Abstract
In tumors, alternative translation and posttranslational proteolytic cleavage of full-length cyclin E (EL) produces tumorigenic low molecular weight cyclin E (LMW-E) isoforms that lack a portion of the EL amino-terminus containing a nuclear localization sequence. Therefore, we hypothesized that LMW-E isoforms have altered subcellular localization. To explore our hypothesis, we compared EL versus LMW-E localization in cell lysates and in vivo using fractionation and protein complementation assays. Our results reveal that LMW-E isoforms preferentially accumulate in the cytoplasm where they bind the cyclin E kinase partner, cyclin-dependent kinase 2 (Cdk2), and have associated kinase activity. The nuclear ubiquitin ligase Fbw7 targets Cdk2-bound cyclin E for degradation; thus, we examined if altered subcellular localization affected LMW-E degradation. We found that cytoplasmic LMW-E/Cdk2 was less susceptible to Fbw7-mediated degradation. One implication of our findings is that altered LMW-E and LMW-E/Cdk2 subcellular localization may lead to aberrant LMW-E protein interactions, regulation, and activity, ultimately contributing to LMW-E tumorigenicity.
Introduction
Cyclin E is a protein that regulates the cell cycle by promoting G1-S transition and regulating S phase progression (1). As a consequence, cyclin E accumulation is tightly regulated to prevent constitutive cyclin E activity, which can lead to genomic instability and tumorigenesis (2). Deregulation of cyclin E can result from gene amplification (3-6), enhanced gene transcription (7-9), increased protein stability (10-13), and the generation of low molecular weight cyclin E (LMW-E) isoforms (14-16).
LMW-E isoforms, which range in size from 33 to 45 kDa, are generated from NH2-terminal elastase cleavage of 50 kDa, EL protein at amino acids Q40-E45 or A69-D70, or from alternative translation of cyclin E at M46 (16). LMW-E accumulation is tumor specific and these isoforms have been found in multiple tumor types including breast, ovarian and colorectal cancers, and melanomas (5, 17-20). Furthermore, LMW-E proteins are strong correlative biomarkers in breast and ovarian cancer (20, 21).
LMW-E isoforms have a more profound effect on cell cycle deregulation than full-length cyclin E (EL; refs. 16, 18, 19, 22-24), and transgenic mice expressing LMW-E isoforms have more mammary tumor development and metastasis than EL transgenic mice (25). Thus, LMW-E isoforms seem more aggressive than EL in cell cycle abrogation and mammary tumor initiation and maintenance. However, the mechanisms governing LMW-E regulation and function remain unclear.
One approach to uncovering LMW-E regulation and function is to analyze secondary protein structure, as structural properties can affect localization, protein-protein interactions, and enzymatic activity. One noted structural difference between EL and LMW-E proteins is the loss of the canonical NH2-terminal nuclear localization sequence in the LMW-E isoforms. Thus, we hypothesized that LMW-E isoforms have altered subcellular localization. Using protein fractionation and protein complementation, we investigated LMW-E subcellular localization and its effects on LMW-E functional and regulatory protein interactions. Here, we provide evidence that EL and LMW-E subcellular localization differs, thereby affecting interaction with the cyclin E coactivator cyclin-dependent kinase 2 (Cdk2). Furthermore, differential subcellular localization of EL/Cdk2 and LMW-E/Cdk2 complexes influences their respective proteasomal regulation.
Materials and Methods
Cell culture conditions/treatments
Cancer cells and 293T human embryonic kidney cells were cultured in α-MEM and D-MEM media (Hyclone), respectively, supplemented with 10% fetal bovine serum (FBS; Atlanta Biological). Immortalized human mammary epithelial cells were cultured in D-media (26) composed of 1/2 HQ Ham's/F-12K (Hyclone) and 1/2 α-MEM supplemented with 1% FBS. Cycloheximide-treated cells were incubated in growth media containing cycloheximide before harvesting. Cells were maintained in a 37°C, 6.5% CO2 growth chamber.
Plasmids/transfections
To generate cyclin E-IFPN vectors, EL-, T1-, and T2-FLAG sequences (14) were directionally subcloned into the IFPN (VYF099) protein complementation vector (27) using 5′ Hind III and 3′ Cla I restriction sites. Cdk2 cDNA sequence was fused to IFPC (VYF102; ref. 27) using 5′ Cla I and 3′ Hind III restriction sites. Primer sequences (Integrated DNA Technologies) used for subcloning are as follows, restriction sites are italicized: EL-Flag: sense primer, 5′GGAAGCTTATGCCGAGGGAGCGCAGGGA3′; antisense primer, 5′AAGGACGACGATGACAAGATCGATCC3′; T1-Flag: sense primer, 5′GGAAGCTTATGGATCCAGATGAAGAAA3′; antisense primer, 5′AAGGACGACGATGACAAGATCGATCC3′; T2-Flag: sense primer, 5′GGAAGCTTATGGCAGTCTGTGCAGACC3′, antisense primer, 5′AAGGACGACGATGACAAGATCGATCC3′; Cdk2: sense primer, 5′GGATCGATATGGAGAACTTCCAAAAGGTGG; antisense primer, 5′GTACCCATCTTCGACTCTGAAAGCTTCC3′. Intensely fluorescent protein (IFP) cloning vectors and IFPN-AKT1 and PDK1-IFPC vectors were provided from Gordon Mills (27). FLAG-Fbw7 vectors were provided from Wade Harper (28). pCDNA3.1 (Invitrogen) was an empty vector control. To determine transfection efficiency, TRE-luciferase plasmid was cotransfected and luciferase activity measured using the Luciferase Assay II Reagent (Promega). Cell transfections were performed using Gene Juice Transfection Reagent (Novagen) and analyzed 24 to 72 h posttransfection.
Protein isolation
Cyclin E (EL, T1, or T2)-IFPN and IFPC-Cdk2 fusion proteins were generated in vitro using the Promega TnT Quick Coupled Transcription/Translation System. NP40 lysis buffer was used to extract total protein from whole cell lysates. Cell fractionation: Pelleted cells were resuspended in buffer A [20 mmol/L Hepes (pH 7.9), 10 mmol/L KCl, 2.5 mmol/L MgCl, 1 mmol/L EDTA, 1 mmol/L DTT, 0.2% NP40, and protease inhibitors], incubated on ice for 5 min, and centrifuged at 14,000 rpm, 4°C for 1 min. The supernatant is the soluble cytoplasmic fraction. The pellet was resuspended in buffer B (buffer A, 20% glycerol, 20 mmol/L NaCl), incubated on ice for 30 min, and centrifuged at 14,000 rpm, 4°C for 20 min. The supernatant is the soluble nuclear fraction.
Western blot analysis
Western blot analysis was performed as described (14). Primary antibodies used were as follows: cyclin E HE-12 (Santa Cruz Biochemicals), Cdk2 (Transduction Laboratories), GFP (Abcam), growth factor receptor binding protein 2 (BD Transduction), parp (Cell Signaling), and vinculin (Sigma). ImageQuant TL v2005 software (Amersham Biosciences) was used for densitometry.
Histone HI kinase
Cyclin E-associated kinase activity was assayed as described (14).
Microscopy
To determine the percentage of cells with nuclear or cytoplasmic/perinuclear IFP fluorescence, fluorescent interphase cells were counted in 5 to 10 microscopic fields per biological replicate. Cells expressing IFP in both the nucleus and cytoplasm were tallied as a positive for each subcellular compartment. Images were captured using ×10 or ×20 objectives on a Leica DM4000B fluorescent microscope (Leica Microsystems) with a RT KE/SE SPOT camera (Diagnostics Instruments, Inc.). The SPOT 4.6 software (Diagnostic Instruments, Inc.) was used to process the images.
Statistical analysis
Results shown as mean ± SD were compared by the Student's t test with a significant P value level of <0.05. The nonparametric Spearman correlation was used to quantify the relationship between protein expression and kinase activity.
Results
LMW-E isoforms preferentially accumulate in the cytoplasm
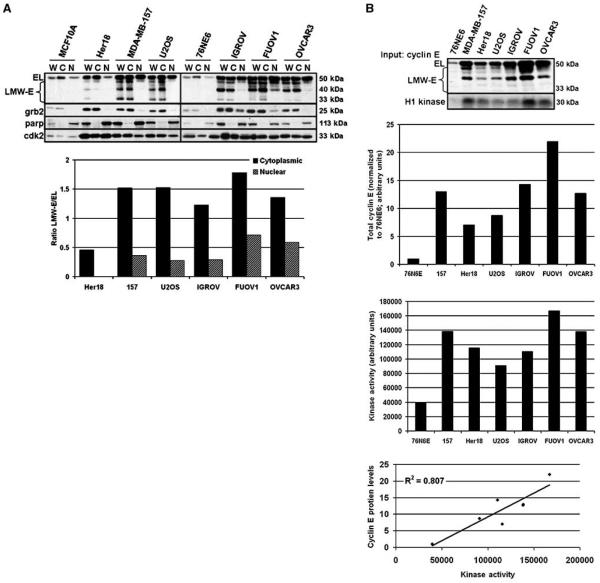
The LMW-E isoforms lack the first 40, 45, or 70 amino acids of EL (16). EL contains a canonical nuclear localization sequence, RSRKRK (29), located at amino acids 27 to 32, which prompted us to investigate LMW-E subcellular localization. We examined cyclin E distribution in multiple cell lines of normal and tumor origin, including two immortalized human mammary epithelial cell lines (76NE6 and MCF10A) and in several LMW-E–accumulating breast, osteosarcoma, and ovarian cancer cell lines. We separated the cell lines into whole cell and soluble cytoplasmic and nuclear lysates, which were then subjected to Western blot analysis for cyclin E. As expected, the MCF10A and 76NE6 nontumorigenic cell lines only expressed EL, whereas tumor cells expressed both EL and LMW-E isoforms (Fig. 1A). However, our analysis revealed the novel finding that in all of the cancer types assayed, the LMW-E proteins primarily accumulated in the cytoplasmic fraction (Fig. 1A). To quantify our observation, we used densitometry to measure LMW-E subcellular accumulation within each subcellular fraction and calculated the ratio of total LMW-E to EL. In tumor cells, the ratio of total LMW-E to EL in the cytoplasm was consistently greater than the ratio of total LMW-E to EL in the nucleus (Fig. 1A, graph). In fact, LMW-E isoforms were only detected in the cytoplasmic fraction of Her18 cells (Fig. 1A, graph). Furthermore, the ratio of total LMW-E to EL in the cytoplasmic fraction of MDA-MB-157, U2OS, IGROV, FUOV1, and OVCAR3 cell lines were, respectively, 4.2, 5.6, 4.2, 2.5, and 2.3 times greater than that of the LMW-E/EL ratio in the nuclear fraction (Fig. 1A, graph). Thus, LMW-E isoforms, which are tumor specific, show a marked subcellular localization to the cytoplasm.
Figure 1. LMW-E isoforms are cytoplasmic.
A, normal immortalized mammary epithelial (MCF10A, 76NE6) cell lines and breast (Her18, MDA-MB-157), osteosarcoma (U2OS), and ovarian (IGROV, FUOV1, OVCAR3) cancer cell lines were separated into whole cell (W), cytoplasmic (C), and nuclear (N) fractions and analyzed for cyclin E and Cdk2 accumulation by Western blot. Grb2 and parp antibodies were used to indicate the cytoplasmic and nuclear fractions, respectively. Western blots were subject to densitometry and the ratio of total LMW-E to EL was graphed. B, the Western blot shows the relative levels of cytoplasmic cyclin E (Input: cyclin E) in each cell line that was assayed for cyclin E–associated kinase activity using histone H1 as a substrate (H1 kinase). Bar graphs, densitometric values for total cytoplasmic cyclin E (top bar graph; normalized to 76N6E) and H1 phosphorylation (bottom bar graph; raw values). Linear regression graph, correlation between cyclin E levels and cyclin E–associated kinase activity for each cell line; R2 = 0.807; P = 0.0060.
Cdk2 is the primary cyclin E coactivator (30). We previously reported that LMW-E proteins also bind Cdk2 (16, 22, 24). Because most LMW-E protein is cytoplasmic, we wanted to examine the subcellular localization of Cdk2. Western blot analysis showed that Cdk2 protein accumulates in both the cytoplasmic and nuclear fractions (Fig. 1A), suggesting that LMW-E isoforms might form complexes with Cdk2 in the cytoplasm.
Cytoplasmic cyclin E has associated kinase activity
Cyclin EL/Cdk2 complexes localize to the nucleus to phosphorylate target proteins (1). However, LMW-E proteins, which are found primarily in the cytoplasm (Fig. 1A), also have associated kinase activity in whole cell extracts (14, 16, 22, 24). This prompted us to investigate if cytoplasmic cyclin E has associated kinase activity in tumor cells that accumulate LMW-E isoforms. We immunoprecipitated cyclin E immune complexes from cytoplasmic lysates of 76NE6 normal immortalized cells and breast, ovarian, and osteosarcoma cancer cells and assayed the cyclin E immune complexes for kinase activity in vitro using histone H1 as a substrate. The Western blot in Fig. 1B shows the relative levels of cytoplasmic LMW-E and/or EL protein present in each cell line. Histone H1 phosphorylation was higher in all tumor cell lines, which accumulate both EL and LMW-E protein, compared with 76NE6 cells, which only accumulate EL (Fig. 1B, bottom). We used densitometry to quantify the total cyclin E protein levels (EL and LMW-E) and cyclin E–associated kinase activity in the nontumorigenic and tumor cells (Fig. 1B, bar graphs). Correlative analysis of these results revealed that there is a positive correlation (R2 = 0.807; P = 0.0060) between cyclin E protein levels and cyclin E–associated kinase activity (Fig. 1B, line graph). Thus, our results suggest that cytoplasmic cyclin E, including the LMW-E isoforms, form active kinase complexes in the cytoplasm.
Protein complementation assay as a tool to examine cyclin E/Cdk2 subcellular localization
Based on our subcellular fractionation and kinase data, we hypothesized that the kinase activity associated with LMW-E proteins in the cytoplasm is through its interaction with Cdk2. To directly address this question, we used protein complementation technology, which unlike immunoprecipitation and immunoblotting, is not limited by input protein concentration and antibody efficacy. Furthermore, by using protein complementation, the formation and localization of cyclin E/Cdk2 interaction can be observed in vivo for each specific cyclin E isoform.
We used the IFP complementation method (27). IFP is an improved variant of green fluorescent protein (31) that has been rationally fragmented into NH2 terminus (IFPN) and COOH terminus (IFPC) peptides (32). When proteins fused to the IFPN and IFPC peptides interact, the IFP fragments are brought into proximity and emit green fluorescence, thereby enabling visualization of the localization of protein-protein interactions in vivo (32). Supplementary Fig. S1A shows a schematic of the cyclin E and Cdk2 protein complementation constructs used in this study, which we show make fusion proteins (Supplementary Fig. S1B) and interact to form functionally active complexes (Supplementary Fig. S1C).
LMW-E isoforms bind to Cdk2 in the cytoplasm, perinuclear membrane, and nucleus
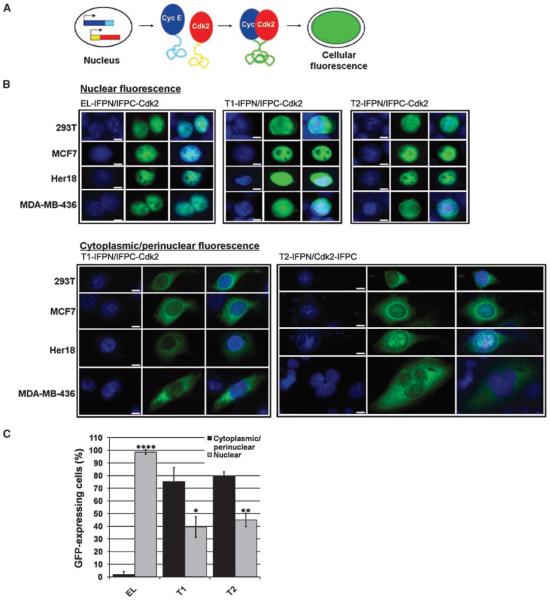
To determine the localization of cyclin E-IFPN/IFPC-Cdk2 complexes in vivo, we coexpressed the fusion proteins in mammalian cells and observed IFP green fluorescence using microscopy as depicted (Fig. 2A). We coexpressed the cyclin E-IFPN and IFPC-Cdk2 fusion proteins in 293T cells as proof-of-principal and in MCF7, Her18, and MDA-MB-436 breast cancer cells, which can readily be transfected and also represent a range of physiologically relevant breast cancer cell lines. MCF7 and Her18 (MCF7-derived, Her2 overexpressing) cells are p53, Rb, and estrogen receptor positive, whereas MDA-MB-436 cells are p53, Rb, and estrogen receptor negative (33). In addition, unlike MCF7 cells, Her18 (Fig. 1A) and MDA-MB-436 cells accumulate LMW-E protein (34).
Figure 2. LMW-E/Cdk2 complexes are cytoplasmic.
A, the schematic shows the principle of protein complementation. Cyclin E/Cdk2 binding brings the IFP fragments into proximity, thereby emitting green fluorescence. B, cyclin E (EL, T1, or T2)-IFPN and IFPC-Cdk2 were coexpressed in various cell lines. Cells expressing nuclear (top) or cytoplasmic/perinuclear (bottom) green fluorescence are shown. Nuclei are 4′,6-diamidino-2-phenylindole(DAPI)–stained blue. Images were taken using the ×20 objective and merged. Scale bar, 5 μm. C, the bar graph shows the percentage of green fluorescent 293T cells that show cyclin E-IFPN/IFPC-Cdk2 signal in the cytoplasm/perinuclear membrane and the percentage of cells that show signal in the nucleus. A total of 606 ≤ n ≤ 767 green fluorescent cells over 5 to 10 microscopic fields were counted for each condition. The SD was calculated for three biological replicates. Student's t test was performed for cytoplasmic/perinuclear versus nuclear localization for each condition; P < 0.05 (*), P < 0.01 (**), and P < 0.0001 (****).
Upon cotransfection, we found that in each cell line, EL-IFPN and IFPC-Cdk2 bound together and emitted green fluorescence at interphase nuclei (Fig. 2B). EL-IFPN/IFPC-Cdk2 localization served as a positive control for the IFP protein complementation method because cyclin E/Cdk2 nuclear localization has been previously reported (35). To examine whether the fluorescence observed for cyclin E-IFPN/IFPC-Cdk2 was specifically due to cyclin E/Cdk2 binding, we used four different approaches. First, we compared the localization patterns of EL-IFPN/IFPC-Cdk2 and previously reported IFPN-AKT1/PDK1-IFPC (27). Results revealed that the localization patterns for these two sets of fusion proteins are distinct (Supplementary Fig. S2), suggesting that the IFP fragments are not influencing localization and that the patterns we see indeed represent the localization of endogenous cyclin E/Cdk2 interaction. Second, we cotransfected 293T cells with EL-IFPN and PDK1-IFPC, which does not reportedly bind cyclin E, and found only minimal nonspecific fluorescence (Supplementary Fig. S2). Third, we cotransfected 293T cells with the IFP cloning vectors and, again, observed negligible fluorescence (Supplementary Fig. S2). Finally, expression of any one of the IFP cloning vectors or fusion proteins alone did not produce fluorescence (data not shown). Taken together, these data suggest that the IFP localization patterns displayed in cells coexpressing cyclin E-IFPN and IFPC-Cdk2 are specific to cyclin E binding to Cdk2.
Despite lack of the nuclear localization sequence in T1 and T2, T1-IFPN/IFPC-Cdk2 and T2-IFPN/IFPC-Cdk2 complexes also localized to interphase nuclei (Fig. 2B). We observed nuclear localization even at 10-fold lower plasmid concentrations (data not shown), suggesting that T1- and T2-IFPN/IFPC-Cdk2 nuclear localization was not due nonspecifically to overexpression. However, unlike EL-IFPN/IFPC-Cdk2, T1- and T2-IFPN/IFPCCdk2 complexes were also readily detectable in the cytoplasm and at the perinuclear membrane of interphase cells (Fig. 2B). Thus, the protein complementation results imply that the endogenous cytoplasmic LMW-E isoforms we detect by Western blot may indeed form complexes with Cdk2 in the cytoplasm.
Given the LMW-E proteins accumulate in the cytoplasm (Fig. 1A), we were interested in whether the T1- or T2-IFPN/IFPC-Cdk2 complexes might also favor cytoplasmic localization. Therefore, we calculated the percentage of interphase cells emitting IFP fluorescence in the cytoplasm and/or perinuclear membrane versus those cells emitting fluorescence in the nucleus. We performed these experiments using 293T cells because of their high transfection efficiency. Fluorescent EL-IFPN/IFPC-Cdk2–expressing cells showed IFP signal almost exclusively in the nucleus of interphase cells (P < 0.0001; Fig. 2C). On the other hand, the T1- and T2-IFPN/Cdk2–expressing cells emitted fluorescence in the cytoplasm/perinuclear membrane nearly twice more often than in the nucleus (P < 0.05 and P < 0.01; Fig. 2C). Thus, whereas ELIFPN/IFPC-Cdk2 complexes preferentially localize to the nucleus, T1- and T2-IFPN/IFPC-Cdk2 complexes preferentially localize to the cytoplasm and/or perinuclear membrane.
These protein complementation results correlate with endogenous LMW-E subcellular distribution and suggest that LMW-E localization affects the formation and localization of LMW-E protein interactions. To investigate whether LMW-E subcellular distribution might also affect its regulation, we compared EL and LMW-E proteasomal degradation.
LMW-E isoforms are susceptible to proteasomal degradation
Cyclin E proteasomal degradation is regulated, in part, by its subcellular localization and interaction with Cdk2. For example, cyclin E/Cdk2 complexes reside in the nucleus (35) where they are targeted for ubiquitination by the nuclear isoforms of the E3 ubiquitin ligase, Fbw7 (36, 37).
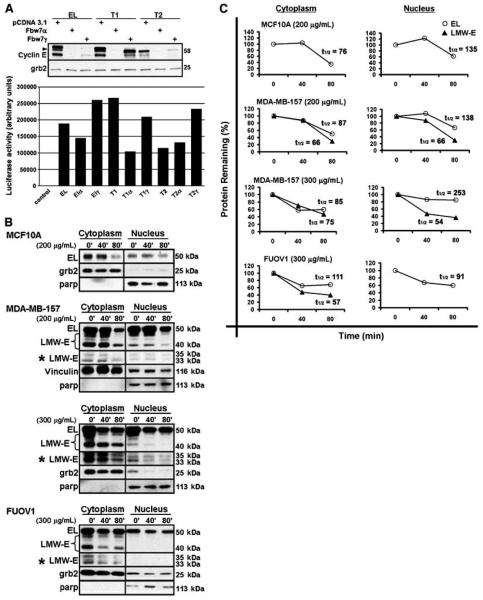
Fbw7-mediated proteasomal degradation of cyclin E requires that Cdk2 bind and phosphorylate cyclin E (38). Given that cyclin E/Cdk2 resides in the nucleus (Fig. 2B; ref. 35), it is not surprising that the nuclear Fbw7α and Fbw7γ isoforms target cyclin E, whereas cytoplasmic Fbw7β is not required for cyclin E degradation (36). However, because our results suggest that in cancer cells, LMW-E/Cdk2 favors nonnuclear accumulation (Fig. 2C), we questioned if LMW-E isoforms would be susceptible to Fbw7-mediated degradation. To begin to address this question, we analyzed the effect of Fbw7α and -γ on T1- and T2-IFPN protein levels. We cotransfected MDA-MB-436 breast cancer cells with EL-, T1-, or T2-IFPN and IFPC-Cdk2 plasmids in the absence or presence of Fbw7α or -γ expression vectors. Our vector transfections were efficient as measured by the luciferase activity of a cotransfected TRE-luciferase plasmid (Fig. 3A, bar graph). Cells were collected 48 hours after transfection and whole cell lysates were analyzed by Western blot for cyclin E accumulation. The results revealed that EL-, T1-, and T2-IFPN protein levels were significantly reduced in the presence of Fbw7 (Fig. 3A), suggesting that Fbw7 does in fact regulate both EL and LMW-E protein stability.
Figure 3. LMW-E isoforms are sensitive to proteasomal degradation.
A, MDA-MB-436 cells were transfected with cyclin E-IFPN in the absence (pCDNA3.1) or presence of Fbw7. Whole cell lysates were analyzed by Western blot for cyclin E. Arrow, the M46 cyclin E isoform. Grb2 is a loading control. The activity of a cotransfected TRE-luciferase vector served as a transfection efficiency control (bar graph; raw values). B, nontumorigenic and tumor cells were treated with cycloheximide. Control cells were left untreated (0 min). Cells were fractionated and analyzed for cyclin E accumulation by Western blot. *, longer exposed film. C, densitometric values were used to plot the percent degradation and calculate the half-life (t1/2, min) for full-length (EL) and total LMW-E cyclin E.
To confirm the proteasomal regulation of endogenous LMW-E, we treated cells with cycloheximide, which blocks new protein synthesis thereby allowing us to determine the degradation rate of existing cyclin E protein. To directly compare the stability of each cyclin E isoform, we used MDA-MB-157 breast and FUOV1 ovarian cancer cell lines. MCF10A normal immortalized cells served as a nontumorigenic control for experimental efficacy. These three different cell lines were treated with different but equitoxic doses of cycloheximide, as there is normal/tumor differential sensitivity to cycloheximide. After treatment of cells with cycloheximide, cytoplasmic and nuclear lysates were isolated and analyzed by Western blot for cyclin E (Fig. 3B) and quantified for degradation rate and half-life using densitometry (Fig. 3C). The results showed that EL levels were reduced in the cycloheximide-treated nontumorigenic and cancer cells (Fig. 3B and C). LMW-E levels also declined in the cycloheximide-treated cancer cells and seemed slightly less stable than EL (Fig. 3B and C). Thus, our data indicate that like EL, endogenous LMW-E isoforms are susceptible to degradation by the proteasome in cancer cells.
Cytoplasmic LMW-E/Cdk2 complexes have reduced sensitivity to Fbw7-mediated degradation
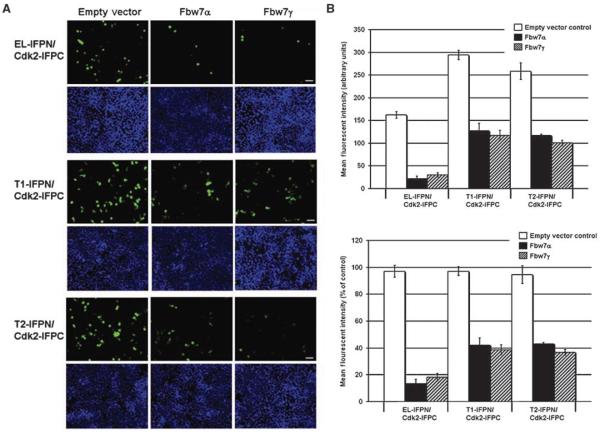
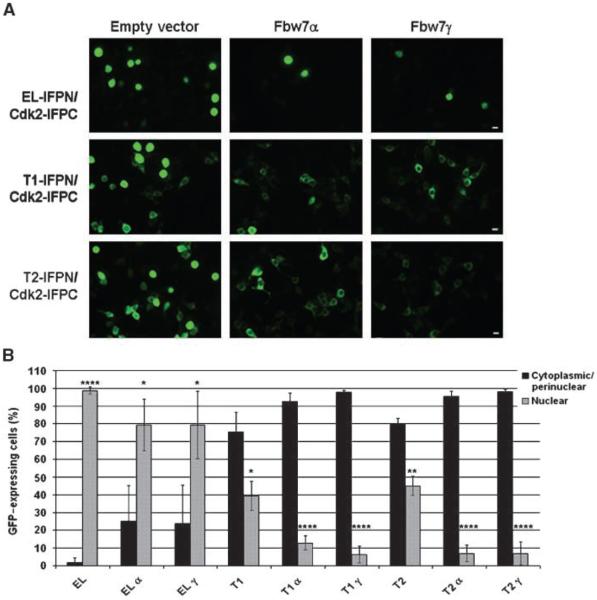
Although LMW-E proteins (i.e., T1 and T2) are sensitive to Fbw7-mediated proteasomal degradation, we questioned how Fbw7-mediated degradation affects subcellular distribution of LMW-E/Cdk2 complexes. This is an important question because LMW-E tumorigenic potential is not expected to exclusively correlate with its protein levels, but other properties as well, such as its spatial and temporal localization and biochemical interaction with other proteins. To determine the effect of Fbw7-mediated degradation on cyclin E subcellular distribution, we used the protein complementation assay to examine the effect of Fbw7 on cyclin E-IFPN/IFPC-Cdk2 localization using fluorescence microscopy. We coexpressed cyclin E-IFPN (EL, T1, or T2) and IFPC-Cdk2 in 293T cells in the absence or presence of Fbw7α or -γ and analyzed the cells 24 hours after transfection. Representative images of the cotransfected cells are shown in Fig. 4A. In the absence of Fbw7, cyclin E and Cdk2 IFP fusion proteins formed numerous and intense fluorescent complexes (Fig. 4A, empty vector). In the presence of Fbw7α or Fbw7γ, cyclin E-IFPN/IFPC-Cdk2 fluorescent signal was significantly reduced (Fig. 4A). Fluorescence activated cell sorting (FACS) was then used to quantify cyclin E-IFPN/IFPC-Cdk2 IFP signal in the absence or presence of Fbw7 (Fig. 4B). The mean fluorescence (Fig. 4B, top graph) and the normalized percent fluorescence (Fig. 4B, bottom graph) were quantified. The results revealed that EL-IFPN/IFPC-Cdk2 signal was reduced by 80% and T1- and T2-IFPC/Cdk2 signal were reduced by 60% in the presence of Fbw7 (Fig. 4B, bottom graph). T1- and T2-IFPC/Cdk2 mean fluorescence in the control cells was higher than EL-IFPN/IFPC-Cdk2 (Fig. 4B, top graph), which may explain why Fbw7 had less effect on T1- and T2-associated IFP signal than that of EL. Nevertheless, our results indicate that Fbw7 can reduce cyclin E/Cdk2 formation.
Figure 4. Fbw7 reduces cyclin E-IFPN/IFPC-Cdk2 complex formation.
A, 293T cells were cotransfected with cyclin E-IFPN and IFPC-Cdk2 in the absence or presence of Fbw7. Top, cells expressing green fluorescence. Blue DAPI-stained nuclei (bottom) are shown to show that each field has a comparable number of cells. Images were taken using the ×10 objective. Scale bar, 50 μm. B, FACS was used to measure IFP fluorescence in 293T cells. Top graph, mean fluorescent intensity; bottom graph, normalized to the empty vector controls. Columns, mean of three biological replicates, n = 30,000 cells per replicate analyzed by FACS; bars, SD.
To address the effect of Fbw7 on cyclin E-IFPN/IFPC-Cdk2 subcellular distribution, we examined the localization of these complexes in vivo using microscopy. We discovered a striking trend for cyclin E-IFPN/Cdk2-IFPN subcellular accumulation in the presence of Fbw7. Specifically, Fbw7 interfered with nuclear cyclin E-IFPN/IFPC-Cdk2 complex formation significantly more than cytoplasmic and/or perinuclear complex formation (Fig. 5A). To quantify our observation, we determined the percentage of cells expressing IFP in the cytoplasm/perinuclear membrane versus the nucleus in the absence or presence of Fbw7 (Fig. 5B). The total percentages exceeded 100% because any cell that showed signal in both the cytoplasm/perinuclear membrane and in the nucleus was included in both subcellular calculations. A graph showing the breakdown of cells expressing IFP signal in the cytoplasmic/perinuclear membrane, the nucleus, or in both subcellular compartments is shown (Supplementary Fig. S3). There was an increase in the percentage of cells showing cytoplasmic/perinuclear IFP signal in the presence of Fbw7 over the absence of Fbw7 for EL-, T1-, and T2-IFPN/IFPC-Cdk2 (Fig. 5B), resulting in a greater ratio of cytoplasmic/perinuclear to nuclear IFP expression for each cyclin E-IFPN/IFPC-Cdk2 complex in the presence versus the absence of Fbw7. In particular, the subcellular distribution of T1- and T2-IFPN/IFPC-Cdk2 complexes was significantly more stratified toward the cytoplasm in the presence than in the absence of Fbw7 (P < 0.0001; Fig. 5B). Taken together, these data suggest that tumor cells that accumulate LMW-E isoforms have acquired a growth advantage due to LMW-E subcellular localization. In particular, the physiologically relevant nuclear Fbw7 isoforms seem to preferentially target nuclear cyclin E/Cdk2 complexes and stratify the subcellular distribution of cyclin E/Cdk2 complexes to favor cytoplasmic accumulation.
Figure 5. Cytoplasmic cyclin E-IFPN/IFPC-Cdk2 complexes are less susceptible to Fbw7.
A, the images show the localization of cyclin E-IFPN/IFPC-Cdk2 in 293T cells cotransfected with cyclin E-IFPN and IFPC-Cdk2 in the absence or presence of Fbw7. Images were using ×20 objective. Scare bar, 10 μm. B, the bar graph shows the percentage of green fluorescent 293T cells that show cyclin E-IFPN/IFPC-Cdk2 signal in the cytoplasm/perinuclear membrane and the percentage of cells that show signal in the nucleus. A total of 386 ≤ n ≤ 767 green fluorescent cells over 5 to 10 microscopic fields were counted for each condition. Columns, mean of three biological replicates; bars, SD; P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), and P < 0.0001 (****).
Discussion
Our findings suggest that the loss of the NH2 terminus affects LMW-E subcellular localization, the localization of cyclin E–binding proteins, and LMW-E regulation by the proteasome. Thus, deregulated localization of cyclin E through the generation of LMW-E isoforms may be a mechanism by which cancer cells alter cyclin E regulation and function, imparting a growth advantage over normal cells. Indeed, tumor cells have higher cyclin E–associated kinase activity in the cytoplasm than do normal cells because tumor cells accumulate EL/Cdk2 and tumor-specific LMW-E/Cdk2 complexes in the cytoplasm. It is important to note that although preferentially accumulating in the cytoplasm, we also found LMW-E isoforms and LMW-E/Cdk2 complexes in the nucleus of cancer cells. Previous studies have shown that the nuclear localization sequence motif of cyclin E is not required for cyclin E nuclear localization (16, 39, 40). Furthermore, it is possible that LMW-Es are shuttled into the nucleus via protein-protein interactions. We are now exploring what additional proteins might interact with LMW-E isoforms in the cytoplasm and how these proteins may be involved in the tumorigenic process.
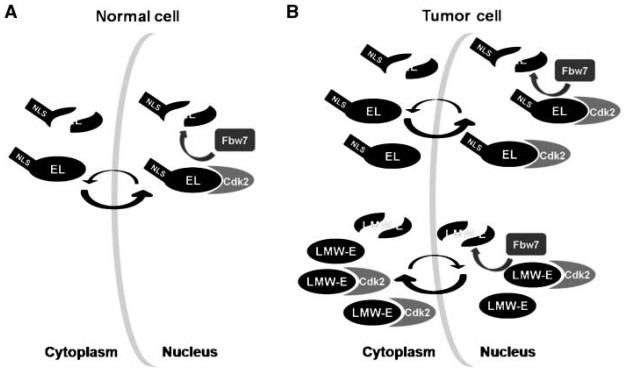
Cyclin E and cyclin E/Cdk2 subcellular localization is not a static process and, in fact, cyclin E/Cdk2 complexes are known to shuttle between the cytoplasm and nucleus (35). Based on our results, we propose a model where, in tumor cells, EL and LMW-E proteins have differing nucleocytoplasmic shuttling dynamics that cause LMW-E/Cdk2 to be imported into the nucleus at a slower rate and/or exported more efficiently than EL/Cdk2 (Fig. 6). We also propose that these spatio-temporal differences affect EL and LMW-E proteasomal regulation in cancer cells, where the physiologically relevant, nuclear-localized Fbw7 isoforms target nuclear cyclin E/Cdk2 complexes (Fig. 6). Of note, as reported for cyclin E (38, 41), we also observed that ectopic expression of cytoplasmically localized Fbw7β is sufficient to mediate degradation of EL-, T1-, and T2-IFPN and we found that Fbw7β targets nuclear cyclin E-IFPN/IFPC-Cdk2 complexes (data not shown). Fbw7β and cyclin E can form an immune complex (41); thus, it is possible that Fbw7β is shuttled into the nucleus to mediate cyclin E degradation via cyclin E interaction.
Figure 6. Proposed model of the effect of subcellular localization on cyclin E stability and activity in normal versus tumor cells.
A, in normal cells, EL shuttles between the nucleus and cytoplasm (arrows). Nuclear import is faster than export causing EL/Cdk2 to accumulate in the nucleus where nuclear Fbw7 can target EL for degradation. EL shuttling results in a net loss of protein from both subcellular compartments. B, EL shows similar subcellular distribution patterns and targeting by Fbw7 in tumor cells as outlined for normal cells. LMW-E and LMW-E/Cdk2 preferentially accumulate in the cytoplasm due to lack of the nuclear localization sequence and altered shuttling dynamics (arrows) and have reduced susceptibility to nuclear Fbw7. Finally, tumor cells have higher cyclin E–associated kinase activity in the cytoplasm than do normal cells because tumor cells accumulate EL/Cdk2 and tumor-specific LMW-E/Cdk2 in the cytoplasm.
In summary, we show that EL and LMW-E isoforms are differentially regulated at the level of subcellular accumulation in cancer cells. Altered localization of LMW-E and LMW-E/Cdk2 complexes seems to affect the regulation of LMW-E isoforms by cellular regulatory proteins and has implications for LMW-E function. Thus, it is our goal to further uncover differences in the mechanisms of EL and LMW-E regulation and function, so that ultimately, we can design novel treatment strategies for patients that accumulate the tumorigenic LMW-E proteins.
Supplementary Material
Acknowledgments
Grant support: NIH CA87548 (K. Keyomarsi), CA87548S1 (N.A. Delk), 1F32CA128296-01A1 (N.A. Delk); National Cancer Institute P50CA116199 (K. Keyomarsi); and Clayton Foundation (K. Keyomarsi).
We thank the Keyomarsi and Hunt laboratories, including Drs. Said Akli, Rozita Yarmand, and Hannah Wingate, Ben Mull, and Curtis Pickering; Dr. Zhiyong Ding (Mills Laboratory), and the MD Anderson FACS Core Facility for intellectual and technical advice and/or critical review of this manuscript; and both Dr. Mills and Dr. Ding for their input in the studies with protein-protein complementation assays. The protein complementation vectors were a gift from Dr. Gordon B. Mills (University of Texas at M. D. Anderson Cancer Center, Houston, TX) and the Fbw7 vectors were a gift from Dr. Wade Harper (Harvard Medical School, Boston, MA).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Moroy T, Geisen C, Cyclin E. Int J Biochem Cell Biol. 2004;36:1424–39. doi: 10.1016/j.biocel.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene. 2005;24:2776–86. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- 3.Akama Y, Yasui W, Yokozaki H, et al. Frequent amplification of the cyclin E gene in human gastric carcinomas. Jpn J Cancer Res. 1995;86:617–21. doi: 10.1111/j.1349-7006.1995.tb02442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Courjal F, Louason G, Speiser P, Katsaros D, Zeillinger R, Theillet C. Cyclin gene amplification and over-expression in breast and ovarian cancers: evidence for the selection of cyclin D1 in breast and cyclin E in ovarian tumors. Int J Cancer. 1996;69:247–53. doi: 10.1002/(SICI)1097-0215(19960822)69:4<247::AID-IJC1>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 5.Keyomarsi K, Pardee AB. Redundant cyclin over-expression and gene amplification in breast cancer cells. Proc Natl Acad Sci U S A. 1993;90:1112–6. doi: 10.1073/pnas.90.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitahara K, Yasui W, Kuniyasu H, et al. Concurrent amplification of cyclin E and CDK2 genes in colorectal carcinomas. Int J Cancer. 1995;62:25–8. doi: 10.1002/ijc.2910620107. [DOI] [PubMed] [Google Scholar]
- 7.McElroy AK, Dwarakanath RS, Spector DH. Dys-regulation of cyclin E gene expression in human cytomegalovirus-infected cells requires viral early gene expression and is associated with changes in the Rb-related protein p130. J Virol. 2000;74:4192–206. doi: 10.1128/jvi.74.9.4192-4206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salon C, Merdzhanova G, Brambilla C, Brambilla E, Gazzeri S, Eymin B. E2F-1, Skp2 and cyclin E oncoproteins are upregulated and directly correlated in high-grade neuroendocrine lung tumors. Oncogene. 2007;26:6927–36. doi: 10.1038/sj.onc.1210499. [DOI] [PubMed] [Google Scholar]
- 9.Soucek T, Pusch O, Hengstschlager-Ottnad E, Adams PD, Hengstschlager M. Deregulated expression of E2F-1 induces cyclin A- and E-associated kinase activities independently from cell cycle position. Oncogene. 1997;14:2251–7. doi: 10.1038/sj.onc.1201061. [DOI] [PubMed] [Google Scholar]
- 10.Akhoondi S, Sun D, von der Lehr N, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–12. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 11.Mukherji A, Janbandhu VC, Kumar V. HBx-dependent cell cycle deregulation involves interaction with cyclin E/A-cdk2 complex and destabilization of p27Kip1. Biochem J. 2007;401:247–56. doi: 10.1042/BJ20061091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–22. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 13.Welcker M, Clurman BE. The SV40 large T antigen contains a decoy phosphodegron that mediates its interactions with Fbw7/hCdc4. J Biol Chem. 2005;280:7654–8. doi: 10.1074/jbc.M413377200. [DOI] [PubMed] [Google Scholar]
- 14.Harwell RM, Porter DC, Danes C, Keyomarsi K. Processing of cyclin E differs between normal and tumor breast cells. Cancer Res. 2000;60:481–9. [PubMed] [Google Scholar]
- 15.Libertini SJ, Robinson BS, Dhillon NK, et al. Cyclin E both regulates and is regulated by calpain 2, a protease associated with metastatic breast cancer phenotype. Cancer Res. 2005;65:10700–8. doi: 10.1158/0008-5472.CAN-05-1666. [DOI] [PubMed] [Google Scholar]
- 16.Porter DC, Zhang N, Danes C, et al. Tumor-specific proteolytic processing of cyclin E generates hyperactive lower-molecular-weight forms. Mol Cell Biol. 2001;21:6254–69. doi: 10.1128/MCB.21.18.6254-6269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bales E, Mills L, Milam N, et al. The low molecular weight cyclin E isoforms augment angiogenesis and metastasis of human melanoma cells in vivo. Cancer Res. 2005;65:692–7. [PubMed] [Google Scholar]
- 18.Bedrosian I, Lu KH, Verschraegen C, Keyomarsi K. Cyclin E deregulation alters the biologic properties of ovarian cancer cells. Oncogene. 2004;23:2648–57. doi: 10.1038/sj.onc.1207408. [DOI] [PubMed] [Google Scholar]
- 19.Corin I, Di Giacomo MC, Lastella P, Bagnulo R, Guanti G, Simone C. Tumor-specific hyperactive low-molecular-weight cyclin E isoforms detection and characterization in non-metastatic colorectal tumors. Cancer Biol Ther. 2006;5:198–203. doi: 10.4161/cbt.5.2.2356. [DOI] [PubMed] [Google Scholar]
- 20.Davidson B, Skrede M, Silins I, Shih Ie M, Trope CG, Florenes VA. Low-molecular weight forms of cyclin E differentiate ovarian carcinoma from cells of mesothelial origin and are associated with poor survival in ovarian carcinoma. Cancer. 2007;110:1264–71. doi: 10.1002/cncr.22918. [DOI] [PubMed] [Google Scholar]
- 21.Keyomarsi K, Tucker SL, Buchholz TA, et al. Cyclin E and survival in patients with breast cancer. N Engl J Med. 2002;347:1566–75. doi: 10.1056/NEJMoa021153. [DOI] [PubMed] [Google Scholar]
- 22.Akli S, Zheng PJ, Multani AS, et al. Tumor-specific low molecular weight forms of cyclin E induce genomic instability and resistance to p21, p27, and antiestrogens in breast cancer. Cancer Res. 2004;64:3198–208. doi: 10.1158/0008-5472.can-03-3672. [DOI] [PubMed] [Google Scholar]
- 23.Wingate H, Bedrosian I, Akli S, Keyomarsi K. The low molecular weight (LMW) isoforms of cyclin E deregulate the cell cycle of mammary epithelial cells. Cell Cycle. 2003;2:461–6. [PubMed] [Google Scholar]
- 24.Wingate H, Zhang N, McGarhen MJ, Bedrosian I, Harper JW, Keyomarsi K. The tumor-specific hyperactive forms of cyclin E are resistant to inhibition by p21 and p27. J Biol Chem. 2005;280:15148–57. doi: 10.1074/jbc.M409789200. [DOI] [PubMed] [Google Scholar]
- 25.Akli S, Van Pelt CS, Bui T, et al. Overexpression of the low molecular weight cyclin E in transgenic mice induces metastatic mammary carcinomas through the disruption of the ARF-p53 pathway. Cancer Res. 2007;67:7212–22. doi: 10.1158/0008-5472.CAN-07-0599. [DOI] [PubMed] [Google Scholar]
- 26.Band V, Zajchowski D, Kulesa V, Sager R. Human papilloma virus DNAs immortalize normal human mammary epithelial cells and reduce their growth factor requirements. Proc Natl Acad Sci U S A. 1990;87:463–7. doi: 10.1073/pnas.87.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding Z, Liang J, Lu Y. A retrovirus-based protein complementation assay screen reveals functional AKT1-binding partners. Proc Natl Acad Sci U S A. 2006;103:15014–9. doi: 10.1073/pnas.0606917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welcker M, Orian A, Jin J, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101:9085–90. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore JD, Kornbluth S, Hunt T. Identification of the nuclear localization signal in Xenopus cyclin E and analysis of its role in replication and mitosis. Mol Biol Cell. 2002;13:4388–400. doi: 10.1091/mbc.E02-07-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson DG, Walker CL. Cyclins and cell cycle checkpoints. Annu Rev Pharmacol Toxicol. 1999;39:295–312. doi: 10.1146/annurev.pharmtox.39.1.295. [DOI] [PubMed] [Google Scholar]
- 31.Nagai T, Ibata K, Park E, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 32.Remy I, Michnick SW. A cDNAlibrary functional screening strategy based on fluorescent protein complementation assays to identify novel components of signaling pathways. Methods. 2004;32:381–8. doi: 10.1016/j.ymeth.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 33.Gray-Bablin J, Zalvide J, Fox MP, Knickerbocker CJ, DeCaprio JA, Keyomarsi K. Cyclin E, a redundant cyclin in breast cancer. Proc Natl Acad Sci U S A. 1996;93:15215–20. doi: 10.1073/pnas.93.26.15215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koepp DM, Schaefer LK, Ye X, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–7. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 35.Jackman M, Kubota Y, den Elzen N, Hagting A, Pines J. Cyclin A- and cyclin E-Cdk complexes shuttle between the nucleus and the cytoplasm. Mol Biol Cell. 2002;13:1030–45. doi: 10.1091/mbc.01-07-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Drogen F, Sangfelt O, Malyukova A, et al. Ubiquitylation of cyclin E requires the sequential function of SCF complexes containing distinct hCdc4 isoforms. Mol Cell. 2006;23:37–48. doi: 10.1016/j.molcel.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 37.Ye X, Wei Y, Nalepa G, Harper JW. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol Cell Biol. 2003;23:8586–600. doi: 10.1128/MCB.23.23.8586-8600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welcker M, Singer J, Loeb KR, et al. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol Cell. 2003;12:381–92. doi: 10.1016/s1097-2765(03)00287-9. [DOI] [PubMed] [Google Scholar]
- 39.Geisen C, Moroy T. The oncogenic activity of cyclin E is not confined to Cdk2 activation alone but relies on several other, distinct functions of the protein. J Biol Chem. 2002;277:39909–18. doi: 10.1074/jbc.M205919200. [DOI] [PubMed] [Google Scholar]
- 40.Kelly BL, Wolfe KG, Roberts JM. Identification of a substrate-targeting domain in cyclin E necessary for phosphorylation of the retinoblastoma protein. Proc Natl Acad Sci U S A. 1998;95:2535–40. doi: 10.1073/pnas.95.5.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye X, Nalepa G, Welcker M, et al. Recognition of phosphodegron motifs in human cyclin E by the SCF(Fbw7) ubiquitin ligase. J Biol Chem. 2004;279:50110–9. doi: 10.1074/jbc.M409226200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.