Abstract
We have investigated photoabsorption spectra of bovine rhodopsin and its mutants (E122Q and E113Q) by hybrid quantum mechanical/molecular mechanical (QM/MM) calculations as well as retinal in vacuo by pure QM calculations, employing multireference (MR) ab initio and TD-B3LYP methods. The sophisticated MR-SORCI+Q and MRCISD+Q methods extrapolated with respect to adopted approximations can reproduce the experimental absorption maxima of retinal very well. The relatively inexpensive MR-DDCI2+Q method gives absorption maxima blue-shifted by ca. 65 nm from experimental values; however, this error is systematic and thus MR-DDCI2+Q can be used to estimate spectral shifts. In MR calculations, the ground state energy of retinal at B3LYP geometry is significantly lower than that at CASSCF geometry. Therefore, B3LYP geometry is more reliable than CASSCF geometry, which has blue-shift error as large as 100 nm in the gas phase. The effect of ground state geometry on the excitation energies is less critical in the polarizing field of protein environments. At the B3LYP geometry, there is no significant charge transfer upon vertical excitation to the S1 excited state either from Glu113 to retinal or from Schiff-base terminal to β-ionone ring through the polyene chain. All-trans to 11-cis isomerization of retinal in the gas phase has no influence on the calculated S1 absorbing state, in agreement with experiment. The shoulder of the experimental absorption spectrum of retinal in vacuo at the S1 absorbing band appears to be the second electronic transition (S2) in our calculations, contrary to previous tentative assignment to vibrational state of S1 or to the S1 band of a retinal isomer.
Keywords: Spectral Tuning, Retinal, Multireference ab initio methods, QM/MM calculations, Electronic Excitation
I. Introduction
Rhodopsin (Rh) is responsible for the dim-light vision in vertebrate eyes and belongs to the G-protein-coupled receptor family.1,2 It is activated by the light-induced 11-cis to all-trans isomerization of protonated Schiff-base retinal (PSBR). While the retinal in vacuo absorbs light at 610 nm,3 the retinal in bovine Rh absorbs at 500 nm.1,2 Despite extensive mutagenesis and computational analyses in the last two decades (see ref 2 and references therein), the molecular basis of spectral tuning, e.g., how environments of retinal modulate its absorption maximum in the UV-visible spectrum, remains unclear.
In general, computation of the absorption maxima for retinal proteins is a very challenging problem. Our recent ONIOM quantum mechanical/molecular mechanical (QM/MM) calculations on bovine Rh, employing density functional theory (DFT), show that the effects of amino acids other than Glu113 on the first (S1) vertical excitation energy of retinal tend to cancel to each other.5 Analogously, the protein environment of retinal and Glu113 represented with several point charge models contributes up to ±20 nm at CASPT2/ANO level.6 However, the protein environment around retinal and Glu113 red-shifts the S1 excitation energy by 110 nm at (aug-MCQDPT2/cc-pVDZ)/EFP level, where EFP is an effective fragment potential method for calculating QM/MM interaction.7 Glu113 blue-shifts the S1 excitation energy in the gas phase by ca. 60, 150 and 210 nm at TD-B3LYP/6-31G*,4 CASPT2/(ANO or 6-31G*)6,8 and aug-MCQDPT2/cc-pVDZ7 levels, respectively. Although this varying Glu113 effect is well captured at those levels where its atoms are represented with point charges rather than included in the QM model system, SACCI/D95(d) calculations find a significant blue-shift (ca. 100 nm) charge-transfer contribution of Glu113.9
The computed gas-phase excitation energies of all-trans and 11-cis retinals also vary quite a lot.2,5 For example, the errors in the S1 excitation energy of all-trans retinal (experiment:3,4 610 nm) are -100/-50 nm (TD-B3LYP/6-31G* at B3LYP and complete active space self-consistent field [CASSCF] geometries, respectively),10,11 -65 nm (CASPT2/6-31G* at CASSCF geometry),12 -10/-40 nm (OM2-MRCI at B3LYP and CASSCF geometries, respectively),11 25/-20 nm (spectroscopy-oriented configuration interaction [SORCI]/6-31G* at B3LYP and CASSCF geometries, respectively)11 and more than 200 nm (SACCI/D95(d) at B3LYP geometry).14 However, CASPT2 calculations with a large ANO basis set at MP2 geometry (606 nm)6,15 reproduce the experiment (610 nm).
In our previous study,5 we investigated absorption spectra of bovine rhodopsin and its mutants by time-dependent (TD)-DFT QM/MM calculations in ONIOM scheme and reproduced the experimental absorption maxima within 10 nm. In this study, we employ high-level multireference (MR) ab initio QM/MM calculations on retinal to pinpoint origins of differences among various calculations and to provide computational strategies for accurately calculating its absorption spectra in the gas phase as well as in the protein environments of bovine rhodopsin and its mutants.
II. Computational Details
For bovine Rh and its mutants (E122Q and E113Q), ONIOM(B3LYP/6-31G*:AMBER)-optimized structures with electronic embedding (EE) are used in this study (see ref 5). All single-point ab initio QM/MM calculations on these structures were carried out with ORCA 2.6.19 program package16 using an EE scheme,17 in which fixed MM point charges are included in the one-electron QM Hamiltonian and the QM/MM electrostatic interactions are evaluated from the QM electrostatic potential and the MM atomic charges. Although this EE scheme17 is slightly different from the ONIOM-EE,5,18 they result in identical excitation energies at given Rh geometries in our test calculations. Therefore, ONIOM-EE geometries can be used for QM/MM excitation energy calculations with the ORCA program package that has many MR methods implemented. No cutoffs were introduced for the nonbonding MM and QM/MM interactions. Hydrogen link atoms19 were used at the QM/MM boundary along with a charge shift model.20
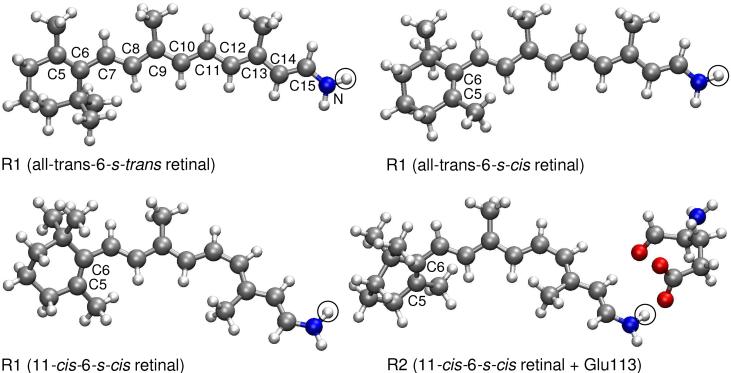
We employed two QM regions shown in Figure 1: (i) R1, including the full retinal along with covalently bound N/NH moiety of Lys296 for deprotonated/protonated Schiff-base (SB) linkage, which results in total of 50/51 atoms. We do not extend this QM region to Cε atom of Lys296 since it red-shifts the S1 excitation energy only 10 nm in TD-DFT5 and CASPT212 studies. (ii) R2, consisting of R1 region plus full Glu113, which results in total of 68 atoms when either retinal or Glu113 side chain is protonated. The gas-phase QM calculations on the bare R1 and R2 regions at the QM/MM geometries will be called as QM-none. Unless stated otherwise, our calculations were performed for PSBR modeled as R1 and using 6-31G* basis set.21 In order to assess the basis set dependence of computed excitation energies, we also used VDZP (known also as SV(P)),22 VTZP,22 TZVP,23 double-ξ {BNANO-DZP: (33s12p3d) → [3s2p1d] for N, (32s12p3d) → [3s2p1d] for C and (4s1p) → [2s1p] for H} and triple-ζ {BNANO-TZ2P: (44s18p6d) → [4s3p2d] for N, (43s18p6d) → [4s3p2d] for C and (5s2p) → [3s2p] for H} type ANO basis sets from the ORCA library.16,24 Auxiliary basis sets used to speed-up the ab initio calculations are SV/C and TZV/C for double-ζ and triple-ζ quality basis sets, respectively.25 The calculated excitation energies are not affected by the use of auxiliary basis sets for the current system.
Figure 1.
QM models and atom numbering of PSBR. SBR corresponds to removing the hydrogen circled or transferring it to Glu113 carboxylic oxygen.
We applied CASSCF26 theory for obtaining zero-order wave function of all MR ab initio calculations. CASSCF captures a major part of static correlation by mixing the active-space configurations at the full configuration interaction (CI) level. The largest 6-root CASSCF(12/12) calculations include all valance π-electrons of the polyene chain of R1 from C5 to the SB terminal. For the extensive post-CASSCF calculations at SORCI and MRCISD levels, we had to use 3-root CASSCF(6/6). For R2 with Glu113 in the QM region, we performed 6-root CASSCF(12/12) calculations including π-electrons of Glu113 in the active space.
Subsequent post-CASSCF MR calculations on the CASSCF reference wave function were performed at several CI levels. As cost-effective post-CASSCF methods, we applied difference dedicated CI (DDCI) methods DDCI2 and DDCI3.27 These methods use a CI expansion aimed at improving energy difference between individual states rather than their absolute correlation energies. Whereas the DDCI2 method includes double excitations which involve either two particles in the inactive empty orbitals or two holes in the inactive occupied orbitals in addition to all single excitations, the DDCI3 method also includes out of the active-space configurations with two holes and one particle or two particles and one hole.27-29 The effects of excitations higher than doubles (i.e., disconnected quadruple excitations) were estimated with the a posteriori MR Davidson +Q term, which corrects the energy but not the expansion coefficients of the correlated wavefunction.30 To assess the effect of size-consistency error in DDCI type methods more accurately, we applied a slightly modified version of MR averaged coupled pair functional ACPF/230 procedure to DDCI2 excitations, leading to the size-consistent DDACPF/2a-2 method implemented in ORCA.
We also applied the SORCI method,28 which performs a DDCI3 calculation in a truncated basis of approximate average natural orbitals (AANOs) obtained from a DDCI2 calculation. In the DDCI3 step of SORCI, AANOs with an occupation number less than a threshold Tnat are rejected while those with an occupation number larger than 2.0-Tnat are frozen.
Finally, we also applied the most straightforward MRCISD approach including all single and double excitations32,33 to benchmark the validity of results obtained with the lower level DDCI2, DDCI3 and SORCI methods. For accounting size-consistency error, +Q correction was also calculated for SORCI and MRCISD results.
To enhance computational efficiency, the ORCA thresholds Tnat (used only for SORCI), Tpre and Tsel were set to 10-6, 10-4 and 10-6 Eh, respectively.11,28,13 The present DDCI2 and SORCI calculations on retinal in vacuo and the previous SORCI calculations on model retinal systems (see Supporting Information of ref 13) show that looser ORCA thresholds of Tsel and Tnat result in the significant loss of accuracy. Improved virtual orbitals were not used. Core orbitals with energies of less than -4 Eh were frozen, and no virtual orbitals were neglected. In all perturbative treatments, a level shift of 0.4 Eh was applied in order to avoid intruder state problems.
III. Results
A. Protonated Schiff-Base Retinal (PSBR) in Vacuo
In this section, we mainly discuss the calculated first (S1) and second (S2) singlet vertical excitation energies of PSBR.
Basis Set Dependence
The improvement of the 6-31G* basis set to aug-cc-pVTZ red-shifts the S1 vertical energy of PSBR by only 10 nm in DFT calculations.5 As the correlated ab initio calculations converge to the basis set limit slower than DFT calculations, it is necessary to investigate the basis set effect on the excitation energies of PSBR for MR ab initio treatments. Indeed, for a small model of PSBR (without β-ionone ring), the S1 and S2 vertical energies were found to red-shift by ca. 20 nm when 6-31G* basis set is improved to aug-cc-pVTZ at the SORCI level (see Supporting Information of ref 13). To assess the basis set effect further on the full all-trans retinal, we performed 6-root DDCI2+Q(12/12) calculations at the gas-phase B3LYP/6-31G* geometry of all-trans-6-s-trans retinal, as shown in Table 1. When the double-ζ quality basis sets were improved to TZVP, the S1 excitation energy red-shifts by ca. 20 nm. However, with the largest basis set considered (BNANO-TZ2P), it comes back and gives only 5 nm blue-shifted result relative to the 6-31G*. This ANO basis set gave slightly better agreement with experiment than the aug-cc-pVTZ basis set in previous test calculations.24 As to the S2 excitation energy, 6-31G* and VDZP basis sets give ca. 20 nm blue-shift compared with the larger basis sets, which is similar to the results on model systems.13 In the following, we will use the cost-effective 6-31G* basis set that contains ca. 5 (20) nm blue-shift error for the S1 (S2) excitation energy of full PSBR.
Table 1.
Basis set dependence of the S1 (S2 in parentheses) vertical excitation energy (nm) of PSB all-trans-6-s-trans retinal in vacuo at the 6-root CASSCF(12/12) and DDCI2+Q(12/12) level at the B3LYP/6-31G*-optimized geometry
| Basis Set | Basis Set Size | CASSCF(12/12) | DDCI2+Q(12/12) |
|---|---|---|---|
| 6-31G* | 354 | 553 (404) | 545 (416) |
| VDZP [or SV(P)] | 444 | 566 (405) | 555 (420) |
| BNANO-DZP | 444 | 559 (405) | 555 (437) |
| TZVP | 579 | 572 (406) | 570 (438) |
| VTZP | 600 | 572 (406) | 563 (441) |
| BNANO-TZ2P | 753 | 552 (404) | 550 (437) |
The Effects of Several Computational Settings
We have investigated the effects of several computational settings (approximations) at the DDCI2+Q/6-31G* level on the excitation energies of PSB all-trans-6-s-trans retinal, as shown in Table 2. The number of roots has no significant effect on the computed excitation energies. Reducing CASSCF(12/12) to CASSCF(6/6) introduces a large blue-shift error of ca. 160 (60) nm in the CASSCF S1 (S2) vertical excitation energy. CI calculations substantially reduces the error due to small active space; the DDCI2+Q S1 (S2) excitation energy error due to the reduction of the active space is ca. 15 (30) nm to red (blue). Tightening Tsel from 10-6 Eh to 10-7 Eh blue-shifts the S1 (S2) DDCI2+Q excitation energy by ca. 15 (5) nm whereas tightening Tpre = 10-4 has no notable effect, a situation similar to SORCI calculations on model systems.13 In an overall assessment, CI calculations with Tsel = 10-6 using CASSCF(6/6) wave function have a net error of ca. +15 (-55) nm in S1 (S2) excitation energy, arising from Tsel [+15 (-5) nm], active space size [+15 (-30) nm], basis set [-5 (-20) nm] and the use of NH2 terminal rather than NHCH3 [-10 (0) nm11].
Table 2.
The effects of active space size, the number of roots and Tsel (Eh) in post-CASSCF/6-31G* calculations on the computed S1 (S2) vertical excitation energy (nm) of PSB all-trans-6-s-trans retinal in vacuo optimized at B3LYP/6-31G* level
| 6-root CAS(12/12) | 3-root CAS(12/12) | 3-root CAS(6/6) | 3-root CAS(6/6) | |
|---|---|---|---|---|
| CASSCF | 553 (404) | 548 (391) | 382 (334) | 382 (334) |
| Tsel | 10-6 | 10-6 | 10-6 | 10-7 |
| DDCI2+Q | 545 (416) | 544 (412) | 559 (385) | 545 (379) |
Accuracy of Several CI Levels
Experimental absorption spectra of PSBR (both all-trans and 11-cis conformations) have absorption bands peaking at 610 nm and ca. 390 nm.3,4 These bands were assigned previously to S1 and S2 transitions, respectively.3,4 The band at 610 nm has a shoulder at around 540 nm.3,4 This shoulder was tentatively assigned either to the S1 state of an all-trans retinal isomer or to a vibrational state of the electronic S1 state.3,4 The results of several CI calculations are given in Table 3. From the error estimates in the previous section, one should subtract 15 nm from S1 and add 55 nm to S2 excitation energies given in Table 3 when CI results using CASSCF(6/6)/6-31G* are compared with experiment. The magnitude of +Q correction for the S1 (S2) excitation energy is sizable and depends on the applied CI level: -26 (-33) nm, 42 (0) nm, 47 (5) nm and -73 (-280) nm at DDCI2, DDCI3, SORCI and MRCISD levels, respectively. Therefore, we consider the results only with Q correction.
Table 3.
S1 and S2 vertical excitation energies (nm) for PSB all-trans-6-s-trans retinal in vacuo at various CI/6-31G* levels along with CASSCF and TD-B3LYP results at the B3LYP/6-31G*-optimized geometry
| Method | S1 | S2 |
|---|---|---|
| TD-B3LYP | 501 | 380 |
| CASSCF(12/12)a | 553 | 404 |
| CASSCF(6/6) | 382 | 334 |
| DDCI2+Q(12/12)a | 545 | 416 |
| DDCI2+Q(6/6) | 559 | 385 |
| DDACPF/2a-2(12/12)a | 527 | 394 |
| DDCI3+Q(6/6) | 640 | 474 |
| SORCI+Q(6/6) | 626 | 491 |
| MRCISD+Q(6/6) | 604 | 458 |
6-root CAS(12/12) results are given for comparison.
As already found in the previous section, CI calculations improve the poor performance of small CASSCF(6/6) and make the results comparable to the CI results based on CASSCF(12/12). Our lowest level post-CASSCF calculation (DDCI2+Q) corrects the TD-B3LYP S1 excitation energy by ca. 50 nm. DDCI2+Q and size-consistent DDACPF/2a-2 results agree within 20 nm. DDCI3+Q, SORCI+Q and MRCISD+Q methods improve DDCI2+Q energies significantly, making the S1 excitation energies agree with experiment within ±20 nm (including 15 nm correction). Natural orbital iteration at SORCI+Q level further improves the S1 DDCI3+Q excitation energy toward experiment by ca. 15 nm.
As to the S2 excitation energy, when the DDCI3+Q, SORCI+Q and MRCISD+Q S2 excitation energies based on CASSCF(6/6) are corrected with our error estimate of 55 nm, we obtain values (513-546 nm) that are close to the 540 nm shoulder of S1 band rather than to the 390 nm absorption band assigned to S2 in the experimental spectrum.3,4 At the B3LYP-optimized geometry, the calculated oscillator strengths f of S1 and S2 excitation for the 6-s-trans/6-s-cis conformation are around 1.8/1.8 and 0.03/0.14, respectively. As these oscillator strengths are significantly different than zero, we assign the shoulder of the S1 band to the S2 band of ground-state 6-s-cis conformation (see Table 4 for the results on this species), rather than a vibrational splitting or S1 of a local minimum. The difference between the S1 and third (S3) excitation energies in DDCI2+Q(12/12)/6-31G* (not listed in Table 3, see Supporting Information) is 237 nm; which agrees reasonably well with the difference (225 nm) in the experimental absorption maxima of 610 and 385 nm bands of all-trans retinal. Therefore, we assign the band peaking at 385 nm in the experimental spectrum as the S3 band, contrary to the previous assigned S2.3,4 The appearance of three absorption bands in the absorption spectrum of PSB all-trans retinal in vacuo is in agreement with two-photon spectroscopy of PSB all-trans retinal in the protein environment of bacteriorhodopsin (568 nm, 488 nm and 410 nm).34
Table 4.
Geometry dependence of the S1 (S2) excitation energy of PSB retinal in vacuo in MR calculations with 6-31G* basis set based on 3-root CAS(6/6) or 6-root CAS(12/12) levels
| CASSCF(12/12) | CASSCF(6/6) | DDCI2+Q(12/12) | SORCI+Q(6/6) | MRCISD+Q(6/6) | |
|---|---|---|---|---|---|
| all-trans-6-s-trans | |||||
| CASSCF geometrya | 526 (345) | 421 (295) | 539 (360) | 565 (395) | 603 (412) |
| B3LYP geometry | 553 (404) | 382 (334) | 545 (416) | 626 (491) | 604 (458) |
| all-trans-6-s-cis | |||||
| CASSCF geometryb,c | 457 (312) | 415 (328) | 470 (361) | 505 (396) | 546 (421) |
| B3LYP geometryd | 545 (391) | 467 (350) | 551 (412) | 610 (475) | 604 (493) |
| 11-cis-6-s-cis | |||||
| B3LYP geometry | 535 (383) | 362 (313) | 541 (403)e | 625 (473) | 601 (454) |
Geometry Effect at Several CI Levels
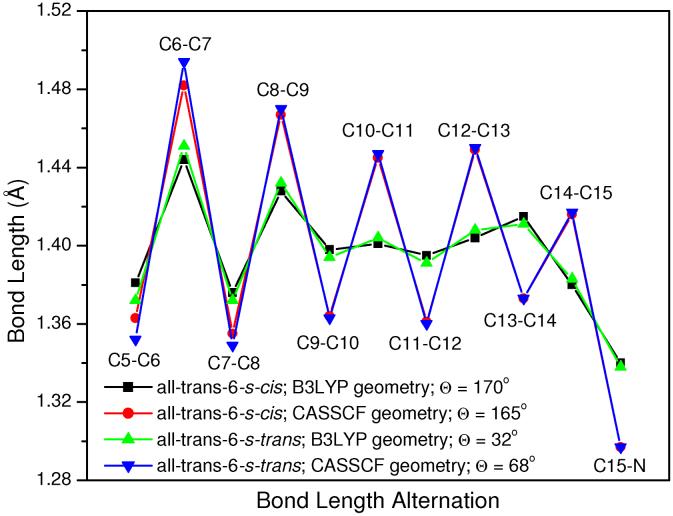
As shown in Figure 2, CASSCF single and double bonds are on average 0.04 Å longer and 0.03 Å shorter than B3LYP distances, respectively. In the SORCI+Q and MRCISD+Q calculations, the B3LYP geometry of PSB all-trans retinal (both 6-s-trans and 6-s-cis) is at least 8 kcal/mol more stable than the CASSCF geometry. Therefore, the B3LYP ground-state geometries (incorporating both static and dynamical correlations) are more appropriate for retinal systems than CASSCF ones (incorporating only static correlation and unbalanced). B3LYP, MP2 and CASPT2 geometries have been shown to be almost the same on small PSBR models, whereas HF (no electron correlation) and CASSCF (incorporating only static correlation) methods overestimate bond length alternation (BLA) of the polyene chain.5,35-37 As discussed earlier, the clear BLA pattern of PSBR in rhodopsins is the result of the polarizing field of protein environments.5 Therefore, it is not expected to have a significant BLA in gas-phase retinal systems.
Figure 2.
Bond length alternation of polyene chain of PSB all-trans retinal at B3LYP/6-31G* and CASSCF/6-31G* levels along with C5-C6-C7-C8 tilt angle of β-ionone ring (Θ)
CASSCF and B3LYP geometries are both used extensively in literature for retinal systems. However, going from CASSCF geometries to B3LYP ones in the present post-CASSCF calculations, the S1 and S2 excitation energies improve up to 100 nm and reach to the range of experimental value at SORCI+Q and MRCISD+Q levels, as shown in Table 4. An analogous geometry effect was also seen in the previous SORCI,13 SACCI calculations9,14 and some lower level calculations.13 For example, going from HF geometries of retinal proteins (being similar to CASSCF geometries) to B3LYP ones (being similar to CASPT2 geometries), the S1 SACCI excitation energy red-shifts by ca. 100 nm.9,14 Remembering the basis set effect on the S1 and S2 excitation energies of PSBR is small, the difference between the previous CASPT2 excitation energies of all-trans-6-s-cis retinal with the 6-31G* basis set at CASSCF geometry (534 nm for S1 and 353 nm for S2)12 and with an ANO basis set at MP2 geometries (606 nm for S1 and 436 nm for S2)6 should also mainly arise from such geometry effects. Actually, these CASPT2 S1 excitation energies are almost the same as our MRCISD+Q results at CASSCF (546 nm) and B3LYP (604 nm) geometries. However, the S2 excitation energy calculated at the CASPT2 level (353 nm12 and 436 nm6) is ca. 60 nm blue-shifted than that at the MRCISD+Q level (421 and 493 nm).
The SORCI+Q and MRCISD+Q S1 excitation energies of 6-s-cis and 6-s-trans conformations at B3LYP geometries (calculated: 604 nm; estimate with 15 nm corrrection: 589 nm) agree with experiment3,4 reasonably well. The calculated S2 excitation energies are more sensitive to the conformational changes. Including the error estimate of 55 nm in our post-CASSCF(6/6) S2 excitation energy calculations, the maximum of the shoulder at the S1 band of experimental spectrum (ca. 540 nm),3,4 e.g., S2 band in our new assignment discussed above, is reproduced at SORCI+Q (530 nm) and MRCISD+Q (548 nm) levels using the B3LYP geometry of 6-s-cis conformation. However, it is ca. 30 nm blue-shifted at the MRCISD+Q level for the 6-s-trans conformation (513 nm). Hence, the absorption spectrum of all-trans retinal recorded in vacuo3 should belong mainly to the 6-s-cis conformation. This conformation is at least 3 kcal/mol more stable than the 6-s-trans one at SORCI+Q and MRCISD+Q levels.
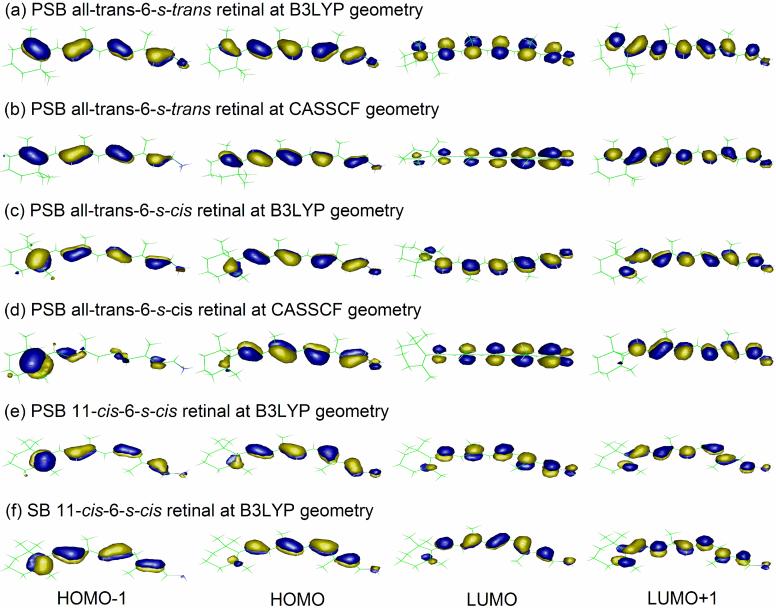
11-cis-retinal with 6-s-cis conformation is ca. 6 kcal/mol above the ground-state all-trans-6-s-cis retinal in all of our MR calculations at the B3LYP geometry. Its frontier orbitals (Figure 3) and calculated excitation energies are very similar to those for all-trans retinal (Table 4). Therefore, 11-cis ↔ all-trans isomerization is not expected to affect the S1 excitation energy much in protein environments.
Figure 3.
The key 6-root CASSCF(12/12) orbitals of retinal in vacuo.
Dominant configurations in S0, S1, and S2 states are the closed-shell singlet, the HOMO to LUMO single excitation, and the HOMO-1 to LUMO single excitation, respectively (see Figure 3).38 If HF or CASSCF geometry with overestimated polyene chain BLA is used, LUMO is artificially destabilized and thus S1 and S2 transitions shift to blue. At the B3LYP geometries, HOMO-1 or HOMO to LUMO double excitation contributes significantly in the S2 transition since LUMO becomes stable enough with the decreased BLA. In other words, the decreased (increased) BLA results in the red (blue) shift in the S1 and S2 excitation energies.
At the CASSCF geometries, half of the positive charge located on the CNH2 moiety moves toward the β-ionone ring upon S1 excitation, consistent with previous studies.8,39 This originates from the nature of the LUMO, whose π-electron density decreases from the SB terminal to β-ionone ring (Figure 3). However, there appears to be no such charge transfer at B3LYP geometries. Therefore, the charge transfer effect seen at CASSCF geometries upon the S1 transition is artificial and arises from the overestimation of BLA.
B. Deprotonated SBR in Vacuo
The conjugation scheme in the frontier orbitals of the charged PSBR (Figure 3a-e) and neutral SBR (Figure 3f) is different from each other. The effects of computational approximations used for reducing computational cost in CI calculations may thus be different between the two systems. This leads us to investigate several computational factors in accessing the error in post-CASSCF(6/6)/6-31G* calculations of deprotonated SB 11-cis-retinal with Tsel = 10-6 Eh, as shown in Table 5. These analyses are especially important for estimating the error in ab initio QM/MM calculations of visual pigments with deprotonated SBR, such as E113Q mutant of rhodopsins.
Table 5.
The effect of active space size, basis set and Tsel (Eh) in CASSCF and CI calculations on the computed S1 and S2 (in parentheses) vertical excitation energy (nm) of deprotonated SB 11-cis-6-s-cis retinal in vacuo optimized at B3LYP/6-31G* level
| Number of Roots |
6 |
6 |
6 |
3 |
3 |
3 |
|---|---|---|---|---|---|---|
| CAS Size |
CAS(12/12) |
CAS(10/10) |
CAS(10/10) |
CAS(6/6) |
CAS(6/6) |
CAS(6/6) |
| Basis Set | 6-31G* | 6-31G* | BNANO-TZ2P | 6-31G* | BNANO-TZ2P | 6-31G* |
| CASSCF | 285 (231) | 285 (230) | 285 (230) | 246 (231) | 247 (235) | 246 (231) |
| Tsel | 10-6 | 10-6 | 10-6 | 10-6 | 10-6 | 10-7 |
| DDCI2+Q | 310 (309) | 299 (292) | 320 (294) | 283 (274) | 298 (278) | 282 (269) |
Reducing the CAS(12/12) to CAS(6/6) blue-shifts the CASSCF S1 excitation energy of SBR by ca. 40 nm, whereas the CASSCF S2 excitation energy is not affected. It also has 27 (35) nm blue-shift effect for the S1 (S2) DDCI2+Q excitation energy. Tightening Tsel from 10-6 to 10-7 Eh does not have much influence on the S1 or S2 excitation energies. Although the S1 and S2 CASSCF as well as the S2 DDCI2+Q excitation energies are not affected much by the basis set extension from 6-31G* to BNANO-TZ2P, the S1 DDCI2+Q excitation energy blue-shifts by 21 and 15 nm at CAS(10/10) and CAS(6/6) levels, respectively. With the summation of these error estimates, our practical DDCI2+Q(6/6)/6-31+G* calculations are expected to have a blue-shift error of ca. 50 (35) nm for the S1 (S2) excitation energy of SBR relative to the DDCI2+Q(12/12)/BNANO-TZ2P.
C. Bovine Rhodopsin and its Mutants
Bovine Rh and its mutants with PSBR have two absorption maxima at around 500 nm and 380 nm. In our SORCI+Q/MM and MRCISD+Q/MM calculations, S1 is an absorbing state (f = 1.2-2.0) whereas S2 is a nonabsorbing state with f ~ 0 due to structural distortions in the protein environment. Therefore, the S2 state in these calculations cannot be assigned to the experimental absorption band at around 380 nm. As the spectral tuning of visual pigments is mainly discussed on the S1 absorbing state, we will not address the tuning of the second absorbing state, which will require an extension of the present 3-root calculations. For deprotonated SBR, the S1 and S2 states are very close in energy (within 10-20 nm), and S2 may become absorbing rather than S1 depending on the computational settings; here we only discuss the absorbing state (f = 1.4-2.0) in relating the calculations to the experiments.
The Effects of Protein Environment
The QM part used in the QM/MM calculation in this section is R1, the full 11-cis-retinal along with covalently bound N/NH moiety of Lys296 for deprotonated/protonated SB linkage. Geometries were obtained by ONIOM(B3LYP:AMBER)-EE optimization. QM/MM excitation energy calculations with TD-B3LYP, SORCI+Q and MRCISD+Q reproduce experimental absorption maximum of WT bovine Rh perfectly (experiment: 500 nm), as shown in Table 6. In the present QM/MM calculations with TD-B3LYP, DDCI2+Q, SORCI+Q and MRCISD+Q, the experimental spectral shift of -20 nm with E122Q mutation2,5 is also reproduced within 10 nm. Hence, all of these methods can be used in evaluating QM/MM spectral shifts of PSBR with mutations.
Table 6.
The calculated excitation energy (nm) of the absorbing state for the WT bovine Rh and its E122Q and E113Q mutants at protein geometries in the absence (QM-none) and presence (QM/MM) of protein environments (QM region: R1; basis set: 6-31G*)a
| Structure [Experiment] |
WT Rh [500 (0)] |
E122Q [480 (-20)] |
E113Q[384 (-116)] |
|||
|---|---|---|---|---|---|---|
| Retinal Protonation |
PSBR |
PSBR |
SBR |
|||
| QM Method | QM-none | QM/MM | QM-none | QM/MM | QM-none | QM/MM |
| TD-B3LYP | 560 (0) | 503 (0) | 566 (6) | 492 (-11) | 398 (-162) | 409 (-94) |
| DDCI2+Q(12/12) | 527 (0) | 435 (0) | 522 (-5) | 427 (-8) | 310 (-217) | 320 (-115) |
| SORCI+Q(6/6) | 616 (0) | 495 (0) | 586 (-30) | 462 (-33) | 327 (-289) | 330 (-165) |
| MRCISD+Q(6/6) | 626 (0) | 499 (0) | 584 (-42) | 490 (-9) | 323 (-303) | 322 (-177) |
Numbers (nm) in parentheses are spectral shifts relative to the WT bovine Rh. The calculated QM and QM/MM shifts are relative to the QM-none and QM/MM results of WT Rh.
The polarizing protein environment induces an increased BLA for the polyene chain.5 This BLA increase is more significant with B3LYP (ca. 0.03 Å5) than with CASSCF (ca. 0.01 Å8). Thus, CASSCF geometries in protein environments are more similar to B3LYP ones and more reliable than those in the gas phase. Actually, previous CASPT2/MM S1 excitation energies at CASSCF/AMBER and DFTB (being similar to B3LYP one, see ref 5) geometries are very similar to each other (480 and 506 nm, respectively) for bovine Rh.8,39
In our previous TD-B3LYP/MM study,5 we reproduced experimental spectral shifts due to mutations on bovine Rh within 10 nm except for E113Q mutant. Here we reinvestigate the E113Q mutant with SBR by using MR methods. As our post-CASSCF calculations are expected to have ca. 15 (50) nm red (blue) shift error for the absorbing state of PSBR (SBR), our MR calculations with present settings should overestimate the spectral shift going from WT Rh to E113Q mutant by ca. 65 nm. As expected, SORCI+Q(6/6)/MM and MRCISD+Q(6/6)/MM shifts are ca. 65 nm larger than the experimental shift.40 Whereas DDCI2+Q/MM reproduces the SORCI+Q/MM and MRCISD+Q/MM excitation energies of SBR (Table 6), its result for S1 of PSBR is ca. 65 nm blue-shifted since some contributing double excitations are ignored. Therefore, going from PSBR to SBR, the error arising from our computational settings (ca. 50 nm red shift with CAS(12/12)) and the intrinsic error of DDCI2 for S1 of PSBR (ca. 65 nm blue shift) tend to cancel to each other. Hence, DDCI2+Q(12/12)/MM reproduces the experimental shift due to error cancellation without applying any correction.
The Effects of Glu113 Counterion and Retinal Protonation
DDCI2+Q reproduces the experimental shifts reasonably well with the current settings as a result of error cancellation. Therefore, we will assess the effect of Glu113 counterion on the absorbing state of 11-cis-retinal with 6-root DDCI2+Q(12/12)/6-31G* calculations, as shown in Table 7.
Table 7.
Gas-phase QM-none (e.g., QM at protein geometry) and QM/MM excitation energies (nm) for the absorbing state of bovine Rh in the absence (R1) and presence (R2) of Glu113 in the QM region and the effect retinal protonation (basis set: 6-31G*) along with QM and QM/MM spectral shifts with deprotonation of retinal (in parentheses) and with inclusion of Glu113 in the QM model (△)
| Protonation State | Method | QM region = R1 (without Glu113) | QM region = R2 (with Glu113) | △ = E(R2) - E(R1) |
|---|---|---|---|---|
| QM=TD-B3LYP | ||||
| PSBR (deprotonated Glu113) | QM-none | 560 (0) | 500 (0) | -60 |
| QM/MM | 503 (0) | 499 (0) | -4 | |
| SBR with protonated Glu113 | QM-none | 408 (152) | 423 (77) | 15 |
| QM/MM | 429 (74) | 435 (64) | 6 | |
| SBR with deprotonated Glu113 | QM-none | 396 (164) | 380 (120) | -16 |
| QM/MM | 380 (123) | 382 (117) | -2 | |
| QM=DDCI2+Q(12/12) | ||||
| PSBR (deprotonated Glu113) | QM-none | 527 (0) | 395 (0) | -133 |
| QM/MM | 435 (0) | 432 (0) | -2 | |
| SBR with protonated Glu113 | QM-none | 318 (209) | 309 (86) | -9 |
| QM/MM | 336 (99) | 337 (95) | 1 | |
| SBR with deprotonated Glu113 | QM-none | 313 (214) | 298 (97) | -15 |
| QM/MM | 309 (126) | 294 (138) | -15 |
When Glu113 is included in the gas-phase QM model of PSBR at the protein geometry, the S1 excitation energy blue-shifts at DDCI2+Q level by ca. 130 nm. This shift agrees with previous CASPT2 results within 30 nm.6,8,39,41,42 The remaining protein environment red-shifts the S1 excitation energy by ca. 35 nm, similar to a previous CASPT2/ANO study at DFTB geometry.6 This shift is significantly larger (ca. 100 nm) in CASPT2 studies at CASSCF gas phase geometry,8,39 which should not be used because of overestimated BLA. The calculated S1 excitation energy of PSBR at TD-B3LYP/MM and DDCI2+Q/MM levels does not depend on the presence or absence of Glu113 in the QM region (R1 vs R2). Hence, Glu113 contributes to the S0 and S1 states equally and the Glu113 point-charge model can be safely used in calculating the S1 excitation energy of retinal.
Although previous SACCI/AMBER9 and aug-MCQDPT2/EFP7 calculations reproduce the experimental absorption maximum of bovine Rh, their energy contributions are somewhat different to each other and from the other studies. First, concerning the SACCI/AMBER result, inclusion of Glu113 in the QM region, e.g. R2, is necessary to reproduce experimental S1 absorption maximum. It blue-shifts the SACCI/AMBER S1 excitation energy by ca. 100 nm.9 However, such a charge transfer effect from Glu113 to retinal has not been found in the present ab initio MR-QM/MM and the previous TD-B3LYP/MM calculations. Furthermore, the SACCI S1 excitation energy with R1 has a large error of more than 200 nm in the gas phase. The source of errors in the SACCI and SACCI/MM excitation energies with R1 remains a puzzle.9,13 Next, concerning the aug-MCQDPT2/EFP7 result, the protein environment around retinal and Glu113 (an R2-like QM region) red-shifts S1 excitation energy (QM-none = 388 nm vs. QM/MM = 515 nm) more than 100 nm.7 However, all previous calculations with R2 (TD-B3LYP,5 CASPT26 and the present DDCI2+Q) find a small contribution from the protein environment. It seems that this discrepancy comes from significant blue-shift error in the gas-phase aug-MCQDPT2 result.7
TD-B3LYP has a significant error of 110 nm for the S1 excited state of protonated R1 in the gas phase (Table 3) as a result of the use of nonlocal exchange functionals.13 The gas phase TD-B3LYP S1 excitation energy of protonated R1 at the protein geometry has a smaller error of ca. 60 nm, i.e. the difference between TD-B3LYP gas-phase spectral shifts relating to R1 with the corresponding DDCI2+Q shifts in Table 7. The inclusion of the effect of Glu113 fixes the error in TD-B3LYP calculations significantly. Hence, TD-B3LYP can be useful for estimating excitation energies in protein environments both with R1 and R2 QM regions as well as in the gas phase with R2 (but not with R1). This means that the conclusions of the previous TD-B3LYP/AMBER study remain true although the gas phase results with R1 are blue-shifted.5
Deprotonation of retinal blue-shifts the absorption maximum by ca. 90 nm (Table 7). The inclusion of Glu113 in the QM region and/or of the protein environment does not affect the SBR excitation energies much in both TD-B3LYP and DDCI2+Q calculations (less than 20 nm, see ref 5 for detailed discussions). When the interaction between SBR and its counterion is weakened, the absorbing state blue-shifts, as seen from the comparison of the results with the protonated and deprotonated Glu113.
IV. Discussion and Conclusions
We have investigated vertical excitation energies of retinal in vacuo and bovine Rh along with its E122Q and E113Q mutants by using MR ab initio methods in combination with the QM/MM method in order to clarify the origins of differences among various previously published calculations and to provide computational strategies for accurately calculating absorption spectra. MR excitation energy calculations are too demanding with sufficiently large QM regions, active spaces, basis sets and configuration selections. Therefore, we need to adjust these computational settings looser than desired, to a certain extent. Taking each of the settings at a desired level while all of the remaining ones are fixed at lower levels, we roughly estimated the errors in reducing computational accuracy. For example, the post-CASSCF(6/6)/6-31G* calculations with Tsel = 10-6 Eh has a red-shift (blue-shift) error of ca. 15 (50) nm for the S1 absorbing state of PSBR (SBR). Applying these error corrections to SORCI+Q(6/6)/6-31G* and MRCISD+Q(6/6)/6-31G* excitation energies with Tsel = 10-6 Eh, the experimental absorption maxima are reproduced quite well.
The relatively inexpensive DDCI2+Q method has an intrinsic blue-shift error of ca. 65 nm in calculating the S1 excitation energy of PSBR in the gas-phase and protein environment as a result of the ignored double excitations. As this error is constant, (a) absolute values of excitation energies of PSBR can be estimated from the calculated DDCI2+Q excitation energies and (b) DDCI2+Q reproduces experimental spectral shifts of PSBR due to mutation reasonably well. The absorbing excitation energy of SBR calculated with DDCI2+Q is almost the same as that with higher level SORCI+Q and MRCISD+Q calculations, e.g. ca. 60 nm blue-shifted compared with experiment due to computational settings. As this error in SBR and the intrinsic error in PSBR DDCI2+Q excitation energies are almost the same, the experimental spectral shift due to deprotonation of retinal can be reproduced well by using the bare DDCI2+Q results.
The TD-B3LYP S1 excitation energy of retinal in the gas phase has a significant blue-shift error arising from the use of nonlocal exchange functionals.13 The net error in the excitation energy calculations with TD-B3LYP diminishes significantly when including the effect of H-bonding counterion (Glu113) of retinal.
The level of ground-state geometry optimization has a large impact on the computed vertical excitation energies. In the post-CASSCF excitation energy calculations, B3LYP geometries give results that are consistent with experiment both in the gas phase and in the protein environment. S1 may have up to 100 nm blue-shift error in the gas phase if the CASSCF structure with overestimated BLA is used. However, at CASSCF/MM geometries, the error in the post-CASSCF/MM excitation energies is small since polarizing field of protein environment makes the CASSCF structure closer to the B3LYP structure. Our calculations indicate that the large error in the previous CASPT2 S1 excitation energy in vacuo12 arises mainly from the use of an inappropriate CASSCF geometry.
The previous8,39 and present calculations at CASSCF geometries find that half of the positive charge located on the SB terminal moves toward β-ionone ring upon S0 to S1 excitation. However, there appears to be no such charge transfer at the more appropriate B3LYP geometries.
All-trans to 11-cis isomerization of retinal has no significant effect on the S1 excitation energy in our MR calculations, in agreement with experiments,3,4 showing almost the same absorption spectra for both isomers. The 6-s-cis conformer of all-trans retinal is more stable than the 6-s-trans one in the present MR calculations. Moreover, its calculated S2 excitation energy is more consistent with the experimental value. Therefore, experimental spectra of all-trans retinal3,4 belong mainly to the 6-s-cis conformer, in agreement with ground state energetics. Our calculations also suggest that the shoulder of the experimental absorption spectrum of retinal in vacuo at the S1 absorbing band belongs to the second electronic transition (S2), contrary to previous tentative assignment to vibrational state of S1 or to the S1 band of a retinal isomar.
Glu113 has a blue-shift effect of ca. 130 nm on the S1 excitation energy of PSBR whereas it has almost no effect on that of SBR. The net Glu113 effect for PSBR is mainly electrostatic in the present MR and the previous TD-B3LYP5 calculations. In other words, S0 to S1 excitation does not involve any charge transfer from Glu113 to retinal. Thus, it is appropriate to place Glu113 in the MM part of QM/MM calculations. The protein environment of retinal other than Glu113 has small effect on the S1 excitation energy. Deprotonation of retinal may play an important role in the UV vision of vertebrates. It blue-shifts the S1 excitation energy by ca. 90 nm.
In summary, the present calculations pinpoint the origin of most of the conflicting issues in literature and thus enhance significantly our understanding on spectral tuning mechanism in retinal systems. The relatively inexpensive DDCI2+Q method can be used in calculating spectral shifts. TD-B3LYP is also useful in calculating the S1 excitation energy shifts in protein environments.
Supplementary Material
Acknowledgment
We thank Dr. Frank Neese for supplying his ORCA program package and for invaluable discussions. We also thank Drs. Hiroshi Nakatsuji and Jun-ya Hasegawa for discussions. This work at Emory is supported by a grant from the National Institutes of Health (R01EY016400-03), and the work at Kyoto is in part supported by Japan Science and Technology Agency (JST) with a Core Research for Evolutional Science and Technology (CREST) grant in the Area of High Performance Computing for Multiscale and Multiphysics Phenomena.
Footnotes
Supporting Information Available. All calculated vertical excitation energies along with their oscillator strengths. This information is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- (1).Yokoyama S. Prog. Ret. Eye Res. 2000;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- (2).Altun A, Yokoyama S, Morokuma K. PhotoChem. PhotoBiol. 2008;84:845–854. doi: 10.1111/j.1751-1097.2008.00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Andersen LH, Nielsen IB, Kristensen MB, El Ghazaly MOA, Haacke S, Nielsen MB, Petersen MA. J. Am. Chem. Soc. 2005;127:12347–12350. doi: 10.1021/ja051638j. [DOI] [PubMed] [Google Scholar]
- (4).Nielsen IB, Lammich L, Andersen LH. Phys. Rev. Lett. 2006;96:018304/1–4. doi: 10.1103/PhysRevLett.96.018304. [DOI] [PubMed] [Google Scholar]
- (5).Altun A, Yokoyama S, Morokuma K. J. Phys. Chem. B. 2008;112:6814–6827. doi: 10.1021/jp709730b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Sekharan S, Sugihara M, Buss V. Angew. Chem. Int. Ed. 2007;46:269–271. doi: 10.1002/anie.200603306. [DOI] [PubMed] [Google Scholar]
- (7).Bravaya K, Bochenkova A, Granowsky A, Nemukhin A. J. Am. Chem. Soc. 2007;129:13035–13042. doi: 10.1021/ja0732126. [DOI] [PubMed] [Google Scholar]
- (8).Coto PB, Strambi A, Ferre N, Olivucci M. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17154–17159. doi: 10.1073/pnas.0604048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Fujimoto K, Hayashi S, Hasegawa J, Nakatsuji H. J. Chem. Theo. Comp. 2007;3:605–618. doi: 10.1021/ct6002687. [DOI] [PubMed] [Google Scholar]
- (10).Vreven T, Morokuma K. Theor. Chem. Acc. 2003;109:125–132. [Google Scholar]
- (11).Hoffmann M, Wanko M, Strodel P, König PH, Frauenheim T, Schulten K, Thiel W, Tajkhorshid E, Elstner M. J. Am. Chem. Soc. 2006;128:10808–10818. doi: 10.1021/ja062082i. [DOI] [PubMed] [Google Scholar]
- (12).Cembran A, Gonzalez-Luque R, Altoe P, Merchan M, Bernardi F, Olivucci M, Garavelli M. J. Phys. Chem. A. 2005;109:6597–6605. doi: 10.1021/jp052068c. [DOI] [PubMed] [Google Scholar]
- (13).Wanko M, Hoffmann M, Strodel P, Koslowski A, Thiel W, Neese F, Frauenheim T, Elstner M. J. Phys. Chem. B. 2005;109:3606–3615. doi: 10.1021/jp0463060. [DOI] [PubMed] [Google Scholar]
- (14).Fujimoto K, Hasegawa J, Hayashi S, Kato S, Nakatsuji H. Chem. Phys. Lett. 2005;414:239–242. [Google Scholar]
- (15).Sekharan S, Weingart O, Buss V. Biophys. J. 2006;91:L07–L09. doi: 10.1529/biophysj.106.087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).(a) Neese F. ORCA-an ab initio, DFT and semiempirical electronic structure package. version 2.6. Institut für Physikalische und Theoretische Chemie, Universität Bonn; Germany: 2007. revision 19. [Google Scholar]; (b) Latest version available from http://www.thch.uni-bonn.de/tc/orca.
- (17).Bakowies D, Thiel W. J. Phys. Chem. 1996;100:10580–10594. [Google Scholar]
- (18).Vreven T, Byun KS, Komáromi I, Dapprich S, Montgomery JA, Jr., Morokuma K, Frisch MJ. J. Chem. Theo. Comp. 2006;2:815–826. doi: 10.1021/ct050289g. [DOI] [PubMed] [Google Scholar]
- (19).Antes I, Thiel W. In: Hybrid Quantum Mechanical and Molecular Mechanical Methods. Gao J, editor. ACS Symposium Series 712, American Chemical Society; Washington DC: 1998. pp. 50–65. [Google Scholar]
- (20).de Vries AH, Sherwood P, Collins SJ, Rigby AM, Rigutto M, Kramer GJ. J. Phys. Chem. B. 1999;103:6133–6141. [Google Scholar]
- (21).(a) Ditchfield R, Hehre WJ, Pople JA. J. Chem. Phys. 1971;54:724–728. [Google Scholar]; (b) Hehre WJ, Ditchfield R, Pople JA. J. Chem. Phys. 1972;56:2257–2261. [Google Scholar]; (c) Hariharan PC, Pople JA. Theor. Chim. Acta. 1973;28:213–222. [Google Scholar]; (d) Clark T, Chandrasekhar J, Spitznagel GW, Schleyer P. v. R. J. Comp. Chem. 1983;4:294–301. [Google Scholar]
- (22).Schäfer A, Horn H, Ahlrichs R. J. Chem. Phys. 1992;97:2571–2577. [Google Scholar]
- (23).(a) Schäfer A, Huber C, Ahlrichs R. J. Chem. Phys. 1994;100:5829–5835. [Google Scholar]; (b) Ahlrichs R, May K. Phys. Chem. Chem. Phys. 2000;2:943–945. [Google Scholar]
- (24).(a) Neese F. 2008. unpublished basis sets.; (c) Berry JF, George SD, Neese F. Phys. Chem. Chem. Phys. 2008;10:4361–4374. doi: 10.1039/b801803k. [DOI] [PubMed] [Google Scholar]; (b) These ANO basis sets give energies that are intermediate in quality between those obtained with the correlation consistent triple-ζ (cc-pVTZ) and the much larger correlation consistent quadruple-ζ (cc-pVQZ) basis sets in test calculations.
- (25).(b) Eichkorn K, Weigend F, Treutler O, Ahlrichs R. Theor. Chem. Acc. 1997;97:119–124. [Google Scholar]; (c) Weigend F, Häser M. Theor. Chem. Acc. 1997;97:331–340. [Google Scholar]; (a) The auxiliary basis sets were obtained from the TURBOMOLE basis set library under ftp.chemie.uni-karlsruhe.de/pub/cbasen.
- (26).Roos BO, Taylor PR. Chem. Phys. 1980;48:157–173. [Google Scholar]
- (27).Miralles J, Castell O, Caballol R, Malrieu JP. Chem. Phys. 1993;172:33–43. [Google Scholar]
- (28).Neese F. J. Chem. Phys. 2003;119:9428–9443. [Google Scholar]
- (29).Altun A, Kumar D, Neese F, Thiel W. J. Phys. Chem. A. 2008 doi: 10.1021/jp802092w. DOI: 10.1021/jp802092w. [DOI] [PubMed] [Google Scholar]
- (30).Hirsch G, Bruna PJ, Peyerimhoff SD, Buenker RJ. Chem. Phys. Lett. 1977;52:442–448. [Google Scholar]
- (31).Gdanitz RJ. Int. J. Quant. Chem. 2001;85:281–300. [Google Scholar]
- (32).Siegbahn PEM. J. Chem. Phys. 1980;72:1647–1656. [Google Scholar]
- (33).Sharp SB, Gellene GI. J. Chem. Phys. 2000;113:6122–6131. [Google Scholar]
- (34).Birge RR, Zhang C-F. J. Chem. Phys. 1990;92:7178–7195. [Google Scholar]
- (35).Page CS, Olivucci M. J. Comput. Chem. 2003;24:298–309. doi: 10.1002/jcc.10145. [DOI] [PubMed] [Google Scholar]
- (36).Geskin VM, Bredas JL. Int. J. Quantum Chem. 2003;91:303–310. [Google Scholar]
- (37).Blomgren F, Larsson S. J. Comput. Chem. 2005;26:738–742. doi: 10.1002/jcc.20210. [DOI] [PubMed] [Google Scholar]
- (38).The main characters of S1 and S2 states are inverted at CASSCF(6/6) level artificially. Our post-CASSCF(6/6) calculations repair such problems in using small active spaces.
- (39).Andruniow T, Ferre N, Olivucci M. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17908–17913. doi: 10.1073/pnas.0407997101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Lewis JW, Szundi I, Fu W-Y, Sakmar TP, Kliger DS. Biochemistry. 2000;39:599–606. doi: 10.1021/bi991860z. [DOI] [PubMed] [Google Scholar]
- (41).Ferre N, Olivucci M. J. Am. Chem. Soc. 2003;125:6868–6869. doi: 10.1021/ja035087d. [DOI] [PubMed] [Google Scholar]
- (42).Hufen J, Sugihara M, Buss V. J. Phys. Chem. B. 2004;108:20419–20426. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.