Abstract
Motivation
Many experimental and algorithmic approaches in biology generate groups of genes that need to be examined for related functional properties. For example, gene expression profiles are frequently organized into clusters of genes that may share functional properties. We evaluate a method, neighbor divergence per gene (NDPG), that uses scientific literature to assess whether a group of genes are functionally related. The method requires only a corpus of documents and an index connecting the documents to genes.
Results
We evaluate NDPG on 2796 functional groups generated by the Gene Ontology consortium in four organisms: mouse, fly, worm and yeast. NDPG finds functional coherence in 96, 92, 82 and 45% of the groups (at 99.9% specificity) in yeast, mouse, fly and worm respectively.
INTRODUCTION
The increasing application of genome scale approaches to biology is shifting the focus of data analysis from individual genes to systems of genes participating in a common biological process. Many experimental protocols result in the definition of gene groups. For example, gene expression data can be used to cluster genes into groups (Eisen et al., 1998) and protein or amino acid sequences can be used to find other related sequences (Altschul et al., 1990, 1997). Rapidly recognizing whether a set of genes share a common function is useful for assessing the significance of experimentally derived gene sets and prioritizing those sets that deserve follow-up.
We have developed a novel computational method, neighbor divergence per gene (NDPG), that assesses whether a set of genes share a common biological function by automatic analysis of scientific text. The scientific literature often contains the relevant information to assess whether a group of genes share function. The published literature is accessible in electronic form—often as full text, and almost always in abstract form (http://www.ncbi.nlm.nih.gov/PubMed/). Our method uses statistical natural language processing (NLP) methods (Manning and Schütze, 1999; Rosenfeld, 2000) to mine the literature and assign a functional coherence score to the group of genes.
Our method can be used to do a literature-based evaluation of gene groups produced by analytical algorithms or experimental protocols. For example, NDPG scoring can be used to detect gene expression clusters that are functionally relevant. NDPG performs a literature-based assessment of whether the genes in each cluster have related functional properties.
NDPG requires only a corpus of articles relevant to all of the genes being studied (e.g. all genes within an organism) and an index associating the articles to appropriate genes. Such reference lists are often available from sequence databases such as SWISS-PROT (Bairoch and Apweiler, 1999); genomic databases such as SGD (Cherry et al., 1998), MGD (Blake et al., 2002), FlyBase (Gelbart et al., 1997), and WormBase (Stein et al., 2001); or can be compiled automatically by scanning titles and abstracts of articles for gene names (Jenssen et al., 2001). In a small-scale, preliminary evaluation conducted on 19 yeast functional groups NDPG achieved 95% sensitivity at 100% specificity, whereas a naïve method based on consistent article word usage in gene references achieved only 10.5% sensitivity at 98.9% specificity (Raychaudhuri et al., 2002b; Raychaudhuri, Schütze and Altman, 2003).
Here we conduct a large-scale evaluation of the NDPG method in four organisms across many different functions. Gene ontology (GO) is a large hierarchical vocabulary devised to describe genetic function in a standard way across many types of organisms (Ashburner et al., 2000). It has three major branches: biological process, molecular function, and cellular compartment. One of the purposes of the development of this vocabulary is for effective gene annotation in a standard way. Its hierarchical nature allows specific annotations (for example triosephosphate isomerase) to be generalized to other terms up the hierarchy (such as isomerase or enzyme).
To evaluate our method we assembled functional groups of genes from four species: Saccharomyces cerevisiae (yeast), Mus muscularis (mouse), Drosophila melanogaster (fly), and Caenorhabditas elegans (worm). Reference indices were assembled for each species by collecting gene references from the same online resources. We used the GO annotations on genes to generate functional groups. Each GO term generates a species-specific gene group consisting of genes assigned that term. The assignments are either explicitly indicated, or are inferred from assignments to more specific terms. Each group with six or more genes was scored with our method. If the NDPG method is successful these functional groups of genes should receive very high scores. Random groups of genes were assembled also. Ideally we expect that these groups of genes will receive low scores.
METHODS
We applied the NDPG method to four different organisms. For application of this method to each of the four different organisms, a different set of genes and articles referring to them were assumed.
Evaluation
Obtaining gene ontology code assignments
For each of the four organisms, Gene Ontology assignments were obtained from the genome databases for each organism (SGD for yeast (Cherry et al., 1998), MGD for mouse (Blake et al., 2002), FlyBase for fly (Gelbart et al., 1997), and WormBase for worm (Stein et al., 2001)) between 25th March and 5th April 2002. We assigned the listed GO codes by the databases to the genes (explicit annotations). Also, we assigned all parent codes in the ontology of listed codes to the gene also (inferred annotations). All ontology files were downloaded from the GO consortium web site (http://www.geneontology.org) on 25th March. For each species, a gene ontology term that was assigned to six or more genes defined a functional group. There were a total of 2796 such functional groups across all four species.
Obtaining gene references indices and articles
A gene reference list was obtained from each of the genomic databases. A reference list is a table that connects PubMed abstracts to genes in the species; each line in the table has a PubMed ID and an appropriate gene identifier. The reference lists were either obtained from the database website directly (SGD), or obtained after a database query made on the web site (WormBase), or with assistance from the site curators (MGI and FlyBase). The reference lists were obtained between 25 March and 8 April, and were used to make the reference indices for the NDPG method. Only genes that had at least one article reference were considered; others were altogether eliminated from analysis. All relevant articles were obtained from PubMed.
Evaluating NDPG
For each organism 200 random groups were devised of each size: 6, 12, 24, 48 and 96 genes, for a total of 1000 random groups per organism. The NDPG method was used to score all 1000 random groups; the 99.9th percentile of scores for each organism was identified as a cutoff.
The functional groups derived from GO were scored with NDPG. The percentage of functional groups within the organism scoring above the cutoff was tabulated. The percentage of groups above the cutoff is the sensitivity of our method at 99.9% specificity. The median score and the percentage of scores exceeding the cutoff were computed for each GO branch in each organism.
Comparing annotation quality to NDPG performance
For each branch of GO and each organism, we calculated the percent of explicit and inferred annotations that could be attributed to either the TAS (‘traceable author statement’) or IDA (‘inferred from direct assay’) annotation. These annotations are generally regarded as reliable, high quality annotations. The resulting percentages were plotted against the percent of functional groups that NDPG was able to successfully identify in that GO branch and organism.
Neighbor divergence per gene (NDPG) method
The NDPG method used here will be described in detail elsewhere, but can be summarized here (Raychaudhuri, Schütze, and Altman, 2003).
Data types: document corpus and reference index
NDPG calculation of a gene group requires a corpus of documents relevant to all genes in the organism, and a reference index indicating the articles that are germane to each gene. Here, the documents are the title and abstract fields of PubMed records.
Identifying semantic neighbors for corpus articles
For each document, the 19 most similar documents (not including the document itself) are pre-computed. To quantify the similarity between two documents, we calculate the cosine of the angle between the inverse document frequency weighted word vectors of the documents (Manning and Schütze, 1999). In the selection of the 19 most similar documents for each document, those documents are excluded that refer only to genes contained in the set of genes referred to in the seed document.
Scoring article relative to gene groups
Given a gene group, NDPG then assigns a score to each document. The score is the count of semantic neighbors that refer to group genes.
A theoretical distribution of scores
If the gene group has no coherent functional structure, the semantic neighbors of any given document should refer to group genes independently with a probability q. If q is small, a Poisson distribution estimates the distribution of scores. In this case:
where λ = 19 * q, S is the document score, and n ranges from 0 to 19. For a given gene group we estimate q, the fraction of documents referring to group genes, by counting documents referring to the gene group and dividing by the number of documents.
Quantifying the difference between the empirical score distribution for a gene and the theoretical one
For each gene in the group, an empirical distribution of the scores of documents referring to it is computed. If the group contains no functional coherence, all of the distributions of scores should be similar to the Poisson distribution. The difference between the empirical distribution of scores for each group gene and the theoretical Poisson distribution is quantified with the KL-divergence (Manning and Schütze, 1999). Given two distributions, a theoretical one, h, and an observed one, g, we calculate KL-divergence:
If two distributions are the same, the divergence is zero; the more disparate the two distributions the larger the divergence.
Functional coherence score of a group of genes
The functional coherence score assigned to a gene subgroup is the average KL-divergence for all genes in the subgroup.
RESULTS
Table 1 contains descriptive statistics about the literature index for each organism and the GO annotations also. Mouse has the most number of genes with references, while worm has the fewest. For the total number of references per article and references per gene, the mean exceeds the median in all organisms. For each organism there are a few outlier genes with many article references, and there are a few outlier articles with many gene references.
Table 1.
Summary of primary data for four organisms For each of the four organisms we have listed summary statistics for the reference lists obtained from the genome databases and GO annotations.
| Yeast | Mouse | Fly | Worm | ||
|---|---|---|---|---|---|
| Summary of reference lists obtained from the genome centersa | |||||
| Genes with references | 5 151 | 26 148 | 14 732 | 2 289 | |
| Article | 22 934 | 41 669 | 15 495 | 2 144 | |
| References | 62 473 | 113 738 | 97 117 | 13 659 | |
| References/article | Median | 2 | 1 | 3 | 4 |
| Mean | 2.73 | 2.73 | 6.27 | 6.37 | |
| References/gene | Median | 4 | 1 | 1 | 2 |
| Mean | 12.12 | 4.35 | 6.59 | 5.97 | |
| Summary of gene ontology annotations obtained from the GO consortiumb | |||||
| Genes with codes assigned | 4 276 | 6 148 | 4 042 | 523 | |
| GO codes | Process | 874 | 904 | 1 019 | 196 |
| Component | 251 | 233 | 347 | 42 | |
| Function | 1 132 | 1 432 | 1 458 | 246 | |
| Total | 2 257 | 2 569 | 2 824 | 484 | |
| Explicit GO annotations | 13 770 | 27 122 | 14 405 | 2 235 | |
| Inferred GO annotations | 49 781 | 68 075 | 47 801 | 5 017 | |
| Ratio explict/implicit | 3.62 | 2.51 | 3.32 | 2.24 | |
| Annotations/gene | Median | 14 | 15 | 14 | 13 |
| Mean | 14.86 | 15.48 | 15.39 | 13.87 | |
| Annotations/code | Median | 3 | 3 | 2 | 3 |
| Mean | 28.15 | 37.06 | 22.02 | 14.98 | |
References are the total number of times a gene is referred to by a document.
Most organisms use only a few of the 4773 process, 977 functions, and 5015 function GO codes. Explicit GO annotations are ones assigned by curators while inferred annotations are more general annotations that are parents in the ontology to the explicit codes. The ratio indicates the average number of inferred parents terms generated by each explicit term annotation
All of the different organisms used about the same number of GO codes for annotation, about 2500 codes, except worm which used fewer, about 500 codes. For each term assigned explicitly to a gene as an annotation, many more annotations that are parents of the term are also implied. The more specific the term, the more inferred terms apply to the same gene. In general the yeast and fly annotations are very specific, and the ratio of inferred annotations to explicit annotations is large (3.6 and 3.3) compared to mouse and worm, where the annotations are more general (2.5 and 2.2).
Table 2 contains the results of the analysis on the GO functional groups. A separate cutoff was generated for each organism; the cutoffs are listed in the table. The cutoff was selected as a 99.9% specificity threshold. Alternately, the cutoff can be thought of as the score above which there is a probability less than 0.001 that the group of genes is random. The results are presented separately for each of the three branches of GO. For all GO branches yeast has the greatest percentage of groups exceeding the cutoff, followed by mouse, then fly, then finally worm. When all worm functional groups are combined, the median group score of worm functional groups is less than the cutoff.
Table 2.
Sensitivity of NDPG in four organisms. For each branch of GO in each organism, the functional coherence of each of the annotations with six or more genes is computed. The total number of such groups is listed, followed by the median number of genes in the group. Also listed is the median NDPG functional coherence score for the annotation, and the percent of groups exceeding the 99.9% specificity cutoff. The cutoff score is listed at the bottom of the table for each organism. Also in the table, is the overall performance across all branches of the GO ontology for each organism
| Yeast | Mouse | Fly | Worm | ||
|---|---|---|---|---|---|
| Process GO codes | Number of groups | 429 | 354 | 349 | 71 |
| Median group size | 21 | 20 | 16 | 17 | |
| Median NDPG score | 15.32 | 10.20 | 5.21 | 1.42 | |
| % of groups exceeding cutoff | 97.44% | 87.85% | 86.82% | 46.48% | |
| Component GO codes | Number of groups | 148 | 111 | 151 | 18 |
| Median group size | 20 | 18 | 16 | 16 | |
| Median NDPG score | 18.63 | 11.73 | 5.59 | 2.40 | |
| % of groups exceeding cutoff | 94.59% | 90.99% | 81.46% | 77.78% | |
| Function GO codes | Number of groups | 264 | 435 | 382 | 84 |
| Median group size | 17 | 16 | 17 | 15 | |
| Median NDPG score | 11.35 | 13.39 | 3.58 | 1.53 | |
| % of groups exceeding cutoff | 93.56% | 96.09% | 78.27% | 36.90% | |
| All GO codes | Number of groups | 841 | 900 | 882 | 173 |
| Median group size | 20 | 18 | 16 | 16 | |
| Median NDPG score | 15.11 | 11.84 | 4.47 | 1.58 | |
| % of groups exceeding cutoff | 95.72% | 92.22% | 82.20% | 45.09% | |
| 99.9% specificity cutoff | 3.43 | 3.19 | 1.34 | 1.63 |
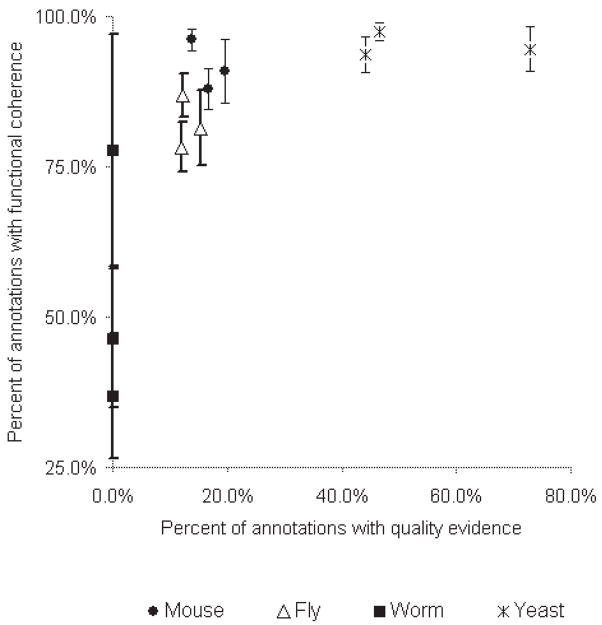
In Figure 1 we have plotted the percentage of annotations attributable to the most reliable evidence codes versus the percentage of functional groups in each GO branch of each organism exceeding the cutoff. In general, annotated sets where many of the annotations were derived from high quality evidence had a greater percentage of annotated groups exceeding the cutoff.
Fig. 1.
Coherence of an annotated group corresponds to evidence quality in yeast. Functional annotations of genes were separated by organism and GO branch into twelve sets. Along the y-axis, the percentage of functional groups with six or more genes in that set exceeding the cutoff is plotted; also indicated in the plot is a 95% confidence interval for that percentage. Along the x-axis the percent of annotations in the set for which the attributed evidence codes are either TAS or IDA is plotted.
DISCUSSION
The challenge of designing a literature-based method that assesses the functional coherence of a gene group is in compensating for the great disparities in biological literature. Some functions are thoroughly represented in the literature while others may not be; some genes are heavily studied while others are newly discovered.
The NDPG method is effective at identifying groups of genes that are functionally coherent in multiple organisms. In Table 2 we have listed the sensitivity of the NDPG method for four different organisms. The method is best able to identify functionally coherent groups in yeast, and performs poorest in worm. The method achieves 96, 92, 82 and 45% sensitivity at 99.9% specificity in yeast, mouse, fly and worm respectively.
The variable performance in the four different organisms can be accounted for by different factors. The first is the quality of the references in the reference indices; in a good reference index, genes should be connected to the appropriate articles. This is difficult to objectively assess. It is possible that different literature indices have more or less appropriate references depending on the articles available and the level of care placed in putting the literature resource together.
A second issue is the abundance of available articles in the reference index. Yeast has the strongest performance; it has over 20 000 articles and a 4:1 ratio of articles to genes. Worm on the other hand has the smallest corpus with one-tenth the number of articles that the yeast reference index has and a ratio less than 1:1 of articles to genes; our method is less than half as sensitive for worm functional groups.
An additional contributing factor may be the quality of the GO annotations themselves. Gene Ontology is a massive effort, and remains a work in progress. Since this is the case, it is perhaps not an ideal gold standard yet. Currently annotation of genes with GO codes remains an active area with many groups experimenting with different strategies involving manual and computational analysis of literature, sequence, and experimental data (Hill et al., 2001; Hvidsten et al., 2001; Dwight et al., 2002; Raychaudhuri et al., 2002a; Schug et al., 2002; Xie et al., 2002). The online resources for the different organisms rely more heavily on different strategies of annotation. The strategy used to make a specific annotation is listed as an ‘evidence code’. We considered IDA (‘inferred from direct assay’) and TAS (‘traceable author statement’) as the two highest quality and most reliable evidence codes. We determined the percentage of inferred and explicit annotations that could be attributed to each of these two evidence codes in the three GO branches of the four organisms. In Figure 1 it is evident that there is a relationship between the percent of high quality annotations and the performance of our methods. The percent of high quality annotations is an indication of the amount of manual effort involved in that organism’s GO annotation. We reason that the more effort, the better the quality of the annotation, and the more reliable of a gold standard it is, and consequently the better our performance.
A functional group may not be identified as functionally coherent if the shared function is not represented in the corpus of scientific literature. This may be the case if the function has not yet been described in the literature, or if the function has not been well studied in that organism. For example the tricarboxylic acid cycle (TCA) functional group in yeast receives a significant score of 15.43, whereas in mouse the same functional group receives an insignificant score of 1.97. The subject of TCA genetics has not been well described in the mouse literature. A Medline query for articles assigned the MeSH subject headings of ‘tricarboxylic acid cycle’ and ‘genetics’ yielded 365 articles on 5 July 2002. Only 13 of these articles had the ‘mouse’ MeSH heading also, and none of those 13 references were listed in the mouse reference index. In contrast, 52 had the ‘yeast’ MeSH heading and of those 32 were listed in the yeast reference index. The TCA GO annotations in mouse were made without direct reference to the literature. Eight of the nine genes that were assigned the TCA function because of the presence of an appropriate keyword in the SWISS-PROT sequence entry for the gene; these annotations were assigned the evidence code IEA (‘inferred from electronic annotation’). The other gene was assigned the TCA function by sequence similarity search; this annotation was assigned the evidence code ‘ISS’ (‘inferred from sequence similarity’). Since NDPG is a literature-based approach, this functional group is missed altogether. This issue might be mitigated if references to homologous genes in other organisms were included as additional references to genes. Undiscovered functions would still not be discernible.
In this implementation of the algorithm we used 19 semantic neighbors, although we have informally tested the method with 199 semantic neighbors. Our preliminary analysis revealed that the performance is not very sensitive to the number of neighbors used. Here we used a smaller number of neighbors since the reference index of one for one of the organisms, worm, had few articles.
Since the goal of many bioinformatics analyses is to define groups of genes that are in some way similar, NDPG provides a means to incorporate a literature-based component into these analyses. For example, we have also developed an algorithm that uses scientific literature to direct a search for groups of genes that share function and gene expression properties. This algorithm finds a plane in gene expression data that separates out a group of genes from the remainder that has a high NDPG score (Raychaudhuri, Schütze and Altman, 2003). NDPG is used to assess whether the separated genes share a common function. Another practical application of NDPG to bring the content of literature to bear on a bioinformatics problem might include using literature in sequence-based homology modeling or motif finding (MacCallum et al., 2000; Chang et al., 2001). These results suggest the potential for NDPG in the same sorts of analyses across different species.
Acknowledgments
RBA is supported by NIH LM06244, GM61374, NSF DBI-9600637, and a grant from the Burroughs–Wellcome Foundation; SR is supported by NIH GM-07365. The authors also thank SGD, MGI, Flybase, and Wormbase for making gene reference lists available to us.
Footnotes
Availability: Contact authors.
Contact: russ.altman@stanford.edu
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1999. Nucleic Acids Res. 1999;27:49–54. doi: 10.1093/nar/27.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA, Richardson JE, Bult CJ, Kadin JA, Eppig JT. The Mouse Genome Database (MGD): the model organism database for the laboratory mouse. Nucleic Acids Res. 2002;30:113–115. doi: 10.1093/nar/30.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Raychaudhuri S, Altman RB. Including biological literature improves homology search. Pac Symp Biocomput. 2001;14:374–383. doi: 10.1142/9789814447362_0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, et al. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwight SS, Harris MA, Dolinski K, Ball CA, Binkley G, Christie KR, Fisk DG, Issel-Tarver L, Schroeder M, Sherlock G, et al. Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO) Nucleic Acids Res. 2002;30:69–72. doi: 10.1093/nar/30.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FlyBase. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 2002;30:106–108. doi: 10.1093/nar/30.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart WM, Crosby M, Matthews B, Rindone WP, Chillemi J, Russo Twombly S, Emmert D, Ashburner M, Drysdale RA, Whitfield E, et al. FlyBase: a Drosophila database. The FlyBase consortium. Nucleic Acids Res. 1997;25:63–66. doi: 10.1093/nar/25.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DP, Davis AP, Richardson JE, Corradi JP, Ringwald M, Eppig JT, Blake JA. Program description: strategies for biological annotation of mammalian systems: implementing gene ontologies in mouse genome informatics. Genomics. 2001;74:121–128. doi: 10.1006/geno.2001.6513. [DOI] [PubMed] [Google Scholar]
- Hvidsten TR, Komorowski J, Sandvik AK, Laegreid A. Predicting gene function from gene expressions and ontologies. Pac Symp Biocomput. 2001:299–310. doi: 10.1142/9789814447362_0030. [DOI] [PubMed] [Google Scholar]
- Jenssen TK, Laegreid A, Komorowski J, Hovig E. A literature network of human genes for high-throughput analysis of gene expression. Nat Genet. 2001;28:21–28. doi: 10.1038/ng0501-21. [DOI] [PubMed] [Google Scholar]
- MacCallum RM, Kelley LA, Sternberg MJ. SAWTED: structure assignment with text description–enhanced detection of remote homologues with automated SWISS-PROT annotation comparisons. Bioinformatics. 2000;16:125–129. doi: 10.1093/bioinformatics/16.2.125. [DOI] [PubMed] [Google Scholar]
- Manning CM, Schütze H. Foundations of Statistical Natural Language Processing. MIT Press; Cambridge, MA: 1999. [Google Scholar]
- Raychaudhuri S, Chang JT, Sutphin PD, Altman RB. Associating genes with gene ontology codes using a maximum entropy analysis of biomedical literature. Genome Res. 2002a;12:203–214. doi: 10.1101/gr.199701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Schütze H, Altman RB. Using text analysis to identify functionally coherent gene groups. Genome Res. 2002b;12:1582–1590. doi: 10.1101/gr.116402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Schütze H, Altman RB. Inclusion of textual documents in the analysis of multi-dimensional data sets: application to gene expression data. Machine Learning. 2003 in press. [Google Scholar]
- Rosenfeld R. Two decades of statistical language modeling: where do we go form here? Proc IEEE. 2000;88:1270–1278. [Google Scholar]
- Schug J, Diskin S, Mazzarelli J, Brunk BP, Stoeckert CJ., Jr Predicting gene ontology functions from ProDom and CDD protein domains. Genome Res. 2002;12:648–655. doi: 10.1101/gr.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L, Sternberg P, Durbin R, Thierry-Mieg J, Spieth J. WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res. 2001;29:82–86. doi: 10.1093/nar/29.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Wasserman A, Levine Z, Novik A, Grebinskiy V, Shoshan A, Mintz L. Large-scale protein annotation through gene ontology. Genome Res. 2002;12:785–794. doi: 10.1101/gr.86902. [DOI] [PMC free article] [PubMed] [Google Scholar]