in vivo (28). In ovarian cancer, DACH1 is overexpressed and may promote tumor progression by antagonizing TGFβ signaling (29, 30).
The role of DACH1 gene expression in prostate cancer was unknown. In the current studies DACH1 abundance was reduced in human prostate cancer. Re-expression of DACH1 inhibited prostate cancer cellular growth and colony formation in soft agar assays. DACH1 inhibited DHT-dependent AR activity requiring the conserved DS domain. DACH1 inhibited the activity of mutant AR derived from patient samples. The DACH1 gene is a novel inhibitor of prostate cancer cellular growth, inhibiting ligand-dependent AR activity.
MATERIALS AND METHODS
Plasmid construction, reporter genes, expression vectors, reagents and kits
The expression plasmids encoding an N-terminal FLAG peptide linked to DACH1, DACH1 DS-domain alone (DS) or DACH1 DS-domain deleted (ΔDS), the tet-inducible DACH1 expression vector and DACH1-VP16 fusion protein were previously described (27, 30). The Ski cDNA was subcloned into the 3x FLAG-CMV7.1 vector (Sigma, St. Louis, MO). Expression vectors for human TIP-60, SRC-1, ARA70 and SIRT1 were previously described (10, 15, 16). The androgen-responsive synthetic reporter constructs (MMTV-LUC, PSA-LUC, Sc-ARE, (ARE)4LUC, pG5LUC) and the expression vectors (pCMVHA-p300, pcDNA3AR-GFP, hARwt, the AR carboxyl terminal ligand binding domain mutants and the AR point mutants ARK630T, ARK630Q) were previously described (10, 11, 15–17, 31–34). DHT, R1181, TSA, and Nicotinamide were purchased from Sigma, St. Louis, MO. Free PSA in the culture medium was measured by Free-PSA ELISA kit from BioQuant (San Diego, CA, Cat. BQ092F).
Cell culture, DNA transfection, and luciferase assays
Cell culture, DNA transfection, and luciferase assays were performed as previously described (15–17). The CV1, DU145, LNCaP, and HEK293T cell lines were cultured as previously described (10). Transfection and data analyses were as described in supplemental information.
DNA synthesis analysis and colony formation assay
DNA synthesis was analyzed by 3H-TdR incorporation. 1 × 105 cells were plated into 24-well plate and cultured for 36 h. 1 μCi of 3H-TdR was added to each well and culture continued for 2 h. Cells were washed and fixed with incorporation of 3H-TdR measured by liquid scintillation. For contact-dependent growth, 4 × 103 LNCaP cells were plated in triplicate in the presence or absence of 2 μg/ml doxycycline into 6-cm dish and medium was changed every 3 days. The colonies were visualized after 2-weeks of growing by staining with 0.04% crystal violet in methanol for 1 h. For contact-independent growth assays, 4 × 103 LNCaP cells were plated in triplicate in 2 ml of 0.3% agarose in complete growth medium in the presence or absence of 2 μg/ml doxycycline overlaid on a 0.5% agarose base, also in complete growth medium. 2 weeks after incubation, colonies greater than 50 μm in diameter were counted using an Omnicon 3600 image analysis system.
Western blot analysis, tissue microarrays, and immunohistochemistry
Western blot analysis using antibodies to FLAG, AR (Santa Cruz Biotechnology, Santa Cruz, CA), and the loading control β–tubulin was conducted as described (10). Immunohistochemistry for DACH1 was performed as previously described (29, 35). A human prostate tissue array consisting of 32 paired prostate cancer tissues with corresponding normal tissues was from AccuMax Array (MD, USA, ISU Abxis). The immunostaining intensity was scored as negative: 0, minimally positive: 1, positive: 2, strong positive: 3, respectively. The DACH1 positive cells were counted and recorded by a pathologist.
RNA extraction from FFPE human prostate tissue specimens, Real-time quantitative PCR
RNA was extracted from formalin fixed paraffin embedded (FFPE) archival human prostate tissue blocks containing 10 normal human prostates and 10 matched prostate tumor tissue specimen, using RecoverAll™ Total Nucleic Acid Isolation Kit for FFPE Tissues (Applied Biosystems, Foster City, CA). Subsequent analysis was as described in supplemental information (28).
Chromatin immuno-precipitation assays (ChIP)
Endogenous PSA promoter ChIP assays using antibodies directed to the FLAG epitope of DACH1, AR, Sirt1, NCoR, HDAC3, and control IgG were performed as previously described (9, 23, 28, 36). (see supplemental information)
RESULTS
DACH1 expression is reduced in human prostate cancer
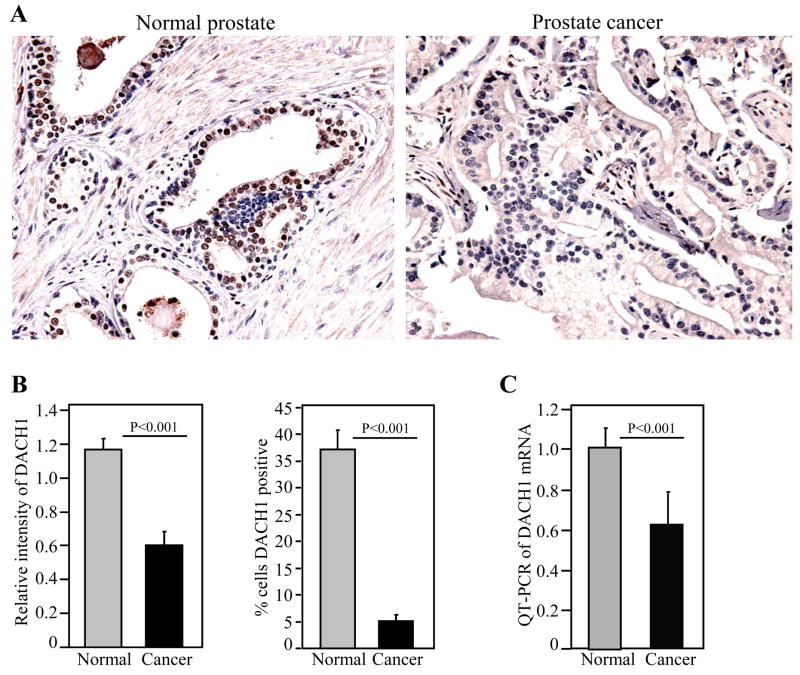
In order to determine the relative abundance of DACH1 in normal and prostate cancer samples, immunohistochemical staining was performed on paired normal and prostate cancer tissues, using a previously well-characterized antibody (23). DACH1 staining was identified in the nucleus and cytoplasm of normal prostate epithelial cells (Fig. 1A). Approximately 35% of epithelial cells stained positively for DACH1 in the normal prostate. The relative intensity of the immunohistochemical staining was quantitated for each sample. Relative mean intensity was compared between those individual samples staining positively for DACH1 in the normal and tumorous tissues. The relative intensity of immunostaining was significantly reduced (~ 2-fold) in the prostate tumors, compared to the corresponding normal prostate epithelium (Fig. 1B). The percentage of cells staining for DACH1 was also significantly reduced (~ 7-fold) (Fig. 1B). The relative abundance of DACH1 mRNA was assessed in matched prostate tissue comparing normal and prostate cancer samples from each patient (Fig. 1C). The relative abundance was normalized for epithelial cells using KLK3 (PSA) mRNA. DACH1 mRNA was reduced in prostate cancer samples compared with the normal prostate tissue of the individual. Thus, the relative abundance of DACH1 is reduced in prostate cancer epithelium compared with normal prostate epithelium.
Figure 1. Immunohistochemical staining and mRNA expression of DACH1 in human prostate cancer.
(A) Representative example of DACH immunostaining of normal prostate and prostate cancer (400 ×). (B) Quantification of DACH1 relative immune intensity and percent of cells staining for DACH1. (C). DACH1 mRNA determined by quantitative PCR. Comparison was made between matched normal and tumorous prostate samples.
DACH1 inhibits human prostate cancer cellular growth
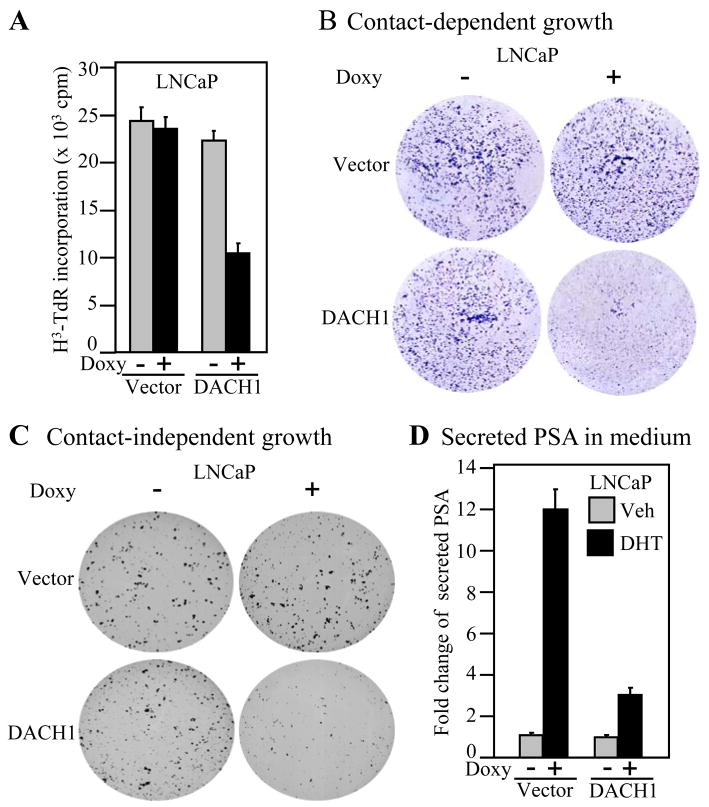
In order to determine the role of DACH1 in prostate cancer cellular growth, the LNCaP cell line was used to create tetracycline inducible DACH1 gene expression. The addition of deoxycycline induced DACH1 expression as characterized by Western blot analysis to the FLAG epitope of the DACH1 protein (Supplementary Fig. S1A). β-tubulin, used as a loading control for protein abundance, showed similar protein levels. Doxycycline did not affect 3H thymidine uptake in LNCaP cells. DACH1 expression inhibited LNCaP cell proliferation by greater than 60% (Fig. 2A). Contact-dependent growth of LNCaP cells was characterized by colony formation assay. LNCaP cells formed colonies in tissue culture (Fig. 2B). The number and size of colonies were reduced by > 85% upon expression of DACH1. The induction of DACH1 expression upon doxycycline treatment reduced colony formation in soft agar by >85% (Fig. 2C). Dihydrotestosterone (DHT, 10−8 M) induced PSA secretion ~9.5 fold (Fig. 2D). Expression of DACH1 reduced DHT-dependent induction of PSA by approximately 60% (Fig. 2D). Collectively, these studies demonstrate DACH1 inhibits LNCaP cell contact-dependent growth and DNA synthesis.
Figure 2. DACH1 inhibits prostate cancer cellular proliferation and growth.
(A) DNA synthesis analysis of LNCaP DACH1-doxy-inducible cell line. Data are mean ± SEM for N>5 separate experiments. The Doxycycline (Doxy) inducible DACH1 LNCaP cell line were assessed for (B) colony formation (C) contact-independent growth in soft agar and (D) PSA secretion in the media determined by ELISA. Data are mean ± SEM for N≥5 throughout.
DACH1 repression of ligand-dependent AR activity involves the DS domain
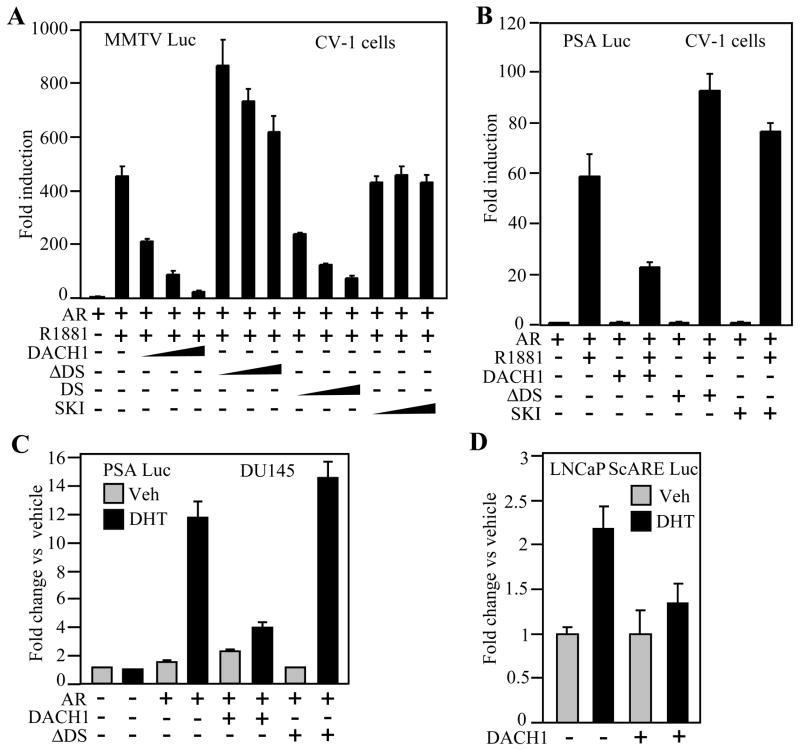
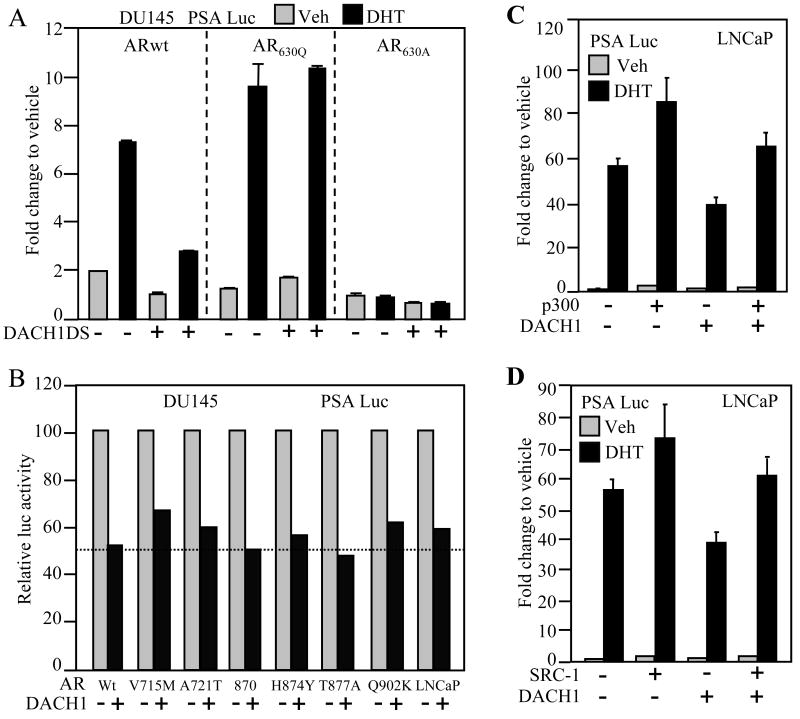
As DACH1 expression had inhibited PSA secretion of LNCaP cells, we considered the possibility that DACH1 may directly inhibit AR transcriptional activity. To determine whether DACH1 inhibited AR transcriptional activity, CV1 cells were used and androgen responsive reporter genes encoding Androgen Response cis Elements (ARE) were co-introduced with an expression vector encoding the wild type AR. In the presence of DHT and the AR, ARE activity was induced approximately ~ 450-fold. The transfection of increasing doses of DACH1 expression vector reduced repression of ligand-dependent AR activity assay using the ARE reporter gene (Fig. 3A). In order to determine the specificity of transcriptional repression of AR signaling by DACH1, comparison was made with a deletion mutant of the conserved DS domain of DACH1 (DACH1 ΔDS). Tranfection of equal amounts of the DACH1 ΔDS expressing vector failed to repress ligand-dependent activity of the AR (Fig. 3A). Expression of the DS domain alone however was sufficient for repression of ligand-dependent AR activity (Fig. 3A). As the DS domain shares identity with the Ski protein at the amino acid level, we examined the effect of Ski expression on ligand-dependent AR activity. In contrast with DACH1, expression of Ski failed to repress ligand-dependent activation of the AR (Fig. 3A). Similar experiments were conducted using the PSA promoter luciferase reporter gene. Addition of the AR agonist R1881 induced PSA luciferase reporter activity ~60 fold (Fig. 3B). Expression of DACH1 reduced PSA-Luc activity ~40 fold. The deletion of the DS domain abrogated DACH1 repression and, as with a simple ARE, Ski failed to repress AR activity assessed using the PSA promoter (Fig. 3B). As the transcriptional repression function of co-repressors may vary by cell type, we examined the repression function of DACH1 in the AR-deficient prostate cancer cells DU145. The addition of DHT with the AR induced PSA luciferase activity ~12 fold (Fig. 3C). DHT-induced AR activity was reduced ~8 fold upon expression of DACH1. Deletion of the DS domain abrogated DACH1-dependent repression of ligand-dependent activity of the PSA promoter (Fig. 3C). Similarly, in HEK293T cells, the ligand-dependent activation of the PSA promoter was repressed from 6- to 3- fold by DACH1 expression. The effect of DACH1 was abrogated by deletion of the DS domain and the DS domain alone was sufficient for repression of ligand-dependent AR activity (Supplementary Fig. S1B). We examined the effect of DACH1 in regulating ligand-dependent AR activity at distinct AREs. The secretory component (SC) of the polymeric immunoglobulin receptor (pIgR) gene is an androgen responsive gene responsible for transepithelial transport of IgA and IgM. The scARE was linked to the luciferase reporter gene and transduced into the LNCaP and HEK293T cells. DHT induced scARE activity ~2.5 fold and this activity was abrogated by expression of DACH1 (Fig. 3D, Supplementary Fig. S1C). Thus, DACH1 inhibits ligand-dependent AR activity requiring the DS domain assessed using multiple distinct AREs in both DU145 and CV-1 cells.
Figure 3. DACH1 regulation of AR activity.
Androgen-response luciferase reporter genes were assessed for DHT-mediated activity in transient expression studies. Cells were co-transfected with expression vectors encoding DACH1, mutant DACH1 (ΔDS) or Ski. Cells were treated with AR agonist (either R1881-10nM or DHT-10nM). Luciferase reporter gene assays were conducted in distinct cell lines as indicated (A; B; CV1) (C; DU145) (D; LNCaP). Data are mean ± SEM of N≥5 separate transfections.
DACH1 recruits NCoR and SirT1 to endogenous Androgen Response Elements (AREs)
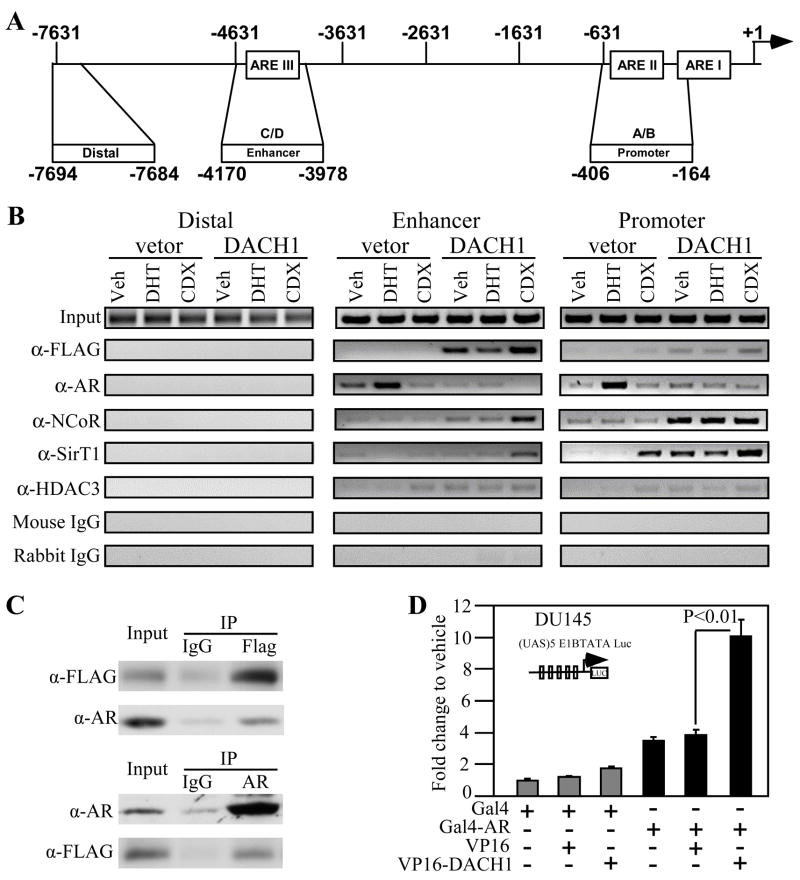
The AR associates in the context of local chromatin at Androgen Response Elements (AREs) within the proximal PSA promoter as assessed by chromatin immuno-precipitation (ChIP) assays. In order to determine whether DACH1 mediated AR repression involved direct association of DACH1 at an endogenous ARE, ChIP studies were conducted in LNCaP cells. A doxycycline (Doxy) inducible retrovirus expression vector encoding FLAG-tagged DACH1 was used to transduce LNCaP cells. ChIP analyses were conducted in the presence of vehicle, DHT (10nM) or the DHT antagonist Casodex (CDX, 15μM). Comparison was made between the PSA proximal promoter (ARE I-II), the enhancer (ARE III) and a distal region of the PSA promoter that does not bind the AR as a form of negative control (Fig. 4A). ChIP conducted to DACH1 demonstrated Tet-induced DACH1 expression correlated with DACH1 recruitment to the PSA ARE I-II and ARE III, but not the control distal promoter region (Fig. 4B). DHT reduced and Casodex enhanced DACH1 recruitment to ARE I-II/III. In the presence of DHT, the AR abundance was increased at ARE I-II/III, which was reduced by Casodex or DACH1 expression (Fig. 4B). NCoR recruitment was enhanced by DACH1 expression at ARE I-II/III with further recruitment by Casodex at ARE III. NCoR and DACH1 associate with class II TSA-sensitive HDACs. Recent studies demonstrated SirT1 represses the AR through deacetylation and is recruited to the ARE (10, 37). Consistent with previous studies, Casodex enhanced SirT1 abundance at the ARE I-II. DACH1 expression enhanced SirT1 recruitment in the basal state and in the presence of DHT at ARE I-II. DACH1 increased SirT1 recruitment in the presence of Casodex to ARE III. Immunoprecipation-wetsern blot analysis detected the increased association of AR with DACH1 in the presence of DHT and recruitment of the co-repressor protein HDAC1 and NCoR (Fig. 4B). Mammalian two hybrid analysis using Gal4-AR and VP16-DACH1 demonstrated their association (Fig. 4C).
Figure 4. DACH1 associates with the AR and is co-recruited to an endogenous ARE.
(A) Schematic representation of the human PSA promoter indicating the ARE (I-III) (B) ChIP analysis of LNCaP cells stably transduced with a Doxy-regulated DACH1 expression vector. Cells were treated for 12 hours with either DHT (10nM), Casodex (CDX, 15μM), or vehicle control. ChIP assays were conducted with the antibodies indicated. (C) immunoprecipation-western blot assay in LNCaP cell. (D) Association of DACH1 and AR in mammalian two-hybridization assay.
DACH1 DS domain-mediated AR repression involves the AR acetylation site
In order to determine the AR domains required for DACH1-dependent repression, a series of point mutants of the AR were examined in co-transfection experiment. In some patients with prostate cancer, point mutations arise in the carboxyl terminal ligand binding domain and contribute to promiscuous activation of the AR by different ligands. It has been considered that these mutations may thereby contribute to androgen antagonist failure (11). Using the androgen-responsive reporter PSA-luc in DU145 cells, comparison was made with point mutations of the AR acetylation site, as mutation of this site has been identified in prostate cancer and this site is known to be a key determinant of androgen-dependent prostate tumor cellular growth (22, 36, 38). As DACH1 repression of the wild type AR was reduced by deletion of the DS domain and the DS domain was sufficient for repression of the wild type AR (Fig. 3A), we tested the effect of DACH1 on the AR acetylation site point mutants using the DACH1 DS domain. Point mutation of the AR acetylation site enhanced ligand-induced transactivation (Fig. 5A) as previously described (15) inducing PSA activity ~10 fold. In contrast with AR wt, expression of the DACH1 DS domain failed to repress the gain-of-function AR acetylation point mutant. These studies suggest the involvement of AR acetylation site in AR repression by the DACH1 DS domain. An acetylation site dead mutant (ARK630A) failed to induce AR activity. No significant interaction was observed upon the expression of DACH1 (Fig. 5A).
Figure 5. DACH1 inhibition of AR reporter gene activity via the AR acetylation cite and overcome by p300.
(A). Luciferase reporter gene assays were conducted using the AR-responsive PSA-Luc reporter gene. Cells were co-transfected with the AR wt or AR mutants and DACH1 DS domain as indicated. The AR K630 site is acetylated in response to ligand. ARK630Q represents a gain-of-function mutant, ARK630A, an acetylation dead mutant. (B) AR wt or carboxyl terminal ligand binding mutants were assessed for DACH1 repression. (C) p300 and (D) SRC-1 enhanced AR activation and overcome DACH1 repression. Data are mean ± SEM of N≥5 separate transfections.
The role of the AR carboxyl (C)-terminus was next examined. A series of point mutations in the AR C-terminus were examined for DACH1 repression. The AR wt and ligand-binding domain point mutants each activated PSA activity (Fig. 5B). Ligand-dependent activity was normalized to 100% to enable an assessment of DACH1 repression. DACH1 inhibited ligand-dependent activation of the PSA promoter assessed using either the wildtype AR or each of the ligand binding domain AR point mutants (Fig. 5B). Collectively these studies suggest the interaction between DACH1 and the AR does not involve the C-terminal ligand binding domain and may involve the AR acetylation site.
DACH1 repression of AR signaling involves class III deacetylases
AR activity is determined by co-activators including p300, SRC-1, TIP60, and ARA-70. Reporter gene assays were conducted to examine the effect of these co-activator proteins on DACH1-mediated repression of the AR. Ligand-dependent augmentation of AR activity was induced by p300 and SRC-1, and the expression of p300 and SRC-1 overcame the effect of DACH1 repression on PSA luciferase activity in the presence of DHT in both LNCaP and DU145 cells (Fig. 5C, D and Supplementary Fig. S2C, lanes 6 vs. 8). Similarly, expression of pCAF, TIP60, or ARA-70 was capable of overcoming DACH1 repression of AR activity in the presence of ligand in both LNCaP and DU145 cells (Supplementary Fig. S2A-F, lanes 6 vs. 8). Collectively, these studies demonstrate AR co-activators may either partially or completely rescue DACH1-dependent AR repression.
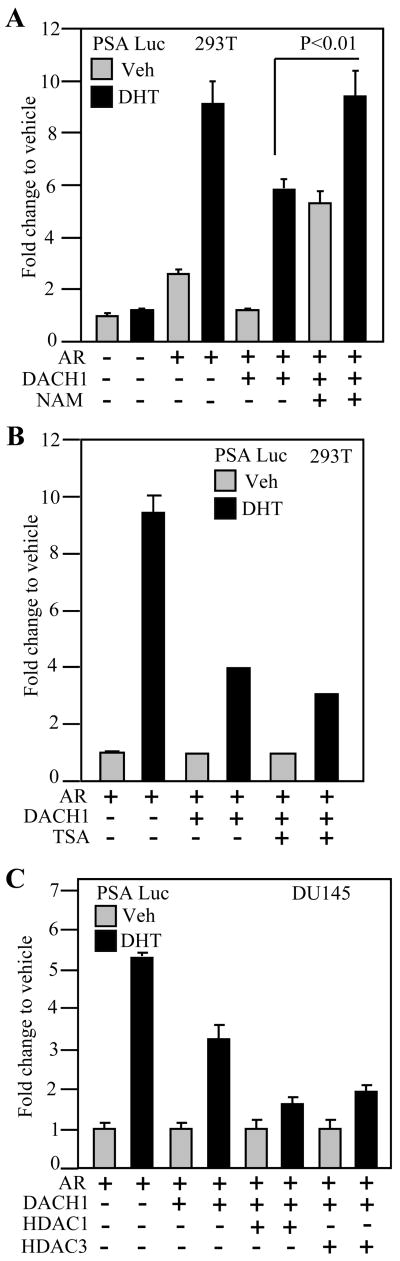
The AR acetylation site gain-of-function mutant ARK630Q evaded repression by the DACH1 DS domain. Endogenous SIRT1, a class III histone deacetylase, constitutively represses AR activity in prostate cancer cells (10, 37). We examined the role of endogenous Sirt1 in DACH1 repression using Sirt1 inhibitors (nictotinamide (NAM), Sirtinol, splitomycin). AR activity was assayed using the PSA promoter in HEK293T cells. AR activity was induced ~ 4.5 fold by DHT and repressed 50% by DACH1. The addition of Nicotinamide (5μM) reversed the DACH1-dependent repression in the presence of ligand (Fig. 6A). DACH1 repression of DHT-dependent AR activity was not effected by the addition of TSA (Fig. 6B). Co-expression of either HDAC1 or HDAC3 augmented DACH1-dependent repression of the AR responsive PSA promoter (Fig. 6C). In addition to the known association of class I/II HDACs with DACH1, the current studies suggest class III HDACs are involved in DACH1-repression of the AR.
Figure 6. DACH1 inhibition of AR reporter gene activity was mediated through sirt1 dependent.
Luciferase reporter gene assays were conducted using the AR-responsive PSA-Luc reporter gene. Cells were co-transfected with the AR and DACH1 as indicated and treated with (A) NAM or (B) TSA, or (C) co-transfected with HDAC. Data are mean ± SEM of N≥5 separate transfections.
DISCUSSION
The current studies demonstrate that DACH1 inhibits DHT-dependent AR signaling. DACH1 inhibited DHT-dependent DNA synthesis, cellular proliferation, and contact-dependent growth. DACH1 repression of the AR was dependent upon the ARE, the presence of the AR, and the presence of the AR ligand DHT. Repression of AR activity required the conserved DS domain of DACH1. This region of DACH1 is conserved with the Sno/Ski oncogenes. However, neither Ski, nor Sno repressed ligand-dependent AR activity. The DS domain alone was sufficient for repression of ligand-dependent AR activity. The DS domain is known to physically associate with NCoR/HDAC1 and HDAC3 (23, 24). Androgen response element activity is determined by the relative abundance of co-activators and co-repressor proteins which either encode or recruit HDAC activity. The AR co-repressors, including NCoR, SMRT, HDAC1/3 and SIRT1 (8, 9, 19, 39, 40) play a key role in regulating ligand-dependent activity (41). The current studies are consistent with a model in which DACH1 recruits known AR co-repressors to maintain the AR in a repressed state.
The DS domain of DACH1 was sufficient for repression of AR activity. Point mutation of the AR within the carboxyl terminus did not affect the magnitude of repression by DACH1. Thus DACH1, like SIRT1 (41), inhibits the activity of mutant AR that arise in patients that are resistant to androgen ablation therapy. Point mutation of the AR acetylation site, however, created an AR mutant that was defective in repression by the DS domain. The AR is modified by phosphorylation, acetylation, ubiquitination, and sumoylation (41). The AR acetylation at lysine residues 630, 632, and 633 is known to enhance AR activity and gain-of-function mutations of the AR acetylation site (ARK630Q, ARK630T) convey enhanced growth and reduced apoptosis in human prostate cancer cells in culture and in vivo in nude mice (41). Molecular analysis of the gain-of-function AR acetylation site mutants demonstrated enhanced binding of co-activator proteins (p300) and reduced binding of the co-repressor complex including NCoR, HDAC1, HDAC3, SMAD6 (10). The finding that the DACH1 DS domain failed to repress the gain-of-function AR acetylation site mutant is consistent with previous studies showing DACH1 binds NCoR, HDAC1/3 (23, 27) and studies showing HDAC1/3 and NCoR (15, 16) have reduced binding affinity to this AR acetylation site mutant.
Herein the AR was recruited to an ARE in the presence of ligand. In the current studies, DACH1 was recruited to both the proximal promoter (ARE I-II) and the distal enhancer (ARE III) and augmented NCoR recruitment. DACH1 enhanced Casodex-mediated recruitment of NCoR to the distal enhancer ARE III. The mechanisms governing NCoR recruitment are important as transcriptional repression by the AR induced by androgen antagonist is dependent upon recruitment of a co-repressor complex involving NCoR (10). Furthermore, NCoR appears to be required for transcriptional repression by androgen antagonists (10) and Casodex (bicalutamide) may function as an agonist in the absence of this co-repressor (10). Previous studies have been consistent with a model in which the AR binds to and represses target gene transcription upon binding an androgen antagonist. Antagonist bound AR undergoes a conformational change rendering it competent to bind transcriptional co-repressors. The current studies demonstrate DACH1 expression is associated with the recruitment of NCoR in the absence of androgen antagonist. DACH1 thus functions to recruit the NCoR co-repressor in the absence of antagonist.
The repression of AR activity was dependent upon Nicotinamide, adenosine-dinucleotide (NAD). The class III HDAC family encoded by the Sirtuin proteins, which regulate diverse biological processes involved in the stress response, metabolism, apoptosis, aging and nuclear receptor function (22). Sirt1 regulates transcription factors including p53, Foxo proteins, MyoD, NF-κB, and the p300 and PGC1α co-activators (22). Consistent with a prior study (22), SirT1 was recruited to ARE I-II upon Casodex treatment. SirT1 inhibits ligand-dependent AR-transactivation (22) and is required for AR antagonist induced transcriptional repression (37). Sirt1 deacetylates and represses AR activity (10). In a previous study, SirT1 was recruited in the context of local chromatin to AREs (37). Herein DACH1 enhanced Sirt1 recruitment to the endogenous ARE of the PSA gene. SirT1 is expressed in prostate cancer and represses AR activity and AR-dependent cellular proliferation (37). DACH1 like Casodex recruits SirT1. DACH1 loss in prostate cancer may uncouple Sirt1 repression. The inhibitions of DACH1 repression by the SIRT1 inhibitors (Sirtinol, nicotinamide) are consistent with the participation of class III HDAC in DACH1 function. Thus, DACH1 repression of the liganded AR involves Type I/II and Type III HDAC activity.
The Dachshund gene encodes a protein of the helix-turn-helix family with a winged Forkhead structure (26). Initially described as a dominant repressor of the EGF receptor, ellipse mutation (25). Drosophila Dachshund (Dac) encodes one of seven genes required for compound eye specification. The current studies have implications for cross-talk between the RDGN signaling pathway and hormonal signaling through the AR. Drosophila gonadal development is dependent upon Dac gene expression and Dac mutants flies have defective external male genitalia and defective ovarian duct formation (42, 43).
Herein, DACH1 expression inhibited DHT-dependent cellular growth, DNA synthesis and proliferation. Although the molecular mechanisms by which DACH1 regulated DHT-dependent cellular proliferation remains to be determined the inhibition of AR transcriptional activity is likely to be the key mechanism. DHT induced LNCaP cellular proliferation is AR-dependent and DACH1 expression inhibited transcriptional activity of the AR mutant identified in LNCaP cells. DACH1 may inhibit proliferation via additional targets of the AR. DACH1 can inhibit the transcriptional activity of c-Jun and repress expression of cyclin D1 in fibroblasts (23, 27). Given the role of Dachshund in both the RDGN network and AR signaling, it will be of interest to determine the role of the RDGN genes in cell fate determination and prostate tumor progression.
Supplementary Material
Acknowledgments
This work was supported in part by R01CA70896, R01CA75503, R01CA86072, (R.G.P.), and the Kimmel Cancer Center is supported by the NIH Cancer Center Core grant P30CA56036 (R.G.P.). This project is a generous grant from the Dr. Ralph and Marian C. Falk Medical Research Trust (R.G.P.), Margaret Q. Landenberger Research Foundation (K.W.) and supported in part by a grant from the Pennsylvania Department of Health (R.G.P).
References
- 1.Boring CC, Squires TS, Tong T, Montgomery S. Cancer statistics, 1994. A Cancer J Clin. 1994;44:7–26. doi: 10.3322/canjclin.44.1.7. [DOI] [PubMed] [Google Scholar]
- 2.Deutsch E, Maggiorella L, Eschwege P, Bourhis J, Soria JC, Abdulkarim B. Environmental, genetic, and molecular features of prostate cancer. Lancet Oncol. 2004;5:303–13. doi: 10.1016/S1470-2045(04)01468-8. [DOI] [PubMed] [Google Scholar]
- 3.Chen CD, Welsbie DS, Tran C, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 4.Sharifi N, Farrar WL. Androgen receptor as a therapeutic target for androgen independent prostate cancer. Am J Ther. 2006;13:166–70. doi: 10.1097/00045391-200603000-00013. [DOI] [PubMed] [Google Scholar]
- 5.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 6.Onate SA, Tsai SY, Tsai MJ, O’Malley BM. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–57. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 7.Shen HC, Buchanan G, Butler LM, et al. GRIP1 mediates the interaction between the amino- and carboxyl-termini of the androgen receptor. Biol Chem. 2005;386:69–74. doi: 10.1515/BC.2005.009. [DOI] [PubMed] [Google Scholar]
- 8.Zhu P, Baek SH, Bourk EM, et al. Macrophage/cancer cell interactions mediate hormone resistance by a nuclear receptor derepression pathway. Cell. 2006;124:615–29. doi: 10.1016/j.cell.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 9.Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol Cell. 2002;9:601–10. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 10.Fu M, Liu M, Sauve AA, et al. Hormonal control of androgen receptor function through SIRT1. Mol Cell Biol. 2006;26:8122–35. doi: 10.1128/MCB.00289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taplin ME, Bubley GJ, Shuster TD, et al. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N Engl J Med. 1995;332:1393–8. doi: 10.1056/NEJM199505253322101. [DOI] [PubMed] [Google Scholar]
- 12.Tilley WD, Buchanan G, Hickey TE, Bentel JM. Mutations in the androgen receptor gene are associated with progression of human prostate cancer to androgen independence. Clin Cancer Res. 1996;2:277–85. [PubMed] [Google Scholar]
- 13.Wang C, Powell MJ, Popov VM, Pestell RG. Acetylation in nuclear receptor signaling and the role of sirtuins. Mol Endocrinol. 2008;22:539–45. doi: 10.1210/me.2007-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C, Fu M, Angeletti RH, et al. Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity. J Biol Chem. 2001;276:18375–83. doi: 10.1074/jbc.M100800200. [DOI] [PubMed] [Google Scholar]
- 15.Fu M, Wang C, Wang J, et al. Acetylation of the androgen receptor enhances coactivator binding and promotes prostate cancer cell growth. Mol Cell Biol. 2003;23:8563–75. doi: 10.1128/MCB.23.23.8563-8575.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu M, Wang C, Wang J, et al. The Androgen Receptor Acetylation governs transactivation and MEKK1-induced apoptosis without affecting in vitro sumoylation and transrepression function. Mol Cell Biol. 2002;22:3373–88. doi: 10.1128/MCB.22.10.3373-3388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu M, Wang C, Reutens AT, et al. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J Biol Chem. 2000;275:20853–60. doi: 10.1074/jbc.M000660200. [DOI] [PubMed] [Google Scholar]
- 18.Gong J, Zhu J, Goodman OB, et al. Activation of p300 histone acetyltransferase activity and acetylation of the androgen receptor by bombesin in prostate cancer cells. Oncogene. 2006;25:2011–21. doi: 10.1038/sj.onc.1209231. [DOI] [PubMed] [Google Scholar]
- 19.Hodgson MC, Astapova I, Cheng S, et al. The androgen receptor recruits nuclear receptor CoRepressor (N-CoR) in the presence of mifepristone via its N and C termini revealing a novel molecular mechanism for androgen receptor antagonists. J Biol Chem. 2005;280:6511–9. doi: 10.1074/jbc.M408972200. [DOI] [PubMed] [Google Scholar]
- 20.Cheng S, Brzostek S, Lee SR, Hollenberg AN, Balk SP. Inhibition of the dihydrotestosterone-activated androgen receptor by nuclear receptor corepressor. Mol Endocrinol. 2002;16:1492–501. doi: 10.1210/mend.16.7.0870. [DOI] [PubMed] [Google Scholar]
- 21.Kang Z, Janne OA, Palvimo JJ. Coregulator recruitment and histone modifications in transcriptional regulation by the androgen receptor. Mol Endocrinol. 2004;18:2633–48. doi: 10.1210/me.2004-0245. [DOI] [PubMed] [Google Scholar]
- 22.Whittle JR, Powell MJ, Popov VM, Shirley LA, Wang C, Pestell RG. Sirtuins, nuclear hormone receptor acetylation and transcriptional regulation. Trends Endocrinol Metab. 2007:356–64. doi: 10.1016/j.tem.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Wu K, Li A, Rao M, et al. DACH1 is a cell fate determination factor that inhibits Cyclin D1 and breast tumor growth. Mol Cell Biol. 2006;26:7116–29. doi: 10.1128/MCB.00268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Perissi V, Liu F, Rose DW, Rosenfeld MG. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science. 2002;297:1180–3. doi: 10.1126/science.1073263. [DOI] [PubMed] [Google Scholar]
- 25.Mardon G, Solomon NM, Rubin GM. Dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–86. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- 26.Kim SS, Zhang RG, Braunstein SE, Joachimiak A, Cvekl A, Hegde RS. Structure of the retinal determination protein Dachshund reveals a DNA binding motif. Structure. 2002;10:787–95. doi: 10.1016/s0969-2126(02)00769-4. [DOI] [PubMed] [Google Scholar]
- 27.Wu K, Liu M, Li A, et al. The Cell Fate Determination Factor DACH1 Inhibits c-Jun Induced Contact-Independent Growth. Mol Biol Cell. 2007;18:755–67. doi: 10.1091/mbc.E06-09-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu K, Katiyar S, Li A, et al. Dachshund inhibits oncogene-induced breast cancer cellular migration and invasion through suppression of interleukin-8. Proc Natl Acad Sci U S A. 2008;105(19):6924–9. doi: 10.1073/pnas.0802085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sunde JS, Donninger H, Wu K, et al. Expression Profiling Identifies Altered Expression of Genes That Contribute to the Inhibition of Transforming Growth Factor-β Signaling in Ovarian Cancer. Cancer Res. 2006;66:8404–12. doi: 10.1158/0008-5472.CAN-06-0683. [DOI] [PubMed] [Google Scholar]
- 30.Wu K, Yang Y, Wang C, et al. DACH1 inhibits transforming growth factor-beta signaling through binding Smad4. J Biol Chem. 2003;278:51673–84. doi: 10.1074/jbc.M310021200. [DOI] [PubMed] [Google Scholar]
- 31.Gingras S, Moriggl R, Groner B, Simard J. Induction of beta-hydroxysteroid dehydrogenase/delta5-delta4 isomerase type 1 gene transcription in human breast cancer cell lines and in normal mammary epithelial cells by interleukin-4 and interleukin-13. Mol Endocrinol. 1999;13:66–81. doi: 10.1210/mend.13.1.0221. [DOI] [PubMed] [Google Scholar]
- 32.Sathya G, Chang CY, Kazmin D, Cook CE, McDonnell DP. Pharmacological uncoupling of androgen receptor-mediated prostate cancer cell proliferation and prostate-specific antigen secretion. Cancer Res. 2003;63:8029–36. [PubMed] [Google Scholar]
- 33.Reutens AT, Fu M, Watanabe G, et al. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol Endocrinol. 2001;15:797–811. doi: 10.1210/mend.15.5.0641. [DOI] [PubMed] [Google Scholar]
- 34.Matsumura I, Kitamura T, Wakao H, et al. Transcriptional regulation of cyclin D1 promoter by STAT5: its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J. 1999;18:1367–77. doi: 10.1093/emboj/18.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee RJ, Albanese C, Fu M, et al. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol Cell Biol. 2000;20:672–83. doi: 10.1128/mcb.20.2.672-683.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu M, Rao M, Bouras T, et al. Cyclin D1 inhibits PPARgamma -mediated adipogenesis through HDAC recruitment. J Biol Chem. 2005;280:16934–41. doi: 10.1074/jbc.M500403200. [DOI] [PubMed] [Google Scholar]
- 37.Dai Y, Ngo D, Forman LW, et al. Sirtuin 1 is required for antagonist-induced transcriptional repression of androgen-responsive genes by the androgen receptor. Mol Endocrinol. 2007;21:1807–21. doi: 10.1210/me.2006-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu M, Wang C, Zhang X, Pestell RG. Nuclear receptor modifications and endocrine cell proliferation. J Steroid Biochem Mol Biol. 2003;85:133–8. doi: 10.1016/s0960-0760(03)00223-1. [DOI] [PubMed] [Google Scholar]
- 39.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–7. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 40.Liao G, Chen LY, Zhang A, et al. Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. J Biol Chem. 2003;278:5052–61. doi: 10.1074/jbc.M206374200. [DOI] [PubMed] [Google Scholar]
- 41.Wang L, Hsu CL, Chang C. Androgen receptor corepressors: an overview. Prostate. 2005;63:117–30. doi: 10.1002/pros.20170. [DOI] [PubMed] [Google Scholar]
- 42.Keisman EL, Baker BS. The Drosophila sex determination hierarchy modulates wingless and decapentaplegic signaling to deploy dachshund sex-specifically in the genital imaginal disc. Development. 2001;128:1643–56. doi: 10.1242/dev.128.9.1643. [DOI] [PubMed] [Google Scholar]
- 43.Sanchez L, Gorfinkiel N, Guerrero I. Sex determination genes control the development of the Drosophila genital disc, modulating the response to Hedgehog, Wingless and Decapentaplegic signals. Development. 2001;128:1033–43. doi: 10.1242/dev.128.7.1033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.