Abstract
In eukaryotic cells, the ubiquitin–proteasome pathway is the major mechanism for the targeted degradation of proteins with short half-lives. The covalent attachment of ubiquitin to lysine residues of targeted proteins is a signal for the recognition and rapid degradation by the proteasome, a large multi-subunit protease. In this report, we demonstrate that the human estrogen receptor (ER) protein is rapidly degraded in mammalian cells in an estradiol-dependent manner. The treatment of mammalian cells with the proteasome inhibitor MG132 inhibits activity of the proteasome and blocks ER degradation, suggesting that ER protein is turned over through the ubiquitin–proteasome pathway. In addition, we show that in vitro ER degradation depends on ubiquitin-activating E1 enzyme (UBA) and ubiquitin-conjugating E2 enzymes (UBCs), and the proteasome inhibitors MG132 and lactacystin block ER protein degradation in vitro. Furthermore, the UBA/UBCs and proteasome inhibitors promote the accumulation of higher molecular weight forms of ER. The UBA and UBCs, which promote ER degradation in vitro, have no significant effect on human progesterone receptor and human thyroid hormone receptor β proteins.
The ubiquitin–proteasome pathway is the major system in the eukaryotic cell for the selective degradation of short-lived regulatory proteins (1, 2). A common feature of proteasome-mediated protein degradation is the covalent attachment of ubiquitin, a highly conserved 8.6-kDa protein, to lysine residues of proteins targeted for degradation followed by the formation of polyubiquitin chains attached covalently to the targeted protein. Ubiquitinated proteins are recognized and degraded by the multi-subunit protease complex, the 26S-proteasome (3–6). In addition to the role it plays in protein degradation, ubiquitination may serve regulatory functions such as directing the subcellular localization of proteins (3, 4). The ubiquitin–proteasome pathway also plays an important role in various cellular processes such as cell-cycle regulation, signal transduction, differentiation, antigen processing, and degradation of tumor suppressors (3, 4, 7–11).
Protein ubiquitination involves three classes of enzymes, namely the E1 ubiquitin-activating enzyme (UBA), E2 ubiquitin-conjugating enzymes (UBCs), and E3 ubiquitin–protein ligases. The UBA first activates ubiquitin in an ATP-dependent manner. The activated ubiquitin then forms a thioester bond between the carboxyl-terminal glycine residue of ubiquitin and a cysteine residue of the UBA. Next, ubiquitin is transferred from the E1 to one of the several E2s (UBCs), preserving the high-energy thioester bond (1, 2, 5, 11). In some cases, ubiquitin is transferred directly from the E2 to the target protein through an isopeptide bond between the ɛ-amino group of lysine residues of the target protein and the carboxy terminus of ubiquitin. In other instances, the transfer of ubiquitin from UBCs to target proteins proceeds through an E3 ubiquitin–protein ligase intermediate (12, 13). It has been proposed that the biological specificity of the ubiquitin pathway is modulated by the selective combination of UBCs and E3 proteins. To date, more than 30 UBCs and 25 E3 proteins have been identified (7, 14).
Recent studies from our laboratory and others suggest that the ubiquitin-conjugating enzyme, UBC9, and the E3 ubiquitin–protein ligases, E6-associated protein and RPF1/RSP5, interact with members of the nuclear hormone receptor superfamily and modulate their transactivation functions (15–18). Similarly, yeast SUG1, an ATPase subunit of the 26S-proteasome complex, also interacts with and modulates nuclear hormone receptor functions (19–22). These studies suggest a possible regulatory role for the ubiquitin–proteasome pathway in nuclear hormone receptor-mediated gene activation.
The stability of the human estrogen receptor (ER) is modulated by its ligand, estradiol. In the absence of estradiol, the half-life of ER is ≈5 days, but only 3–4 hr in the presence of estradiol (23, 24). Because the ER protein has a short half-life in the presence of ligand (24), it is possible that the receptor itself would be a target of the ubiquitin–proteasome degradation pathway. In fact, a previously published study suggests that the ER protein in uterus may be ubiquitinated (23). However, not all members of the steroid hormone receptor superfamily are similarly regulated. For example, the progesterone receptor (PR) and glucocorticoid receptor are reported to have longer half-lives (≈20–25 hr) regardless of the presence of ligand (25, 26).
In this report, we now show that ER is degraded in a hormone-dependent manner and the proteasome inhibitor, MG132, promotes the in vivo accumulation of ER and blocks hormone-induced receptor degradation. We demonstrate that ER is degraded in vitro and that this degradation depends on UBA and UBC enzymes of the ubiquitin pathway and the proteasome inhibitors, MG132 and lactacystin, block ER degradation in vitro. Furthermore, the UBA/UBCs, regardless of the presence of proteasome inhibitors, promote the accumulation of higher molecular weight forms of ER. Our data also indicate that the ubiquitin pathway enzymes that facilitate ER degradation are unable to promote the degradation of PR and human thyroid hormone receptor β (TR) under similar experimental conditions and suggest that specific complexes of UBA and UBCs may target different nuclear receptors for degradation.
MATERIALS AND METHODS
Plasmid Constructs.
The mammalian expression plasmid for ER (27), the in vitro expression plasmids for ER, PR, and TR (27, 28), the bacterial expression plasmids of Arabidopsis thaliana UBA1 (29), and expression plasmids of various UBCs, [UbcH5B (30) and UbcH7 (31)] have been described previously. The estrogen-responsive reporter plasmid, pERE.E1b.LUC, was constructed by ligating a PvuII–SmaI fragment of pERE.E1B.CAT into the SmaI site of the pGL3 Basic plasmid (Promega).
Transfections.
HeLa cells were maintained in DMEM supplemented with 10% fetal bovine serum. Twenty-four hours before transfection, 3 × 105 cells per well were plated in six-well Falcon dishes in phenol red-free DMEM containing 5% dextran-coated charcoal-stripped serum. Cells were transfected with 4 ng of ER expression plasmid and 750 ng of the estrogen-responsive reporter plasmid by using Lipofectamine (Life Technologies, Grand Island, NY), according to the manufacturer’s recommended guidelines. Cells were washed and fed with phenol-red free DMEM containing 5% charcoal-stripped serum and subsequently treated with 10−9 M estradiol (E2) and 1 μM proteasome inhibitor, MG132 (Sigma). As a control, cells were treated with dimethyl sulfoxide (DMSO) both in the absence and presence of estradiol. After 24 hr, cells were harvested and cell extracts were prepared for ER protein analysis.
Analysis of ER Protein Levels.
To analyze the ER protein levels, transfected cells were harvested and lysed in ER extraction buffer [50 mM Tris⋅HCl (pH8.0)/5 mM EDTA/1% Nonidet P-40/0.2% Sarkosyl/0.4 M NaCl/100 μM sodium vanadate/10 mM sodium molybdate/20 mM NaF]. Subsequently, 40 μg of protein extracts was loaded and resolved by 7.5% SDS/PAGE and then transferred to a nitrocellulose membrane. The nitrocellulose membrane was incubated in a blocking buffer [50 mM Tris⋅HCl (pH7.5)/150 mM NaCl/0.5% Tween 20/1% dried nonfat milk] for 1 hr at room temperature. Then the membrane was incubated with the H222 antibody, which specifically recognizes the ER protein. After extensive washing, the membrane was first incubated with rabbit anti-rat antibody and then with horseradish peroxidase-conjugated goat-anti-rabbit IgG, and ER protein levels were visualized with the ECL+Plus Western blotting detection system (Amersham).
Bacterial Expression of Ubiquitin Pathway Enzymes.
A. thaliana UBA1/E1 and ubiquitin-conjugating enzymes (UbcH5B and UbcH7) were expressed in Escherichia coli BL21 (λDE3) by using the pET expression system (Novagen) (32). Bacterial cells harboring appropriate expression plasmids were grown overnight in 400 ml cultures at 25°C. The next morning, expression of proteins was induced with 1 mM isopropyl-d-thiogalactoside for 3–4 hr. Subsequently, cells were lysed in sonication buffer [10 mM Tris⋅HCl (pH 7.9)/10% glycerol/0.5 M NaCl/0.1% Nonidet P-40/5 mM β-mercaptoethanol and protease inhibitors (100 μg/ml phenylmethylfulfonyl fluoride/2 μg/ml leupeptin/2 μg/ml aprotinin/1 μg/ml pepstatin)]. Coomassie blue staining of an aliquot of each lysate separated by 7.5% SDS/PAGE was used to determine the relative amounts of each protein.
In Vitro Expression of ER, PR, and TR.
In vitro expression of radiolabeled ER, PR, and TR proteins was performed by using in vitro transcription and translation (TNT)-coupled rabbit reticulocyte extracts in the presence of [35S]methionine, according to manufacturer’s recommended conditions (Promega).
Protein Degradation and Ubiquitination Assays.
35S-labeled ER was incubated with and without UBA1/E1 (≈5–10 ng) and UBCs/E2s (≈100 ng) in reaction mixtures containing 20 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 4 mM ATP, 10 mM MgCl2, 0.2 mM DTT, and 4 μg of ubiquitin (Sigma) for 2–3 hr at 25°C. Reactions were terminated by boiling samples in the presence of SDS-loading buffer [100 mM Tris⋅HCl (pH 8.0)/200 mM DTT/4% SDS/20% glycerol/0.2% bromophenol blue]. The reaction mixtures were resolved by 7.5% SDS/PAGE, and radiolabeled bands were visualized by autoradiography. However, in Fig. 3B the ER protein was analyzed by Western blot analysis using H222 antibody.
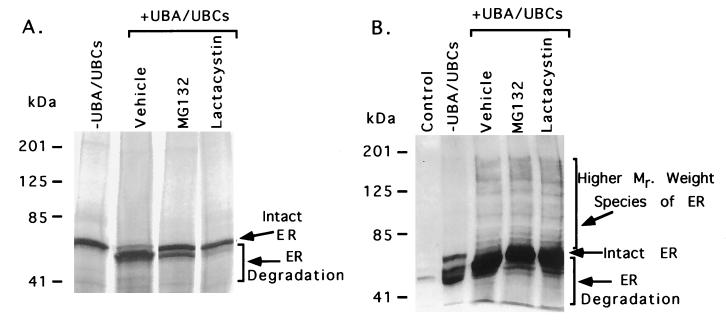
Figure 3.
The proteasome inhibitors, MG132 and lactacystin, block ER degradation in vitro. (A) 35S-labeled ER protein was synthesized in vitro in the presence of either vehicle only, 33 μM MG132 or 33 μM lactacystin with TNT-coupled rabbit reticulocyte extracts. The labeled ER protein was then incubated with ATP and ubiquitin either in the absence of UBA/UBCs or in the presence of UBA and UBCs (UbcH5B and UbcH7). Arrows indicate the position of intact and degraded ER protein. (B) The UBA/UBCs and proteasome inhibitors, MG132 and lactacystin, promote the accumulation of slower migrating forms of ER (shown by a bracket). 35S-labeled ER protein was synthesized in vitro in the presence of either vehicle only, 33 μM MG132 or 33 μM lactacystin with TNT-coupled rabbit reticulocyte extracts. The labeled ER protein was then incubated with ATP and ubiquitin either in the absence of UBA/UBCs or in the presence of UBA and UBCs (UbcH5B and UbcH7). Then the ER protein was analyzed by Western blot analysis using H222 antibody that specifically recognizes ER. The control lane contains reticulocyte extract only. Arrows indicate the position of intact, degraded, and slower migrating forms of ER protein.
RESULTS AND DISCUSSION
It has been reported that ER in the uterus is ubiquitinated and exhibits a short half-life in other estrogen target tissues in the presence of estradiol (23, 24). To determine whether down-regulation of the ER protein is mediated by the ubiquitin–proteasome pathway, we performed transient cotransfection assays in the presence or absence of the proteasome inhibitor, MG132. HeLa cells were cotransfected with an expression plasmid for ER and a reporter plasmid containing an estrogen response element and subsequently incubated with either DMSO (vehicle) or MG132 both in the absence and presence of estradiol. The effect of hormone and MG132 on ER protein levels was analyzed by Western blot analysis of cell extracts from these cells. As shown in Fig. 1, the DMSO-treated control cells exhibit lower levels of ER protein compared with that of MG132-treated cells. Addition of estradiol to the control cells reduces the level of ER protein compared with cells that were not treated with hormone (lane 1 vs. lane 2). However, MG132 blocks the estradiol-induced degradation of the ER protein (lane 3 vs. lane 4). The small molecular weight (<66 kDa) bands apparent in the MG132-treated cells likely are the result of nonproteasomal degradation of overexpressed ER. These data are consistent with the previously published report that indicates that estradiol induces down-regulation of the ER protein (23, 24). Our results also suggest that estrogen-dependent down-regulation of ER proceeds through the proteasome.
Figure 1.
ER degradation depends on the ubiquitin–proteasome pathway. HeLa cells were transiently transfected with 4 ng of pCMV5hER and 750 ng of pERE.E1b.LUC. The cells were treated with either vehicle (DMSO) or proteasome inhibitor (1 μM MG132) both in the absence (−) and presence of 10−9 M estradiol (E2). The ER expression was analyzed by Western blot by using an anti-ER antibody, H222.
To further investigate whether hormone-dependent ER down-regulation was through the ubiquitin–proteasome pathway, we performed in vitro protein degradation and ubiquitin assays. 35S-labeled ER protein was synthesized in vitro by using TNT-coupled rabbit reticulocyte extracts in the presence of radiolabeled methionine. The 35S-labeled ER protein was then incubated with ATP and ubiquitin either in the absence of UBA/UBCs or in the presence of bacterially expressed UBA and UbcH5B and UbcH7 (UBCs), and reactions were terminated at varying times. In vitro, UBA and UBC enzymes promoted the degradation of ER protein compared with a control that lacks UBA and UBCs (Fig. 2). Furthermore, ER is degraded in a time-dependent manner. As shown in Fig. 2, most of the full-length receptor protein is degraded into a smaller form within 2 hr. This receptor degradation is not complete. The ER undergoes limited proteolysis that results in a slightly smaller form of ER. This restricted pattern of ER degradation is analogous to that of tramtrack and vitamin D receptor degradation, which are also degraded into slightly smaller forms through a proteasome pathway (22, 33). It is likely that more complete degradation of ER does not occur in these in vitro assays because of limiting amounts of proteasome pathway components. Furthermore, addition of hormone in in vitro assays did not change the ER degradation pattern (data not shown). These data support the hypothesis that the proteasome pathway is involved in ER protein degradation.
Figure 2.
In vitro ER degradation depends on ubiquitin pathway enzymes, UBA and UBCs. 35S-labeled ER protein was synthesized in vitro with TNT-coupled rabbit reticulocyte extracts. The labeled ER protein was incubated with ATP and ubiquitin either in the absence of UBA/UBCs (for 120 min) or presence of bacterially expressed UBA and UBCs (UbcH5B and UbcH7). Reactions were terminated at varying times by adding SDS-loading buffer and analyzed by SDS/PAGE and autoradiography. Arrows indicate the position of intact and degraded ER protein.
Next, we asked whether inhibitors of the proteasome pathway were able also to reduce the in vitro degradation of ER. A control reaction in which ER was incubated with vehicle exhibited UBA- and UBCs-dependent ER protein degradation (Fig. 3A). However, the proteasome inhibitors MG132 or lactacystin significantly inhibited the UBA/UBC-mediated degradation of ER. These data are consistent with our intact cell data (Fig. 1), which indicate that the ER protein is degraded through the proteasome pathway, and that inhibitors of this pathway inhibit ER degradation. The in vitro inhibition of ER protein degradation by MG132 is less effective than inhibition by lactacystin. The weaker effect of MG132 may be caused by the fact that MG132 binds to the proteasome in a reversible manner, and that ubiquitinated ER effectively competes for binding to the proteasome because of a higher affinity for the proteasome. In contrast, lactacystin binds to the proteasome in an irreversible manner.
Because the UBA/UBCs promote ER degradation, and proteasome inhibitors decrease the degradation of ER protein both in vivo and in vitro, we asked whether the UBA/UBCs, MG132, and lactacystin-treated reactions promote the accumulation of higher molecular weight forms of ER. Because ubiquitin is conjugated to multiple lysine residues of target proteins and forms polyubiquitin chains, ubiquitin-tagged proteins can be seen as a ladder of higher molecular weight species on SDS/PAGE gels (11, 23, 31). As shown in Fig. 3B, the Western blot analysis of ER protein reveals that the control reaction without proteasome inhibitors (vehicle) exhibited UBA- and UBCs-dependent degradation of ER. Addition of MG132 and lactacystin to the reaction decreased ER degradation. Furthermore, a ladder of higher molecular weight species of ER is visible only in the reactions treated with UBA/UBCs regardless of the presence of proteasome inhibitors compared with that of the −UBA/UBCs reaction. Similarly, higher molecular weight species of ER can be seen in Fig. 3A after exposing the gel ≈10 times longer than the one shown in Fig. 3A (data not shown). It is possible that the ER degradation pattern seen in Fig. 3B is slightly different from that of Fig. 3A because of increased sensitivity in the Western blot, which preferentially amplifies the signal of some minor ER species. The high molecular weight species of ER presumably represent the ubiquitinated form of ER since the −UBA/UBCs reaction did not exhibit the higher molecular weight species ER protein (Fig. 3B). These data are similar to the previously published report that indicates that ubiquitinated forms of ER exhibit a ladder of higher molecular weight species (23). Taken together, these data are highly suggestive that ER is degraded through the ubiquitin–proteasome pathway.
To determine whether the ubiquitin–proteasome pathway also promotes the degradation of other members of the nuclear receptor superfamily, we performed in vitro protein degradation and ubiquitin assays on the PR and TR proteins. The 35S-labeled PR and TR proteins were synthesized by TNT-coupled rabbit reticulocyte extracts in the presence of radiolabeled methionine. The 35S-labeled PR and TR proteins were then incubated with ATP and ubiquitin either in the absence of UBA and UBCs or in the presence of bacterially expressed UBA, UbcH5B, and UbcH7. As shown in Fig. 4A, the addition of ubiquitin pathway enzymes, UBA and UBCs, has no significant effect on PR protein levels. Furthermore, addition of MG132 also exhibited no significant effect on the level of PR protein (Fig. 4A). Data from our transfection studies also suggest that PR protein levels are not significantly altered by either hormone or protease inhibitors in mammalian cells (data not shown).
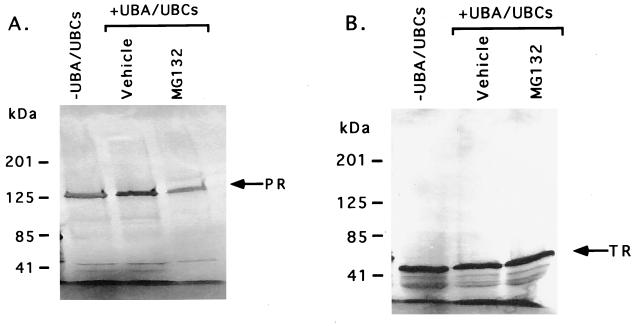
Figure 4.
PR and TR proteins are not the target of the ubiquitin–proteasome pathway. (A) 35S-labeled PR protein was incubated with ATP and ubiquitin either in the absence of UBA/UBCs or in the presence of UBA and UBCs for 120 min (UbcH5B and UbcH7). In the presence of UBA/UBCs, the reaction mixtures were treated with either vehicle or 33 μM MG132. The position of intact PR is indicated by the arrow. (B) 35S-labeled TR was incubated with ATP and ubiquitin either in the absence of UBA/UBCs or in the presence of UBA and UBCs for 120 min (UbcH5B and UbcH7). The reaction mixtures containing UBA and UBCs were treated with either vehicle or 33 μM MG132. The position of intact TR is indicated by the arrow.
Like PR, ubiquitin–proteasome pathway enzymes have no significant effect on the level of TR. The TR is intact both in the absence and presence of ubiquitin pathway enzymes (Fig. 4B). Similarly, MG132 exhibited no significant effect on TR. These data suggest that PR and TR are not the target of the ubiquitin–proteasome pathway in this assay system. Our PR data appear to be in contrast with a previously published study that reports that chicken PR protein is ubiquitinated (34). Several reasons may account for this difference. Our study used human PR protein instead of chicken PR protein, and it is possible that each protein possesses intrinsic differences that may account for this discrepancy. Alternatively, if the human PR protein is degraded by means of the ubiquitin pathway, it may require a different set of UBC enzymes than those used to demonstrate degradation of ER in our in vitro assays.
In eukaryotic cells, several different ubiquitin and ubiquitin-like pathways exist that are mediated by ubiquitin itself and by two other ubiquitin-like proteins, neural precursor cell-expressed developmentally down-regulated (NEDD8) and sentrin (2, 14, 35–37). To date, only the ubiquitin pathway has been clearly implicated in targeted protein degradation. NEDD8 and sentrin are both small (≈8 kDa) proteins that share significant homology to ubiquitin and also are covalently attached to target proteins. The role of the NEDD8 and sentrin pathways in protein degradation is not clear and the significance of modification by NEDD8 and sentrin is unknown (2, 14, 35–37). However, yeast RUB1 and SMT3 proteins, which are closely related to ubiquitin and the mammalian ubiquitin-related factor SUMO1, have been shown to be required for survival and appropriate cell-cycle progression in yeast (38).
The importance of the ubiquitin–proteasome pathway in higher eukaryotes has been well established in cell-cycle regulation, signal transduction, and cell differentiation. Recently, the ubiquitin–proteasome pathway has been linked to transcriptional machinery, and it has been demonstrated that the carboxyl-terminal tail of RNA polymerase II itself is a target of the ubiquitin–proteasome pathway (2–4, 7–11, 39). The involvement of the ubiquitin–proteasome pathway in eukaryotic transcription is further strengthened by the observation that UBCs and E3 ubiquitin–protein ligases interact with steroid hormone receptors and several other transcription factors and coactivate their transactivation functions (15–18). Because the coactivation and ubiquitination activities are distinct, this raises the question as to why ubiquitin pathway enzymes are linked to steroid receptor activation.
Eukaryotic cells exhibit rigorous control over gene expression, and one possible mechanism to control gene expression is to modulate the concentrations of transcriptional regulators in the cell by proteasome-mediated protein degradation. This possibility has been reported for the regulation of protein levels of transcription factors such as STAT5a and tramtrack (4, 33). In this manuscript, we present data that suggest that the ubiquitin–proteasome pathway modulates the concentration of ER protein in mammalian cells by promoting its degradation. Considering that the transcriptionally active ER protein is associated with a diverse group of proteins and forms a preinitiation complex, it is possible that subsequent to receptor activation of transcription, proteasome-mediated degradation of the receptor may be a mechanism that dissociates the preinitiation complex. It could be necessary to dissociate the preinitiation complex through targeted protein degradation, since the reinforcing interactions of multiple transcription factors may make passive dissociation of ligand and coactivators impossible. Additionally, it is possible that hormone-induced ER degradation serves to control physiological responses in estrogen target tissues by down-regulating ER, which ultimately serves to limit the expression of estrogen-responsive genes.
Acknowledgments
We thank Peter Howley and Sushant Kumar for the UbcH7 expression plasmids and Allan Weissman for A. thaliana UBA1 and UbcH5B expression plasmids. The H222 antibody was the generous gift of Abbot Laboratories. We also thank Neil McKenna for critical reading of the manuscript. This work was partially supported by an award (Flaming/Davenport) to Z.N. and by National Institutes of Health grants to C.L.S. and B.W.O.
ABBREVIATIONS
- ER
human estrogen receptor
- UBA
ubiquitin-activating E1 enzyme
- UBCs
ubiquitin-conjugating E2 enzymes
- PR
human progesterone receptor
- TR
human thyroid hormone receptor β
- DMSO
dimethyl sulfoxide
- NEDD8
neural precursor cell-expressed developmentally down-regulated
- TNT
in vitro transcription and translation
Footnotes
To whom reprint requests should be addressed. e-mail: berto@bcm.tmc.edu.
References
- 1.Pickart C M. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 2.Haas A L, Siepmann T J. FASEB J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- 3.Chen Z J, Parent L, Maniatis T. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 4.Kim T K, Maniatis T. Science. 1996;273:1717–1719. doi: 10.1126/science.273.5282.1717. [DOI] [PubMed] [Google Scholar]
- 5.Ciechanover A, Schwartz A L. FASEB J. 1994;8:182–191. doi: 10.1096/fasebj.8.2.8119489. [DOI] [PubMed] [Google Scholar]
- 6.Ciechanover A. Biol Chem Hoppe-Seyler. 1994;375:565–581. doi: 10.1515/bchm3.1994.375.9.565. [DOI] [PubMed] [Google Scholar]
- 7.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 8.Cenciarelli C, Wilhelm K G, Jr, Guo A, Weissman A M. J Biol Chem. 1996;271:8709–8713. doi: 10.1074/jbc.271.15.8709. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt M L, Murray J, Lee V M, Hill W D, Wertkin A, Trojanowski J Q. Am J Pathol. 1991;139:53–65. [PMC free article] [PubMed] [Google Scholar]
- 10.Murray A. Cell. 1995;81:149–152. doi: 10.1016/0092-8674(95)90322-4. [DOI] [PubMed] [Google Scholar]
- 11.Murray E J, Bentley G V, Grisanti M S, Murray S S. Exp Cell Res. 1998;242:460–469. doi: 10.1006/excr.1998.4090. [DOI] [PubMed] [Google Scholar]
- 12.Huibregtse J M, Scheffner M, Beaudenon S, Howley P M. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huibregtse J M, Scheffner M, Beaudenon S, Howley P M. Proc Natl Acad Sci USA. 1995;92:5249. doi: 10.1073/pnas.92.11.5249-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamitani T, Kito K, Nguyen H P, Yeh E T. J Biol Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 15.Saltzman A, Searfoss G, Marcireau C, Stone M, Ressner R, Munro R, Franks C, D’Alonzo J, Tocque B, Jaye M, et al. FEBS Lett. 1998;425:431–435. doi: 10.1016/s0014-5793(98)00287-7. [DOI] [PubMed] [Google Scholar]
- 16.Imhof M O, McDonnell D P. Mol Cell Biol. 1996;16:2594–2605. doi: 10.1128/mcb.16.6.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna N J, Nawaz Z, Tsai S Y, Tsai M J, O’Malley B W. Proc Natl Acad Sci USA. 1998;95:11697–11702. doi: 10.1073/pnas.95.20.11697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nawaz Z, Lonard D M, Smith C L, Lehman E L, Tsai S L, Tsai M-J, O’Malley B W. Mol Cell Biol. 1999;19:1182–1189. doi: 10.1128/mcb.19.2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masuyama H, Brownfield C M, St-Arnaud R, MacDonald P N. Mol Endocrinol. 1997;11:1507–1517. doi: 10.1210/mend.11.10.9990. [DOI] [PubMed] [Google Scholar]
- 20.Makino Y, Yamano K, Kanemaki M, Morikawa K, Kishimoto T, Shimbara N, Tanaka K, Tamura T. J Biol Chem. 1997;272:23201–23205. doi: 10.1074/jbc.272.37.23201. [DOI] [PubMed] [Google Scholar]
- 21.Fraser R A, Rossignol M, Heard D J, Egly J M, Chambon P. J Biol Chem. 1997;272:7122–7126. doi: 10.1074/jbc.272.11.7122. [DOI] [PubMed] [Google Scholar]
- 22.Masuyama H, MacDonald P N. J Cell Biochem. 1998;71:429–440. [PubMed] [Google Scholar]
- 23.Nirmala P B, Thampan R V. Biochem Biophys Res Commun. 1995;213:24–31. doi: 10.1006/bbrc.1995.2093. [DOI] [PubMed] [Google Scholar]
- 24.Pakdel F, Le Goff P, Katzenellenbogen B S. J Steroid Biochem Mol Biol. 1993;46:663–672. doi: 10.1016/0960-0760(93)90307-i. [DOI] [PubMed] [Google Scholar]
- 25.Burnstein K L, Cidlowski J A. Mol Cell Endocrinol. 1992;83:C1–8. doi: 10.1016/0303-7207(92)90187-b. [DOI] [PubMed] [Google Scholar]
- 26.Nardulli A M, Greene G L, O’Malley B W, Katzenellenbogen B S. Endocrinology. 1988;122:935–944. doi: 10.1210/endo-122-3-935. [DOI] [PubMed] [Google Scholar]
- 27.Burris T P, Nawaz Z, Tsai M J, O’Malley B W. Proc Natl Acad Sci USA. 1995;92:9525–9529. doi: 10.1073/pnas.92.21.9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beekman J M, Allan G F, Tsai S Y, Tsai M J, O’Malley B W. Mol Endocrinol. 1993;7:1266–1274. doi: 10.1210/mend.7.10.8264659. [DOI] [PubMed] [Google Scholar]
- 29.Hatfield P M, Gosink M M, Carpenter T B, Vierstra R D. Plant J. 1997;11:213–226. doi: 10.1046/j.1365-313x.1997.11020213.x. [DOI] [PubMed] [Google Scholar]
- 30.Hatakeyama S, Jensen J P, Weissman A M. J Biol Chem. 1997;272:15085–15092. doi: 10.1074/jbc.272.24.15085. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S, Kao W H, Howley P M. J Biol Chem. 1997;272:13548–13554. doi: 10.1074/jbc.272.21.13548. [DOI] [PubMed] [Google Scholar]
- 32.Hatfield P M, Vierstra R D. J Biol Chem. 1992;267:14799–14803. [PubMed] [Google Scholar]
- 33.Li S, Li Y, Carthew R W, Lai Z C. Cell. 1997;90:469–478. doi: 10.1016/s0092-8674(00)80507-3. [DOI] [PubMed] [Google Scholar]
- 34.Syvala H, Vienonen A, Zhuang Y H, Kivineva M, Ylikomi T, Tuohimaa P. Life Sci. 1998;63:1505–1512. doi: 10.1016/s0024-3205(98)00417-2. [DOI] [PubMed] [Google Scholar]
- 35.Gong L, Kamitani T, Fujise K, Caskey L S, Yeh E T. J Biol Chem. 1997;272:28198–28201. doi: 10.1074/jbc.272.45.28198. [DOI] [PubMed] [Google Scholar]
- 36.Kamitani T, Kito K, Nguyen H P, Wada H, Fukuda-Kamitani T, Yeh E T H. J Biol Chem. 1998;273:26675–26682. doi: 10.1074/jbc.273.41.26675. [DOI] [PubMed] [Google Scholar]
- 37.Kamitani T, Nguyen H P, Kito K, Fukuda-Kamitani T, Yeh E T. J Biol Chem. 1998;273:3117–3120. doi: 10.1074/jbc.273.6.3117. [DOI] [PubMed] [Google Scholar]
- 38.Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glotzer M, Murray A W, Kirschner M W. Nature (London) 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]