Abstract
Ska1 and Ska2 form a complex at the kinetochore–microtubule (KT–MT) interface and are required for timely progression from metaphase to anaphase. Here, we use mass spectrometry to search for additional components of the Ska complex. We identify C13Orf3 (now termed Ska3) as a novel member of this complex and map the interaction domains among the three known components. Ska3 displays similar characteristics as Ska1 and Ska2: it localizes to the spindle and KT throughout mitosis and its depletion markedly delays anaphase transition. Interestingly, a more complete removal of the Ska complex by concomitant depletion of Ska1 and Ska3 results in a chromosome congression failure followed by cell death. This severe phenotype reflects a destabilization of KT–MT interactions, as demonstrated by reduced cold stability of KT fibres. Yet, the depletion of the Ska complex only marginally impairs KT localization of the KMN network responsible for MT attachment. We propose that the Ska complex functionally complements the KMN, providing an additional layer of stability to KT–MT attachment and possibly signalling completion of attachment to the spindle checkpoint.
Keywords: chromosome congression, kinetochore, microtubule-attachment, mitosis
Introduction
The mitotic spindle, formed at prophase by microtubules (MTs) originating from two spindle poles, generates the forces required for both chromosome congression to a metaphase plate, and the subsequent sister chromatid separation during anaphase. Essential to its function is the spindles' ability to attach to kinetochores (KTs), proteinaceous structures that assemble at the centromere of each sister chromatid (Cleveland et al, 2003; Maiato et al, 2004). Erroneous KT–MT attachment, with either both sister KTs attached to the same pole (syntelic attachment), or both poles attached to the same KT (merotelic attachment), can result in inaccurate segregation of the sister chromatids and subsequent aneuploidy (Pinsky and Biggins, 2005). To prevent aneuploidy, the cell generates a ‘wait-anaphase' signal known as the spindle assembly checkpoint (SAC) that inhibits anaphase onset until all KTs achieve biorientation, and tension is established between sister KTs (Musacchio and Salmon, 2007).
Electron microscopy has revealed KT as a trilaminar structure: the inner KT, situated most proximal to the centromeric chromatin, the outer KT that provides the binding surface for spindle MTs, and a central linker region (Cheeseman and Desai, 2008). In addition, unattached KTs also show a fibrous corona, a dense array of fibres extending from the outer KT plate (McEwen et al, 1998). Binding of MTs to KTs is highly dynamic, and multiple protein complexes at the KT have been implicated in the process (Cheeseman and Desai, 2008). Of particular importance is the so-called KMN network (named after its key components KNL-1, Mis12 and Ndc80) that comprises three functionally linked complexes (Cheeseman et al, 2004, 2006; Obuse et al, 2004; Kline et al, 2006; Kiyomitsu et al, 2007; Ciferri et al, 2008). Individually, both Ndc80 and KNL-1 bind weakly to MTs, but the interaction of the Ndc80 complex is enhanced synergistically in the presence of the other KMN complex proteins (Cheeseman et al, 2006). Removal of any of the components of the KMN network disrupts both the structure of the outer KT plate and its ability to bind to MTs (DeLuca et al, 2005; Kline et al, 2006).
Although the KMN is thought to provide the core binding sites for MTs at the KT, several other protein complexes also function at this critical interphase. These include the minus-end directed motor protein complex dynein/dynactin and proteins involved in its KT recruitment such as the RZZ complex (Karess, 2005; Yang et al, 2007), Spindly (Griffis et al, 2007; Yamamoto et al, 2008) and NudE and NudEL (Stehman et al, 2007). Moreover, the plus-end directed motor protein Cenp-E and its binding partner BubR1 are required for the maintenance of the KT–MT interaction (Lampson and Kapoor, 2005; Elowe et al, 2007; Matsumura et al, 2007). Finally, MT plus-end binding proteins (+Tips) such as EB1, Clip170 and Clasp (Maiato et al, 2004), and MT-bundling proteins like RanGAP1 (Joseph et al, 2004) have also been implicated in KT–MT binding and stabilization.
Recently, we described the novel spindle KT-associated (Ska) complex, of which we identified two members, Ska1 and Ska2 (Hanisch et al, 2006). Ska1 was originally found in a proteomic survey of the human mitotic spindle (Sauer et al, 2005), whereas Ska2 was identified as a Ska1 binding partner in a yeast two-hybrid screen (Hanisch et al, 2006). In accordance with its identification as a spindle component, Ska1 was concentrated at the outer KT in a MT-dependent manner during prometaphase, where it remained until anaphase. Moreover, this localization was lost upon disruption of the outer KT plate by depletion of the Ndc80 component Hec1 (Hanisch et al, 2006). Interestingly, siRNA-mediated depletion of either Ska1 or Ska2 induced a prolonged metaphase arrest that could be overridden by the co-depletion of the SAC component Mad2 (Hanisch et al, 2006). Eventually, however, cells overcame the metaphase arrest, progressed through a normal anaphase and exited mitosis.
Here, we describe a third member of the Ska complex, C13Orf3 (now termed Ska3), which we identified as a novel binding partner of Ska2 by mass spectrometry. Biochemical and immunocytochemical data confirm that Ska3 represents a bona fide member of the Ska complex, and its depletion induces a metaphase delay similar to that described for Ska1 and Ska2. Intriguingly, we show that a more stringent depletion of the entire Ska complex results in a severe chromosome congression defect that leads to cell death. In accordance with this phenotype, KT–MT attachment was severely impaired in Ska-depleted cells, although, remarkably, the KMN network was barely affected. Thus, we propose that the Ska complex is an essential component of the KT–MT interface, functioning in cooperation to the KMN network and possibly signalling the completion of successful attachment.
Results and discussion
Identification of C13Orf3 as a new component of the Ska complex
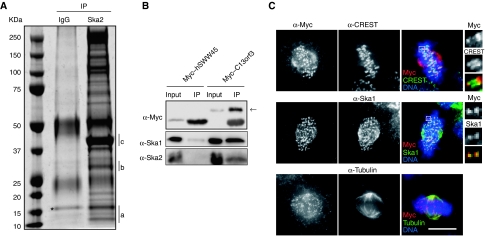
To search for binding partners of the Ska complex (Hanisch et al, 2006), HeLa S3 cells were synchronized in mitosis and Ska2 was immunoprecipitated. Analysis of the immunoprecipitates by mass spectrometry (Figure 1A; Supplementary Figure 1 available online) revealed not only Ska2 and Ska1, but also the earlier uncharacterized protein C13Orf3. Neither of these proteins could be detected in control immunoprecipitates produced by rabbit IgG. To confirm the interaction between C13Orf3 and the Ska complex, Myc-tagged C13Orf3 was overexpressed and immunoprecipitated from HeLa S3 (Figure 1B) and HEK293T cells (data not shown). For control, parallel immunoprecipitations were carried out with the unrelated protein Myc–hSWW45. Out of both cell lines, endogenous Ska1 and Ska2 could readily be co-immunoprecipitated with Myc–C13Orf3, but not with Myc–hSWW45, corroborating the mass spectrometry data. From this result and from the data shown below, we conclude that C13Orf3 is a bona fide member of the Ska complex and we hereafter refer to it as Ska3.
Figure 1.
Identification of C13Orf3 as a new member of the Ska complex. (A) HeLa S3 cells were collected by shake-off in M-phase after treatment with nocodazole followed by 40 min release. Lysates were pre-cleared with rabbit IgG coupled to sepharose A-beads and Ska2 was immunoprecipitated using affinity-purified anti-Ska2 antibody coupled to sepharose A-beads. Proteins were identified by MS analysis. Bands containing Ska2, Ska1 and C13Orf3 are indicated by a, b and c, respectively. Asterisk indicates nonspecific interacting proteins. (B) Myc–hSWW45 (for control) or Myc–C13Orf3 were expressed for 48 h in cells that were subsequently exposed to monastrol for 16 h before immunoprecipitations were performed using 9E10 anti-Myc antibody. All samples were then probed by western blotting with anti-Myc, anti-Ska1 and anti-Ska2 antibodies. Arrow indicates Myc–C13Orf3. Inputs represent 10% of the immunoprecipitated protein. (C) Cells were transfected with a Myc-tagged C13Orf3 construct and then fixed with PTEMF. Cells were stained with 9E10 anti-Myc antibody (red) and with CREST (upper panels), anti-Ska1 (middle panels) or anti-α-Tubulin (lower panels) (green). DNA was visualized using DAPI (blue). Bar=10 μm.
Ska3 has a predicted molecular weight of 45 kDa but, as shown below, migrates at about 55 kDa (Figure 2B; Supplementary Figure 4B), presumably due to its acidic isoelectric point (pI=5). The protein is conserved amongst vertebrates but cannot be identified with confidence below bony fish. According to secondary structure prediction, Ska3 is expected to display a globular N-terminal domain followed by a long unstructured C-terminal region. Furthermore, the Jpred 3 algorithm (Cole et al, 2008) predicts a motif with remote resemblance to a Gle2-binding sequence (GLEBS domain) within the N terminus of Ska3 (aa 17–51, Supplementary Figure 2A), and a second potential GLEBS domain may exist in the C-terminal region (aa 345–382, F Buchholz, personal communication). GLEBS domains, as identified in scMad3, scBub1, hsBub1, hsBubR1 and hsNup98 (Pritchard et al, 1999; Wang et al, 2001; Larsen et al, 2007), are thought to provide interfaces for interactions with β-propeller regions on partner proteins (Larsen et al, 2007).
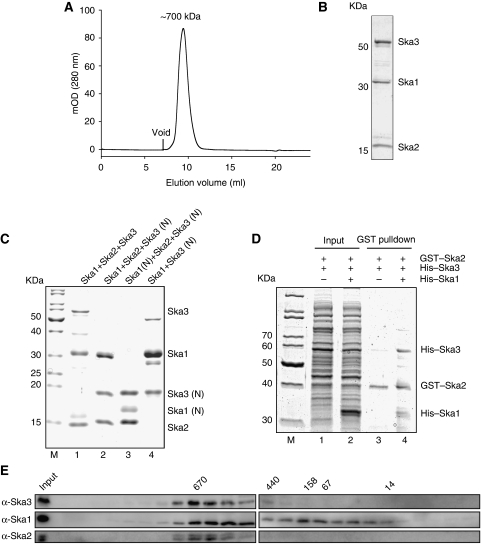
Figure 2.
Characterization of Ska complex formation. (A) Gel filtration was carried out on purified full-length ternary complex reconstituted by co-lysis, where His–Ska1 and GST–Ska2 were co-expressed and His–Ska3 was expressed separately. The predicted MW of the peak fraction as based from the migration in the gel filtration is indicated. (B) Representative Coomassie-stained SDS–PAGE gel from the peak fraction of the Ska ternary complex as determined in (A). (C) Representative fractions of purified full-length ternary complex reconstituted by co-lysis (lane 1), where GST–Ska1 and untagged Ska2 were co-expressed and His–Ska3 was expressed separately. (Lane 2) Ternary complex with C-terminally truncated Ska3 (Ska3 (N)); and (lane 3) ternary complex with C-terminally truncated Ska1 (Ska1 (N)) and Ska3 (N)). In the case of lanes 2 and 3, all three Ska components were co-expressed and were tagged as follows: His–Ska1 (lane 2) or His–Ska1 (N) (lane 3), His–Ska3 (N) and GST–Ska2 (lanes 2 and 3). (Lane 4) Ska1–Ska3 (N) subcomplex between GST–Ska1 and His–Ska3 (N). In all cases, affinity tags are cleaved after GST-affinity chromatography. More details on purification are given in Materials and methods. (D) GST pulldown of Ska2 on cell lysate, where GST–Ska2 and His–Ska3 are co-expressed in the presence or absence of His–Ska1. (Lanes 1 and 2) Input for Ska3 and Ska2 and ternary complex (Ska3, Ska2 and Ska1), respectively. (Lane 3) GST–Ska2 pulldown in the presence of His–Ska3 but absence of His–Ska1. (Lane 4) GST–Ska2 pulldown from the ternary complex. (E) Western blot from SDS–PAGE gel of mitotic HeLa S3 cell lysates fractionated by gel filtration. Membranes were probed with anti-Ska3, anti-Ska1 and anti-Ska2 antibodies. The migrations of MW markers are indicated above the blot (kDa).
The localization of Myc-tagged Ska3 in HeLa S3 cells was analyzed by indirect immunofluoresence microscopy (Figure 1C; Supplementary Figure 3). During interphase, Myc–Ska3 displayed a diffuse cytoplasmic distribution without obvious association with any definable structure (Supplementary Figure 3A). However, beginning in prometaphase, Myc–Ska3 localized to KTs, where it remained throughout metaphase and anaphase, before it associated with the midbody during late telophase (Supplementary Figure 3B). A more detailed examination of the KT association of Ska3 revealed the corresponding signal to lie outside of the centromeric CREST signal (Figure 1C, upper panels), indicating that the KT-bound pool of Ska3 is situated at the outer plate of the KT, similar to the localizations described for Ska1 and Ska2 (Hanisch et al, 2006). A proportion of Ska3 was also seen to associate with the spindle, particularly in the vicinity of the poles during metaphase (Figure 1C, bottom panels). Co-staining of cells for Myc–Ska3 and Ska1 revealed that the two proteins co-localized throughout all stages of mitosis (Figure 1C, middle panels; Supplementary Figure 3B).
Next, we raised a rabbit polyclonal antibody against Ska3. When used for immunofluorescence microscopy, the anti-Ska3 serum produced a specific spindle and KT staining (Supplementary Figure 4A), indistinguishable from that seen with overexpressed Myc-tagged Ska3. By western blotting, the anti-Ska3 antibody detected a protein at a molecular weight of approximately 55 kDa in asynchronously growing cells (Supplementary Figure 4B). In addition, a band of even lower electrophoretic mobility was seen in mitotically arrested cells, suggesting that Ska3 is modified during mitosis (Figure 3A; Supplementary Figure 4B–D). Importantly, both immunoreactive bands were sensitive to Ska3 siRNA (Figure 3A; Supplementary Figure 4B), attesting to their specificity.
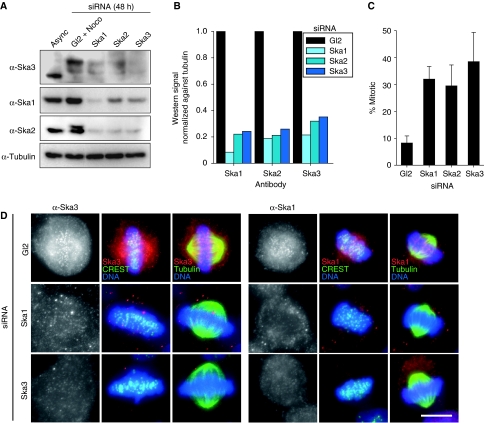
Figure 3.
Interdependence between the Ska complex. (A) HeLa S3 cells were treated for 48 h with control (Gl2) and Ska1, Ska2 and Ska3-specific siRNAs, respectively. Mitotic cells from Gl2-treated cells arrested with nocodazole or Ska1, Ska2 and Ska3-depleted cells were collected by shake-off. Equal amounts of cell extracts were separated by SDS–PAGE and probed by western blotting with anti-Ska3, anti-Ska1, anti-Ska2 and anti-α-Tubulin antibodies. (B) Bar graph of corresponding intensity profile for western signal of (A), normalized against signal for anti-α-Tubulin. (C) Asynchronously growing cells were treated for 48 h with control (Gl2) and Ska1, Ska2 and Ska3-specific siRNAs, respectively. The mitotic indices were determined by light microscopy. Histograms show averages from four independent experiments. Error bars represent standard deviation (s.d.). (D) Cells were treated for 48 h with control (Gl2), Ska1- and Ska3-specific siRNAs, respectively, then fixed with PTEMF and stained with anti-Ska3 (left panels) or anti-Ska1 (right panels) antibodies (red), CREST serum (far red, depicted in green) or anti-α-Tubulin (green). DNA was visualized using DAPI (blue). Bar=10 μm.
To confirm the ability of Ska proteins to directly interact with each other, the three Ska components were expressed in Escherichia coli and used for in vitro reconstitution of a ternary complex (Figure 2A–D). Gel filtration revealed a single homogeneous complex, containing all three Ska components that behaved like a 700-kDa globular protein (Figure 2A and B). At present, we have no definitive information on the exact stoichiometry of the complex. Given that the combined molecular weight of Ska1 (33 kDa), Ska2 (15 kDa) and Ska3 (45 kDa) is <100 kDa, the complex possibly comprises multiple copies of each protein and/or possesses an elongated structure. Interaction mapping using limited proteolysis revealed that the N-terminal regions of Ska3 (aa 1–156) and Ska1 (aa 1–130) were sufficient for stable complex formation (Figure 2C, lanes 2 and 3; Supplementary Figure 2A). Furthermore, Ska3 and Ska1 formed a binary complex in the absence of Ska2 (Figure 2C, lane 4), whereas the ability of Ska3 to bind Ska2 was limited (Figure 2D, lane 3). Interestingly though, the addition of Ska1 enhanced the interaction between Ska3 and Ska2 (Figure 2D, lane 4). This suggests that Ska1 provides a scaffold for complex formation by interacting with both Ska3 and Ska2 through its N-terminal region (Supplementary Figure 2A).
To determine whether a similarly large Ska complex could be identified in mitotic cells, lysates from nocodazole-arrested HeLa S3 cells were subjected to gel filtration and probed by western blotting for Ska components. As shown in Figure 2E, all three Ska proteins comigrated in several fractions that peaked around 720 kDa, in excellent agreement with the in vitro reconstitution data. In addition, Ska1 could be detected in several fractions of lower molecular weight, suggesting the existence of a subpopulation of Ska1 that is not in a complex with Ska2 and Ska3.
Ska3 is required for Ska complex stability and function
Earlier it has been shown that siRNA-mediated depletion of either Ska1 or Ska2 resulted in an increased mitotic index (Hanisch et al, 2006; Rines et al, 2008). Furthermore, the two proteins were found to influence each other's stability and localization (Hanisch et al, 2006). To determine whether Ska3 is also critical for the stability of the entire Ska complex, we examined the effect of Ska3 depletion on Ska1 and Ska2 in HeLa S3 cells. When compared with cells transfected with the control Gl2 oligonucleotide, the Ska3-specific siRNA oligonucleotide effectively depleted the protein, as shown by both western blotting (Figure 3A and B; Supplementary Figure 5A and B) and immunofluorescence microscopy (Figure 3D; Supplementary Figure 5C). Moreover, Ska3 depletion clearly reduced the levels of both Ska1 and Ska2, and likewise, depletion of any other single component of the Ska complex affected the remaining members, albeit to variable extents (Figure 3A, B and D; Supplementary Figure 5A–C). Most importantly, Ska3 depletion effectively blocked mitotic progression in asynchronously growing HeLa S3 cells, producing a 4–5-fold increase in the percentage of mitotic cells above that of the Gl2 control (Figure 3C), with most cells arrested in a metaphase-like state characterized by relatively normal-looking bipolar spindles (Figure 3D; Supplementary Figure 5C). These results are highly reminiscent of earlier data obtained upon depletion of Ska1 and Ska2 (Hanisch et al, 2006).
As mentioned above, Ska3 was clearly modified in mitosis, resulting in its reduced electrophoretic mobility. Analysis by mass spectrometry revealed that Ska3 can be phosphorylated on at least 18 different Ser/Thr residues and on at least one Tyr residue (Supplementary Figure 2A and B) (Brill et al, 2004; Rush et al, 2005; Nousiainen et al, 2006; Daub et al, 2008; Dephoure et al, 2008; Sui et al, 2008; Malik et al, in preparation; Santamaria et al, unpublished result). We, therefore, asked whether the upshift observed in western blots performed on mitotic lysates was due to an M-phase specific phosphorylation on Ska3. Treatment of lysates from mitotically arrested cells with λ phosphatase resulted in the complete loss of the slower migrating band and a concomitant increase in the faster migrating form, indicating that the observed retardation in electrophoretic mobility was indeed dependent on phosphorylation (Supplementary Figure 4C). Interestingly, exposure to calf intestinal phosphatase (CIP) resulted in an intermediate band, suggesting that different phosphorylation events on Ska3 give rise to distinct upshifts. Furthermore, when samples were collected at different times after release from a nocodazole block and examined by western blotting, we observed a step-wise increase in Ska3 mobility concomitant with securin degredation, suggesting that Ska3 is progressively dephosphorylated as cells exit mitosis (Supplementary Figure 4D). The M-phase specificity of Ska3 phosphorylation was further confirmed by demonstrating that Ska3 exists solely in its upshifted (phosphorylated) state when cells are blocked at the metaphase-to-anaphase transition by the addition of the proteosome inhibitor MG132 (Supplementary Figure 4D). The upshifted form of Ska3 was also detected in mitotic cells collected by shake-off 10 h after thymidine release (data not shown), indicating that it is not an artefact caused by prolonged exposure to an anti-mitotic drug. Taken together, these results clearly show that Ska3 is highly phosphorylated during M-phase and then dephosphorylated after SAC inactivation and anaphase onset. In future, it will be interesting to elucidate the role of Ska3 phosphorylation during mitotic progression. One possibility would be that phosphorylation regulates Ska complex formation, but given that interactions among the three Ska proteins can readily be detected amongst recombinant proteins purified from E. coli (Figure 2) it cannot be essential. Thus, it appears more attractive to speculate that Ska3 phosphorylation is required for the localization or function of the complex at the KT. In any event, inspection of the consensus sequences of the phosphorylation sites (Supplementary Figure 2B) suggests that multiple mitotic kinases contribute to control Ska3 function.
Extensive depletion of the Ska complex causes a chromosome congression defect
To explore the function of Ska3, the consequences of depleting individual Ska proteins were examined by live cell imaging performed on HeLa S3 cells stably expressing Histone H2B-GFP. When compared with Gl2-treated control cells, depletion of Ska3, Ska2 or Ska1 resulted in a striking delay between metaphase plate formation and anaphase onset (Figure 4A; Supplementary Movies 1, 2 and 3 and data not shown), consistent with previous results (Hanisch et al, 2006). Eventually, however, cells underwent anaphase and completed mitosis.
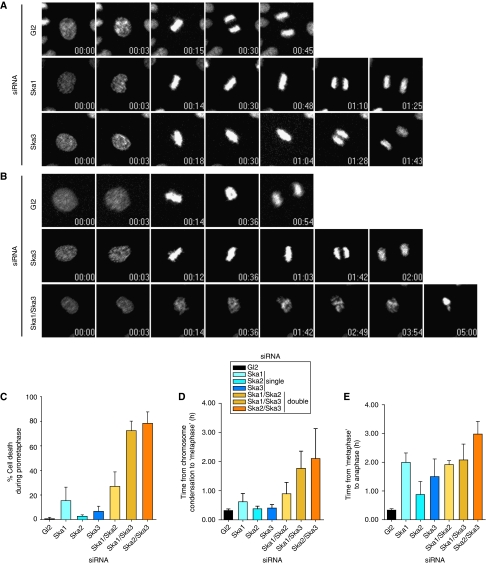
Figure 4.
Time-lapse microscopy analysis of Ska-depleted cells. (A) Stills of representative movies of HeLa S3 cells stably expressing H2B-GFP treated with control (Gl2), Ska1 and Ska3 siRNAs for 36 h before filming. T=0 was defined as the time point one frame before chromosome condensation became evident (prophase). N=3 independent experiments with at least 40 cells counted in each condition. Time points are indicated in h:min. (B) Stills of representative movies of H2B-GFP expressing HeLa S3 cells treated with control (Gl2), Ska3 and Ska1/Ska3 siRNAs for 36 h before filming. T=0 was defined as the time point one frame before chromosome condensation became evident (prophase). N=3 independent experiments with at least 40 cells counted in each condition. Time points are indicated in h:min. (C) The percentage of cells dying in prometaphase calculated from time-lapse movies (Supplementary Movies S4, S5, S6 and S7 and data not shown) of at least 40 siRNA-treated cells per condition per experiment. Histograms show averages from three independent experiments. Error bars represent s.d. The duration from prophase to anaphase onset was calculated from time-lapse movies (Supplementary Movies S4, S5, S6 and S7 and data not shown) of at least 40 siRNA-treated cells per condition per experiment. Time 0 is defined as the first frame where chromosome condensation was apparent. Metaphase was defined as the first frame displaying complete alignment, and anaphase onset was calculated from the first frame at which chromosome segregation was visible. Histograms show the time for progression from chromosome condensation to first complete alignment (D) and from first metaphase until anaphase onset (E). Data are presented as averages from three independent experiments. Error bars represent s.d.
The fact that knock-down of individual Ska proteins delays cells in a metaphase-like state but does not ultimately prevent the completion of mitosis could be interpreted to imply that this complex is not strictly required for mitotic progression. Alternatively, however, the transient nature of the arrest may reflect incomplete depletion of the Ska complex. To explore this latter possibility, we pooled siRNA oligonucleotides targeting two Ska components simultaneously, in the hope that this would achieve a more complete depletion of the Ska complex than any ‘single' knock-down. Quantitative analysis of western blotting revealed that double-siRNA treatment resulted in a more extensive depletion of Ska complex components than single-siRNA treatment, particularly when depletion efficiency was assessed at relatively early time points (Supplementary Figure 5A and B). As shown by time-lapse microscopy, the single depletion of Ska components produced a marked metaphase delay (Figure 4B and E), consistent with earlier results (Hanisch et al, 2006). In contrast, the more complete removal of the complex resulted in a striking chromosome congression failure (Figure 4B and D; Supplementary Figure 5C; Supplementary Movies 4, 5, 6 and 7), followed by a large proportion of cells (up to 80%) dying without ever reaching metaphase (Figure 4C). Remarkably, this phenotype was particularly pronounced whenever Ska3 was one of the two proteins being depleted.
We presume that some cells survived the double-siRNA treatment because they suffered a less complete depletion of the Ska complex. These latter cells eventually reached a stage resembling metaphase, but they required substantially more time (105±35 min for Ska1/Ska3 double depletion) than either cells depleted of Ska3 alone (24±7 min) or Gl2 controls (19±3 min). The exact same Ska-depleted cells then also required much more time to progress to anaphase than Gl2 controls, but differences between singly and doubly siRNA-treated cells were less pronounced (89±36 min for Ska3 single depletion, compared with 124±33 min for Ska1/Ska3 double depletion) (see Figure 4C–E for complete data on all siRNA combinations). Taken together, these data thus suggest that chromosome congression is particularly sensitive to the precise levels of Ska proteins.
Removal of the Ska complex compromises MT–KT attachment
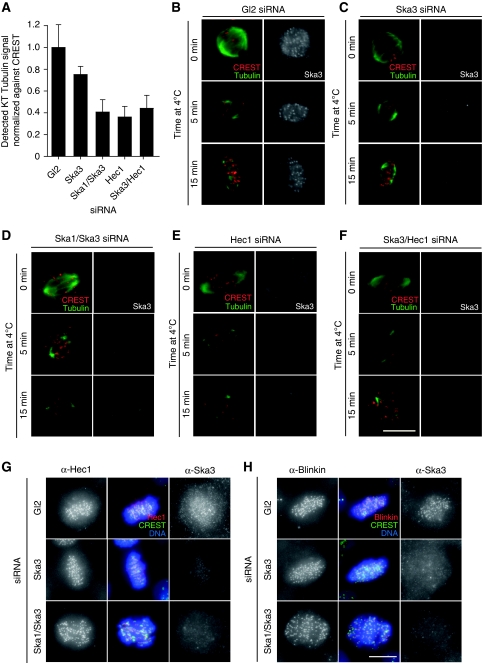
The inability of chromosomes to form a metaphase plate after extensive removal of the Ska complex indicated a possible role of this complex in MT capture and/or maintenance of KT attachment. To explore this possibility, we first confirmed that Ska3 recruitment to KTs was dependent on MTs (Supplementary Figure 6A) and required the presence of the Ndc80 complex (Supplementary Figure 6B), similar to results obtained earlier for Ska1 and Ska2 (Hanisch et al, 2006). Next, we adapted a recently described assay (Toso et al, 2009) to examine the extent of MT interactions with KTs. Specifically, the α-Tubulin signal of MTs was measured and normalized to the CREST signal at KTs (see Materials and methods). As illustrated in Figure 5A, co-depletion of either Ska1/Ska3 or Ska3/Hec1 produced a marked reduction in the KT-associated Tubulin signal, similar to that seen after depletion of Hec1 alone, whereas depletion of only Ska3 produced a minor decrease. This suggested that co-depletion of Ska1/Ska3 interfered with the establishment and/or maintenance of MT–KT attachment to a similar extent as (hypomorphic) Hec1 depletion (DeLuca et al, 2002; Martin-Lluesma et al, 2002). To corroborate this conclusion, we also assayed the stability of KT–MTs (k-fibres) after exposure of Ska-depleted cells to low temperature for either 5 or 15 min (Rieder, 1981). Again, we compared the consequences of both single- and double-Ska depletions with the earlier described Hec1-depletion phenotype (DeLuca et al, 2002; Martin-Lluesma et al, 2002), and we also examined the consequences of Ska3/Hec1 double depletion. Detailed analysis by immunofluorescence microscopy revealed that both the Gl2 control (Figure 5B) and the cells depleted of only Ska3 (Figure 5C) retained many cold-stable k-fibres. In contrast, cells depleted of both Ska1 and Ska3 almost completely lacked k-fibres (Figure 5D), very similar to the cells depleted of Hec1 (Figure 5E). Double depletion of Ska3 and Hec1 did not produce any obvious additional phenotype (Figure 5F), consistent with the observation that the depletion of Hec1 results in the loss of the Ska complex from KTs (Supplementary Figure 6B). Taken together, the above data show that the Ska complex is required for the formation of stable MT–KT attachments. Upon severe depletion of this complex, chromosomes fail to congress successfully and progression through mitosis is compromised.
Figure 5.
Stable MT–KT interactions are dependent on the Ska complex. (A) Histogram represents extent of KT–MT attachment by the level of α-Tubulin signal within the vicinity of the KT (normalized to the CREST signal) in HeLa S3 cells treated for 48 h with control (Gl2), Ska3, Ska1/Ska3, Hec1 or Ska3/Hec1 siRNAs. A significant decrease in α-Tubulin was detected between Gl2 and all other conditions (P<0.01, Student's t-test), and Ska3 and all other combinations (P<0.01). No significant difference was detected among Ska1/Ska3, Hec1 and Ska3/Hec1 siRNA treatment (P>0.01). The average intensity of 30 KT per cell was measured, and bars represent mean and s.d. from five cells per condition. (B–F) HeLa S3 cells were treated with the same combination of siRNA treatments as in (A) and either fixed directly (upper panels) or incubated at 4°C for either 5 min (middle panels) or 15 min (lower panels) before fixation. Cells were stained with anti-α-Tubulin (green), CREST serum (red) and anti-Ska3. Bar=10 μm. Cells were treated for 48 h with control (Gl2), or Ska3 and Ska1/Ska3 specific siRNAs. Cells were fixed with PTEMF and stained with either anti-Hec1 (G) or anti-Blinkin (H) antibodies (red), CREST serum (green) and Ska3 (right columns, respectively). DNA was visualized using DAPI (blue). Bar=10 μm.
In a final series of experiments, we asked whether the extensive removal of the Ska complex affected the localization of KMN components required for KT–MT stabilization (DeLuca et al, 2002; Cheeseman et al, 2006; Kline et al, 2006). Co-depletion of Ska1 and Ska3 did not detectably reduce the overall cellular levels of either Hec1 or Blinkin, the human homologue of KNL-1 (Kiyomitsu et al, 2007) (Supplementary Figure 7A), nor did they produce a major effect on the localization of these proteins (Figure 5G and H). Precise quantifications did suggest slight reductions of the corresponding immunofluorescence signals at KTs (Supplementary Figure 7B and C), but this effect cannot account for the phenotype upon Ska double depletion, as there was no further reduction in the level of KT-bound KMN components when compared with cells only treated with a single Ska siRNA oligonucleotide. The localization of Mis12 could not be monitored directly, but the corresponding complex is unlikely to be affected by Ska depletion, as inferred from the near-normal localization of Hec1 and the known interdependence between the Ndc80 and the Mis12 complexes (Kline et al, 2006). The localizations of other proteins involved in KT–MT attachment, in particular CenpE, the dynein/dynactin complex member p150glued, and the RZZ complex member ZW10 (Lampson and Kapoor, 2005; Yang et al, 2007), were also not noticeably affected by co-depletion of Ska1 and Ska3 in HeLa S3 cells (data not shown). Likewise, the SAC components Mad1, Mad2 and BubR1 could readily be detected at KTs after Ska depletion, in line with persistent activation of the SAC (Hanisch et al, 2006 and data not shown).
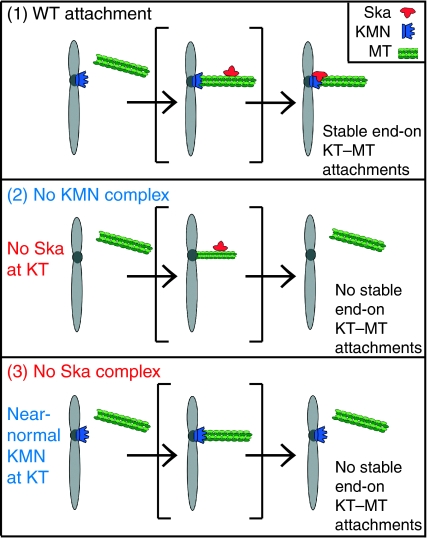
These results argue that the absence of the Ska complex does not significantly disrupt the core KT structure. However, the fact that k-fibre stability is lost upon Ska depletion despite the presence of the KMN network indicates that the Ska complex has an important function in stabilizing KT–MT attachments (Figure 6). The depletion of Hec1 results in a loss of Ska proteins from KTs (Hanisch et al, 2006, this study), implying that a functional KT is required for Ska localization. Although this may suggest a direct interaction between these complexes, it is important to bear in mind that the Ndc80 complex is required for the overall structure of the outer KT (DeLuca et al, 2005). Thus, the loss of Ska proteins upon Hec1 depletion may well reflect the loss of an as yet unidentified anchoring protein. So far, we have been unable to show a direct interaction between the Ndc80 and the Ska complexes, and several focused screens for binding partners of the Ska complex amongst KT components, MT +Tip-binding proteins, or MT-bundling proteins, have failed to reveal interaction partners of interest (Hanisch et al, 2006 and data not shown).
Figure 6.
Model for the requirement of Ska for stable KT–MT interactions. As summarized in this schematic, we propose that the KMN network and the Ska complex cooperate to ensure stable end-on MT attachments to KTs (top panel). Stable attachment fails upon depletion of either the KMN or the Ska complex. In the absence of KMN (middle panel), the Ska complex fails to localize to KTs, whereas, conversely, the KT localization of the KMN does not strictly depend on the Ska complex (bottom panel). This implies that the KMN alone is not sufficient for stable MT attachment but requires cooperation with the Ska complex for full functionality. Finally, we emphasize that the role in chromosome congression shown here for the Ska complex does not rigorously exclude a second, additional role for this complex in SAC silencing (Hanisch et al, 2006).
In summary, we describe the novel phosphoprotein Ska3 (formerly C13Orf3). We show that this protein is a core component of the Ska complex, required for both complex stability and function. Furthermore, through double-depletion siRNA experiments resulting in the near-complete removal of the entire Ska complex, we have been able to describe an essential role for the Ska complex in MT–KT attachment and chromosome congression. Thus, the Ska complex has a more central function in mitotic progression than surmised earlier. We propose that the Ska complex cooperates with the KMN to create the dynamic MT–KT interface that is required for MT attachment, chromosome congression and subsequent segregation. In particular, we speculate that the Ska complex may function as a clamp to stabilize the MT–KT attachments established through the KMN, perhaps providing an analogous function to the Dash–Duo–Dam complex of budding yeast (Cheeseman et al, 2001; Westermann et al, 2007). Additionally, it would be premature to exclude an additional signalling function in the silencing of the SAC (Hanisch et al, 2006). Perhaps, the functions provided by the Ska complex are particularly critical in organisms in which multiple MTs attach to a single KT. This would readily explain the apparent absence of genes coding for Ska proteins from the published yeast genomes.
Materials and methods
Cloning procedures
For cloning of Ska3 (C13Orf3), a cDNA clone (IRATp970D0864D) was obtained from the ‘Deutsches Ressourzentrum für Genomforschung'. This cDNA was cloned in-frame into a pCDNA3.1 vector (Invitrogen, Carlsbad, CA) encoding an N-terminal 3xMyc tag.
Protein purification and antibody production
Antibodies against Ska3 were generated by immunization of rabbits with four injections of 250 μg of N-terminally His6-tagged full-length protein produced in E. coli (Charles River Laboratories, Romans, France). Anti-Ska3 antibodies were affinity purified by applying 1.5 ml immune serum onto nitrocellulose filters containing 400 μg of the antigen used for immunization. The filters were extensively washed with PBST, and anti-Ska3 antibodies were subsequently eluted from the filters with glycine buffer (100 mM pH 2.8) and dialyzed against PBST.
Cell culture and synchronization
HeLa S3, HeLa S3 H2B-GFP and HEK293T cells were cultured in a 5% CO2 atmosphere in Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 10% heat-inactivated fetal calf serum (FCS) and penicillin streptomycin (100 IU/ml and 100 μg/ml, respectively). To arrest cells at prometaphase, they were treated with 50 ng/ml nocodazole (Sigma, St Louis, MO) for 16 h, and mitotic cells were collected by mechanical shake-off, washed twice with PBS, and released into normal growth medium. Samples were taken either from arrested cells or after release for stated time intervals. Alternatively, cells were treated with 45 μg/ml monastrol (Sigma) for 16 h.
Transient transfections and siRNA
Plasmid transfections were performed using TransIT®-LT1 reagent (Mirus Bio Corporation, Madison, WI) according to the manufacturer's instructions. siRNA duplexes were transfected using Oligofectamine (Invitrogen) as described elsewhere (Elbashir et al, 2001). The sequence of the siRNA duplex for targeting Ska3 was: 5′-AGACAAACAUGAACAUUAA-3′ (Qiagen, Hilden, Germany). Ska1 and Ska2 were depleted using established siRNAs targeting published sequences (Hanisch et al, 2006), and Hec1 was depleted using established siRNA targeting published sequence (Martin-Lluesma et al, 2002). As control, a duplex (Gl2) targeting luciferase was used (Elbashir et al, 2001). In experiments comparing single- and double-siRNA treatments, the total concentration of oligonucleotides administered to the cells was kept constant by the addition of Gl2 where applicable.
Cell extracts, immunoprecipitation, western blotting analysis and gel filtration
Lysates were prepared using Hepes lysis buffer (50 mM Hepes pH 7.4, 150 mM NaCl, 0.5% Triton X-100) containing 30 μg/ml RNAse A, 30 μg/ml DNase, 1 μM Okadaic acid, phosphatase inhibitors and complete mini protease inhibitor tablets (1/10 ml) (Roche Diagnostics, Indianapolis, IN). Protein concentrations were determined using the Dc protein assay (Bio-Rad Laboratories, Hercules, CA). Lysates were incubated for 2–4 h at 4°C with 9E10 anti-Myc antibodies (1:10, 9E10 tissue culture supernatant). In each case, 1 μg of antibody was coupled to 1 μl sepharose-A beads (20 μl beads in total) (Pierce, Rockford, IL). After protein capture, beads were washed 4 × with Hepes lysis buffer and resuspended in gel sample buffer and proteins were analysed by SDS–PAGE and immunoblotting. Membranes were probed with the following antibodies: affinity-purified rabbit anti-Ska1 and anti-Ska2 and anti-Ska3 (1 μg/ml), mouse mAb anti-Securin (1:1000, Abcam, Cambridge, UK), mouse mAb anti-Hec1 (1:1000, Abcam), mouse mAb anti-Blinkin (1:200, gift from M Yanaguida), and mouse mAb anti-α-Tubulin (Sigma).
Gel filtration was carried out on mitotic HeLa S3 cells collected by shake-off after synchronization by a sequential thymidine/nocodazole block release protocol, and lysed in NP40-lysis buffer (PBS with 0.5% NP40, complete mini protease inhibitor tablets (1/10 ml) (Roche), and Phosphatase Inhibitors Cocktails 1 and 2 (Sigma)). Before gel filtration, lysates were cleared by sequential centrifugation (4°C) at 16 000 and 200 000 g. Protein extract (6 mg) was then loaded on a Superose 6 10/300 GL column (GE Healthcare, Piscataway, NJ), and isocratic elution was performed at 0.4 ml/min at 4°C with PBS, collecting fractions every 0.65 ml. The protein content of each fraction was precipitated by addition of TCA (13%) followed by centrifugation at 4°C. Protein pellets were washed twice with acetone, dried and resuspended in 60 μl of Laemmli sample buffer. Fractions (10 μl) were loaded on SDS–PAGE and analysed by western blotting.
Ska2 immunoprecipitation and mass spectrometry analysis
For Ska2 immunoprecipitation, mitotic HeLa S3 cells were harvested by shake-off after 16 h nocodazole treatment and release for 40 min. Lysates were prepared using Hepes lysis buffer (see above) and were pre-cleared for 6 h at 4°C with rabbit IgG and Ska2 was subsequently immunoprecipitated overnight with anti-Ska2 antibodies. In each case, 40 μg of antibody was coupled to 40 μl sepharose-A beads (Pierce). After protein capture, beads were washed 4 × with Hepes lysis buffer, resuspended in gel sample loading buffer and resolved by NuPAGE gel (Invitrogen). The gel was processed for analysis by mass spectrometry. This involved in-gel digestion of Coomassie-stained protein bands by trypsin (Promega, sequencing grade) (Shevchenko et al, 1996) and desalting using home-made miniaturized reversed-phase columns (Rappsilber et al, 2003). Peptide mixtures were chromatographically separated with a nanoACQUITY ultra performance liquid chromatography system (Waters, Hertfordshire, UK) connected to a hybrid linear ion trap/orbitrap tandem mass spectrometer (Thermo Electron, Waltham, MA). Peptides dissolved in 0.5% formic acid were loaded to a 14 cm pulled fused silica capillary with an inner diameter of 75 μm and a tip of 8 μm (New Objective, Woburn, MA) packed with reversed-phase ReproSil-Pur C18-AQ 3 μm resin (Dr Maisch GmbH, Ammerbuch-Entringen, Germany). Peptides were separated and eluted by a stepwise 60-min gradient of 0–100% between buffer A (0.2% formic acid in water) and buffer B (0.2% formic acid in acetonitrile). The mass spectrometer was operated in data-dependent MS/MS mode. Survey full scan MS spectra (from m/z 300–2000) were acquired in the FT-Orbitrap with a resolution of 60 000 at m/z 400. A maximum of five peptides were sequentially isolated for fragmentation in the linear ion trap using collision-induced dissociation. MASCOT (version 2.2.0, Matrix Science, London, UK) was used for protein identifications from the human International Protein Index database. For searches, the precursor mass tolerance was set to ±5 ppm, whereas an accuracy of ±0.6 Da was used for MS/MS spectra. Carbamidomethylation was set as fixed modification, whereas oxidation, deamidation (NQ) and phosphorylation (STY) were considered as variable modifications.
In vitro characterization of Ska complex
Full-length and truncated versions of Ska1, Ska2 and Ska3 as N-terminal His- or GST-tagged proteins were cloned into pEC series of vectors (Ligation Independent Cloning modified from pET-28a vector) designed in house. C-terminally His-tagged full-length Ska3 cloned into a pET-28 vector, and untagged Ska2 was cloned into a pET-11 vector. Proteins were expressed in E. coli strain BL21Gold (pLysS) with an overnight induction at 18°C (with 0.2 mM IPTG). Cells were lysed in a buffer containing 20 mM Tris pH 7.5, 500 mM NaCl and 1 mM DTT. Full-length and truncated protein complexes were purified by affinity chromatography with glutathione sepharose 4B beads (GE Healthcare). The affinity tags were cleaved using TEV protease after extensive washing, whereas the proteins were still bound to the beads, followed by anion exchange with a final size exclusion chromatography (Superdex 200, GE Healthcare). The pure reconstituted complex was analysed on a 15% SDS–PAGE gel staining with Coomassie. For the GST-pulldown experiment, GST–Ska2 (full length) and His–Ska3 (full length) were co-expressed in conditions as mentioned above.
Phosphatase treatment
HeLa S3 cells were kept asynchronously growing or were arrested with 150 ng/ml nocodazole for 16 h. The corresponding cell lysates were treated with either CIP (New England Biolabs, Ipswich, MA) or λ Phosphatase (Roche, Indianapolis, IN) or left untreated for 2 h at 30°C. The phosphatase reaction was stopped by addition of sample buffer followed by boiling. Equal protein amounts were loaded and separated by SDS–PAGE followed by western blotting analysis.
Immunofluorescence microscopy
Cells were grown on coverslips and fixed and permeabilized as described earlier (Sillje et al, 2006). Primary antibodies used in this study were rabbit anti-Ska1 and rabbit anti-Ska2 (Hanisch et al, 2006), mouse mAb anti-Myc (1:10, 9E10 tissue culture supernatant), human CREST autoimmune serum (1:2000, Immunovision, Springdale, AR), mouse mAb anti-Hec1 (1:1000, Abcam), mouse mAb anti-Blinkin (1:1000, gift from M. Yanaguida). Primary antibodies were detected with Cy2-, Cy3- and Cy5-conjugated donkey anti-mouse, anti-rabbit or anti-human IgGs (1:1000, Dianavo, Hamburg, Germany). DNA was stained with DAPI (2 μg/ml).
Immunofluorescence microscopy was performed with Zeiss Axio Observer Z1 microscope (Visitron Systems GmbH, Puccheim, Germany), equipped with plan-apochromat 63 × objective, and a CoolSNAP-HQ2 CCD camera. Where applicable, z-axis optical sections were recorded every 0.5 μm, images of a single focal plane were processed with a deconvolution algorithm and optical sections were projected into one picture using Metamorph 7.1 software (Molecular Devices, Sunnyvale, CA). Images were cropped in Adobe Photoshop 6.0 and then sized and placed in figures using Adobe Illustrator 10 (Adobe Systems, San Jose, CA).
Image quantification
Immunofluorescent and western blotting images were quantified using ImageJ software (http://rsb.info.nih.gov/ij/). For immunofluorescence, images used for quantification were taken with identical exposure times within each experiment, acquired from z-axis optical sections every 0.3 μm and compressed to a single plane without deconvolution. Quantification of KT intensities was preformed using a method adapted from that described earlier (Elowe et al, 2007). Essentially, a circular region with fixed diameter was centred on each KT, and the average pixel intensity was measured. CREST intensity was measured in the same region and used for normalization after subtraction of background intensity measured within a cytoplasmic, non-specific region of the cell as described earlier (Toso et al, 2009). To measure the extent of MT attachment at the KT, we adapted the method described recently (Toso et al, 2009). Images were acquired and processed as above, and the intensity of the α-Tubulin signal at the KT measured and normalized against the KT CREST signal, after subtraction of cytoplasmic background intensity for each individual cell. Statistical significances of quantification experiments were verified by Student's t-test.
For quantification of western signal, a set area around the band of interest was selected, the average pixel intensity measured, and the average background signal from an area of identical size at a non-specific region of the membrane was subtracted. The results are presented normalized to the signal for α-Tubulin.
Time-lapse microscopy
For time-lapse microscopy, a HeLa S3 cell line stably expressing Histone H2B-GFP was used (Sillje et al, 2006). Cells were treated with siRNAs for 36 h before image acquisition. The culture plate (Ibidi, Martinsried, Germany) was placed onto a sample stage within an incubator chamber (EMBLEM, Heidelberg, Germany) maintained at a temperature of 37°C, humidity 60%, in an atmosphere of 5% CO2. Imaging was performed using a Zeiss Axio Observer Z1 microscope equipped with a Plan Neofluar 20 × objective. Metamorph 7.1 software (Molecular Devices) was used to collect and process data. Images were captured using 10 ms exposure times every 3-min interval for 16 h.
Supplementary Material
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Movie 4
Supplementary Movie 5
Supplementary Movie 6
Supplementary Movie 7
Supplementary Information
Acknowledgments
We thank Mitsuhiro Yanagida (Kyoto University) for the Blinkin antibody. We are thankful to Anja Wehner for excellent technical assistance, Luca Fava for help with gel filtration, Rainer Malik for bioinformatic support and Roman Körner for mass spectrometry guidance. We gratefully acknowledge Stefan Hümmer, Lily Wang and Sabine Elowe for helpful discussions. This work was supported by the Max Planck Society, the Deutsche Forschungsgemeinschaft (SFB 646), and ENFIN, a Network of Excellence funded by the European Commission within its FP6 Programme (contract number LSHG-CT-2005-518254). AS is supported by a postdoctoral fellowship from the Spanish Education and Science Ministry. BW is partially supported by the Chinese Academy of Sciences.
Footnotes
The authors declare that they have no conflict of interest.
References
- Brill LM, Salomon AR, Ficarro SB, Mukherji M, Stettler-Gill M, Peters EC (2004) Robust phosphoproteomic profiling of tyrosine phosphorylation sites from human T cells using immobilized metal affinity chromatography and tandem mass spectrometry. Anal Chem 76: 2763–2772 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A (2006) The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127: 983–997 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A (2008) Molecular architecture of the kinetochore–microtubule interface. Nat Rev Mol Cell Biol 9: 33–46 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Enquist-Newman M, Muller-Reichert T, Drubin DG, Barnes G (2001) Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J Cell Biol 152: 197–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR III, Oegema K, Desai A (2004) A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev 18: 2255–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, Salek M, Rappsilber J, Moores CA, Salmon ED, Musacchio A (2008) Implications for kinetochore–microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421 [DOI] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36: W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M (2008) Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell 31: 438–448 [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF (2005) Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell 16: 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED (2002) hNuf2 inhibition blocks stable kinetochore–microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol 159: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP (2008) A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA 105: 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Elowe S, Hummer S, Uldschmid A, Li X, Nigg EA (2007) Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev 21: 2205–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Stuurman N, Vale RD (2007) Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol 177: 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A, Sillje HH, Nigg EA (2006) Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J 25: 5504–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M (2004) The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol 14: 611–617 [DOI] [PubMed] [Google Scholar]
- Karess R (2005) Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol 15: 386–392 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T, Obuse C, Yanagida M (2007) Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev Cell 13: 663–676 [DOI] [PubMed] [Google Scholar]
- Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A (2006) The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol 173: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, Kapoor TM (2005) The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat Cell Biol 7: 93–98 [DOI] [PubMed] [Google Scholar]
- Larsen NA, Al-Bassam J, Wei RR, Harrison SC (2007) Structural analysis of Bub3 interactions in the mitotic spindle checkpoint. Proc Natl Acad Sci USA 104: 1201–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, DeLuca J, Salmon ED, Earnshaw WC (2004) The dynamic kinetochore–microtubule interface. J Cell Sci 117: 5461–5477 [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma S, Stucke VM, Nigg EA (2002) Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297: 2267–2270 [DOI] [PubMed] [Google Scholar]
- Matsumura S, Toyoshima F, Nishida E (2007) Polo-like kinase 1 facilitates chromosome alignment during prometaphase through BubR1. J Biol Chem 282: 15217–15227 [DOI] [PubMed] [Google Scholar]
- McEwen BF, Hsieh CE, Mattheyses AL, Rieder CL (1998) A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma 107: 366–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED (2007) The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R (2006) Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci USA 103: 5391–5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C, Iwasaki O, Kiyomitsu T, Goshima G, Toyoda Y, Yanagida M (2004) A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat Cell Biol 6: 1135–1141 [DOI] [PubMed] [Google Scholar]
- Pinsky BA, Biggins S (2005) The spindle checkpoint: tension versus attachment. Trends Cell Biol 15: 486–493 [DOI] [PubMed] [Google Scholar]
- Pritchard CE, Fornerod M, Kasper LH, van Deursen JM (1999) RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J Cell Biol 145: 237–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75: 663–670 [DOI] [PubMed] [Google Scholar]
- Rieder CL (1981) The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma 84: 145–158 [DOI] [PubMed] [Google Scholar]
- Rines DR, Gomez-Ferreria MA, Zhou Y, DeJesus P, Grob S, Batalov S, Labow M, Huesken D, Mickanin C, Hall J, Reinhardt M, Natt F, Lange J, Sharp DJ, Chanda SK, Caldwell JS (2008) Whole genome functional analysis identifies novel components required for mitotic spindle integrity in human cells. Genome Biol 9: R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ (2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol 23: 94–101 [DOI] [PubMed] [Google Scholar]
- Sauer G, Korner R, Hanisch A, Ries A, Nigg EA, Sillje HH (2005) Proteome analysis of the human mitotic spindle. Mol Cell Proteomics 4: 35–43 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Sillje HH, Nagel S, Korner R, Nigg EA (2006) HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol 16: 731–742 [DOI] [PubMed] [Google Scholar]
- Stehman SA, Chen Y, McKenney RJ, Vallee RB (2007) NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J Cell Biol 178: 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui S, Wang J, Yang B, Song L, Zhang J, Chen M, Liu J, Lu Z, Cai Y, Chen S, Bi W, Zhu Y, He F, Qian X (2008) Phosphoproteome analysis of the human Chang liver cells using SCX and a complementary mass spectrometric strategy. Proteomics 8: 2024–2034 [DOI] [PubMed] [Google Scholar]
- Toso A, Winter JR, Garrod AJ, Amaro AC, Meraldi P, McAinsh AD (2009) Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J Cell Biol 184: 365–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Babu JR, Harden JM, Jablonski SA, Gazi MH, Lingle WL, de Groen PC, Yen TJ, van Deursen JM (2001) The mitotic checkpoint protein hBUB3 and the mRNA export factor hRAE1 interact with GLE2p-binding sequence (GLEBS)-containing proteins. J Biol Chem 276: 26559–26567 [DOI] [PubMed] [Google Scholar]
- Westermann S, Drubin DG, Barnes G (2007) Structures and functions of yeast kinetochore complexes. Annu Rev Biochem 76: 563–591 [DOI] [PubMed] [Google Scholar]
- Yamamoto TG, Watanabe S, Essex A, Kitagawa R (2008) SPDL-1 functions as a kinetochore receptor for MDF-1 in Caenorhabditis elegans. J Cell Biol 183: 187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Tulu US, Wadsworth P, Rieder CL (2007) Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr Biol 17: 973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Movie 4
Supplementary Movie 5
Supplementary Movie 6
Supplementary Movie 7
Supplementary Information