Abstract
Conjugation of SUMO to target proteins is an essential eukaryotic regulatory pathway. Multiple potential SUMO substrates were identified among nuclear and chromatin proteins by proteomic approaches. However, the functional roles of SUMO-modified pools of individual proteins remain largely obscure, as only a small fraction of a given target is sumoylated and therefore is experimentally inaccessible. To overcome this technical difficulty in case of Topoisomerase II, we employed constitutive SUMO modification, enabling tracking of modified Top2p, not only biochemically but also cytologically and genetically. Top-oisomerase II fused to a critical number of SUMO repeats is concentrated at the specific intranuclear domain, the nucleolus, when more than four SUMO moieties are added, indicating that fused SUMO repeats are biologically active. Further analysis has established that poly-sumoylation of Top2p is required for the stable maintenance of the nucleolar organizer, linking SUMO-mediated targeting to functional maintenance of ribosomal RNA gene cluster.
Introduction
The SUMO protein was first discovered as a modifier of RanGAP1 (Matunis et al. 1996; Mahajan et al. 1997; Saitoh et al. 1998). The sumoylation pathway is organized similarly to ubiquitination: E1, E2, and E3 steps are required to conjugate the activated SUMO molecule to substrates (Johnson 2004). E1 and E2 enzymes are essential for cell viability in S. cerevisiae. SUMO E3 factors are responsible for substrate specificity of conjugation machinery in vivo (Johnson 2004), although they are partially redundant in vitro (Takahashi et al. 2003). Elimination of all three E3 activities (Siz1, Siz2, and Mms21) is synthetically lethal (Reindle et al. 2006). While recent proteomic studies demonstrated that several hundreds of proteins can be modified in vivo (Panse et al. 2004; Wohlschlegel et al. 2004; Zhou et al. 2004; Denison et al. 2005; Hannich et al. 2005; Wykoff and O’Shea 2005), direct evidence of biological significance and/or functional importance of these modifications is lacking for most substrates. Two main obstacles prevent the elucidation of the sumoylation’s role for a given target protein using straightforward biochemical or genetic methods. First, for the absolute majority of proteins, only a tiny fraction of molecules is modified (Johnson 2004), so that purification of the modified protein and a biochemical assessment of its function is extremely difficult. For the same reason, direct physical mapping of sumoylated sites and therefore their mutagenesis are rarely attempted (Johnson and Blobel 1999).
The low abundance of the sumoylated molecules vs the unmodified pool of the same protein (Johnson 2004) is a mystery in itself, as other post-translational modifications usually affect a significant proportion of the cellular pool of a given protein. Thus, this SUMO-specific phenomenon is largely reminiscent of histone modifications (Shilatifard 2006). Therefore, as sumoylation largely affects chromosomal proteins, it is conceivable that SUMO can serve as a targeting/retention signal for a particular nuclear or chromosomal location, similarly to histone modifications. Results of several attempts to fuse a single SUMO molecule directly to the amino termini of target proteins (that are polysumoylated in vivo) are generally consistent with this hypothesis (Ross et al. 2002; Dobreva et al. 2003; Huang et al. 2003; Shiio and Eisenman 2003). However, biological adequacy of such fusions remains questionable because of unnatural positioning of the fused SUMO and its presence in only a single copy. In our pervious work, we employed a more biologically relevant fusion design, by introducing a single SUMO inside the Top2p in the close vicinity of the natural target site (Takahashi et al. 2006). Such a fusion targeted Top2p to yeast centromeres (Takahashi et al. 2006).
Despite the fact that multisumoylation of Topoisomerase II was found from yeast to mammals, the molecular function of these multiple modifications was not identified. Top2p has a number of rather distinct roles in the cell, most notably in deoxyribonucleic acid (DNA) replication, transcription, and chromosome segregation (Wang 2002), making it tempting to test whether polysumoylation is specifically related to one of these functions. In this report, we attempted to track the artificially poly-sumoylated (SUMO-chain-modified) pool of Topoisomerase II in vivo using the fusion principle introduced in Takahashi et al. (2006). In vivo modeling of Top2p polysumoylation allowed us to approach several unresolved questions about Top2p sumoylation. As a result, we found a plausible explanation for the role of polysumoylation (SUMO chains) and/or multisumoylation (sumoylation at several clustered sites) in targeting Top2 protein to a specific chromosomal location. Mimicking of the physiological sumoylated state was achieved by engineering an internal fusion of multiple SUMO repeats to the Top2 protein via peptide bond, in close proximity to the cluster of natural SUMO conjugation sites. We showed that tandemly fused SUMO molecules are biologically active: The fusion’s activity in vivo and in vitro was dependent on the number of SUMO repeats added, and the certain optimum of repeats (mimicking the wild-type sumoylation levels) produced a specific subnucleolar targeting signal.
Materials and methods
Plasmids used in this study are described in Table 1. S. cerevisiae strains were derived from W303-1A or S288c, as indicated in Table 2. Manipulation of yeast cells, microscopy, and biochemical techniques were essentially as described in Takahashi et al. (2006). For microscopic imaging, the diploid strains carrying specific fluorescent markers, i.e., heterozygous fusions of mRFP to NIC96, SIK1, and SPC42 genes, were used for the integration of TOP2:4SMT3:GFP and TOP2:5SMT3:GFP constructs (Tables 1 and 2).
Table 1.
Plasmids
| Name | Backbone | Insert (targeting site) | Makers | Source |
|---|---|---|---|---|
| pYT1014 | pRS316 | pSMT3:GFP:SMT3(SacI + BglII) | LEU2 URA3 | This study |
| pYT1051 | pTS901IU | top2–1SMT3:HA (AvrII) | URA3 | Takahashi et al. (2006) |
| pYT1052 | pTS901IU | top2–2SMT3:HA (AvrII) | URA3 | This study |
| pYT1053 | pTS901IU | top2–3SMT3:HA (AvrII) | URA3 | This study |
| pYT1054 | pTS901IU | top2–4SMT3:HA (AvrII) | URA3 | This study |
| pYT1055 | pTS901IU | top2–5SMT3:HA (AvrII) | URA3 | This study |
| pYT1056 | pTS901IU | top2–6SMT3:HA (AvrII) | URA3 | This study |
| pYT1057 | pTS901IU | top2–7SMT3:HA (AvrII) | URA3 | This study |
| pYT1058 | pTS901IU | top2–8SMT3:HA (AvrII) | URA3 | This study |
| pYT1059 | pTS901IU | top2SNM–1SMT3:HA (AvrII) | URA3 | This study |
| pYT1102 | pTS910IL | top2–4SMT3:GFP (AvrII) | LEU2 | This study |
| pYT1103 | pTS910IL | top2–5SMT3:GFP (AvrII) | LEU2 | This study |
| pYT1108 | – | pGAL: top2(1219–1428)::5SMT3:GFP | URA3 | This study |
Table 2.
Yeast strains
| Relevant genotype | Source | |
|---|---|---|
| Isogenic to W303 | ||
| W303-1A | MATa ade2 ura3 trp1 leu2 his3 can1 ssd1 | Rothstein |
| 1033-W303-1A | MATa TOP2:HA::URA3 | This study |
| 1035-W303-1A | MATa top2 C:HA::URA3 | This study |
| AS611 | MATa/αTOP2/top2–1SMT3:HA::URA3 | This study |
| AS612 | MATa/α TOP2/top2–2SMT3:HA::URA3 | This study |
| AS613 | MATa/α TOP2/top2–3SMT3:HA::URA3 | This study |
| AS614 | MATa/α TOP2/top2–4SMT3:HA::URA3 | This study |
| AS615 | MATa/α TOP2/top2–5SMT3:HA::URA3 | This study |
| AS616 | MATa/α TOP2/top2–6SMT3:HA::URA3 | This study |
| AS617 | MATa/α TOP2/top2–7SMT3:HA::URA3 | This study |
| AS618 | MATa/α TOP2/top2–8SMT3:HA::URA3 | This study |
| AS619 | MATa/αTOP2/top2-SNM-1SMT3:HA::URA3 | This study |
| AS620 | MATa/α TOP2/top2–4SMT3:GFP::LEU2 | This study |
| AS621 | MATa/α TOP2/top2–5SMT3:GFP::LEU2 | This study |
| 1-YT626 | MATa smt4Δ::cgHIS3 GFP:SMT3::LEU2 NOP1:mRFP::URA3 | This study |
| 2-YT626 | MATa GFP:SMT3::LEU2 NOP1:mRFP::URA3 | This study |
| 1-YT14 | MATa siz1Δ::cgHIS3 siz2Δ::LEU2 | This study |
| 2-YT14 | MATα siz1Δ::cgHIS3 siz2Δ::LEU2 | This study |
| Y2226/JBY1087 | MATa top2-snm:HA:: KanMX | Bachant et al. (2002) |
| Isogenic to S288C | ||
| YPH499 | MATa ura3 lys2 ade2 trp1 his3 leu2 | Hieter |
| AS622 | MATa/α SPC42/SPC42:mRFP::kanMX TOP2/top2–4SMT3:GFP::LEU2 | This study |
| AS623 | MATa/α SIK1/SIK1:mRFP::kanMX TOP2/top2–5SMT3:GFP::LEU2 | This study |
| 3a4031697 | MATa top1Δ0::kanMX | This study |
| 4b4031697 | MATα top1Δ0::kanMX | This study |
| 1aYT625 | MATα his3 trp1 ura3 top2ΔC:HA::URA3 top1Δ0::kanMX | This study |
| 1035-BY4729 | MATα his3 trp1 ura3 top2ΔC:HA::URA3 | This study |
| 4bAS399 | MATa siz1Δ::KanMX siz2Δ::KanMX | Takahashi et al. (2006) |
| 12cAS399 | MATα siz1Δ::KanMX siz2Δ::KanMX | Takahashi et al. (2006) |
| NIC96-EY0987 | MATα NIC96::mRFP::KanMX | Huh et al. (2003) |
| BY4733 | MATa his3Δleu2Δmet15Δtrp1Δura3Δ | ATCC |
| BY4729 | MATa his3Δ200 trp1Δ63 ura3Δ0 | ATCC |
| Congenic to S288C | ||
| 3-YT627 | MATa siz1Δ::cgHIS3 siz2Δ::LEU2 top1Δ::KanMX | This study |
| 4-YT627 | MATα siz1Δ::cgHIS3 siz2Δ::LEU2 top1Δ::KanMX | This study |
To insert SMT3-coding sequences into TOP2, two primers (ttgcgcatgctagcTCATCACCATCATCATCACATGCATGACTACAAG and CTAATACGTAactagtCCAATCTGTTCTCTGTGAGCCTCAAT) were used to amplify the poly-histidine and FLAG-tagged SMT3 (HF:SMT3; Takahashi et al. 2006), without the last glycine codon required for Smt3p processing in vivo. The amplified fragment was digested with NheI and SpeI and inserted into the SpeI site of the 5′-truncated TOP2 gene (cloned into pTS901IU or pTS901IL with 5HA or green fluorescent protein (GFP) tags, respectively; Sasaki et al. 2000). The preserved unique SpeI site was sequentially used for inserting additional copies of SMT3. The HA-tagged constructs were targeted (see Table 1) to the native TOP2 locus in the diploid W303 strain.
To replace the wild-type SMT3 gene with the GFP-tagged versions HFG–SMT3 (GFP tag) expressed from the native SMT3 promoter, the targeting constructs were generated on the basis of the HF-SMT3::LEU2 integrative plasmid pAS924 (Takahashi et al. 2006). The polymerase chain reaction (PCR)-generated GFP fragment (SpeI–NheI) was inserted into the SpeI site between the SMT3 promoter and the SMT3 ORF in pAS924 (Takahashi et al. 2006). The resulting plasmid pYT1014 (HFG–SMT3) was digested with SacI/BglII and transformed into Leu2− yeast cells. The NOP1:mRFP::URA3 construct was a kind gift from Y. Kikuchi (The University of Tokyo).
To generate the amino terminus-truncated Top2p fusion, the COOH-terminal part of TOP2–5SMT3 (corresponding to the Top2p amino acid residues 1,219–1,428) was PCR-amplified from pYT1103, and the resulting PCR product was cloned into pGAL:GFP vector to produce pYT1108.
Ribosomal DNA (rDNA) quantification by quantitative real-time PCR (qPCR) was performed essentially as described previously (Wang et al. 2006). Briefly, total yeast DNA was purified from test strains grown asynchronously to the same OD600=0.8, DNA concentrations in different samples were further equalized in accordance with spectrometric reading. Independent primer pairs for measuring rDNA copy number were #9 and #17 from Wang et al. (2005), which both amplify 300-bp fragments. Primers amplifying TUB2 locus, as in Wang et al. (2005), were used for internal control. qPCR was carried out with the MX3000P real-time PCR system (Stratagene). PCR mixtures (50 μl) contained 1 μl of template DNA, 25 μl of 2× SYBR Green Master Mix (Stratagene), and 50 nM primers. PCR cycle parameters were 1 min at 95°C, 30 s at 55°C, and 1 min at 72°C. The cycle threshold values were determined for each sample, and fold change was calculated relatively to the wild type for four replicates for each DNA sample. The relative amount of rDNA was normalized to TUB2 locus values.
Results
We previously found that a single SUMO fused to Top2p induces a notable enrichment of Top2p in pericentromeric chromatin (Takahashi et al. 2006), which is reminiscent of the Ndc10p (Montpetit et al. 2006), while the native Top2 protein is conjugated to four to five SUMO molecules (Bachant et al. 2002; Takahashi et al. 2006). Therefore, to investigate the biological consequences of Top2p polysumoylation, we devised a modified approach allowing the introduction of multiple SUMO repeats into the same location as in Takahashi et al. (2006). As the SUMO-fused protein was expressed ectopically, in addition to the wild-type one, it was possible to create a good approximation of the wild-type situation, where both sumoylated and unmodified forms of the substrate coexist (Fig. 1a). This approach is more adequate than a traditional mutagenesis approach (Fig. 1a), where the whole pool of the substrate needs to be mutated, to reveal a possible phenotype. Using the linear SUMO fusions is also probably more justified than using linear ubiquitin fusions (Saeki et al. 2004; Varshavsky 2005), as the branching lysine residue of the SUMO molecule is more proximal to its amino terminus (Fig. 1b).
Fig. 1.
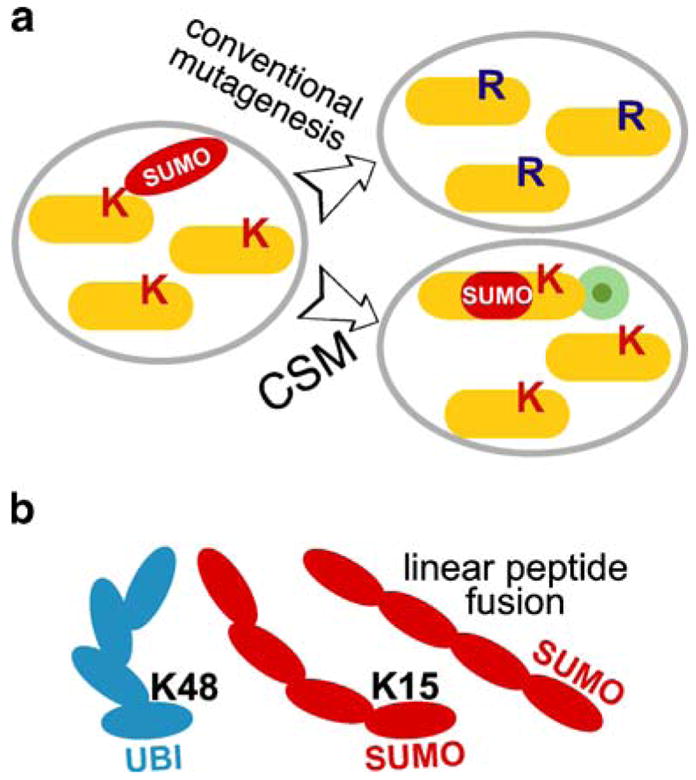
The experimental approach to modeling polysumoylation in vivo. a The advantage of constitutive SUMO modification (CSM) technique over traditional acceptor-site lysine mutagenesis. Ectopic expression of the tagged CSM target (lower right) represents a much better approximation of the wild type situation (left), compared to traditional mutagenesis of the sumoylated sites lysines (K to R; top right). CSM has an added benefit of biochemical/cytological tracking of the CSM pool of a given protein with an epitope tag (green circle). b Tandem peptide fusion of SUMO molecules is a good model of natural polysumoylation because of the proximal NH3-terminal position of the branching site lysine (K15) in SUMO. Branching in a polyubiquitin chain (K48) is shown for comparison
We systematically constructed serial SUMO fusions to the Top2p tail (constitutive SUMO modification, CSM, thereafter). SUMO-encoding sequences were fused in tandem at the internal Top2p site located between the first and the second C-terminal sumoylation sites (Fig. 2), leaving the natural SUMO sites intact. To facilitate detection of the Top2p variants, we also introduced GFP and/or 5× HA tags in all constructs. The presence of polyhistidine and FLAG tags within each SMT3 unit (Johnson and Blobel 1997; Takahashi et al. 2006) allowed single-step purification and robust detection of the given fusion.
Fig. 2.
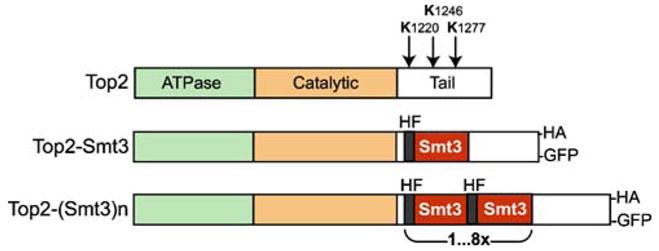
Topoisomerase II CSM constructs. Top panel—schematic domain organization of S. cerevisiae Top2p. Arrows indicate major sumoylation sites. In the middle—the single Smt3p fusion to Top2p, which has been shown to preferentially target pericentromertic chromatin (Takahashi et al. 2006). Lower panel—CSM constructs generated for this work. Each repeated SUMO unit contains polyhistidine/FLAG tags (HF; Takahashi et al. 2006). Most of the CSM constructs also included a COOH-terminal tag (HA or GFP)
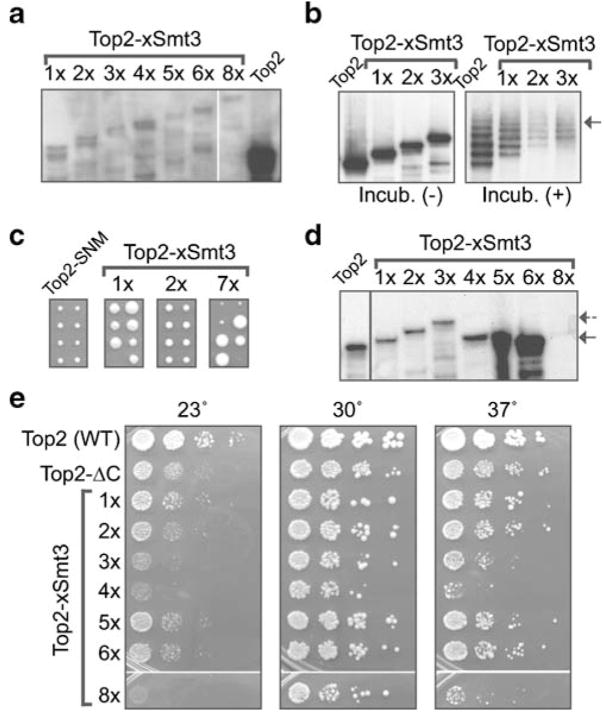
The TOP2::SMT3 fusion constructs were integrated into yeast genome, replacing the wild-type TOP2 locus. Initially, we used diploid yeast cells for transformation, to generate an ectopically expressed fusion (i.e., in the presence of the wild-type TOP2 gene), according to the principles outlined in Fig. 1a. As shown in Fig. 3a, Top2p–nSUMO fusions migrate slower than the backbone form of Top2p, because of the presence of additional SUMO copies. The expression levels of fused constructs appeared to be unaffected by the length of the fusion (Fig. 3a).
Fig. 3.
Poly-SUMO-fused Topoisomerase II variants have distinct biological properties in vivo. a Ectopic expression of Top2p–CSM in diploid cells. Western blotting of Top2p–CSM–HA construct integrated into W303 diploid cells (See Table 2, AS611–AS618). The control lane shows wild-type Top2p–HA in haploid cells (1033-W303-1A; not ectopic). b Fused SUMO molecules are counted toward total Top2p sumoylation in vitro. Sumoylation of chromatin-bound Top2p in vitro is described in detail in Takahashi et al. (2006). Briefly, the purified recombinant components of the conjugation reaction (Takahashi et al. 2003) were used in the reaction mixture containing chromatin-bound Top2p–HA (1033-W303-1A strain), Top2p–1Smt3–HA (AS611 segregant), Top2p–2Smt3–HA (AS612 segregant), and Top2p–3Smt3–HA (AS613 segregant), as substrates (right panel). The mixture kept on ice (Incub. −) was used as a negative control (left panel). Reaction products were analyzed by Western blotting using anti-HA antibody. The arrow points to the upper limit of sumoylation (five to six Smt3p), which is applicable to all tested constructs. c Top2p with long SUMO CSM chains inhibits cell growth. Meiotic progeny tetrads from diploid strains shown in a. Two dissected tetrads (30°C, 3 days) are shown for diploids expressing Top2p–SNM–HA (AS619), control, Top2p–1Smt3 (AS611), Top2p–2Smt3 (AS612), and Top2p–7Smt3 (AS617). The inhibitory effect of Top2p–7Smt3 results in very small colonies (last panel). A similar inhibitory effect is detected starting from 4× SUMO fused (not shown). d Four-SUMO threshold for stable maintenance of Top2p–CSM constructs. Western blotting showing extracts from haploid strains expressing Top2p–CSM constructs as a sole source of Topoisomerase II: Top2–Smt3 (a AS611 segregant), Top2p–2Smt3 (AS612 segregant), Top2p–3Smt3 (AS613 segregant), Top2p–4Smt3 (AS614 segregant), Top2p–5Smt3 (AS615 segregant), Top2p–6Smt3 (AS616 segregant), and Top2p–8Smt3 (AS618 segregant). The complete loss of all tandem SMT3 copies but one is evident for 5× (AS615 segregant) and above constructs, while the 4× (AS614 segregant) construct has only traces of full-length fusion (dashed arrow). The 8× construct in the haploid strain (AS618 segregant) results in the rapid loss of the whole integrated plasmid (including the HA epitope tag). e The necessity to reduce SUMO numbers by recombination results in reduced growth rates for 4–8× Top2p–CSM constructs. Fresh segregants containing Top2p–CSM with various numbers tandem SUMO were spotted onto YPD plates and incubated for 2 days. The growth-inhibitory effects are evident for 4 and 8× constructs, which eliminate excessive CSM SUMO by different recombination pathways. 5 and 6× constructs, which rapidly pop-out excessive CSM SUMO, show little growth inhibition in this analysis
Next, we tested whether the fused SUMO has a potential to be recognized by sumoylation machinery in vitro, before testing them for biologically activity in vivo. Using chromatin extracts from haploid strains (progeny from diploids shown in Fig. 3a), with one to three copies of SUMO-fused Top2p, we tested Top2p modification in vitro according to Takahashi et al. (2003, 2006). Based on this assay, we established that Top2–SUMO fusions can still be modified at several sites, even if reaction efficiency was somewhat decreased, probably as a result of enzyme titration by the fused SUMO (Fig. 3b). Moreover, we uncovered that the total number of SUMO per Top2p remained stable, as if “counting” of both fused and conjugated SUMO moieties has occurred (Fig. 3b, arrow). This indicated that the fused SUMO repeats, if not fully functional, have at least a potential to be recognized in the substrate context.
Some biological activity of fused SUMO could be a reason for the reduced growth rates of diploids with 5× SUMO fusions and higher (not shown). Therefore, we obtained the meiotic progeny of all diploids heterozygous for Top2p–CSM insertions to test Top2p–CSM functionality and localization, as well as the possible phenotypes of these strains, carrying the CSM fusion as a single source of Top2p. However, haploids with more than three copies of SUMO had a severely reduced growth rate and viability (Fig. 3c). Moreover, when the surviving haploids with CSM fusions were analyzed biochemically, we detected the loss of SUMO repeats (Fig. 3d). This loss was prominent beginning with 4× SUMO and resulted in the elimination of all SUMO copies but one (Fig. 3d). In the case of surviving strains with Top2p–8SUMO, recombination apparently removes the whole integrated construct, as indicated by disappearance of the HA tag (Fig. 3d). The comparison of growth rates and viability of different fusions in haploid cells shows that inhibitory effects of the fusion are proportional (Fig. 3e) to the rate of repeat removal (Fig. 3d), indicating that the presence of tandem repeats per se is the source of toxicity. Placing the ectopic Top2p–5SUMO fusion under pGAL promoter showed that such a strain has slower growth even under repressed promoter condition (probably because of the basal expression), and the growth was inhibited in galactose media, with SUMO repeats being lost at both conditions (data not shown). However, expression of the GAL promoter-driven tandem SUMO fusions (4SUMO and 5SUMO) themselves (lacking TOP2 sequences) did not produce any phenotypes in yeast cells (data not shown), suggesting that it is the in vivo Top2p function that is being affected by certain fusion lengths. It is unlikely, however, that long fusions significantly change the enzymatic Top2p activity per se, as the tail region of Top2p is relatively distant from the enzymatic core; therefore, some large molecules, such as GFP, can be harmlessly fused to the C terminus of Top2p (Takahashi et al. 2006).
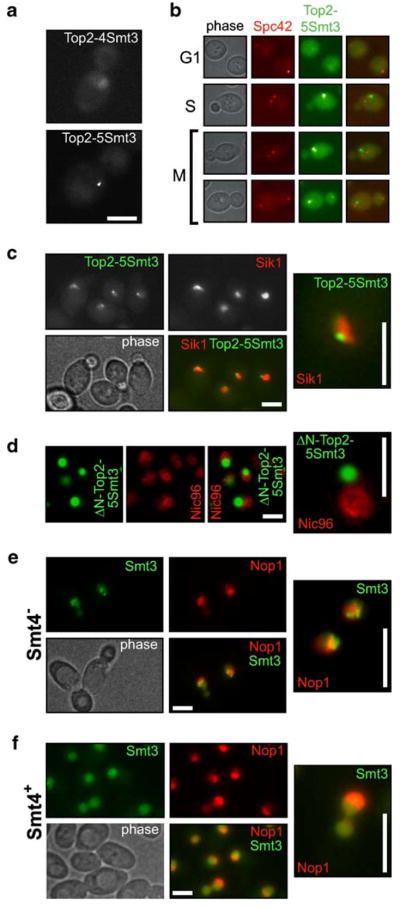
The results in Fig. 3 show that fusing four to five copies of SUMO to the Top2p tail produces a distinct genetic phenotype. Incidentally, this number corresponds to the number of SUMO conjugated to wild-type Top2p in vivo (Bachant et al. 2002; Takahashi et al. 2006). Therefore, we can hypothesize that Top2p with four to five CSM fusions should have some distinct properties in vivo, possibly reflecting the properties of poly- or multisumoylated wild-type Top2p (with the exception of not being a subject of desumoylation). One of the possible roles for such a heavy sumoylation, as noted in “Introduction,” could be localization to a specific chromosomal domain. Therefore, we assessed chromosomal localization of the CSM Top2 proteins, using GFP fusions (Fig. 2). We generated diploid strains carrying heterozygous TOP2:4SMT3 and TOP2:5SMT3 insertions fused to GFP (Table 2) and analyzed localization of the GFP signal. To offset the loss of SMT3 repeats through recombination, we used transformants as soon as they became visible on the transformation plate. As shown in Fig. 4a, typical localization patterns of Top2p–4SUMO and Top2p–5SUMO are distinct both from each other and from the wild-type Top2p–GFP. Unlike the latter, giving a uniform nuclear signal (Takahashi et al. 2006), both 4× CSM and 5× CSM fusions have subnuclear localization, with the 5× fusion showing an extreme concentration of GFP into a dot-like structure (Fig. 4a). Thus, intolerability of 4× CSM (and higher) Top2p in haploid cells (Fig. 3d,e) can be explained by draining the nuclear pool of the CSM Top2p to this concentrated location. We can hypothesize that this compartment corresponds to the natural localization of multisumoylated Top2p. As the 4× CSM fusion is more diffuse and probably represents an intermediate case, based on Western blotting analysis (Fig. 3d), we focused our further attention at the 5× CSM fusion localization.
Fig. 4.
Top2p polysumoylation is a signal for sub-nucleolar localization. a Diploid cells expressing Top2p–4Smt3–GFP (AS620) and Top2p–5Smt3–GFP (AS621). Cells were analyzed by microscopy as soon as transformants appeared on the plates, to avoid progressive loss of CSM SMT3 copies. The 4× construct (AS620) shows some enrichment of subnuclear structures, while the 5× construct (AS621) results in a compact dot-like staining in the nucleus. Scale bar (here and everywhere)=5 μm. b Top2p–5Smt3–GFP does not colocalize with kinetochores. Unlike Top2p–1Smt3–GFP (Takahashi et al. 2006), Top2p–5Smt3–GFP does not show colocalization with Spc42p-mRFP (Huh et al. 2003; AS622 strain). c Top2p–5Smt3–GFP localizes to nucleolar periphery. Diploid strain coexpressing Sik1p–mRFP (a nucleolar marker) and Top2p–5Smt3–GFP (AS623) was analyzed by fluorescent microscopy. For more than 100 GFP+ cells counted, a complete coincidence was observed between the GFP dot (Top2p–5Smt3–GFP) and the nucleolus. Higher magnification reveals that Top2p–5Smt3–GFP is in a subnucleolar domain close to the edge of the nucleolus (the insert). d The activity of Top2p core is required for subnucleolar localization of the Top2p–5Smt3 fusion. The COOH-tail of Top2p–5Smt3 lacking the enzymatic and DNA-binding domains of Top2p (Δ1-1218 TOP2) was expressed from GAL1 promoter with N-terminal fusion of GFP (pYT1108) in a Nic96p-mRFP strain (Nic96-EY0987; Huh et al. 2003). The Nic96p-mRFP marker was used to visualize boundaries of the nucleus. e The GFP–Smt3 protein is concentrated in the subnucleolar area in smt4Δ cells. The smt4Δ cells expressing GFP–Smt3p as a single source of SUMO (from the native SMT3 promoter) show accumulation of SUMO signal in a subnucleolar structure, Nop1p-mRFP was used as a nucleolar marker (1-YT626). f The bulk of GFP–Smt3p is not concentrated in the nucleolus in wild type cells. This control experiment for the e was done with the similarly marked Smt4+ wild-type strain (2-YT627)
Colocalization analysis of Top2–5× SUMO–GFP with a nuclear membrane protein Nic96p-mRFP (Huh et al. 2003) shows that Top2–5× SUMO dots indeed localize inside the nucleus (data not shown). Coexpression of Top2p–5SUMO–GFP with the SPB marker Spc42p-mRFP shows that unlike the single fusion of SUMO to Top2p, which enriches the pericentromeric area (Takahashi et al. 2006), the 5× CSM Top2p fusion localization does not coincide with Spc42p-mRFP (Fig. 4b). 5× CSM Top2p shows, however, some cell-cycle dependency, trailing the Spc42p-mRFP marker during chromosome segregation, both in its intracellular position and in its separation timing (Fig. 4b), suggesting that it is chromosomal.
The nucleolar organizer (or rDNA) is another prominent locus in the yeast genome that has a special Top2p requirement (Brill et al. 1987; Christman et al. 1988; Kim and Wang 1989), probably because of active transcription of rDNA throughout the cell cycle (Wang et al. 2006). Therefore, we compared Top2p–5SUMO–GFP localization with that of the nucleolar protein Sik1-mRFP. As shown in Fig. 4c, the 5× CSM Top2p fusion localizes within the nucleolar area, closer to its periphery. As the CSM Top2p cannot be demodified, it apparently cannot turn over to a SUMO-free form and thus accumulates in the nucleolus causing lethality. This localization analysis indicates that the multisumoylated pool of Top2p could be targeted and/or restricted to the nucleolus, where it performs its specific enzymatic function in rDNA transcription, replication, and segregation.
We conducted two additional control experiments to verify that the observed Top2p–5SUMO–GFP nucleolar concentration is relevant to the SUMO biology and the Top2p activity. First, we tested whether the DNA-binding and the enzymatic core of Top2p are required for nucleolar localization of Top2p–5SUMO. We expressed the amino terminus-truncated Top2p–5SUMO–GFP under the control of the GAL promoter. Such a fusion failed to localize to the nucleolus and was accumulating in the cytoplasmic compartment (Fig. 4d), indicating that some core Top2p activity is involved in the nucleolar targeting of the Top2p–CSM fusion.
In another control experiment, we tested whether a deletion of SMT4/ULP2, the major nuclear SUMO isopepti-dase-encoding gene (Li and Hochstrasser 2000; Strunnikov et al. 2001), would mimic the accumulation of sumoylated substrates in the nucleolus, similarly to Top2p–5SUMO. We constructed the Smt4+ (wild-type) and Smt4− (smt4Δ) strains, which expressed the GFP–Smt3p fusion from the native SMT3 promoter (see “Materials and methods”), as a sole source of SUMO. Both strains were analyzed for GFP–SUMO localization in the presence of a nucleolar RFP marker (Nop1p–RFP). The GFP–Smt3p signal was largely concentrated in the subnucleolar area, structurally reminiscent of rDNA chromatin (Fig. 4e), while in most wild-type SMT4 cells, the GFP–Smt3p signal was evenly distributed in the whole nucleus (Fig. 4f). This result suggests that subnucleolar accumulation of Top2p–5SUMO is not an artifact, but it is likely a reflection of the natural SUMO dynamics in wild type, which loses its transient nature if the SUMO removal is blocked (either by the covalent fusion in Top2p–5SUMO or by isopeptidase inactivation in smt4Δ).
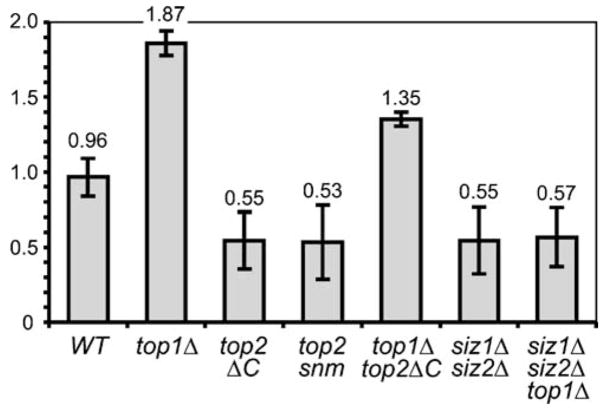
The finding that Top2p–5SUMO nucleolar accumulation likely reflects a general cellular phenomenon—transient targeting of sumoylated proteins to the nucleolus; we tested whether sumoylation of Top2p is required for nucleolar maintenance, particularly the rDNA locus stability. As the top2ΔC-encoded mutant protein lacks any sumoylation in vivo and cannot be modified in vitro (Takahashi et al. 2006), we re-examined the phenotype of this mutant, as well as some additional mutations in the critical top-oisomerase and sumoylation activities. Initial analysis of rDNA content by Southern blotting was inconclusive, as we found that the top2ΔC and especially topΔ1 mutations result in loss of some chromosomal rDNA repeats with the attendant gain of extrachromosomal rDNA, which was difficult to quantify. Therefore, we analyzed the relative rDNA content by qPCR and revealed that top2ΔC and top2–snm cells have reduced levels of rDNA (Fig. 5). However, when Topoisomerase I was inactivated by TOP1 deletion, the combined top1Δ top2ΔC double mutant showed a reproducible increase in rDNA content relatively to top2ΔC, probably a result of extrachromosomal replication (Southern blotting data, not shown). The rDNA content of the top1Δ top2ΔC double mutant was still lower compared to the single top1Δ mutation, indicating that Top2p sumoylation is likely also required to maintain extrachromosomal rDNA induced by top1Δ. Similar to top2ΔC and top2–snm, the rDNA loss was also associated with the siz1Δ siz2Δ double mutation, which blocks the bulk of sumoylation in the cells, including the Top2p modification. It is interesting to note that the top1Δ mutation was unable to restore the rDNA content in siz1Δ siz2Δ even partially (as with top2ΔC), indicating that other factors involved in the rDNA maintenance may need to be SUMO modified, in addition to Top2p. These data confirm that sumoylation of Top2p and probably some additional factors is required for stable rDNA maintenance.
Fig. 5.
Relative rDNA dosage is decreased in cells lacking the sumoylated Topoisomerase II. For most mutants rDNA quantification by qPCR (see “Materials and methods”) was done for at least two independent strains (including divergent backgrounds) of the same relevant genotype: wild type (W303-1A, BY4733, BY4729), top1Δ (3a4031697, 4b4031697), top2ΔC (1035-BY4729, 1035-W303), top1Δ top2ΔC (1aYT625), top2-snm (Y2226/JBY1087), siz1Δ siz2Δ (4bAS399, 12cAS399, 1-YT14, 2-YT14) and siz1Δ siz2Δ top1Δ (3-YT627, 4-YT627) strains. Two rDNA primer pairs were used for each experiment, and multiple independent qPCR analyses are averaged. All values were normalized to the TUB2 (a single-copy gene) PCR product (analyzed in each experiment in parallel) and then to the wild-type (W303) signal
Discussion
In this report, we modeled sumoylated forms of Top-oisomerase II in vivo by fusing an optimal number of SUMO in-frame within the target protein. Without using CSM, it would be extremely problematic to determine the function of poly-sumoylation because sumoylation and desumoylation of a given substrate in vivo is highly dynamic. The presented approach has several distinct advantages: (1) Ectopic expression is not only allowable but is desirable, to mimic the natural balance of the modified/unmodified substrate; (2) addition of specific epitope tags allows both biochemical purification and cytological detection of CSM targets; (3) physical mapping of individual sumoylated sites is not always necessary, as clusters of consensus SUMO conjugation sites provide a reliable prediction of multiple sumoylations (Xue et al. 2006); (4) the approach is not limited to yeast. The toxicity of ectopic constitutive SUMO fusions (as in Fig. 3) could be an unavoidable limitation, as the CSM SUMO is not removed in a normal regulatory fashion by isopeptidase because the COOH-terminal glycine residue of Smt3p is absent from the fusions. While such an approach is also obviously limited to the targets that have sumoylation sites clustered outside of the vital protein parts, e.g., active centers of enzymes, it is unlikely that natural sumoylation sites with a positive regulatory effect would frequently occur in such critical regions because of the bulkiness of conjugated SUMO.
The discovery of nucleolar targeting of Topoisomerase II by poly-(multi-)sumoylation suggests that heavily sumoylated topoisomerase has a specialized function in rDNA. The localization of Topoisomerase II to a subnucleolar compartment has been also described in the metazoan system: The C-terminal domain of mammalian Topoisomerase II beta localizes to a subnucleolar structure (Sakaguchi et al. 2001). One of the nucleolus-specific functions of Top2p could be rDNA transcription by RNA polymerase I (Brill et al. 1987), especially prominent in yeast where rDNA is constantly transcribed (Wang et al. 2006). However, we may have uncovered a more general role of heavy sumoylation in nucleolar maintenance, by showing that the bulk of SUMO is concentrated in the nucleolus in the smt4Δ mutant. While some reports showed that increase in sumoylation correlates with proteins leaving the nucleolus (Mo et al. 2002; D’Amours et al. 2004; Torres-Rosell et al. 2007), in neither case, the localization of the sumoylated pool was tracked.
Based on our findings, the general role of (poly-) sumoylation could be to target subpopulations of chromatin proteins with a general nuclear function, to the nucleolus. Substantial deregulation of such targeting, e.g., by over- or undersumoylation of several substrates, may result in some nucleolar structure and segregation defects (Li and Hochstrasser 2000; Strunnikov et al. 2001). Our analysis of rDNA content in the top2ΔC mutant indicates that the absence of sumoylated Top2p alone leads to the loss of tandem rDNA repeats (Fig. 5), characteristic to much more severe top2 mutations (Brill et al. 1987; Christman et al. 1988; Kim and Wang 1989). However, in the case of top2ΔC and top2–snm, viabilities of mutant strains are not affected, consistent with an rDNA-specific defect, e.g., elevated recombination resulting in increased turnover and loss of repeats (Kim and Wang 1989). The fact that the siz1Δ siz2Δ double mutant essentially phenocopies this trait of top2ΔC and top2–snm indicates that Top2p sumoylation is a key player in rDNA maintenance, although some additional sumoylated factors must be involved (as suggested by the different mode of siz1Δ siz2Δ interaction with top1Δ (Fig. 5).
The nucleolar role of multi-SUMO modification in the unchallenged cell cycle is indirectly supported by other experimental results. Among them are the existence of a nucleolus-resident E3 SUMO ligase Mms21p (Zhao and Blobel 2005), the localization of two mammalian SUMO-isopeptidases to the nucleolus (Mukhopadhyay and Dasso 2007), and especially by proteomic data in budding yeast (Wohlschlegel et al. 2004; Hannich et al. 2005). Indeed, among the 196 nuclear sumoylated proteins that have at least two sumoylated sites in vivo (Wohlschlegel et al. 2004), 39 are nucleolar (according to GO), 41 are confirmed to be partially nucleolar, 90 are chromatin components that can potentially have a nucleolar subset, six are SUMO pathway components, and only 20 are uncharacterized or unlikely to be nucleolar. Although the hypothesis of polysumoylation’s role in nucleolar targeting needs additional extensive validation, its emergence highlights the usefulness of the improved SUMO fusion approach (CSM), both for practical application to a given SUMO target and for advancement of general knowledge about SUMO biology. Based on our observations, we can infer the existence of a mechanisms that controls nucleolar localization of a subset of given nuclear proteins by their targeting to the nucleolus via transient polysumoylation and their nucleolar retention via recognition by some SUMO-binding protein(s) residing in the nucleolus. As putative SUMO-binding domains have recently been identified (Hecker et al. 2006; Lin et al. 2006; Kerscher 2007; Sun et al. 2007; Uzunova 2007), it is conceivable that some nucleolar proteins may have an ability to serve as receptors for sumoylated proteins, which hence acquire a nucleolar-specific function through such interactions.
Acknowledgments
Authors thank Y. Kikuchi for plasmids and strains, V. Yong-Gonzalez, P. Butylin, S. Dulev, A. Samoshkin, and X. Strunnikova for fruitful discussion, and S. Malhotra and Y. Ejamo for technical assistance. This work was supported by the NICHD Intramural Program.
Footnotes
Communicated by E.A. Nigg
References
- Bachant J, Alcasabas A, et al. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell. 2002;9(6):1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- Brill SJ, DiNardo S, et al. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326(6111):414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- Christman MF, Dietrich FS, et al. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell. 1988;55(3):413–425. doi: 10.1016/0092-8674(88)90027-x. [DOI] [PubMed] [Google Scholar]
- D’Amours D, Stegmeier F, et al. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117(4):455–469. doi: 10.1016/s0092-8674(04)00413-1. [DOI] [PubMed] [Google Scholar]
- Denison C, Rudner AD, et al. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol Cell Proteomics. 2005;4(3):246–254. doi: 10.1074/mcp.M400154-MCP200. [DOI] [PubMed] [Google Scholar]
- Dobreva G, Dambacher J, et al. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev. 2003;17(24):3048–3061. doi: 10.1101/gad.1153003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannich JT, Lewis A, et al. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem. 2005;280(6):4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- Hecker CM, Rabiller M, et al. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem. 2006;281(23):16117–16127. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- Huang TT, Wuerzberger-Davis SM, et al. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell. 2003;115(5):565–576. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272(43):26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147(5):981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O. SUMO junction—what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8(6):550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RA, Wang JC. A subthreshold level of DNA topoisomerases leads to the excision of yeast rDNA as extrachromosomal rings. Cell. 1989;57(6):975–985. doi: 10.1016/0092-8674(89)90336-x. [DOI] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol. 2000;20(7):2367–2377. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DY, Huang YS, et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell. 2006;24(3):341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, et al. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88(1):97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, et al. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135(6 Pt 1):1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo YY, Yu Y, et al. Nucleolar delocalization of human topoisomerase I in response to topotecan correlates with sumoylation of the protein. J Biol Chem. 2002;277(4):2958–2964. doi: 10.1074/jbc.M108263200. [DOI] [PubMed] [Google Scholar]
- Montpetit B, Hazbun TR, et al. Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J Cell Biol. 2006;174(5):653–663. doi: 10.1083/jcb.200605019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32(6):286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Panse VG, Hardeland U, et al. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem. 2004;279(40):41346–41351. doi: 10.1074/jbc.M407950200. [DOI] [PubMed] [Google Scholar]
- Reindle A, Belichenko I, et al. Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J Cell Sci. 2006;119(Pt 22):4749–4757. doi: 10.1242/jcs.03243. [DOI] [PubMed] [Google Scholar]
- Ross S, Best JL, et al. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol Cell. 2002;10(4):831–842. doi: 10.1016/s1097-2765(02)00682-2. [DOI] [PubMed] [Google Scholar]
- Saeki Y, Isono E, et al. Intracellularly inducible, ubiquitin hydrolase-insensitive tandem ubiquitins inhibit the 26S proteasome activity and cell division. Genes Genet Syst. 2004;79(2):77–86. doi: 10.1266/ggs.79.77. [DOI] [PubMed] [Google Scholar]
- Saitoh H, Sparrow DB, et al. Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr Biol. 1998;8(2):121–124. doi: 10.1016/s0960-9822(98)70044-2. [DOI] [PubMed] [Google Scholar]
- Sakaguchi A, Akashi T, et al. A distinct subnuclear localization of mammalian DNA topoisomerase IIbeta in yeast. Biochem Biophys Res Commun. 2001;283(4):876–882. doi: 10.1006/bbrc.2001.4856. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Toh EA, et al. Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40S ribosome biogenesis in the nucleolus. Mol Cell Biol. 2000;20(21):7971–7979. doi: 10.1128/mcb.20.21.7971-7979.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA. 2003;100(23):13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Aravind L, et al. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics. 2001;158(1):95–107. doi: 10.1093/genetics/158.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Leverson JD, et al. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOy-lated proteins. Embo J. 2007;26:4102–4112. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Toh EA, et al. Comparative analysis of yeast PIAS-type SUMO ligases in vivo and in vitro. J Biochem (Tokyo) 2003;133(4):415–422. doi: 10.1093/jb/mvg054. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Yong-Gonzalez V, et al. SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II. Genetics. 2006;172(2):783–794. doi: 10.1534/genetics.105.047167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J, Sunjevaric I, et al. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol. 2007;9(8):923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- Uzunova K, Gottsche K, et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem. 2007;282:34167–34175. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. Ubiquitin fusion technique and related methods. Methods Enzymol. 2005;399:777–799. doi: 10.1016/S0076-6879(05)99051-4. [DOI] [PubMed] [Google Scholar]
- Wang BD, Butylin P, et al. Condensin function in mitotic nucleolar segregation is regulated by rDNA transcription. Cell Cycle. 2006;5(19):2260–2267. doi: 10.4161/cc.5.19.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BD, Eyre D, et al. Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol Cell Biol. 2005;25(16):7216–7225. doi: 10.1128/MCB.25.16.7216-7225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3(6):430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Johnson ES, et al. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J Biol Chem. 2004;279(44):45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- Wykoff DD, O’Shea EK. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol Cell Proteomics. 2005;4(1):73–83. doi: 10.1074/mcp.M400166-MCP200. [DOI] [PubMed] [Google Scholar]
- Xue Y, Zhou F, et al. SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res. 2006;34(Web Server issue):W254–W257. doi: 10.1093/nar/gkl207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA. 2005;102(13):4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Ryan JJ, et al. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem. 2004;279(31):32262–32268. doi: 10.1074/jbc.M404173200. [DOI] [PMC free article] [PubMed] [Google Scholar]