Abstract
Although neuronal nitric oxide synthase (nNOS) plays a substantial role in skeletal muscle physiology, nNOS-knockout mice manifest an only mild phenotypic malfunction in this tissue. To identify proteins that might be involved in adaptive responses in skeletal muscle of knockout mice lacking nNOS, 2D-PAGE with silver-staining and subsequent tandem mass spectrometry (LC-MS/MS) was performed using extracts of extensor digitorum longus muscle (EDL) derived from nNOS-knockout mice in comparison to C57Bl/6 control mice. Six proteins were significantly (P≤ 0.05) more highly expressed in EDL of nNOS-knockout mice than in that of C57 control mice, all of which are involved in the metabolism of reactive oxygen species (ROS). These included prohibitin (2.0-fold increase), peroxiredoxin-3 (1.9-fold increase), Cu2+/Zn2+-dependent superoxide dismutase (SOD; 1.9-fold increase), heat shock protein β-1 (HSP25; 1.7-fold increase) and nucleoside diphosphate kinase B (2.6-fold increase). A significantly higher expression (4.1-fold increase) and a pI shift from 6.5 to 5.9 of peroxiredoxin-6 in the EDL of nNOS-knockout mice were confirmed by quantitative immunoblotting. The concentrations of the mRNA encoding five of these proteins (the exception being prohibitin) were likewise significantly (P≤ 0.05) higher in the EDL of nNOS-knockout mice. A higher intrinsic hydrogen peroxidase activity (P≤ 0.05) was demonstrated in EDL of nNOS-knockout mice than C57 control mice, which was related to the presence of peroxiredoxin-6. The treatment of mice with the chemical NOS inhibitor l-NAME for3 days induced a significant 3.4-fold up-regulation of peroxiredoxin-6 in the EDL of C57 control mice (P≤ 0.05), but did not alter its expression in EDL of nNOS-knockout mice. ESR spectrometry demonstrated the levels of superoxide to be 2.5-times higher (P≤ 0.05) in EDL of nNOS-knockout mice than in C57 control mice while an in vitro assay based on the emission of 2,7-dichlorofluorescein fluorescence disclosed the concentration of ROS to be similar in both strains of mice. We suggest that the up-regulation of proteins that are implicated in the metabolism of ROS, particularly of peroxiredoxin-6, within skeletal muscles of nNOS-knockout mice functionally compensates for the absence of nNOS in scavenging of superoxide.
Neuronal nitric oxide synthase (nNOS), by whose action the gaseous signalling molecule nitric oxide (NO) is produced, exists at high concentrations in the skeletal muscles of mammals, including mice, rats and humans (reviewed in Reid, 1998; Stamler & Meissner, 2001). Various isoforms of nNOS have been characterized at the protein level. The two major species include a 160 kDa α-form and a 140 kDa β-form, which have different N-terminal domains. The α-form uniquely manifests a 237 amino acid long stretch, which is encoded by exon 2 (Brenman et al. 1996). In skeletal muscle, both α-and β-nNOS isoforms contain an alternatively spliced 34 amino acid long μ-exon in the central portion of their tertiary structures, which has no significant bearing on their catalytic properties (Silvagno et al. 1996).
Expressed as a dimer, nNOS is found preferentially in fast twitch-oxidative (type IIa) muscle fibres of rats (Kobzik et al. 1994; Planitzer et al. 2001) and is thus enriched in skeletal muscle with a high proportion of such fibres (e.g. extensor digitorum longus muscle (EDL) of the lower limb). In contrast, nNOS is preferentially expressed in type I muscle fibres in humans (Frandsen et al. 2000). Targeting to the sarcolemma is mediated mainly by its N-terminal PDZ domain, which facilitates the integration of nNOS into the dystrophin glycoprotein complex (Brenman et al. 1995). This large protein aggregate is down-regulated in patients suffering from Duchenne's muscular dystrophy (DMD), but it is unclear whether nNOS is involved in the aetiology of this disease (reviewed in Campbell & Stull, 2003).
Many functions have been attributed to nNOS within skeletal muscle (reviewed in Reid, 1998; Stamler & Meissner, 2001). Oxidative metabolism is inhibited by the activity of nNOS, which reduces the depletion of energy stores and supports anaplerotic reactions (Kapur et al. 2000). In contracting skeletal muscles of the rat limb, the raised levels of nNOS-derived NO promote relaxation (Kobzik et al. 1994). Furthermore, NO supports the uptake of glucose by muscle fibres during contraction although the contribution of nNOS and the corresponding molecular mechanisms underlying this modulation is controversial (Roberts et al. 1997; Higaki et al. 2001). Other investigations indicate that the nNOS-mediated production of NO plays a role in paracrine signalling, facilitating the interaction between skeletal muscle fibres and the adjacent microvasculature (Thomas et al. 2003; Percival et al. 2008). In particular, nNOS has been shown to control the cGMP-dependent relaxation of smooth muscle cells in skeletal muscle (Grange et al. 2001), thereby aiding eNOS-facilitated vasodilation. Furthermore, NO produced by the activity of nNOS also enhances angiogenesis within skeletal muscle (Hudlicka et al. 2000; Williams et al. 2006a).
Although nNOS is involved in pivotal physiological processes in many tissues, nNOS-knockout mice with deleted exon 2 are vital (Huang et al. 1993). The most remarkable phenotypic change in these mice is an increase in the size of the pylorus, owing to the absence of a functional plexus myentericus (Huang et al. 1993). Ageing nNOS-knockout mice manifest hypertrophy of the heart ventricles, but do not suffer from hypertension (Barouch et al. 2002). The peripheral organs of nNOS-knockout mice are less sensitive to insulin than those of their wild-type counterparts (Shankar et al. 2000). As a consequence of NO dependency, both the primary discharge of urine and the re-absorption of bicarbonate are reduced in the kidneys of nNOS-knockout mice (Wang et al. 2000).
So far, a loss of tissue weight is the only skeletal muscle-associated alteration that has been observed in nNOS-knockout mice (Huang, 1999; Wadley et al. 2007; Kobayashi et al. 2008). To our opinion, this change in macroscopic phenotypic appears to be rather mild with respect to the various important functions assigned to nNOS in skeletal muscle. Thus, we hypothesize the existence of adaptive mechanisms that functionally compensate for the absence of nNOS in skeletal muscle of nNOS-knockout mice.
To search for proteins that could be involved in such a molecular adaptation, we performed proteomics analysis (which included two dimensional polyacrylamide electrophoresis (2D-PAGE) and subsequent liquid chromatography–mass spectrometry/mass spectrometry (LC-MS/MS). Six proteins involved in the metabolism of reactive oxygen species (ROS) were identified that exhibit differential expression patterns in EDL between nNOS-knockout mice and C57/Bl6 control mice.
Methods
Ethical approval
All animal experiments were approved by the Freie Universität Berlin and by the state authorities for animal welfare (LAGETSI Berlin).
Animal treatment and tissue preparation
C57Bl/6 control and nNOS-knockout mice were bred in our animal care facility under standard conditions (food (Sniff, Soest, Germany) and water ad libitum; 22°C room temperature; 60% air humidity; 12 h light and 12 h dark cycle; one animal per cage). The knockout strain was purchased from Jackson Laboratories (Bar Harbour, ME, USA) and originally produced by Huang and colleagues (Huang et al. 1993). In contrast to an earlier report (Brenman et al. 1996), the nNOS-knockout mice used in this study do not express any residual nNOS isoform (β, γ) in skeletal muscles at the protein level (Baum et al. 2002). Healthy, 3-to 5-month-old male mice weighing 25–30 g with no bias in age were used for this study. To our experience, the skeletal muscle phenotype of mice does not alter significantly within this age, as e.g. the fibre types of the skeletal muscle fibres and the microvasculature are already established/remain unchanged. They were killed in a generally anaesthetized state induced with Narcoren (Merial, Halbergmoos, Germany) by excision of the heart.
N-Nitro-l-arginine methyl ester (l-NAME), which is a specific inhibitor for all NOS forms, was obtained from Sigma (Taufkirchen, Germany) and freshly dissolved in 150 mm NaCl as previously described (Baum et al. 2004). In one group of mice (six C57 control mice and six nNOS-knockout mice), l-NAME was injected i.p. in a concentration of 30 mg (kg body weight)−1 while the second group of mice (six C57 control mice and six nNOS-knockout mice) received the same volume of saline.
Endothelial cell fractions were prepared as described before (Da Silva-Azevedo et al. 2002). Tissue sections 10 μm thick were obtained as previously reported (Baum et al. 2002).
Two-dimensional polyacrylamide gel electrophoresis (2D-PAGE)
Each extensor digitorum longus muscle (EDL) was ground in a liquid nitrogen-frozen condition and then homogenized in 250 μl of isoelectric focusing (IEF) sample buffer (7 m urea, 2 m thiourea, 2% (v/v) ampholytes (Amersham/GE Healthcare, Munich, Germany), 1% (w/v) dithiothreitol (DTT) and 2% (w/v) CHAPS). Insoluble material was removed by centrifugation at 20 000 g for 15 min. Proteins were precipitated by using the 2D Clean Up kit (Amersham) and re-dissolved in IEF sample buffer. The protein content was estimated using the Ethan protein 2D quant determination kit (Amersham). Sixty micrograms of protein per 300 μl of IEF sample buffer was used for rehydration of 18 cm long immobilized pH-gradient (IPG) strips covering the non-linear pH range 3–10 (Amersham). After overnight incubation, IEF was performed using a Multiphor II chamber (Pharmacia) at 20°C (5 h at 500 V and 14.5 h at 3500 V).
After IEF, IPG strips were either stored at –70°C or directly transferred to equilibration buffer (6 m urea, 30% (v/v) glycerol, 2% (w/v) SDS in 0.05 m Tris-HCl, pH 8.8) for 30 min. During the first 15 min, the equilibration buffer was supplemented with 1% (w/v) DTT, which was replaced with 4% (w/v) iodoacetamide during the second half of the incubation period.
Electrophoretic separation of the proteins in the second dimension (16 mA/gel) at 20°C in the Ettan six multichamber (Amersham) was achieved using 7–14% gradient SDS-polyacrylamide gels, which contained 1,4-bis(acryloyl)-piperazine as cross-linker.
The gels were stained with silver nitrate (Shevchenko et al. 1996). For staining with Coomassie Blue, the gels were fixed overnight in 2% (v/v) phosphoric acid and 50% (v/v) methanol and then rinsed with distilled water for 10 min. They were then incubated for 1 h in a solution containing 2% (v/v) phosphoric acid, 17% (w/v) ammonium sulphate and 34% (v/v) methanol prior to addition of 0.066% (w/v) Coomassie Blue G-250. The gels were stained for 3 days on a shaking platform and then de-stained for 5 min in water.
The molecular weights of the proteins were determined from co-separated standard markers. The isoelectric points (pI) of the proteins were deduced by a software-supported (PD Quest, Biorad) assignment of prominent spots on the master gels to defined ones on 2D maps in the Expasy data base (http://www.expasy.org/cgi-bin/map2/def?_MOUSE). Gels were scanned with a flatbed scanner (ScanMaker 9800 XL, Microtek, Willich, Germany) and images were processed with PD Quest software.
The absolute density of each spot was determined on silver-stained gel duplicates of five C57Bl/6 control mice and nNOS-knockout mice. To identify differentially expressed proteins, the 20 densitometric values assigned to each of the spots were subjected to statistic analysis.
LC-MS/MS
Spots of interest were excised from the gels, cut into five cubes of 1 mm2 size and incubated in 50 mm ammonium bicarbonate, pH 8.0 (Fluka, Buchs, Switzerland) containing 50% (v/v) acetonitrile (Chromasolve LC-MS grade, Fluka) for 15 min. Trypsin digestion, peptide extraction and tandem mass spectrometry (LC-MS/MS) were performed as described before (Stalder et al. 2008). The collision-induced dissociation (CID) spectra were analysed using PHENYX software in conjunction with Uniprot mammalian protein database release 48.8 (GeneBio, Geneva, Switzerland). As minimal criteria for peptide identification, a PHENYX P value ≤ 10−8 together with trypsin-specific cleavage at the N-and C-termini was mandatory. Protein identification was accepted only if two peptides registered positive in each of two independent analyses.
Immunoblotting
Tissue homogenates were extracted in 20 mm sodium phosphate buffer (pH 7.8) containing 150 mm NaCl, 1% (w/v) Triton X-100, 50 mm sodium fluoride, 1 mm sodium vanadate, 2.5 mm EDTA, 1 mm phenylmethylsulphonylfluoride, 2 μg ml−1 leupeptin, 2 μg ml−1 aprotinin and 2 μg ml−1 pepstatin A. Immunoblotting was performed after the electrophoretic separation of 50 μg of protein per lane after SDS-PAGE on 12% gels with an anti-peroxiredoxin-6 antiserum (diluted 1: 10 000), which was provided by Drs A. Fisher and Y. Manevich (Institute for Environmental Medicine, University of Pennsylvania School of Medicine, Philadelphia, PA, USA). For additional immunoblotting, monospecific antibodies recognizing the following proteins were used: prohibitin (Biolegend, San Diego, CA, USA, in a 1: 2000 dilution); peroxiredoxin-3 (Ab Frontier, Seoul, South Korea); SOD1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA, in a 1: 400 dilution); HSP25 (Calbiochem; Darmstadt, Germany, in a 1: 5000 dilution); NDP kinase B (Santa Cruz, in a 1: 400 dilution); eNOS (BD Biosciences Pharmingen; Heidelberg, Germany, in a 1: 500 dilution) and iNOS (BD Biosciences Pharmingen, in a 2000 dilution).
The development of the immunoblots, the scanning procedure and the densitometric analysis were performed as previously described (Da Silva-Azevedo et al. 2002).
Quantitative real-time PCR
Total RNA was prepared from EDL of mice using the RNeasy Mini kit (Qiagen, Hilden, Germany) in accordance with the manufacturer's protocol. RNA concentrations were determined photometrically. cDNA was synthesized by reverse transcription for 1 h at 42°C using 2 μg of total RNA, 1 μg of oligo-dT primers and 200 U of M-MLV reverse transcriptase (Promega, Mannheim, Germany). The cDNA samples thereby obtained were analysed by quantitative real-time PCR using gene-specific pairs of primers that had been selected with primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) applying a protocol that has been described before (Chlench et al. 2007). The absolute mRNA copy numbers of the genes per microgram of RNA were determined as previously described (Williams et al. 2006b). Non-RT and non-template reactions were run as negative controls.
In vitro DCF fluorescence assay
A 2 mm solution of 2,7-dichlorofluorescein (DCFH) was prepared from 2,7-dichlorofluorescein diacetate (DCFH-DA, Invitrogen, Eugene, OR, USA) as described by Silveira et al. (2003). Therefore, 10 mg of DCFH-DA was dissolved in 2 ml of methanol prior to mixing with 2 ml of 0.05 m NaOH (pH 7.0) at ambient temperature. After 30 min, the stock solution was neutralized with 5 ml of 50 mm sodium dihydrogen phosphate (pH 7.4) and stored at −80°C.
The in vitro DCF fluorescence assay was performed to quantify both the hydrogen peroxidase activity and the intrinsic production of ROS within the EDL. The assay was carried out as described (Silveira et al. 2003) with minor modifications. It is based on the oxidation of xanthine by xanthine oxidase to yield uric acid and hydrogen peroxide, which then oxidizes the non-fluorescent DCFH to the fluorescent 2,7-dichlorofluorescin (DCF). The reaction mixture consisted of 0.1 mm of xanthine (Merck, Darmstadt, Germany), 1 mU of xanthine oxidase (Roche, Mannheim, Germany) and 1 μm DCFH in a total volume of 1 ml. The oxidation reaction was initiated by the addition of xanthine and conducted at 37°C for up to 25 min. The oxidation of DCFH was quantified in a fluorescence spectrophotometer (Perkin Elmer 650-10S, Rodgau-Juegesheim, Germany) using an excitation wavelength set at 488 nm and an emission wavelength at 525 nm.
In order to evaluate the specificity of the assay, the in vitro DCF fluorescence assay was carried out with SOD (7 U) and catalase (35 U) as external superoxide anion and hydrogen peroxide scavengers in accordance to Silveira et al. (2003).
For the quantification of hydrogen peroxidase activity within EDL, 50 μg of the proteins from the muscle homogenates isolated without the use of detergents (or, alternatively, 50 μg of bovine serum albumin as control) was added to the reaction mixture. To deplete the EDL homogenates of peroxiredoxin-6, 50 μg of the proteins from EDL homogenate was incubated with 2 μl of anti-peroxiredoxin-6 antiserum and 5 mg of protein A-sepharose (Sigma) for 2 h at 4°C under agitation. The anti-peroxiredoxin-6 antiserum was exchanged by 2 μl of preimmune serum to conduct control incubations. After centrifugation at 800 g for 20 s, the supernatant was withdrawn and used for the assay.
To quantify the intrinsic production of ROS within the EDL, 200 μg of protein from the EDL homogenates was incubated with DCFH in the absence of xanthine/xanthine oxidase.
Electron spin resonance spectrometry
The levels of free radicals within the EDL were determined by electron spin resonance (ESR) spectrometry using the cell-permeant 1-hydroxy-3-methoxycarbonyl-2,2,5,5-tetramethyl-pyrrolidine (CMH) as spin probe, which preferentially detects superoxide (Dikalov et al. 2007). ESR spectra were obtained and analysed by Dr Bruno Fink (Noxygen, Elzach, Germany) as described (Dikalov & Fink, 2005). For sample collection, the EDL were minced with scissors. After weighing of the tissue, samples of 20–50 mg was re-suspended in ice-cold Krebs-Hepes buffer containing 25 μm of deferoxamine and 5 μm of sodium diethyldithiocarbamate. Subsequently, CMH was added to a final concentration of 500 μm. Blank values without CMH or tissue were collected in parallel. After incubated at 37°C for 15 min, the reaction was stopped by cooling on ice and then treated as described (Dikalov & Fink, 2005).
Statistics
Numerical data are expressed as mean values together with the standard deviation. The statistical significance of differences was evaluated using Student's t test for unpaired data or a two-way analysis of variance (ANOVA) with Tukey's post hoc analysis (hydrogen peroxide reductase activity in the DCF fluorescence assay; peroxredoxin-6 expression after l-NAME application) with the P-value being set at less than 0.05.
Results
Comparison of the total body weight of age-matched male animals revealed C57Bl/6 control mice to be 18% heavier than nNOS-knockout mice (n= 14, data not shown). EDL and TA muscles of C57 control mice weighed significantly more than those of nNOS-knockout mice (EDL: 8.8 ± 1.5 mg versus 6.5 ± 1.5 mg and TA: 39.4 ± 8.8 mg versus 31.2 ± 6.6 mg; data not shown).
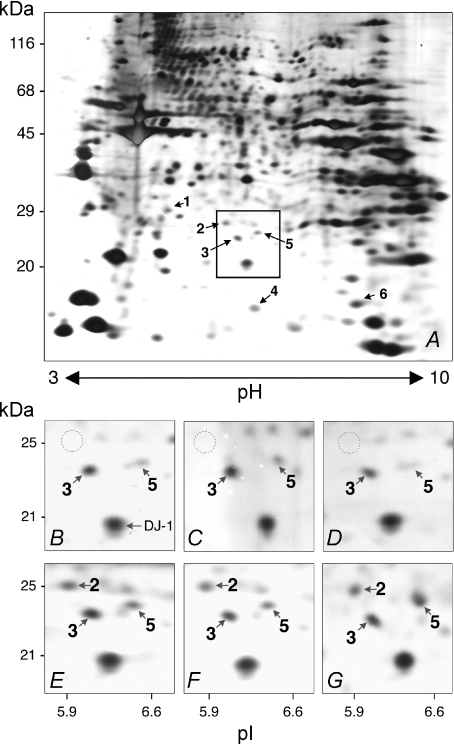
To identify differences in the protein expression patterns between C57 control mice and nNOS-knockout mice, EDL extracts were subjected to 2D-PAGE. Approximately 800 proteins were detected on silver-stained 2D-gels (Fig. 1). Six proteins were expressed at significantly higher levels in the EDL of the nNOS-knockout mice than in that of the C57 control mice. These six differentially expressed proteins were identified by LC-MS/MS (Table 1). Peroxiredoxin-6 was subject to the greatest fluctuations as it was apparently undetected in the EDL of C57 control mice but always prominent in EDL of nNOS-knockout mice with a molecular mass of 25 kDa and pI of 5.9 (Fig. 1). The other five proteins that were significantly up-regulated in EDL of nNOS-knockout mice compared to C57 control mice were prohibitin (2.0-fold), peroxiredoxin-3 (1.9-fold), Cu2+/Zn2+-dependent superoxide dismutase (SOD1; 1.9-fold), heat shock protein β-1 (HSP25; 1.7-fold) and nucleoside diphosphate kinase B (NDP kinase B; 2.6-fold).
Figure 1. 2D-PAGE of proteins isolated from EDL of nNOS-knockout mice and C57 control mice.
A, 60 μg of protein derived from the urea extract isolated from the EDL of a nNOS-knockout mouse was subjected to 2D-PAGE after IEF on IPG strips covering the non-linear pH range 3–10. Numbered arrows point to spots that were significantly more intense in the EDL extracts of nNOS-knockout mice than in the co-analysed ones of C57 control mice on the silver-stained gel after densitometric analysis as listed in Table 1. The box indicates the gel area that is shown in higher magnification in panels B–G. B–G, spot 2 (molecular mass 25 kDa; pI 5.9) was present only in nNOS-knockout mice (E–G), not C57 control mice (B–D), as exemplified individually for three animals of each strain. LC-MS/MS revealed this protein to be peroxiredoxin-6. The spot marked as ‘DJ-1’ in B denotes a protein whose levels were not influenced by nNOS-deficiency. The areas corresponding to spot 2 are encircled in B–D.
Table 1.
Proteins whose expression levels were higher in the EDL of nNOS-knockout mice than in that of C57Bl/6 control mice as determined by 2D-PAGE
| Spot | Estimated mol. mass (kDa) | Estimated pI | Protein | Swissprot accession number | Sequence coverage | Theoretical mol. mass (kDa) | Theoretical pI | Fold change in expression nNOS-knockout/C57 control | P |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 30 | 5.4 | Prohibitin | P67778 | 24.3% | 30 | 5.6 | 2.0 | 0.002 |
| 6 peptides | |||||||||
| 2 | 25 | 5.9 | Peroxiredoxin 6 | O08709 | 52.5% | 25 | 5.7 | Not quantifiable | < 0.0001 |
| 8 peptides | |||||||||
| 3 | 22 | 6.1 | Peroxiredoxin 3 | P20108 | 27.8% | 22 | 5.7 | 1.9 | 0.030 |
| 5 peptides | |||||||||
| 4 | 16 | 6.4 | Cu2+/Zn2+-superoxide dismutase (SOD1) | P08228 | 32.7% | 16 | 6.0 | 1.9 | 0.047 |
| 4 peptides | |||||||||
| 5 | 23 | 6.4 | Heat-shock protein | P14602 | 33.9% | 23 | 6.1 | 1.7 | 0.007 |
| β-1 (HSP25) | 7 peptides | ||||||||
| 6 | 17 | 7.6 | Nucleoside diphosphate | Q01768 | 66.5% | 17 | 7.0 | 2.6 | 0.002 |
| kinase B (NDP kinase B) | 8 peptides |
The positions of the spots in the 2D-gels were established by comparison using protein markers of known molecular mass and pI. Quantitative expression data (‘Fold change’) were based on the densitometric analysis of gel duplicates of 5 animals of each strain. The 6 proteins were identified by LC-MS/MS. As peroxiredoxin-6 was not detectable in EDL of C57 control mice, its ‘fold exchange’ was not quantifiable.
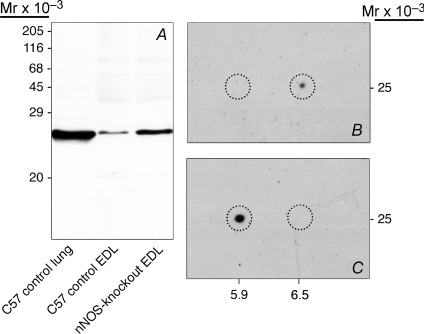
Using anti-peroxiredoxin-6 antibodies, a single 25 kDa band was detected by immunoblotting in EDL and lung of C57 control mice as well as in EDL of nNOS-knockout mice (Fig. 2A). The size of this band corresponds to the established molecular mass of peroxiredoxin-6. The densitometric analysis demonstrated peroxiredoxin-6 to be expressed at a 4.1-fold significantly higher expression level within the EDL of nNOS-knockout mice than within that of C57 control mice while its concentration was 18.5-fold significantly higher in the lung than in the EDL of C57 control mice (n= 6; data not shown). 2D-immunoblotting furthermore showed the peroxiredoxin-6-immunoreactive 25 kDa protein to have a pI of 6.5 in EDL of C57 control mice, while it exhibited a pI of 5.9 in EDL of nNOS-knockout mice (Figs 2B and C).
Figure 2. Immunoblotting confirms that peroxiredoxin-6 is markedly up-regulated in the EDL of nNOS-knockout mice.
A, after SDS-PAGE, immunoblotting with anti-peroxiredoxin-6 antibodies revealed a single band (molecular mass 25 kDa) in the EDL extracts of nNOS-knockout mice. This band was only slightly detectable in the EDL extracts but prominent in lung extracts of C57 control mice. B and C, after 2D-PAGE, immunoblotting with anti-peroxiredoxin-6 antibodies demonstrated the presence of a 25 kDa protein with a pI of 6.5 in the EDL extracts of C57 control mice (B) and with pI of 5.9 in the EDL extracts of nNOS-knockout-mice (C). The corresponding areas (molecular mass of 25 kDa and pI of 5.9 or 6.5) in B and C are encircled.
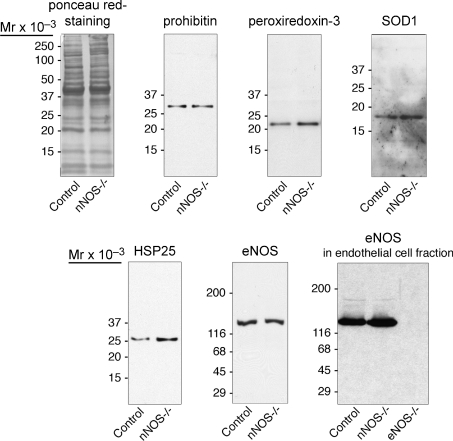
Additional immunoblotting was performed with monospecific antibodies against the other five proteins, which were found to be up-regulated in nNOS-knockout-mice (Fig. 3). Densitometric analysis of samples (n= 4) revealed HSP25 (4.6-fold), peroxiredoxin-3 (1.7-fold) and SOD1 (1.7-fold) to be significantly more highly expressed in the EDL of nNOS-knockout mice than in that of C57 control mice (data not shown). The protein levels of prohibitin (0.7-fold decrease) did not differ significantly between the two mice strains. The antibodies against NDP kinase B that we tested did not specifically recognize this protein (data not shown). Similar levels of eNOS were detected in homogenate and endothelial cell fraction of EDL in C57 control mice and nNOS-knockout mice (Fig. 3), while iNOS protein was not noticeable (data not shown).
Figure 3. Quantitative immunoblotting analysis.
Fifty micrograms of protein derived from the EDL homogenates of C57 control mice and nNOS-knockout mice were subjected to immunoblotting with antibodies against four proteins being present in the 2D-PAGE analysis at higher concentrations in the EDL of nNOS-knockout mice than in that of C57 control mice and against eNOS. The levels of peroxiredoxin-3, SOD1 and HSP25 were higher in the EDL of nNOS-knockout-mice than in that of C57 control mice, while those of prohibitin and eNOS (determined also in the vascular endothelial cell fraction) did not vary between the two strains. As loading control, a nitrocellulose matrix stained with ponceau red is shown. Representative examples of n= 4 immunoblots for each protein and mouse strain.
The mRNA expression levels of the six modulated proteins were determined by quantitative RT-PCR (Table 2). This analysis revealed significantly higher mRNA concentrations of peroxiredoxin-6 (2.7-fold), peroxiredoxin-3 (2.3-fold), HSP25 (1.6-fold), NDP kinase B (1.5-fold) and SOD1 (1.4-fold) in the EDL of nNOS-knockout mice than in that of C57 control mice. The mRNA levels of prohibitin (1.5-fold increase) did not differ significantly between the two strains.
Table 2.
Quantitative RT-PCR analysis for determination of mRNA concentrations of the six proteins being present at higher concentrations in the EDL of nNOS-knockout mice than in that of C57 control mice
| Copy no. in EDL (per μg of mRNA) |
||||
|---|---|---|---|---|
| Gene | C57 control | nNOS-knockout | Fold mRNA change* | P |
| Prohibitin | 141 633 ± 97 407 | 80 613 ± 68 178 | 0.57 | 0.424 |
| Peroxiredoxin-6 | 5 300 000 ± 2 671 573 | 22 640 000 ± 5 980 100 | 4.27 | 0.010 |
| Peroxiredoxin-3 | 1 396 333 ± 742 856 | 3 729 250 ± 1 664 531 | 2.67 | 0.049 |
| Cu2+/Zn2+-superoxide dismutase (SOD1) | 347 000 ± 122 233 | 656 250 ± 162 133 | 1.89 | 0.031 |
| Heat-shock protein β-1 (HSP25) | 400 167 ± 86 314 | 805 800 ± 490 732 | 2.01 | 0.026 |
| Nucleoside diphosphate kinase B (NDP kinase B) | 508 333 ± 138 564 | 968 500 ± 310 724 | 1.91 | 0.045 |
Fold change in mRNA expression nNOS-knockout/C57 control. The mRNA levels were quantified by RT-PCR. The copy numbers of each gene given per μg of RNA were determined using external standards. Mean values for 5 animals of each strain are represented together with the standard deviation.
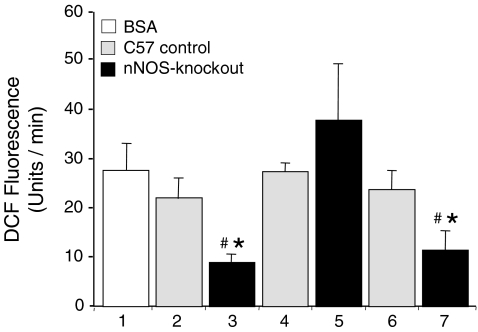
Peroxiredoxin-6-dependent hydrogen peroxidase activity within the EDL of nNOS-knockout mice and C57 control mice was quantified using the DCF fluorescence assay, which is based on the incubation of xanthine with xanthine oxidase to produce hydrogen peroxide which then oxidizes DCFH. As observed in preliminary experiments, the incubation with catalase lowered the DCF fluorescence by almost 40%, while incubation with SOD had no effect suggesting that particularly hydrogen peroxide and not superoxide is generated and quantified by this approach (data not shown). As shown in Fig. 4, a strong increase of DCF fluorescence was measured in the control experiment with 50 μg of bovine serum albumin (BSA) in the incubation buffer (28 ± 6 U min−1). When the assay was performed in the presence of 50 μg of protein that was derived from EDL homogenates of C57 control mice, the DCF fluorescence was non-significantly lower (22 ± 5 U min−1). In contrast, in the presence of a similar amount of protein that was derived from EDL homogenates of nNOS-knockout mice, the DCF fluorescence increase was significantly dropped (9 ± 2 U min−1). If the EDL homogenates had been subjected to immunoprecipitation with anti-peroxiredoxin-6 antibodies prior to performing the assay, then the DCF fluorescence in samples of C57 control mice (28 ± 2 U min−1) and nNOS-knockout mice (37 ± 13 U min−1) did not differ significantly from that in the control incubations with BSA. These data suggest that the hydrogen peroxide reductase activity that was present in the EDL extracts of nNOS-knockout mice is mainly associated with peroxiredoxin-6. If the EDL homogenates had been exposed to antibodies of a preimmune serum prior to performance of the assay, then DCF fluorescence values were similar to those registered for the EDL homogenates not subjected to immunoprecipitation, namely, 24 ± 4 U min−1 and 11 ± 5 U min−1 for C57 control mice and nNOS-knockout mice, respectively.
Figure 4. The peroxiredoxin-6-dependent hydrogen peroxide-degrading activity of the EDL is higher in nNOS-knockout than in C57 control mice.
The oxidation of xanthine by xanthine oxidase generates hydrogen peroxide, which oxidizes the non-fluorescent DCFH to the fluorescent DCF. The fluorescence signal emitted at a wavelength of 525 nm was measured after a 5 min incubation period at 37°C. The assay was performed in the presence of 50 μg aliquots of bovine serum albumin (1: control) and of proteins from EDL homogenates that were derived from C57 control (2) and nNOS-knockout mice (3). Prior to the assay, some of the EDL homogenates were subjected to immunoprecipitation with antibodies against peroxiredoxin-6 (4: C57 control; 5: nNOS-knockout) or a pre-immune serum (6: C57 control; 7: nNOS-knockout). Values represent mean ± standard deviation of 6 independent experiments. *Values in nNOS-knockout mice differ significantly from values in C57 control mice. #Values differ significantly from values of controls with BSA.
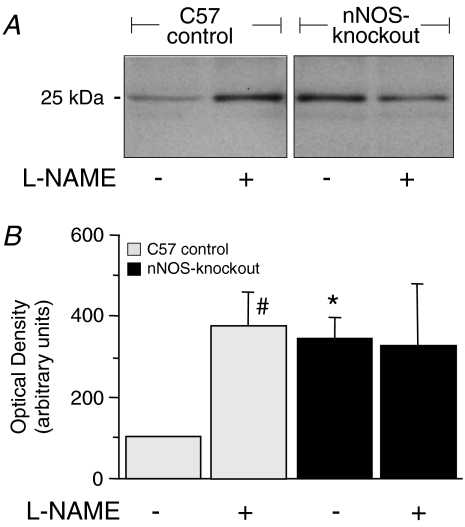
The acute regulation of peroxiredoxin-6 expression in the EDL was analysed after administrating the mice for 3 days with the NOS inhibitor l-NAME. Such treatment induced a significant increase of peroxiredoxin-6 in EDL of C57 control mice (3.7-fold at the protein level (immunoblotting, Fig. 5) and 1.5-fold at the mRNA level (qRT-PCR; 8 761 800–12 984 000 copies per μg RNA; n= 4; data not shown), but no significant change in that for the nNOS-knockout strain. The expression of the five other genes/proteins found to be differentially expressed in the 2D-PAGE analysis was not significantly altered in EDL of either C57 control mice or nNOS-knockout mice after l-NAME treatment for 3 days at both the protein (prohibitin: 0.8-fold; peroxiredoxin-3: 0.8-fold; SOD1: 1.4-fold; HSP25: 0.9-fold; NDP kinase B: 1.3-fold; n= 4; data not shown) and mRNA level (prohibitin: 0.7-fold; peroxiredoxin-3: 0.9-fold; SOD1: 1.4-fold; HSP25: 1.5-fold; NDP kinase B: 1.1-fold; n= 4; data not shown).
Figure 5. Chemical inhibition of NOS for three days in vivo induces the expression of peroxiredoxin-6 within the EDL.
A, C57Bl/6 control and nNOS-knockout mice were treated for 3 days with the NOS-inhibitor l-NAME. EDL extracts were then subjected to immunoblotting with anti-peroxiredoxin-6 antibodies. B, densitometric quantification of the 25 kDa peroxiredoxin-6 band. The density value for the EDL in untreated control mice was set at 100%. Mean values for 6 independent experiments are represented together with the standard deviation. *Values in nNOS-knockout mice differ significantly from values in C57 control mice with the same treatment. #Values in treated mice differ significantly from values in untreated mice.
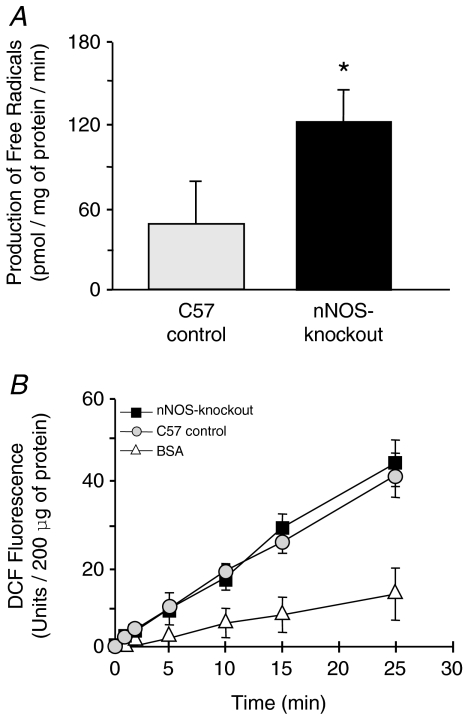
The concentrations of the various ROS were quantified in the EDL using two different approaches, namely ESR spectrometry (Fig. 6A) and the DCF fluorescence assay (Fig. 6B). Using the first approach, significant 2.5 times higher levels of free radicals were found in nNOS-knockout mice (123 ± 25 pmol (mg protein)−1 min−1) than in C57 control mice (49 ± 35 pmol (mg protein)−1 min−1). Using the second approach, the concentration of ROS was found to be similar in the EDL homogenates of nNOS-knockout mice (44 ± 6 U (200 μg protein)−1 (25 min)−1) and C57 control mice (41 ± 3 U (200 μg protein)−1 (25 min)−1). A similar increase in DCF fluorescence was measured if tissue sections instead of homogenates were introduced into the assay (data not shown). Co-incubation of the homogenates with catalase as external hydrogen peroxide scavenger significantly reduced the DCF fluorescence in both mouse strains (C57 control: 32 ± 4 U (200 μg protein)−1 (25 min)−1); nNOS-knockout: 34 ± 6 U (200 μg protein)−1 (25 min)−1); n= 3; data not shown). These results suggest that hydrogen peroxide contributes to approximately 20% of the DCFH oxidation in EDL homogenates of C57 control mice and nNOS-knockout mice as determined in our in vitro DCF fluorescence assay.
Figure 6. The production of free radicals but not of hydrogen peroxide is higher in the EDL of nNOS-knockout mice than in that of C57 control mice.
A, to quantify the concentration of free radicals, EDL homogenates from C57 control mice and nNOS-knockout mice were subjected to electron spin resonance (ESR) spectrometry using 1-hydroxy-3-methoxycarbonyl-2,2,5,5-tetramethyl-pyrrolidine (CMH) as spin probe. Mean values for 8 independent experiments are represented together with the standard deviation. *Values in nNOS-knockout mice differ significantly from those in C57 control mice. B, 200 μg aliquots of bovine serum albumin (BSA) and of EDL homogenates that were derived from C57 control and nNOS-knockout mice were incubated with DCFH at 37°C. The fluorescence signal emitted at a wavelength of 525 nm was monitored for 25 min incubation. Mean values for three independent experiments are represented together with the standard deviation.
Discussion
To identify the proteins that are differentially expressed within the EDL of nNOS-knockout mice and C57/Bl6 control mice and might therefore be involved in an adaptive response for the absence of nNOS at the molecular and physiological level, we conducted a proteomics analysis. This involved two dimensional polyacrylamide gel electrophoresis (2D-PAGE) and LC-MS/MS. Six proteins (peroxiredoxin-6, nucleoside diphosphate kinase B (NDP kinase B), prohibitin; peroxiredoxin-3; Cu2+/Zn2+-dependent superoxide dismutase (SOD1) and heat shock protein β-1 (HSP25/HSP27)) were expressed at higher levels within the EDL of nNOS-knockout mice than within that of C57 control mice. With the exception of prohibitin (slightly down-regulated) and NDP kinase B (no working antibody available), the significant up-regulation of these proteins within the EDL of nNOS-knockout mice compared to C57 control mice was confirmed by quantitative immunoblotting. The levels of mRNA encoding for all of these proteins but prohibitin were likewise significantly higher within the EDL of the nNOS-knockout mice.
At both the gene and the protein-level, peroxiredoxin-6 exhibited the most distinct differences in concentration between the two mouse strains. In addition, peroxiredoxin-6 exhibited a pI of 6.5 in the EDL of C57 control mice, which varied from the pI of 5.9 detected in the EDL of nNOS-knockout mice by immunoblotting after 2D-PAGE. The lower sensitivity of silver staining protocols compared to immunoblotting detection could explain why we have not observed an up-regulated spot at pI of 6.5 in the EDL of C57 control mice in silver-stained 2D-gels. A similar pI shift was previously attributed to different states of oxidation of peroxiredoxin-6 (Wagner et al. 2002; Cullingford et al. 2006; Power et al. 2008). According to these studies, the more acidic protein reflects the protein form of peroxiredoxin-6 that has been oxidized by superoxide-derived hydrogen peroxide, while the more basic protein represents a reduced peroxiredoxin-6 form. The data of our ESR spectroscopy support this conclusion as we found a more oxidative milieu with higher levels of superoxide in skeletal muscles of nNOS-knockout mice than in those of C57 control mice.
Only peroxiredoxin-6 expression within the EDL was up-regulated in C57 control mice after the acute inhibition of NOS activity with l-NAME at the mRNA and protein level. This observation is consistent with the results of the 2D-PAGE analysis. Both findings performed under acute and chronic conditions confirm that peroxiredoxin-6 in skeletal muscle is negatively correlated with the availability of NO. In macrophages, on the other hand, peroxiredoxin-6 and NOS levels are uniformly adjusted (Diet et al. 2007). This apparent contradiction could arise from different functions of NO (generated, e.g. by nNOS) and ROS (scavenged, e.g. by peroxiredoxin-6) in the two cells/tissues: while both systems of radicals contribute in concert to the immune response of macrophages, they exhibit antagonistic effects in skeletal muscle (see below).
Although peroxiredoxin-6 is expressed in many organs and tissues, the highest concentrations are encountered in the lung (Mo et al. 2003; Wang et al. 2003). In skeletal muscle, the physiological levels of peroxiredoxin-6 are known to be low (Wang et al. 2003) as confirmed by our immunoblotting data for C57 control mice. Peroxiredoxin-6 exhibits two functions (for review, see Manevich & Fisher, 2005). (1) The enzyme reduces hydrogen peroxide to water and other hydroperoxides including phospholipid hydroperoxides to their corresponding alcohols during the oxidation of glutathione (Phelan et al. 2003). Accordingly, the intrinsic hydrogen peroxidase activity of EDL homogenates, which was found to be higher in nNOS-knockout mice than in C57 control mice, could be mainly ascribed to peroxiredoxin-6 and not to other peroxidases, e.g. catalase or glutathione peroxidase (see below). (2) Unique amongst the peroxiredoxins, peroxiredoxin-6 exhibits as second function a Ca2+-independent phospholipase A2 activity (Manevich et al. 2007).
The other five proteins that were more expressed within the EDL of nNOS-knockout mice than control mice in the 2D-PAGE analysis were not significantly modulated under conditions of acute NOS inhibition (l-NAME administration) suggesting that they contribute to adaptation to lifelong lack of nNOS. These five proteins are likewise known to have an impact on the metabolism of ROS and other oxygen derivatives. Mitochondrial peroxiredoxin-3 is involved in the regulation of redox reactions (Chang et al. 2004) and protects radical-sensitive enzymes from oxidative damage (Wood et al. 2003). Sarcoplasmic SOD1 destroys free radicals by quenching superoxide (Zelko et al. 2002). HSP25 (identical to HSP27), which is abundant within contractile cells, is implicated in the consolidation of the intracellular redox homeostasis (Escobedo et al. 2004; Arrigo et al. 2005). Sarcoplasmic NDP kinase B catalyses the reversible conversion of nucleoside diphosphates to nucleoside triphosphates and thus fulfills a homeostatic function in energy metabolism. However, it also protects BAF3 cells against oxidative stress (Arnaud-Dabernat et al. 2004). In skeletal muscle, mitochondrial prohibitin regulates cellular respirative activity (Theiss et al. 2007). Because the up-regulation of prohibitin was not confirmed by immunoblotting and qRT-PCR, it, however, remains elusive whether prohibitin is actually up-regulated in EDL of nNOS-knockout mice. Although the striking up-regulation does not necessarily imply a higher activity of these proteins in ROS scavenging, we suggest that the nNOS-deficiency within skeletal muscle of nNOS-knockout mice is associated with changes in the metabolism of ROS. Accordingly, ESR spectroscopy revealed higher levels of free radicals in EDL of nNOS-knockout mice than those within C57 control mice. Because CMH was used as the spin trap in ESR spectroscopy, which preferentially detects superoxide (Dikalov et al. 2007), we suggest that the concentration of this free radical in particular is elevated within the EDL of nNOS-knockout mice.
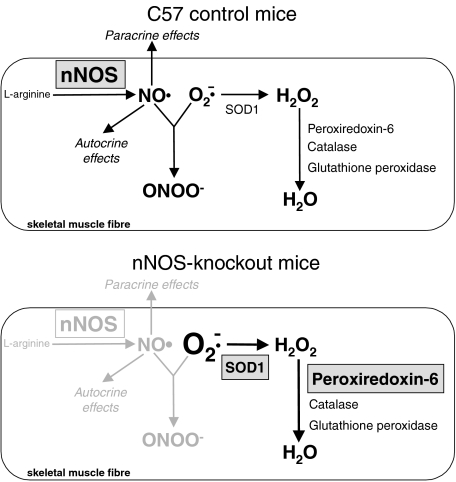
In skeletal muscle, the existence of a dynamic balance of ROS and NO is relevant in the control of contraction (for review see Smith & Reid, 2006). Low levels of ROS are required for the generation of normal forces while NO limits the force. If the redox homeostasis is perturbed, e.g. by increased levels of superoxide, then the contractile function of skeletal muscle is compromised, resulting in muscle weakness and fatigue (for review see Smith & Reid, 2006). It seems reasonable that the concentration of both radical cascades in skeletal muscle is controlled to counterbalance the activity of one system in the case that the other system is deregulated. Deduced from the experimental data, our hypothesis of how the counterbalance between NO and ROS is accomplished in skeletal muscle of nNOS-knockout-mice is schematically summarized in Fig. 7.
Figure 7. Schematic summary of the altered ROS metabolism in skeletal muscle fibres of nNOS-knockout-mice.
Under wild-type conditions (e.g. in C57 control mice), nitric oxide (NO) acts in skeletal muscle fibres as paracrine and autocrine signalling molecule but also might react with superoxide (O2−) to generate peroxynitrite (ONOO−). Scavenging of superoxide is furthermore achieved by enzymatic catalysis of superoxide dismutases (e.g. the cytosolic SOD1) to yield hydrogen peroxide (H2O2), which is subsequently reduced to water by catalase, glutathione peroxidase or peroxiredoxin-6 activity. In nNOS-knockout mice, up-regulated SOD1 and peroxiredoxin-6 enable the scavenging of higher superoxide levels. Other proteins with impact on the ROS metabolism that were found to be up-regulated in skeletal muscle fibres of nNOS-knockout mice in the 2D-PAGE analysis (prohibitin, peroxiredoxin-3, HSP25 and NDP kinase B) are not included in the scheme. Grey down-toning or gradation of reactions or enzymes, respectively, indicates their absence or down-regulation. For further details, see Discussion.
The superoxide (anion) represents the top of the ROS cascade, which is generated mainly by the activity of NADPH oxidase and as a byproduct by the respiratory chain complexes I and III (for review, see Jackson et al. 2007). Superoxide is highly toxic as it inactivates enzymes that contain iron–sulphur clusters as prosthetic group (Valko et al. 2007). Therefore, the rapid detoxification of superoxide is of physiological relevance. The reaction of superoxide with NO, which yields peroxynitrite, represents one possible mode of elimination. This reaction is approximately three times more efficient in scavenging superoxide than is that catalysed by the SOD system (Pattwell et al. 2004). Thus it is one function of nNOS in skeletal muscle to ensure the protection from toxic levels of superoxide. Consequently, if other scavengers of superoxide were insufficiently up-regulated in skeletal muscle of nNOS-knockout mice, superoxide would accumulate, as demonstrated in our ESR spectrometry analysis. Furthermore, levels of peroxynitrite would be reduced what was not investigated in this study.
We introduced muscle homogenates (lysis without detergents) in the DCF fluorescence assay to quantify the levels of ROS in the EDL. A similar approach was previously applied to determine ROS concentrations in homogenates of brain (Martinez-Lazcano et al. 2007) and skeletal muscle cells (Silveira et al. 2003). It is however, obvious that this methodology can introduce artifacts in the measurements, e.g. by disruption of the cellular microenvironment (intact mitochondria are an important source of superoxide) or by suppression of catalytic activities (dissociation of enzyme subunits). Three experiments were performed to validate the DCF fluorescence assay: (1) incubation with BSA instead of homogenate protein did not lead to an increase of DCF fluorescence indicating that the DCFH oxidation is specifically achieved by a homogenate component, (2) the successful replacement by tissue sections shows that the ROS producing activity in homogenates is retained despite mechanical cellular destruction, and (3) the coincubation with catalase revealed that hydrogen peroxide contributes to the DCFH oxidation in EDL homogenates of C57 control mice and nNOS-knockout mice to a similar extent suggesting that the production rates of this ROS are similar in both mouse strains.
The SOD protein family comprises three members (CuZn-SOD (SOD1), Mn-SOD (SOD2) and EC-SOD (SOD3)), which oxidize superoxide to hydrogen peroxide (Zelko et al. 2002). Because SOD1 was expressed at higher levels within the EDL of nNOS-knockout mice than within that of the control strain, one would have expected the concentration of ROS to be likewise higher, but this was not the case. It is thus conceivable that higher levels of ROS were rapidly eliminated, e.g. by the up-regulated peroxiredoxin-6. In murine hepatocytes, elevated levels of hydrogen peroxide have been shown to induce the up-regulation of peroxiredoxin-6, which in these cells contributes physiologically to the homeostasis of ROS (Gallagher & Phelan, 2007).
An up-regulation of the enzymes that degrade ROS and other oxygen derivatives is thus necessary for the elimination of the surplus of ROS if the levels of NO are reduced. Consistent with this tenet is the finding that the cerebellum (but not the cortex and the striatum) of nNOS-knockout mice contains lower levels of ROS and is less susceptible to oxidative damage than that of control mice (Martinez-Lazcano et al. 2007). Apparently, the molecular counteraction to scavenge ROS in response to the nNOS-deficiency is in this part of the brain more distinct than in the other cerebral parts or in skeletal muscle.
We postulate that the up-regulation of peroxiredoxin-6 within the EDL of nNOS-knockout mice is induced to eliminate an excess quantity of hydrogen peroxide (Fig. 7). Such a protective function of peroxiredoxin-6 has been proposed for melanoma cells which express low levels of this protein (de Souza et al. 2006) and has been deduced from studies with peroxiredoxin-6-knockout mice (Wang et al. 2003,2008; Nagy et al. 2006). An up-regulation of the peroxiredoxin-6 system as a consequence of the lack/inhibition of nNOS or another NOS form has not been reported so far. Two additional observations furthermore indicate that peroxiredoxin-6 is the main target among the enzymes exerting hydrogen peroxide reductase activity (e.g. glutathione peroxidase-1 and catalase) that is modulated in skeletal muscle of nNOS-knockout-mice: (1) only levels of peroxiredoxin-6 were revealed to be increased in EDL of nNOS-knockout mice as detected in the densitometric analysis after 2D-PAGE, and (2) the main proportion of cellular hydrogen peroxidase activity in nNOS-knockout mice was assigned to peroxiredoxin-6 as determined in the in vitro DCF fluorescence assay after immunodepletion. Alternatively, the metabolism of ROS within skeletal muscles of nNOS-knockout mice might be modulated so as to generate intermediates with a signal transduction function (for review, see Rhee et al. 2005), which would serve as surrogates for nNOS-derived NO. Given that peroxiredoxin-6 functions also as a phospholipase (for review, see Manevich & Fisher, 2005; Winterbourn & Hampton, 2008), its up-regulation within the EDL of nNOS-knockout mice could likewise compensate for the absence of nNOS in signal transduction processes that depend upon nNOS activity in control mice.
An up-regulation of eNOS in EDL of nNOS-knockout mice was not detected as previously reported (Wadley et al. 2007). Thus it still remains controversial whether eNOS contributes to the compensatory response in EDL of nNOS-knockout mice.
In summary, we have identified six proteins that are up-regulated within the EDL of nNOS-knockout mice. Each of these proteins is known to be involved in the metabolism of ROS. However, it is not yet clear whether this up-regulation is a response to nNOS deficiency to exclusively scavenge an excess of ROS, or whether it represents a controlled adaptive reaction of the skeletal muscle fibres to additionally compensate for nNOS function in cellular signalling. It also has to be examined whether our results obtained in EDL of age-matched C57Bl/6 control mice and nNOS-knockout mice with identical genetic background but different offspring can be validated using wild-type and nNOS-knockout littermates.
Further experiments are now needed to establish the functional relevance of the six proteins that are implicated in the metabolism of ROS (particularly that of peroxiredoxin-6) in compensating for nNOS function within the skeletal muscle of nNOS-knockout mice.
Acknowledgments
We would like to thank Prof Aron Fisher and Dr Yefim Manevich (Institute for Environmental Medicine, University of Pennsylvania School of Medicine, Philadelphia, PA, USA) for providing the anti-peroxiredoxin-6 antibody. The encouraging support of Prof Hans Hoppeler and linguistic help of Dr Cery England is gratefully acknowledged. This work was funded by grants from the Deutsche Forschungsgemeinschaft (Pr 271/5-4 and Za 184/3-2).
References
- Arnaud-Dabernat S, Masse K, Smani M, Peuchant E, Landry M, Bourbon PM, Le Floch R, Daniel JY, Larou M. Nm23–M2/NDP kinase B induces endogenous c-myc and nm23–M1/NDP kinase A overexpression in BAF3 cells. Both NDP kinases protect the cells from oxidative stress-induced death. Exp Cell Res. 2004;301:293–304. doi: 10.1016/j.yexcr.2004.07.026. [DOI] [PubMed] [Google Scholar]
- Arrigo AP, Virot S, Chaufour S, Firdaus W, Kretz-Remy C, Diaz-Latoud C. Hsp27 consolidates intracellular redox homeostasis by upholding glutathione in its reduced form and by decreasing iron intracellular levels. Antioxid Redox Signal. 2005;7:414–422. doi: 10.1089/ars.2005.7.414. [DOI] [PubMed] [Google Scholar]
- Barouch LA, Harrison RW, Skaf MW, Rosas GO, Cappola TP, Kobeissi ZA, Hobai IA, Lemmon CA, Burnett AL, O'Rourke B, Rodriguez ER, Huang PL, Lima JA, Berkowitz DE, Hare JM. Nitric oxide regulates the heart by spatial confinement of nitric oxide synthase isoforms. Nature. 2002;416:337–339. doi: 10.1038/416337a. [DOI] [PubMed] [Google Scholar]
- Baum O, Da Silva-Azevedo L, Willerding G, Wockel A, Planitzer G, Gossrau R, Pries AR, Zakrzewicz A. Endothelial NOS is main mediator for shear stress-dependent angiogenesis in skeletal muscle after prazosin administration. Am J Physiol Heart Circ Physiol. 2004;287:H2300–H2308. doi: 10.1152/ajpheart.00065.2004. [DOI] [PubMed] [Google Scholar]
- Baum O, Miethke A, Wockel A, Willerding G, Planitzer G. The specificity of the histochemical NADPH diaphorase reaction for nitric oxide synthase-1 in skeletal muscles is increased in the presence of urea. Acta Histochem. 2002;104:3–14. doi: 10.1078/0065-1281-00625. [DOI] [PubMed] [Google Scholar]
- Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF, Froehner SC, Bredt DS. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and α1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- Brenman JE, Chao DS, Xia H, Aldape K, Bredt DS. Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell. 1995;82:743–752. doi: 10.1016/0092-8674(95)90471-9. [DOI] [PubMed] [Google Scholar]
- Campbell KP, Stull JT. Skeletal muscle basement membrane–sarcolemma–cytoskeleton interaction. J Biol Chem. 2003;278:12599–12600. doi: 10.1074/jbc.R300005200. [DOI] [PubMed] [Google Scholar]
- Chang TS, Cho CS, Park S, Yu S, Kang SW, Rhee SG. Peroxiredoxin III, a mitochondrion-specific peroxidase, regulates apoptotic signaling by mitochondria. J Biol Chem. 2004;279:41975–41984. doi: 10.1074/jbc.M407707200. [DOI] [PubMed] [Google Scholar]
- Chlench S, Mecha Disassa N, Hohberg M, Hoffmann C, Pohlkamp T, Beyer G, Bongrazio M, Da Silva-Azevedo L, Baum O, Pries AR, Zakrzewicz A. Regulation of Foxo-1 and the angiopoietin-2/Tie2 system by shear stress. FEBS Lett. 2007;581:673–680. doi: 10.1016/j.febslet.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Cullingford TE, Wait R, Clerk A, Sugden PH. Effects of oxidative stress on the cardiac myocyte proteome: modifications to peroxiredoxins and small heat shock proteins. J Mol Cell Cardiol. 2006;40:157–172. doi: 10.1016/j.yjmcc.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Da Silva-Azevedo L, Baum O, Zakrzewicz A, Pries A. Vascular endothelial growth factor is expressed in endothelial cells isolated from skeletal muscles of nitric oxide synthase knockout mice during prazosin-induced angiogenesis. Biochem Biophys Res Commun. 2002;297:1270–1276. doi: 10.1016/s0006-291x(02)02370-7. [DOI] [PubMed] [Google Scholar]
- de Souza GA, Godoy LM, Teixeira VR, Otake AH, Sabino A, Rosa JC, Dinarte AR, Pinheiro DG, Silva WA, Jr, Eberlin MN, Chammas R, Greene LJ. Proteomic and SAGE profiling of murine melanoma progression indicates the reduction of proteins responsible for ROS degradation. Proteomics. 2006;6:1460–1470. doi: 10.1002/pmic.200500243. [DOI] [PubMed] [Google Scholar]
- Diet A, Abbas K, Bouton C, Guillon B, Tomasello F, Fourquet S, Toledano MB, Drapier JC. Regulation of peroxiredoxins by nitric oxide in immunostimulated macrophages. J Biol Chem. 2007;282:36199–36205. doi: 10.1074/jbc.M706420200. [DOI] [PubMed] [Google Scholar]
- Dikalov S, Fink B. ESR techniques for the detection of nitric oxide in vivo and in tissues. Methods Enzymol. 2005;396:597–610. doi: 10.1016/S0076-6879(05)96052-7. [DOI] [PubMed] [Google Scholar]
- Dikalov S, Griendling KK, Harrison DG. Measurement of reactive oxygen species in cardiovascular studies. Hypertension. 2007;49:717–727. doi: 10.1161/01.HYP.0000258594.87211.6b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobedo J, Pucci AM, Koh TJ. HSP25 protects skeletal muscle cells against oxidative stress. Free Radic Biol Med. 2004;37:1455–1462. doi: 10.1016/j.freeradbiomed.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Frandsen U, Hoffner L, Betak A, Saltin B, Bangsbo J, Hellsten Y. Endurance training does not alter the level of neuronal nitric oxide synthase in human skeletal muscle. J Appl Physiol. 2000;89:1033–1038. doi: 10.1152/jappl.2000.89.3.1033. [DOI] [PubMed] [Google Scholar]
- Gallagher BM, Phelan SA. Investigating transcriptional regulation of Prdx6 in mouse liver cells. Free Radic Biol Med. 2007;42:1270–1277. doi: 10.1016/j.freeradbiomed.2007.01.023. [DOI] [PubMed] [Google Scholar]
- Grange RW, Isotani E, Lau KS, Kamm KE, Huang PL, Stull JT. Nitric oxide contributes to vascular smooth muscle relaxation in contracting fast-twitch muscles. Physiol Genomics. 2001;5:35–44. doi: 10.1152/physiolgenomics.2001.5.1.35. [DOI] [PubMed] [Google Scholar]
- Higaki Y, Hirshman MF, Fujii N, Goodyear LJ. Nitric oxide increases glucose uptake through a mechanism that is distinct from the insulin and contraction pathways in rat skeletal muscle. Diabetes. 2001;50:241–247. doi: 10.2337/diabetes.50.2.241. [DOI] [PubMed] [Google Scholar]
- Huang PL. Neuronal and endothelial nitric oxide synthase gene knockout mice. Braz J Med Biol Res. 1999;32:1353–1359. doi: 10.1590/s0100-879x1999001100005. [DOI] [PubMed] [Google Scholar]
- Huang PL, Dawson TM, Bredt DS, Snyder SH, Fishman MC. Targeted disruption of the neuronal nitric oxide synthase gene. Cell. 1993;75:1273–1286. doi: 10.1016/0092-8674(93)90615-w. [DOI] [PubMed] [Google Scholar]
- Hudlicka O, Brown MD, Silgram H. Inhibition of capillary growth in chronically stimulated rat muscles by NG-nitro-L-arginine, nitric oxide synthase inhibitor. Microvasc Res. 2000;59:45–51. doi: 10.1006/mvre.1999.2193. [DOI] [PubMed] [Google Scholar]
- Jackson MJ, Pye D, Palomero J. The production of reactive oxygen and nitrogen species by skeletal muscle. J Appl Physiol. 2007;102:1664–1670. doi: 10.1152/japplphysiol.01102.2006. [DOI] [PubMed] [Google Scholar]
- Kapur S, Picard F, Perreault M, Deshaies Y, Marette A. Nitric oxide: a new player in the modulation of energy metabolism. Int J Obes Relat Metab Disord. 2000;24:36–40. doi: 10.1038/sj.ijo.0801502. [DOI] [PubMed] [Google Scholar]
- Kobayashi YM, Rader EP, Crawford RW, Iyengar NK, Thedens DR, Faulkner JA, Parikh SV, Weiss RM, Chamberlain JS, Moore SA, Campbell KP. Sarcolemma-localized nNOS is required to maintain activity after mild exercise. Nature. 2008;456:511–515. doi: 10.1038/nature07414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobzik L, Reid MB, Bredt DS, Stamler JS. Nitric oxide in skeletal muscle. Nature. 1994;372:546–548. doi: 10.1038/372546a0. [DOI] [PubMed] [Google Scholar]
- Manevich Y, Fisher AB. Peroxiredoxin 6, a L-Cys peroxiredoxin, functions in antioxidant defense and lung phospholipid metabolism. Free Radic Biol Med. 2005;38:1422–1432. doi: 10.1016/j.freeradbiomed.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Manevich Y, Reddy KS, Shuvaeva T, Feinstein SI, Fisher AB. Structure and phospholipase function of peroxiredoxin 6: identification of the catalytic triad and its role in phospholipid substrate binding. J Lipid Res. 2007;48:2306–2318. doi: 10.1194/jlr.M700299-JLR200. [DOI] [PubMed] [Google Scholar]
- Martinez-Lazcano JC, Perez-Severiano F, Escalante B, Ramirez-Emiliano J, Vergara P, Gonzalez RO, Segovia J. Selective protection against oxidative damage in brain of mice with a targeted disruption of the neuronal nitric oxide synthase gene. J Neurosci Res. 2007;85:1391–1402. doi: 10.1002/jnr.21261. [DOI] [PubMed] [Google Scholar]
- Mo Y, Feinstein SI, Manevich Y, Zhang Q, Lu L, Ho YS, Fisher AB. L-Cys peroxiredoxin knock-out mice express mRNA but not protein for a highly related intronless gene. FEBS Lett. 2003;555:192–198. doi: 10.1016/s0014-5793(03)01199-2. [DOI] [PubMed] [Google Scholar]
- Nagy N, Malik G, Fisher AB, Das DK. Targeted disruption of peroxiredoxin 6 gene renders the heart vulnerable to ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2006;291:H2636–H2640. doi: 10.1152/ajpheart.00399.2006. [DOI] [PubMed] [Google Scholar]
- Pattwell DM, McArdle A, Morgan JE, Patridge TA, Jackson MJ. Release of reactive oxygen and nitrogen species from contracting skeletal muscle cells. Free Radic Biol Med. 2004;37:1064–1072. doi: 10.1016/j.freeradbiomed.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Percival JM, Anderson KN, Gregorevic P, Chamberlain JS, Froehner SC. Functional deficits in nNOSmu-deficient skeletal muscle: myopathy in nNOS-knockout mice. PLoS ONE. 2008;3:e3387. doi: 10.1371/journal.pone.0003387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan SA, Wang X, Wallbrandt P, Forsman-Semb K, Paigen B. Overexpression of Prdx6 reduces H2O2 but does not prevent diet-induced atherosclerosis in the aortic root. Free Radic Biol Med. 2003;35:1110–1120. doi: 10.1016/s0891-5849(03)00462-3. [DOI] [PubMed] [Google Scholar]
- Planitzer G, Miethke A, Baum O. Nitric oxide synthase-1 is enriched in fast-twitch oxidative myofibers. Cell Tissue Res. 2001;306:325–333. doi: 10.1007/s004410100449. [DOI] [PubMed] [Google Scholar]
- Power JH, Asad S, Chataway TK, Chegini F, Manavis J, Temlett JA, Jensen PH, Blumbergs PC, Gai WP. Peroxiredoxin 6 in human brain: molecular forms, cellular distribution and association with Alzheimer's disease pathology. Acta Neuropathol. 2008;115:611–622. doi: 10.1007/s00401-008-0373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid MB. Role of nitric oxide in skeletal muscle: synthesis, distribution and functional importance. Acta Physiol Scand. 1998;162:401–409. doi: 10.1046/j.1365-201X.1998.0303f.x. [DOI] [PubMed] [Google Scholar]
- Rhee SG, Kang SW, Jeong W, Chang TS, Yang KS, Woo HA. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr Opin Cell Biol. 2005;17:183–189. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Roberts CK, Barnard RJ, Scheck SH, Balon TW. Exercise-stimulated glucose transport in skeletal muscle is nitric oxide dependent. Am J Physiol Endocrinol Metab. 1997;273:E220–E225. doi: 10.1152/ajpendo.1997.273.1.E220. [DOI] [PubMed] [Google Scholar]
- Shankar RR, Wu Y, Shen HQ, Zhu JS, Baron AD. Mice with gene disruption of both endothelial and neuronal nitric oxide synthase exhibit insulin resistance. Diabetes. 2000;49:684–687. doi: 10.2337/diabetes.49.5.684. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Silvagno F, Xia H, Bredt DS. Neuronal nitric-oxide synthase-μ, an alternatively spliced isoform expressed in differentiated skeletal muscle. J Biol Chem. 1996;271:11204–11208. doi: 10.1074/jbc.271.19.11204. [DOI] [PubMed] [Google Scholar]
- Silveira LR, Pereira-Da Silva L, Juel C, Hellsten Y. Formation of hydrogen peroxide and nitric oxide in rat skeletal muscle cells during contractions. Free Radic Biol Med. 2003;35:455–464. doi: 10.1016/s0891-5849(03)00271-5. [DOI] [PubMed] [Google Scholar]
- Smith MA, Reid MB. Redox modulation of contractile function in respiratory and limb skeletal muscle. Respir Physiol Neurobiol. 2006;151:229–241. doi: 10.1016/j.resp.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Stalder D, Haeberli A, Heller M. Evaluation of reproducibility of protein identification results after multidimensional human serum protein separation. Proteomics. 2008;8:414–424. doi: 10.1002/pmic.200700527. [DOI] [PubMed] [Google Scholar]
- Stamler JS, Meissner G. Physiology of nitric oxide in skeletal muscle. Physiol Rev. 2001;81:209–237. doi: 10.1152/physrev.2001.81.1.209. [DOI] [PubMed] [Google Scholar]
- Theiss AL, Idell RD, Srinivasan S, Klapproth JM, Jones DP, Merlin D, Sitaraman SV. Prohibitin protects against oxidative stress in intestinal epithelial cells. FASEB J. 2007;21:197–206. doi: 10.1096/fj.06-6801com. [DOI] [PubMed] [Google Scholar]
- Thomas GD, Shaul PW, Yuhanna IS, Froehner SC, Adams ME. Vasomodulation by skeletal muscle-derived nitric oxide requires α-syntrophin-mediated sarcolemmal localization of neuronal nitric oxide synthase. Circ Res. 2003;92:554–560. doi: 10.1161/01.RES.0000061570.83105.52. [DOI] [PubMed] [Google Scholar]
- Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Wadley GD, Choate J, McConell GK. NOS isoform-specific regulation of basal but not exercise-induced mitochondrial biogenesis in mouse skeletal muscle. J Physiol. 2007;585:253–262. doi: 10.1113/jphysiol.2007.141309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E, Luche S, Penna L, Chevallet M, Van Dorsselaer A, Leize-Wagner E, Rabilloud T. A method for detection of overoxidation of cysteines: peroxiredoxins are oxidized in vivo at the active-site cysteine during oxidative stress. Biochem J. 2002;366:777–785. doi: 10.1042/BJ20020525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Inglis FM, Kalb RG. Defective fluid and HCO3− absorption in proximal tubule of neuronal nitric oxide synthase-knockout mice. Am J Physiol Renal Physiol. 2000;279:F518–F524. doi: 10.1152/ajprenal.2000.279.3.F518. [DOI] [PubMed] [Google Scholar]
- Wang X, Phelan SA, Forsman-Semb K, Taylor EF, Petros C, Brown A, Lerner CP, Paigen B. Mice with targeted mutation of peroxiredoxin 6 develop normally but are susceptible to oxidative stress. J Biol Chem. 2003;278:25179–25190. doi: 10.1074/jbc.M302706200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Feinstein SI, Fisher AB. Peroxiredoxin 6 as an antioxidant enzyme: Protection of lung alveolar epithelial type II cells from H2O2-induced oxidative stress. J Cell Biochem. 2008;104:1274–1285. doi: 10.1002/jcb.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JL, Cartland D, Hussain A, Egginton S. A differential role for nitric oxide in two forms of physiological angiogenesis in mouse. J Physiol. 2006a;570:445–454. doi: 10.1113/jphysiol.2005.095596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JL, Weichert A, Zakrzewicz A, Da Silva-Azevedo L, Pries AR, Baum O, Egginton S. Differential gene and protein expression in abluminal sprouting and intraluminal splitting forms of angiogenesis. Clin Sci (Lond) 2006b;110:587–595. doi: 10.1042/CS20050185. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC, Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Wood ZA, Schroder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33:337–349. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]