Abstract
Residue 225 in serine proteases of the chymotrypsin family is Pro or Tyr in more than 95% of nearly 300 available sequences. Proteases with Y225 (like some blood coagulation and complement factors) are almost exclusively found in vertebrates, whereas proteases with P225 (like degradative enzymes) are present from bacteria to human. Saturation mutagenesis of Y225 in thrombin shows that residue 225 affects ligand recognition up to 60,000-fold. With the exception of Tyr and Phe, all residues are associated with comparable or greatly reduced catalytic activity relative to Pro. The crystal structures of three mutants that differ widely in catalytic activity (Y225F, Y225P, and Y225I) show that although residue 225 makes no contact with substrate, it drastically influences the shape of the water channel around the primary specificity site. The activity profiles obtained for thrombin also suggest that the conversion of Pro to Tyr or Phe documented in the vertebrates occurred through Ser and was driven by a significant gain (up to 50-fold) in catalytic activity. In fact, Ser and Phe are documented in 4% of serine proteases, which together with Pro and Tyr account for almost the entire distribution of residues at position 225. The unexpected crucial role of residue 225 in serine proteases explains the evolutionary selection of residues at this position and shows that the structural determinants of protease activity and specificity are more complex than currently believed. These findings have broad implications in the rational design of enzymes with enhanced catalytic properties.
Keywords: complement, molecular evolution, thrombin, water
Serine proteases of the chymotrypsin family share a common fold and are involved in diverse and important functions, including digestion and degradative processes, blood coagulation, fibrinolysis, cellular and humoral immunity, embryonic development, and fertilization (1, 2). Previous studies have shown that activity and specificity in these enzymes are influenced by residues directly involved in substrate recognition, especially in the primary specificity site, as well as by more distal regions like the surface loops 180 and 220 and three adjacent β-strands (3–5). Recent studies have shown that the nature of residue 225, located immediately downstream to the 220-loop, specifies an important property in serine proteases. Tyr at position 225 enables Na+ binding near the primary specificity site, whereas Pro at the same position abrogates this function (6, 7). Binding of Na+, the most abundant cation in the extracellular fluids where most serine proteases act in vivo, enhances allosterically the catalytic activity of the enzyme and might have been evolutionarily advantageous. Here we show that residue 225 also contributes to the architecture of the primary specificity site and that, by virtue of this unexpected structural role, it has played a pivotal function in the molecular evolution of the entire class of serine proteases.
MATERIALS AND METHODS
Database.
A total of 284 complete sequences from the Swiss-Prot database were analyzed. Pro at position 225 is found in 244 cases, including all enzymes involved in degradation (trypsin, chymotrypsin, and elastase), fibrinolysis (plasmin, tissue plasminogen activator, and urokinase), cellular-mediated immunity (granzymes and mast cell proteases), and embryonic development (Easter and Snake). Tyr is found in 27 cases, including the vitamin K-dependent enzymes of blood coagulation (thrombin, factor Xa, factor IXa, and activated protein C) and complement proteases (C1r, C1s, MASP-1, and MASP-2). Other residues are documented only in a few cases. These are: Ser (five cases), Phe (four cases), Thr (two cases, chymotrypsins from Streptomyces gryseus), Ile (one case, coagulation factor VIIa in mouse), and Val (one case, coagulation factor VIIa in rabbit). The probability of getting such distribution from a random selection of the 20 aa is 1 in ≈10304. The variability of residue 225 in coagulation factor VIIa is exceptional and probably is linked to the optimization of binding of tissue factor (8) in different species.
Protein Expression and Purification.
Saturation mutagenesis of Y225 of human thrombin was carried out by using PCR primers and human thrombin cDNA (hII cDNA) contained in a HPC4-pNUT expression vector (9). The hII cDNA was modified by silent mutations (T1777C and C1843T) to insert the restriction sites KpnI and PmlI and an AscI site after the TAG codon. The recombinant protein was expressed in BHK21 cells and the metothrexate-stabilized expressing cell line was grown in cell factories with yields of up to 5 mg/liter per day. Tissue culture supernatant (≈10 liters) was collected and stored at either −20°C or −80°C. Batch adsorption of the recombinant protein on QAE Sephadex A-50 was followed by binding on a HPC4 Affi-Gel 10 column (1.5 × 20 cm) in the presence of 5 mM CaCl2 and elution with 5 mM EDTA. The purified recombinant prethrombin-1 with the HPC4 epitope fused to the N terminus was activated by using phosphoserine/phosphocholine-bound prothrombinase complex (≈30 min at 37°C) and purified on a MonoS–FPLC column by using a linear gradient of 0.05–0.5 M NaCl in 5 mM Mes, pH 6 at room temperature. The purified enzyme was stored in 30-μl aliquots at −80°C. Each mutant was homogenous on silver-stained reduced SDS gels. The active enzyme concentration was measured by titration with hirudin (7). Mutants with Asp, Asn, or Glu at position 225 repeatedly failed to express in quantities sufficient for characterization. All other mutants and the wild type were studied in their interaction with Na+, chromogenic substrates, fibrinogen, protein C, and antithrombin III as described (6, 7).
Crystallization of the Y225F, Y225P, and Y225I Thrombin Mutants.
Mutants were concentrated to 3.3 mg/ml in 5 mM Mes, 250 mM NaCl, pH 6.0, and inhibited by a 10-fold molar excess of H-D-Phe-Pro-Arg-chloromethylketone (PPACK) for 30 min at room temperature. Crystallization was achieved at 20°C by vapor diffusion against ≈20% polyethylene glycol 8,000, 0.1 M Zn acetate, 0.1 M cacodylate, pH 6.0–6.5. Crystals were cryoprotected in 25% 2-methyl-2,4-pentanediol before flash freezing. Data were collected from a single crystal per mutant at the Advanced Photon Source (beamline ID19, Argonne National Laboratory, Argonne, IL) and processed by using the HKL2000 package (10). The crystals were of space group P212121 and contained one molecule per asymmetric unit. Structures were solved by molecular replacement by using the coordinates of the thrombin/PPACK complex (11) and the program amore (12). Structures were manually rebuilt with the program O (13, 14) into simulated-annealing omit maps (15) by sequentially omitting 10 residues at a time. Refinement was carried out by using the program x-plor (16). Water molecules were added in the final stage of the refinement process. They were subject to visual inspection to check their positioning in electron density and allowed to refine freely. Valence screening of all solvent peaks using the program wasp (17) readily identified the bound Na+ in the Y225F mutant. Coordinates of the structures of the Y225F, Y225P, and Y225I thrombin mutants have been deposited in the Protein Data Bank.
RESULTS
The distribution of residues at position 225 in serine proteases is rather unusual, with Pro and Tyr representing the amino acids of choice in more than 95% of almost 300 documented sequences. The probability of getting such a pattern from a random selection of the 20 aa is less than 1 in 10300. With very few exceptions, Pro is present in all degradative (e.g., trypsin, chymotrypsin, and elastase) and fibrinolytic enzymes, in the kallikreins, granzymes, and mast cell proteases, in enzymes involved in fertilization and embryonic development, and in factors of the alternative pathway of the complement. On the other hand, Tyr is present in the vitamin K-dependent clotting enzymes and factors of the classical and lectin-dependent pathways of the complement.
Residue 225 is highly conserved in proteases from different species. In the trypsins, P225 is conserved from bacteria to human (18, 19). In thrombin, a protease present exclusively in vertebrates, Y225 is conserved from hagfish to human (20). A correlation also exists between residue 225 and the codon usage (TCN or AGY) for the active site S195, which has been used to trace paths of descent in serine proteases (21). The TCN codons for S195 and P225 are found in more ancestral proteases (trypsin and chymotrypsins of bacteria), whereas more specialized proteases found in vertebrates carry Y225 and the AGY codon for S195 (6). The correlation is intriguing because both the TCN→AGY and Pro→Tyr conversions cannot occur by single nucleotide mutations in the genetic code.
The Na+-dependent enhancement of catalytic activity may explain the Pro→Tyr conversion of residue 225 occurring in the vertebrates, but leaves the question about the conservation of these residues unanswered. Saturation mutagenesis of Y225 in thrombin therefore was carried out to elucidate the role of residue 225 in the function and evolution of serine proteases and the peculiar distribution of amino acids at this position. All substitutions (except Asp, Glu, and Asn) expressed in quantities sufficient for characterization with natural substrates and inhibitors.
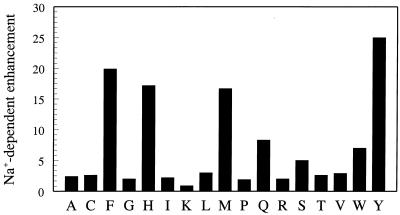
The Na+-dependent enhancement of catalytic activity seen for the wild type is retained to a different extent in the Phe, His, Trp, Gln, Met, and Ser derivatives (Fig. 1). The preference for aromatic residues at 225 (Tyr, Phe, and Trp) suggests a cation-π interaction (22), but the ring of Y225 is more than 5 Å away from the bound Na+ in the crystal structure (23) and makes such interaction unlikely. All other residues at position 225 lead to the loss of Na+ effect, the result of either the lack of binding or failure to transduce binding into enhanced catalytic activity.
Figure 1.
Effect of residue 225 on the Na+ specificity of thrombin, measured as the ratio of the kcat/Km values for the hydrolysis of H-D-Phe-Pro-Arg-p-nitroanilide (FPR) in the presence of 200 mM NaCl or choline chloride, 5 mM Tris, 0.1% polyethylene glycol, pH 8.0 at 25°C (6). Binding of Na+ to wild type (Tyr) enhances kcat/Km nearly 25-fold. A significant enhancement also is observed for Phe, His, Met, Gln, Trp, and Ser, in decreasing order. All other residues show no significant difference between Na+ and choline.
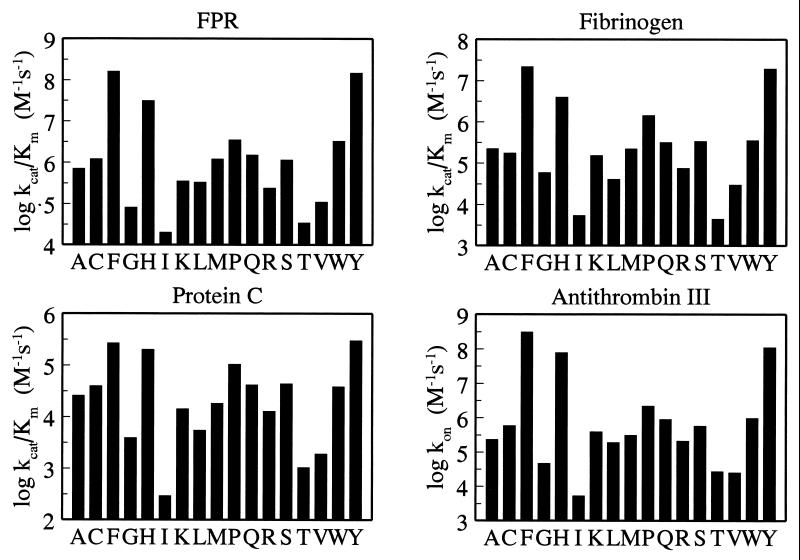
When the mutants were studied in their interaction with chromogenic and natural substrates, it was found that the nature of residue 225 influences the catalytic activity of the enzyme up to 8,000-fold (Fig. 2). This effect is too large to be accounted for solely by the loss of Na+ binding (Fig. 1), which is at most 30-fold for physiological ligands under similar solution conditions (7, 24). The decreased activity primarily is the result of increased Km and compromised substrate binding. Tyr and Phe are associated with the highest specificity (measured as kcat/Km), whereas Pro reduces activity up to 50-fold and reproduces the properties of the Na+-free, anticoagulant slow form of the wild type (7, 24). All other residues show comparable or significantly reduced activity relative to Pro. Short and hydrophobic side chains appear to be especially disruptive at position 225, and so are β-branched residues, whereas bulky ring structures are associated with high catalytic activity. The activity profile observed for a small tripeptide substrate that probes the interior of the active site is remarkably similar to that observed for the natural procoagulant substrate fibrinogen and the anticoagulant substrate protein C, whose cleavage requires binding to additional sites located as much as 20–25 Å away from the active site (24). The ability to interact with the natural inhibitor antithrombin III follows the same pattern as the other ligands and is compromised up to 60,000-fold in the Ile mutant.
Figure 2.
Effect of residue 225 on thrombin interactions. Plotted are the values (in log units) of kcat/Km for the hydrolysis of H-D-Phe-Pro-Arg-p-nitroanilide (FPR), fibrinogen, and protein C (in the presence of 5 mM CaCl2 and 100 nM rabbit thrombomodulin), and the kon values for inhibition of thrombin by antithrombin III (in the presence of 0.5 United States Pharmacopeia units/ml of heparin). Experimental conditions are: 5 mM Tris, 145 mM NaCl, 0.1% polyethylene glycol, pH 7.4 at 37°C (see legend to Fig. 1 for FPR). Measurements of Km and kcat for FPR hydrolysis by several mutants show that the reduced specificity is the result almost entirely of increased Km and compromised substrate binding. The four profiles are quite similar and suggest that perturbation of residue 225 affects the environment of the primary specificity site to which FPR, fibrinogen, protein C, and antithrombin III make contact.
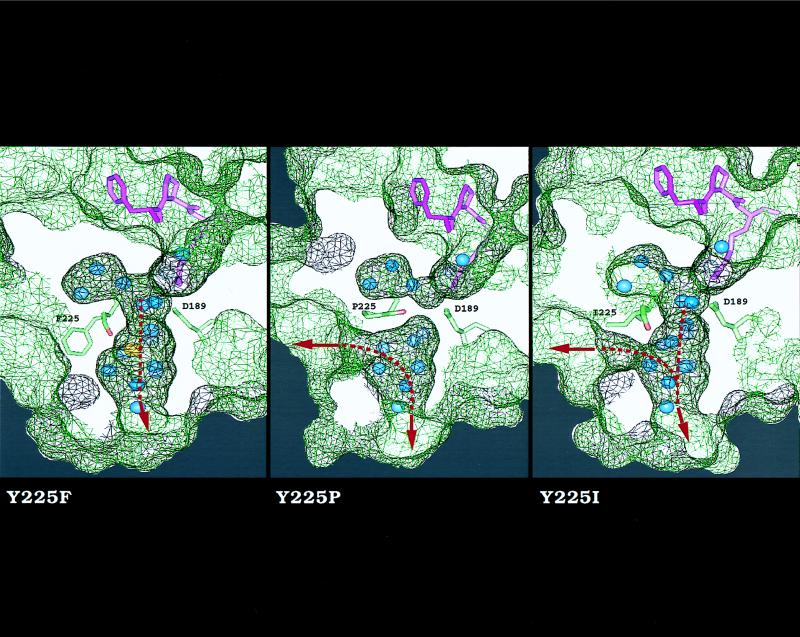
The similarity of the activity profiles in Fig. 2 suggests that the nature of residue 225 influences a structural domain recognized by small chromogenic substrates, natural substrates, and inhibitors alike. The only binding epitope common to all these ligands is the primary specificity site centered around D189. To better understand the molecular origin of the large perturbation in catalytic activity observed on substitution of Tyr at position 225, the mutants Y225F, Y225P, and Y225I were crystallized (Table 1). The Y225F and Y225I mutants span the entire spectrum of activities documented in Fig. 2, whereas the Y225P mutant carries the residue found in all serine proteases devoid of Na+ binding (6). A remarkable difference among these mutants was found on inspection of the water channel embedding the primary specificity site (Fig. 3). The architecture of this channel is highly conserved in serine proteases and is thought to play a key role in substrate recognition and specificity (26–28). In the Y225F mutant, the side chain of F225 is 90% buried in a hydrophobic pocket located on the outer wall of the primary specificity site and is positioned as Y225 in the wild type (11, 24). In this mutant, the channel is shaped as in the wild type (26) and connects the active site to an aperture at the bottom of the molecule delimited by residues of loops 180 and 220 (24). The bound Na+ is coordinated octahedrally as in the wild type (23) and occupies the middle portion of the channel, in close proximity (5 Å) to the side chain of D189 in the primary specificity site. This architecture is common to all proteases with Y225 or F225 (24). Replacement of Y225 with Pro produces a shift in the position of the carbonyl O atom of residue 224, as seen in proteases with P225 (6), rearranges the water molecules in the region, and abrogates Na+ binding. The replacement also causes two changes in the channel. The smaller side chain of Pro unveils a second aperture to the solvent distal to the 220-loop, whereas the reorientation of the carbonyl O atom of residue 224 occludes the channel in its middle, reducing the depth of the primary specificity site around D189. These changes reproduce the architecture of the channel found in all proteases with P225. Interestingly, the depth reduction of the primary specificity site in trypsin, carrying P225, relative to thrombin, carrying Y225, was suggested from functional studies long before crystallographic analysis (29). In the Y225I mutant, the channel is shunted laterally around residue 225 because of the smaller side chain of Ile and is also open in the middle because the carbonyl O atom of residue 224 is oriented as in the wild type.
Table 1.
Crystallographic data for the thrombin mutants Y225F, Y2265P, and Y225I
| Y225F | Y225P | Y225I | |
|---|---|---|---|
| Data collection | |||
| Wavelength, Å | 1.03 | 1.03 | 1.03 |
| Resolution, Å | 30−2.1 | 30−2.1 | 30−2.1 |
| Reflections, observed/unique (I/σ(I) > 1) | 68,774/18,509 | 76,087/17,519 | 72.954/16,825 |
| Completeness*, % | 94.1/89.6 | 88.0/88.2 | 82.9/81.9 |
| Rsym,† % | 4.8/13.1‡ | 4.2/7.1‡ | 4.9/10.4‡ |
| Unit cell dimensions, Å | a = 53.1, b = 75.1, c = 81.4 | a = 53.1, b = 75.1, c = 82.4 | a = 50.6, b = 73.7, c = 89.9 |
| Refinement | |||
| Resolution, Å | 6.0−2.1 | 6.0−2.1 | 6.0−2.1 |
| R factor, % | 19.7 | 19.8 | 22.5 |
| Rfree,§ % | 26.3 | 27.2 | 28.2 |
| Completeness,‡ %, F/σ(F) > 2, working set only§ | 87.5/83.1 | 82.4/81.7 | 78.9/77.6 |
| Number of protein atoms | 2,239 | 2,262 | 2,115 |
| Number of solvent molecules | 141 | 168 | 147 |
| 〈B〉 main chain atoms | 20.9 | 15.7 | 17.8 |
| 〈B〉 side chain atoms and solvent | 24.2 | 20.1 | 20.3 |
| Rmsd bond lengths,¶ Å | 0.010 | 0.010 | 0.011 |
| Rmsd angles,¶ ° | 1.59 | 1.57 | 1.58 |
| Rmsd in B for bonded atoms main chaine | 1.4 | 1.3 | 1.6 |
| Rmsd in B for bonded atoms side chaine | 2.6 | 2.4 | 2.5 |
Percentage of: overall completeness/completeness in the last resolution shell (2.18−2.10 Å).
Defined as Σ |I − 〈I〉|/Σ I, where I is the observed intensity and 〈I〉 is the average intensity from multiple observations of symmetry related reflections.
Rsym for: overall resolution range/last resolution shell (2.18−2.10 Å).
Calculated using 6.5% (Y225F and Y225P) and 4.0% (Y225I) data sets.
rms deviation (Rmsd) from ideal bond lengths and angles (25) and Rmsd in B factors of bonded atoms.
Figure 3.
Effect of residue 225 on the architecture of the water channel around the primary specificity site of thrombin. Shown is a cross section of the enzyme along the water channel that reveals the active site inhibitor PPACK (purple), D189 in the primary specificity site, residue 225 with the carbonyl O atom of residue 224 (red), buried water molecules (blue), and Na+ (yellow). The surface of the enzyme is rendered as a net (black, above the plan of section; green, below it). The side chain of residue 225 points away from D189 and makes no contact with PPACK. The Y225F mutant is practically identical to wild type (11) and shows a bound Na+ coordinated octahedrally by the carbonyl O atoms of K224 and R221a (not shown), and four water molecules (23, 26). In this mutant, the water channel connects the active site to an aperture at the bottom of the molecule (arrow). In the Y225P mutant, there is no evidence of bound Na+; the carbonyl O atom of K224 is shifted 70° toward the interior of the channel and occludes it in the middle. In addition, the channel is shunted laterally (arrow) around residue 225 because of the Y225P replacement. In the Y225I mutant, the channel has three apertures.
The architecture of the water channel around the primary specificity site observed in the three thrombin mutants suggests a possible correlation with the catalytic activity of the enzyme. Optimal activity is observed when the channel is shaped as in the wild-type thrombin or the Y225F mutant and Na+ is bound to its site. A lower, but significant, catalytic activity is observed when the channel is closed in its middle as in the Y225P mutant and all proteases carrying P225. The closure abrogates Na+ binding, but isolates D189 from the apparently detrimental effects of a three-way channel shaped as in the Y225I mutant of thrombin. This correlation is reinforced by the crystal structure of the trypsin from Fusarium oxysporum (30), carrying Ser at position 225. In this structure, the carbonyl O atom of residue 224 is oriented as in the proteases with P225 (6) and the channel is shaped as in the Y225P mutant of thrombin. This finding is consistent with the comparable catalytic activity of the Y225S and Y225P mutants.
The notable changes observed in the architecture of the water channel in the three thrombin mutants translate into minimal perturbations of regions that contact the active site inhibitor PPACK. There is no obvious structural change in the Y225I mutant compared with wild type or the Y225F mutant that can be reconciled immediately with the drastic loss of catalytic activity. It is likely that the perturbation induced by mutation of residue 225 predominantly affects the free form of the enzyme and is corrected by the presence of PPACK. Alternatively, the perturbation may be dynamic in nature and could manifest itself through changes in the mobility of the buried water molecules in the channel and the dielectric environment of the uncompensated charge of D189.
DISCUSSION
The importance of the architecture of the primary specificity site in the catalytic function of serine proteases has been recognized in previous studies (3–5), but the emphasis has been put on residues like G216 and G226, whose mutation obviously would compromise access to this site (2, 3). The importance of residue 225 has been overlooked, because it points outside the primary specificity site and away from D189 or other residues making contact with substrate. The structural determinants of protease activity and specificity therefore are more complex than currently believed (1–3).
In allosteric proteases carrying Tyr or Phe at position 225 (6), the aromatic side chain acts like a lid to close the shunt of the water channel connecting the primary specificity site to the aperture at the bottom of the molecule. This side chain is fixed in its position by optimal van der Waals interactions with the highly conserved residues V163 and V167, that are, however, quite variable in proteases carrying P225. Changes in the position of Y225 can influence Na+ binding and the shunt of the water channel and likely provide the trigger for modulating the catalytic activity of the enzyme as seen in thrombin and other allosteric proteases (24). The presence of a rigid Pro at position 225 makes such modulation impossible.
The information from the present study should facilitate the rational engineering of Na+ binding and enhance catalytic activity in proteases with P225, which would be beneficial in several areas of medical and biotechnological importance. Replacement of P225 with Tyr or Phe is certainly necessary for Na+ binding, but it may not be sufficient (31). Packing of the aromatic side chain in its hydrophobic environment must be optimized to achieve the finely tuned allosteric regulation seen in thrombin.
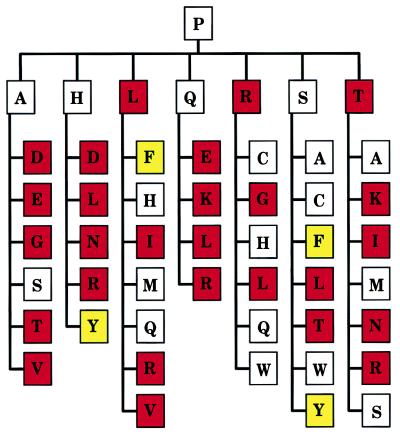
Na+ binding and optimal catalytic activity in serine proteases have emerged during evolution as a result of several changes involving residue 225 and its environment. Remarkably, the activity profiles obtained for thrombin (Fig. 2) recapitulate an important component of this evolutionary transition and help trace the pathway from Pro to Tyr at position 225 (Fig. 4). Starting from Pro in the ancestral serine protease, mutations one step away in the genetic code like Leu, Arg, and Thr would have produced derivatives with significantly lower catalytic activity. Mutations like Ala and Gln would have retained activity, but further mutations from these residues again would have compromised activity in most cases. His might have provided a possible intermediate on the Pro → Tyr pathway, but Ser (with a TCN codon) stands out as a safer and more flexible choice on the pathway leading to the most active derivatives Tyr and Phe. Ser with a TCN codon is present in γ-renin (mouse), granzymes D and E (mouse), and the trypsin from F. oxysporum. On the other hand, there is no example of His at position 225 and the Y225H substitution in coagulation factor IXa is associated with 4% activity compared with wild type and clinical manifestations of hemophilia B (32).
Figure 4.
Evolutionary transitions from P225 in the ancestral serine protease. Shown are the seven residues (in the single-letter code) one mutational step away from Pro in the genetic code (Ala, His, Leu, Gln, Arg, Ser, and Thr), and those one mutational step away from these residues, altogether defining 42 distinct pathways two mutational steps away from Pro. Residues are color-coded based on the associated catalytic activity toward FPR (see Fig. 2). White, residues causing less than 10-fold change in activity compared with Pro. Red, residues causing more than 10-fold loss of activity compared with Pro. Yellow, residues causing more than 10-fold gain in activity compared with Pro. Asn, Asp, and Glu were assumed to significantly compromise catalytic activity and function because the respective mutants of Y225 failed to express in quantities sufficient for characterization. Analysis of trace amounts of Y225D mutant reveals >100-fold reduction in catalytic activity relative to Pro. Furthermore, Asn, Asp, and Glu are not documented at position 225 in serine proteases. If conservation or enhancement of catalytic activity is assumed as an evolutionary criterion for transition, then Ser (with a TCN codon) would have provided the safest and most flexible intermediate on the pathway from Pro to residues with enhanced activity like Tyr and Phe.
We conclude that P225 was retained during evolution in most serine proteases because of its stabilizing structural role on the primary specificity site and the significant loss of activity produced by many alternative residues at position 225. In the vertebrates, or their immediate predecessors, a transition from Pro to Ser and then to Tyr (or Phe) took place most likely in conjunction with the onset of adaptive immunity and blood coagulation. The transition optimized the environment of the primary specificity site through the ability to bind Na+, a property that enhanced significantly the catalytic activity of the enzyme and was retained during evolution. Serine proteases of the complement retain traces of this transition. Pro is present in the enzymes of the alternative pathway (factors D and I), whereas Tyr is present in the enzymes of the lectin-dependent (MASP-1 and MASP-2) and classical (C1r and C1s) pathways that evolved at a later stage (33). Evolutionary pressure to mutate P225 must have occurred in prevertebrates and predated the TCN → AGY conversion of the codon for S195. In fact, a homologue of complement factor B in the sea urchin carries Y225 but a TCN codon for the active site Ser (34). Furthermore, both human MASP-1 and MASP-2 carry Y225, but MASP-1 has a TCN codon for S195 whereas MASP-2 has an AGY codon.
Acknowledgments
Use of the Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Energy Research, under contract No. W-31-109-Eng-38. This work was supported by a grant from CNR Biotecnologia (L.95/95) to S.C., National Institutes of Health Grant GM45948 to G.W., and grants from the National Institutes of Health (HL49413 and HL58141), the American Heart Association, and Monsanto-Searle to E.D.C.
ABBREVIATION
- PPACK
H-D-Phe-Pro-Arg-chloromethylketone
Footnotes
Data deposition: The structures reported in this paper have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID codes 2THF, 1THP, and 1B7X).
References
- 1.Lesk A M, Fordham W D. J Mol Biol. 1996;258:501–537. doi: 10.1006/jmbi.1996.0264. [DOI] [PubMed] [Google Scholar]
- 2.Perona J J, Craik C S. Protein Sci. 1995;4:337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perona J J, Craik C S. J Biol Chem. 1997;272:29987–29990. doi: 10.1074/jbc.272.48.29987. [DOI] [PubMed] [Google Scholar]
- 4.Craik C S, Largman C, Fletcher T, Roczniak S, Barr P J, Fletterick R J, Rutter W J. Science. 1985;228:291–297. doi: 10.1126/science.3838593. [DOI] [PubMed] [Google Scholar]
- 5.Hedstrom L, Szilagyi L, Rutter W J. Science. 1992;255:1249–1253. doi: 10.1126/science.1546324. [DOI] [PubMed] [Google Scholar]
- 6.Dang Q D, Di Cera E. Proc Natl Acad Sci USA. 1996;93:10653–10656. doi: 10.1073/pnas.93.20.10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang Q D, Guinto E R, Di Cera E. Nat Biotechnol. 1997;15:146–149. doi: 10.1038/nbt0297-146. [DOI] [PubMed] [Google Scholar]
- 8.Banner D W, D’Arcy A, Chene C, Winkler F K, Guha A, Konisberg W H, Nemerson Y, Kirchhofer D. Nature (London) 1996;380:41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
- 9.Guinto E R, Vindigni A, Ayala Y M, Dang Q D, Di Cera E. Proc Natl Acad Sci USA. 1995;92:11185–11189. doi: 10.1073/pnas.92.24.11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otwinowski Z. In: Proceedings of the CCP4 Study Weekend: Data Collection and Processing. Sawyers L, Isaacs N, Bailey S, editors. Warrington, U.K.: SERC Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- 11.Bode W, Turk D, Karshikov A. Protein Sci. 1992;1:426–471. doi: 10.1002/pro.5560010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 13.Jones T A, Thirup S. EMBO J. 1986;5:819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 15.Hodel A, Kim S-H, Brünger A T. Acta Crystallogr A. 1992;48:851–858. [Google Scholar]
- 16.Brunger A T. x-plor, version 3.1. New Haven, CT: Yale University; 1992. [Google Scholar]
- 17.Nayal M, Di Cera E. J Mol Biol. 1996;256:228–234. doi: 10.1006/jmbi.1996.0081. [DOI] [PubMed] [Google Scholar]
- 18.Rypniewski W R, Perrakis A, Vorgias C E, Wilson K S. Protein Eng. 1994;7:57–64. doi: 10.1093/protein/7.1.57. [DOI] [PubMed] [Google Scholar]
- 19.Roach J C, Wang K, Gan L, Hood L. J Mol Evol. 1997;45:640–652. doi: 10.1007/pl00006268. [DOI] [PubMed] [Google Scholar]
- 20.Banfield D K, MacGillivray R T A. Proc Natl Acad Sci USA. 1992;89:2779–2783. doi: 10.1073/pnas.89.7.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brenner S. Nature (London) 1988;334:528–530. doi: 10.1038/334528a0. [DOI] [PubMed] [Google Scholar]
- 22.Dougherty D A. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 23.Di Cera E, Guinto E R, Vindigni A, Dang Q D, Ayala Y, Wuyi M, Tulinsky A. J Biol Chem. 1995;270:22089–22092. doi: 10.1074/jbc.270.38.22089. [DOI] [PubMed] [Google Scholar]
- 24.Di Cera E. Trends Cardiovasc Med. 1998;8:340–350. doi: 10.1016/s1050-1738(98)00030-9. [DOI] [PubMed] [Google Scholar]
- 25.Engh R A, Huber R. Acta Crystallogr A. 1991;47:392–400. [Google Scholar]
- 26.Zhang E, Tulinksy A. Biophys Chem. 1997;63:185–200. doi: 10.1016/s0301-4622(96)02227-2. [DOI] [PubMed] [Google Scholar]
- 27.Krem M M, Di Cera E. Proteins. 1998;30:34–42. doi: 10.1002/(sici)1097-0134(19980101)30:1<34::aid-prot3>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 28.Sanschagrin P C, Kuhn L A. Protein Sci. 1998;7:2054–2064. doi: 10.1002/pro.5560071002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berliner L J, Bauer R S, Chang T L, Fenton J W, Shen Y Y. Biochemistry. 1981;20:1831–1837. doi: 10.1021/bi00510a018. [DOI] [PubMed] [Google Scholar]
- 30.Rypniewski W R, Hastrup S, Betzel C, Dauter M, Dauter Z, Papendorf G, Branner S, Wilson K S. Protein Eng. 1993;6:341–348. doi: 10.1093/protein/6.4.341. [DOI] [PubMed] [Google Scholar]
- 31.Vindigni A, Di Cera E. Protein Sci. 1998;7:1728–1737. doi: 10.1002/pro.5560070807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorland E C, Weinshenker B G, Liu J-Z, Ketterling R P, Vielhaber E L, Kasper C K, Ambritz R, Paredes R, Sommer S S. Thromb Haemostasis. 1995;74:1416–1422. [PubMed] [Google Scholar]
- 33.Farries T C, Atkinson J P. Immunol Today. 1991;12:295–300. doi: 10.1016/0167-5699(91)90002-B. [DOI] [PubMed] [Google Scholar]
- 34.Smith L C, Shih C-S, Dachenhausen S G. J Immunol. 1998;161:6784–6793. [PubMed] [Google Scholar]