Abstract
Ras and Rap proteins are closely related small GTPases. Whereas Ras is known for its role in cell proliferation and survival, Rap1 is predominantly involved in cell adhesion and cell junction formation. Ras and Rap are regulated by different sets of guanine nucleotide exchange factors and GTPase-activating proteins, determining one level of specificity. In addition, although the effector domains are highly similar, Rap and Ras interact with largely different sets of effectors, providing a second level of specificity. In this review, we discuss the regulatory proteins and effectors of Ras and Rap, with a focus on those of Rap.
Ras-like small G-proteins are ubiquitously expressed, conserved molecular switches that couple extracellular signals to various cellular responses. Different signals can activate GEFs2 that induce the small G-protein to switch from the inactive, GDP-bound state to the active, GTP-bound state. This induces a conformational change that allows downstream effector proteins to bind specifically to and be activated by the GTP-bound protein to mediate diverse biological responses. Small G-proteins are returned to the GDP-bound state by hydrolyzing GTP with the help of GAPs. Ras (Ha-Ras, Ki-Ras, and N-Ras) and Rap proteins (Rap1A, Rap1B, Rap2A, Rap2B, and Rap2C) have similar effector-binding regions that interact predominantly with RA domains or the structurally similar RBDs present in a variety of different proteins. Both protein families operate in different signaling networks. For instance, Ras is central in a network controlling cell proliferation and cell survival, whereas Rap1 predominantly controls cell adhesion, cell junction formation, cell secretion, and cell polarity. These different functions are reflected in a largely different set of GEFs and GAPs. Also the downstream effector proteins operate in a selective manner in either one of the networks.
GEFs
GEFs for Ras and Rap proteins are usually multidomain proteins that contain a CDC25 homology domain mediating the exchange activity and a REM domain. GEFs for Ras include Sos1 and Sos2, RasGRF, RasGRP1, and RasGRP4. Rap can be activated by C3G, Epac1, Epac2, RasGRP2, PDZ (PSD-95/Dgl/ZO-1)-GEF1, PDZ-GEF2, and PLCε (reviewed in Ref. 1). Within the RasGRP family, RasGRP3 seems to be a more promiscuous GEF, affecting both Rap and Ras (Fig. 1) (2). The general structural basis of nucleotide exchange by CDC25 homology domains was revealed by the crystal structure of Sos in the presence of Ras (3). When the catalytic helix of Sos is inserted into the guanine nucleotide-binding pocket of Ras, affinity for the bound nucleotide is decreased, resulting in its release. Because the concentration of GTP in a cell is higher than that of GDP, GTP will predominantly enter the empty nucleotide-binding pocket and in turn displace the GEF. Recently, the crystal structure of Epac2 with Rap1 was determined, revealing a similar mechanism of nucleotide exchange (4). However, although the interfaces between Sos and Ras and Epac and Rap are both rather extensive, most residues at the interface are different in both GEFs. This shows that although the catalytic mechanism is conserved, the actual interactions are not, allowing the establishment of selectivity or, as in the case of RasGRP3, of promiscuity. The additional domains in the various GEFs are involved in regulating their activation or translocation. For instance, Sos activation involves translocation to tyrosine-phosphorylated proteins, release of autoinhibition, and allosteric regulation of catalytic activity by a distal Ras protein in a positive feedback loop (5). Epac is activated by binding of cAMP, which induces a major conformational change to release its autoinhibition (4). Thus, GEFs are well equipped to selectively regulate the activity of these small G-proteins in time and space in response to a large variety of different stimuli.
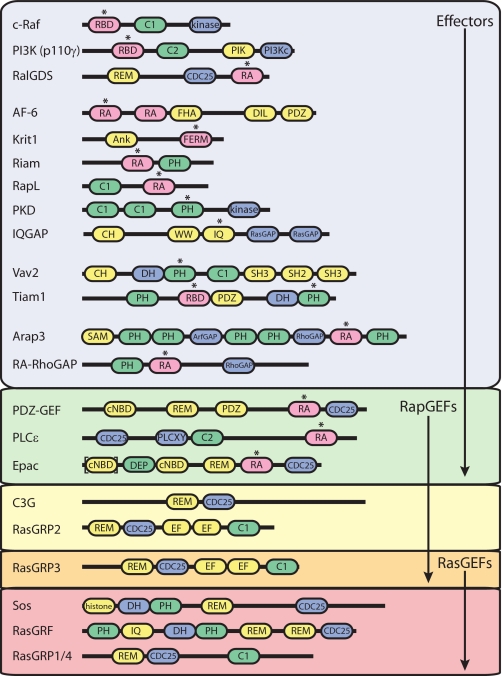
FIGURE 1.
Ras and Rap effector proteins and GEFs. A schematic representation is shown of the domain structures of Ras and Rap GEFs and effector proteins discussed here. RA domains/RBDs are depicted in pink, catalytic domains in blue, lipid-binding domains in green, and other domains in yellow. Asterisks indicate domains required for Ras/Rap binding. C1, protein kinase C conserved region 1; C2, Ca2+-binding motif; PIK, PI3K accessory domain; PI3Kc, PI3K catalytic domain; CDC25, CDC25 homology; FHA, Forkhead-associated domain; DIL, dilute; Ank, ankyrin repeat; CH, calponin homology; SAM, sterile α-motif; cNBD, cyclic nucleotide-binding domain; PLCXY, phospholipase C catalytic regions X and Y; DEP, Dishevelled/Egl-10/pleckstrin; EF, EF-hand; histone, histone domain. 1) The interaction of Tiam1 with Ras has been described for the RBD; for Rap, it was shown to bind to the DH-PH domain. 2) The N-terminal cyclic nucleotide-binding domain is conserved in Epac2 alone, and the RA domain-like sequence in Epac1 is not recognized as such by the SMART Database (smart.embl-heidelberg.de). 3) Although described to be present in PLCε (18, 45), a second RA domain in PLCε is not indicated in the SMART Database.
GAPs
The hydrolysis of GTP in Ras and Rap1 is slow but is accelerated several orders of magnitude by GAPs that insert an additional catalytic side chain into the nucleotide-binding pocket. GAPs acting on Ras include p120RasGAP, neurofibromin, and GAP1. GTP hydrolysis on Rap is catalyzed by Rap1GAP and the Spa-1 family of GAPs (Spa-1, Spa-1-like, and E6TP1) (1). The catalytic domains of RasGAPs and RapGAPs are structurally similar, yet the mode of stimulation of the GTPase reaction is different. RasGAPs use an arginine side chain as a catalytic group, whereas RapGAPs use an asparagine side chain. Catalysis by the arginine side chain involves a glutamine at position 61 of Ras. Indeed, many tumor mutations occur at position 61 to render Ras continuously in its active conformation. Rap proteins do not have a glutamine at position 61, and thus, RapGAPs use a different mode of catalysis, which is provided by the asparagine side chain. There is also a group of GAPs that appears to have a dual specificity for both Ras and Rap, such as several GAP1 (RASAL, CAPRI, and GAPIP4BP) family members and synGAP (6, 7). The isolated GAP domains of these GAPs have low activity for Rap in vitro, but due to allosteric regulation by the additional C2 domain, they can function as RapGAPs in vivo (7). This indicates that there are ways around the selectivity of Ras- and Rap-mediated GTP hydrolysis.
Specificity of Ras Effectors
For Ras, the classic downstream effectors are the three Raf kinases (c-Raf, A-Raf, and B-Raf), various PI3Ks, and RalGDS family members. Raf proteins mediate the Ras-induced activation of the ERK (extracellular signal-regulated kinase)/MAPK (mitogen-activated protein kinase) cascade. They contain an RBD to which Ras binds with high affinity. Rap1 can also bind the Raf RBD in vitro (8) and was proposed to mediate cAMP-mediated effects on Raf kinases in a cell type-dependent manner (9). However, other studies could not reveal a direct connection between Rap1 and Raf kinase signaling (10), and thus, this result remains controversial. Another established Ras effector is PI3K. PI3Ks are heterodimeric proteins with a p110 catalytic and a p85 regulatory subunit. The p110 subunit has an RBD that binds to Ras. This interaction facilitates membrane translocation but in addition allosterically regulates the kinase activity (11). Rap has also been suggested to regulate PI3K. For instance, Rap1 can modestly activate the p110α subunit of PI3K (8), and in B-cells, Rap2 can bind PI3K, although in this system, this correlates with an inhibition of PI3K-mediated protein kinase B activation (12).
RalGDS, Rgl, and Rgl2 (Rlf) contain an RA domain and are GEFs for the small GTPase Ral, downstream of Ras. Their RA domains efficiently bind to both Ras and Rap1 in vitro. Interestingly, Rap1A has a higher affinity for the RalGDS RA domain than Ras and, indeed, also binds in vivo to RalGEFs (8). However, despite high affinity binding to Rap, evidence that endogenous Rap1 activation mediates RalGDS-Ral activation is lacking.
Effectors for Rap Proteins
A large number of proteins have been identified as effectors of Rap proteins, particularly the adapters RAPL, Riam, AF-6, and Krit1; the RacGEFs Tiam1 and Vav2; and the RhoGAPs RA-RhoGAP and Arap3.
AF-6/Afadin—AF-6 is an adapter protein that localizes to cell-cell junctions and contains two RA domains. With its N-terminal RA domain, it binds both Ras and Rap1 with high affinity. Although initial studies showed that AF-6 may participate in Ras-induced junction breakdown (13), others showed that AF-6 binds p120 catenin in a Rap-dependent manner to prevent internalization of E-cadherin (14). However, a longer isoform of AF-6 that regulates E-cadherin in a Rap-independent manner was recently described (15). Thus, a clear picture of the role of AF-6 as an effector of Ras and/or Rap in junctions is still not present. In T-cells, AF-6 is a negative regulator of Rap-induced integrin-mediated cell adhesion (16).
Krit1—Krit1 (Krev interaction trapped 1) is a Rap1-interacting protein that was originally identified in a yeast two-hybrid screen (17). This protein, also called CCM1, is found mutated in cerebral cavernous malformation, a disease associated with defects in brain vasculature. Krit1 has several ankyrin repeats and a FERM (band 4.1/ezrin/radixin/moesin) domain. This domain has a ubiquitin-like fold that is similar to RA domains and RBDs and can bind both Ras and Rap, although the affinity for Rap is higher than for Ras (18). In agreement with this, recent evidence implicates Krit1 as a major effector of Rap1 in the control of endothelial cell-cell junctions (19). In these cells, Krit1 localizes to the junctions, where it associates with junctional proteins and is required for the Rap1-induced reduction in basal and cytokine-induced permeability of the junctions. Krit1 also associates with microtubules, two other CCM proteins (CCM2 and CCM3), and ICAP1 (a protein that binds to and negatively regulates the β1 chain of integrins) (20). The role of these proteins in Rap1/Krit1-mediated regulation of junction permeability is currently unclear.
Riam—Riam contains an N-terminal coiled-coil region, a central RA and PH domain, and a proline-rich C-terminal region, with multiple FPPPP motifs capable of interacting with the EVH1 domains of the actin regulatory proteins Ena and Vasp. Riam interacts with active Rap1 to stimulate the adhesion of Jurkat T-cells through β1 and β2 integrins (21). Ginsberg and co-workers (22) showed that Riam is involved in stimulus-induced, Rap1-mediated recruitment of talin. Talin subsequently binds to and activates the β chain of integrin αIIbβ3. In T-cells, Riam was found in a complex with ADAP (adhesion- and degranulation-promoting adapter protein) and SKAP-55 (Src kinase-associated phosphoprotein of 55 kDa), and this complex is required for Rap1 recruitment to the plasma membrane and T-cell receptor-induced, integrin-mediated cell adhesion to fibronectin and ICAM (intercellular adhesion molecule) (23).
RAPL—RAPL (regulator of adhesion and cell polarity enriched in lymphoid tissues/Nore1B/Rassf5) is another regulator of Rap-induced, integrin-mediated adhesion in T-cells. In vivo, RAPL binds Rap1 after stimulation through the T-cell receptor or by chemokines. In the presence of active Rap1, RAPL binds to the α chain of integrin αLβ2 (LFA-1), resulting in its activation (24). Data from RAPL knock-out mice confirm its role in lymphocyte adhesion. Lymphocytes lacking RAPL are less adherent to ICAM and do not redistribute their integrins after cytokine stimulation. They are defective in cell migration and thus in homing to peripheral lymph nodes (25). Recently, the kinase Mst1 was identified as an effector of RAPL in T-cell adhesion. Activation of Rap1 promotes the binding of RAPL to Mst1 and their relocalization with LFA-1 to the leading edge to induce adhesion (26). Interestingly, whereas Rap1 and RAPL have been shown to regulate both LFA-1 affinity and clustering, Mst1 overexpression enhances only LFA-1 clustering. This suggests that LFA-1 clustering is critical for Rap1-induced T-cell adhesion and that there may also be Mst1-independent mechanisms by which Rap1 regulates LFA-1 affinity. Moreover, in a different T-cell line, RapL was found to interact with Rap2B, regulating random migration and not cell adhesion (27).
PKD1—Also PKD1 may regulate T-cell adhesion through Rap1. Rap1 binds to the PH domain of PKD1, and this interaction facilitates the activation of Rap1 as well as the recruitment of both proteins to the cytoplasmic tail of integrins in the immunological synapse (28). How the interaction with PKD1 induces Rap activation is currently unclear, but it does not depend on the kinase activity of PKD1, indicating that PKD1 functions as a scaffold here.
IQGAP1—IQGAP1 is a scaffold protein that interacts with actin and functions in a number of actin polymerization-driven processes. Recently, Rap1 was found to interact with IQGAP1 in vitro and to colocalize with IQGAP1 at the cell membrane (29). The precise role for Rap1 in IQGAP1 function is currently unclear as both GDP- and GTP-bound Rap1 proteins bind to IQGAP. Thus, the main function of both IQGAP1 and PKD1 appears to be the recruitment of Rap1 rather than being activated by GTP-bound Rap1 to induce biological effects downstream of Rap1.
Interconnectivity with Rac and Rho
In processes controlled by Rap1 signaling, such as junction formation and cell adhesion, Rho family proteins like Rac1, CDC42, and RhoA play a critical role. Rap proteins are directly linked to these proteins through the interaction with the RacGEFs Vav2 and Tiam1 and the RhoGAPs Arap3 and RA-RhoGAP.
RacGEFs—In a search for proteins that could mediate Rap1-induced, Rac-dependent cell adhesion and spreading, Rap1 was found to interact with both Vav2 and Tiam1 (30). Rap1 binds to their catalytic DH-PH domain but does not affect the catalytic activity of the two GEFs. Rather, Rap1 induces the translocation of Vav2 to localize Rac activity to sites of cell spreading. In T-cells, Rap1 binds to Tiam1 as well (31). Here, Rap1 is proposed to recruit Tiam1 and the polarity complex to the future site of polarization. There, by a currently unknown mechanism, Rap1 activates CDC42, which then activates the polarity complex, which in turn activates Rac through Tiam1. Rac then contributes to the induction of T-cell polarity. Because, in contrast to active Rap1, active Rac or CDC42 alone does not induce T-cell polarity, Rap1 may induce a parallel pathway for T-cell polarity, e.g. the Rap-RAPL and/or Rap-Riam pathway. Also in neuronal cells, Rap1B through CDC42 and the polarity complex regulates polarity by defining which of the growing neurites becomes the future axon (32). Another connection between Rap1 and CDC42 was found in junction formation, where Rap1, together with Src, mediates nectin-induced activation of FRG, a GEF for CDC42 (33).
For Tiam1, the interaction of Rap1 with the DH-PH domain is surprising because this protein contains a genuine RBD, which mediates the interaction of Tiam1 with Ras. Indeed, Tiam1 mediates Ras-induced activation of Rac and thus is a genuine effector of Ras as well (34). Rap1 was also reported to bind an ill defined TSS (Tiam-STEF-SIF) homology domain in Tiam2 (STEF) to mediate cAMP-induced, Epac-mediated activation of Rac (35). Because Tiam1 and Tiam2 are rather homologous, these different binding sites are surprising and may require independent confirmation.
RhoGAPs—Arap3 is a dual GAP for both Arf6 and RhoA, with five PH domains and an RA domain that interacts with Rap but not Ras. In vitro, Arap3 GAP activity for Rho, but not Arf6, is stimulated by Rap1. In vivo, activation of Arap3 by Rap1 also requires PI3K activity. In cells, Arap3 seems to regulate growth factor-stimulated formation of lamellipodia. Both overexpression and knockdown of Arap3 in endothelial cells interfered with the normal ruffling response induced by PDGF (36).
RA-RhoGAP was identified in a yeast two-hybrid screen using a human brain cDNA library and active Rap1B as bait. This protein has an RA domain and a RhoGAP domain and, in addition, a PH domain and several annexin-like repeats. Active Rap1 binds to RA-RhoGAP and induces RhoGAP activity. Notably, a mutant lacking the RA domain fails to bind Rap1 but has higher GAP activity for RhoA, strongly suggesting that the RA domain inhibits GAP activity, which is relieved upon Rap1 binding. In undifferentiated neuronal cell lines, Rap1 further enhances RA-RhoGAP-induced neurite outgrowth, whereas small interfering RNA to RA-RhoGAP inhibits Rap-induced neurite outgrowth (37).
Feedback Control and Interconnectivity
RapGEFs with an RA Domain—PDZ-GEF1 and PDZ-GEF2 are multidomain proteins that have, in addition to their catalytic CDC25 homology domains, a PDZ domain, two cAMP-related binding domains, an RA domain, and a C-terminal PDZ domain-binding motif. Both proteins interact with junction proteins of the Magi family, and indeed, PDZ-GEFs have been implicated in the control of cell-cell junctions (38, 39). For PDZ-GEF1, it was shown that Rap1A and Rap2B are the preferred binding partners for its RA domain (40, 41). This suggests that PDZ-GEF is subject to positive feedback by Rap1. PDZ-GEF2 can be activated by M-Ras to regulate Rap1-induced LFA-1 activation in response to tumor necrosis factor-α (42). Epac2 is also a multidomain protein with a regulatory domain consisting of a DEP domain flanked by two cAMP-binding sites and a catalytic region. An RA domain is located between the REM and CDC25 homology domains. This RA domain can interact with active Ha-Ras and may play a role in the translocation of Epac2 to the plasma membrane (43). Also Epac1 has an RA-like sequence between the REM and CDC25 homology domains, but for this RA domain, no interacting proteins have been identified yet.
PLCε—Members of the PLC family are key mediators of many extracellular signals. Upon activation by receptors, PLC converts phosphatidylinositol 4,5-bisphosphate into the protein kinase C-activating lipid diacylglycerol and the second messenger inositol 1,4,5-trisphosphate, which raises the cytosolic calcium concentration. PLCε has, in addition to its lipase catalytic domain, an N-terminal CDC25 homology domain and a two C-terminally located RA domains (44). As an effector, PLCε may be regulated by both Ras and Rap as both proteins bind the C-terminal RA domain with high affinity (18, 44). For instance, after PDGF stimulation, the rapid initial PLCε activation was mediated by Ras, whereas prolonged activation was mediated by Rap1 (45). Originally, the Ras effector function of PLCε was also supported by the observation that in PLCε knock-out mice, Ras-mediated skin tumor formation was reduced (46). However, more recent analysis indicates that this reduction is caused by a non-autonomous effect, i.e. PLCε deficiency resulted in a reduction in 12-O-tetradecanoylphorbol-13-acetate-induced inflammation, suggesting that its role in tumor formation may be due to an increased inflammatory response (47). Recently, Epac and PLCε were shown to mediate β-adrenergic receptor-induced calcium-induced calcium release in cardiomyocytes, suggesting that Rap proteins activated by Epac induce PLCε activity (48). Also in 293 cells expressing the β-adrenergic receptor, Epac mediates the activation of PLCε through Rap2B (49). Thus, both Ras and Rap1 can activate PLCε, and differences in the reported selectivity may depend on the cell context.
As a GEF, PLCε has exchange activity for Rap1 in vitro that is required for sustained PLCε activation upon PDGF stimulation, suggesting that as a GEF, it mediates its own prolonged activation through a positive feedback mechanism (50). Additionally, PLCε may mediate cross-talk between the Ras and Rap signaling networks.
Concluding Remarks
Ras and Rap are highly homologous proteins, each functioning in different but interconnected signaling networks. This specificity of Ras and Rap is achieved by upstream regulatory proteins and downstream effectors (Fig. 1). Most GEFs for Ras or Rap contain a CDC25 homology domain and a REM domain for catalysis, and the mechanism of nucleotide exchange is comparable between RasGEFs and RapGEFs. However, the interaction interfaces between the GEF and its GTPase differ between the different pairs, generally resulting in their tight specificity for each other. In contrast, RasGAPs and RapGAPs do not use the same catalytic mechanism. Whereas RasGAPs provide an arginine side chain as the catalytic group, RapGAPs insert an asparagine side chain in the GTP-binding pocket for catalysis. In addition, GEFs and GAPs are multidomain proteins that regulate these GTPases in time and space, and thus, localization and timing are also important elements in the specificity.
A second way of ensuring specificity in signaling is through effector proteins. Most effectors bind Ras and Rap proteins via an RA domain or an RBD. In vitro, many of these domains bind to both Ras and Rap, albeit with different affinity. However, in vivo, the various effectors are rather specific. In part, this difference in selectivity for the RA domain/RBD may be determined by flanking sequences or by differences in subcellular localization. However, some effectors may be used by both Ras and Rap proteins, e.g. AF-6 and Tiam1. The conclusion that the specificity of Ras and Rap is determined at least at two levels, by both upstream regulators and downstream effectors, implies that results obtained using overexpression of mutant GTPases that are constitutively active lack at least one level of this specificity control. Results obtained with such mutants should therefore be interpreted with care.
For Rap, the list of effectors is rapidly expanding and contains proteins both with and without catalytic activity, which are mostly involved in all aspects of cell adhesion and modulation of the actin cytoskeleton. A number of effectors have been implicated in the control of integrins. For instance, in T-cells, Riam, RAPL, and PKD have all been described as effectors. They may form an “integrin activation complex” consisting of Rap1 and several effectors and perhaps further adapter proteins, required to mediate integrin activation. This complex is then translocated to the integrin upon Rap activation to induce cell adhesion. Also a number of effectors have been identified that regulate the actin cytoskeleton, in particular the GEFs Vav2 and Tiam1 for Rac proteins and the GAPs Arap3 and RA-RhoGAP for Rho proteins. These effectors apparently determine the balance between Rac and Rho signaling and, as such, regulate the dynamics of the actin cytoskeleton. Finally, an interesting aspect in the control of Ras family proteins is the presence of RA domains in GEFs, such as Epac proteins, PDZ-GEFs, and PLCε. These RA domains may be responsible for feedback control or for the connection between signaling networks.
Thus, in the last couple of years, it is truly appreciated that Rap proteins serve in signaling networks that are largely different from Ras signaling networks. However, as with all signaling networks, there is interconnectivity. It is the challenge for the future to understand both networks and the interconnectivity between these and other networks in full detail.
Acknowledgments
We thank Holger Rehmann and Fried Zwartkruis for comments on this manuscript.
This work was supported by the Dutch Cancer Society (to J. H. R.) and by the Netherlands Genomics Initiative through the Cancer Genomics Centre (to J. L. B.). This is the first article of three in the Thematic Minireview Series on Novel Ras Effectors. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
Footnotes
The abbreviations used are: GEF, guanine nucleotide exchange factor; GAP, GTPase-activating protein; RA, Ras association; RBD, Ras-binding domain; REM, Ras exchange motif; PLC, phospholipase C; PI3K, phosphatidylinositol 3-kinase; PH, pleckstrin homology; PKD, protein kinase D; DH, Dbl homology; PDGF, platelet-derived growth factor.
References
- 1.Bos, J. L., Rehmann, H., and Wittinghofer, A. (2007) Cell 129, 865–877 [DOI] [PubMed] [Google Scholar]
- 2.Yamashita, S., Mochizuki, N., Ohba, Y., Tobiume, M., Okada, Y., Sawa, H., Nagashima, K., and Matsuda, M. (2000) J. Biol. Chem. 275, 25488–25493 [DOI] [PubMed] [Google Scholar]
- 3.Boriack-Sjodin, P. A., Margarit, S. M., Bar-Sagi, D., and Kuriyan, J. (1998) Nature 394, 337–343 [DOI] [PubMed] [Google Scholar]
- 4.Rehmann, H., Arias-Palomo, E., Hadders, M. A., Schwede, F., Llorca, O., and Bos, J. L. (2008) Nature 455, 124–127 [DOI] [PubMed] [Google Scholar]
- 5.Margarit, S. M., Sondermann, H., Hall, B. E., Nagar, B., Hoelz, A., Pirruccello, M., Bar-Sagi, D., and Kuriyan, J. (2003) Cell 112, 685–695 [DOI] [PubMed] [Google Scholar]
- 6.Kupzig, S., Deaconescu, D., Bouyoucef, D., Walker, S. A., Liu, Q., Polte, C. L., Daumke, O., Ishizaki, T., Lockyer, P. J., Wittinghofer, A., and Cullen, P. J. (2006) J. Biol. Chem. 281, 9891–9900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pena, V., Hothorn, M., Eberth, A., Kaschau, N., Parret, A., Gremer, L., Bonneau, F., Ahmadian, M. R., and Scheffzek, K. (2008) EMBO Rep. 9, 350–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez-Viciana, P., Sabatier, C., and McCormick, F. (2004) Mol. Cell. Biol. 24, 4943–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stork, P. J., and Schmitt, J. M. (2002) Trends Cell Biol. 12, 258–266 [DOI] [PubMed] [Google Scholar]
- 10.Bos, J. L., de Bruyn, K., Enserink, J., Kuiperij, B., Rangarajan, S., Rehmann, H., Riedl, J., de Rooij, J., van Mansfeld, F., and Zwartkruis, F. (2003) Biochem. Soc Trans 31, 83–86 [DOI] [PubMed] [Google Scholar]
- 11.Pacold, M. E., Suire, S., Perisic, O., Lara-Gonzalez, S., Davis, C. T., Walker, E. H., Hawkins, P. T., Stephens, L., Eccleston, J. F., and Williams, R. L. (2000) Cell 103, 931–943 [DOI] [PubMed] [Google Scholar]
- 12.Christian, S. L., Lee, R. L., McLeod, S. J., Burgess, A. E., Li, A. H., Dang-Lawson, M., Lin, K. B., and Gold, M. R. (2003) J. Biol. Chem. 278, 41756–41767 [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto, T., Harada, N., Kano, K., Taya, S., Canaani, E., Matsuura, Y., Mizoguchi, A., Ide, C., and Kaibuchi, K. (1997) J. Cell Biol. 139, 785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoshino, T., Sakisaka, T., Baba, T., Yamada, T., Kimura, T., and Takai, Y. (2005) J. Biol. Chem. 280, 24095–24103 [DOI] [PubMed] [Google Scholar]
- 15.Lorger, M., and Moelling, K. (2006) J. Cell Sci. 119, 3385–3398 [DOI] [PubMed] [Google Scholar]
- 16.Zhang, Z., Rehmann, H., Price, L. S., Riedl, J., and Bos, J. L. (2005) J. Biol. Chem. 280, 33200–33205 [DOI] [PubMed] [Google Scholar]
- 17.Serebriiskii, I., Estojak, J., Sonoda, G., Testa, J. R., and Golemis, E. A. (1997) Oncogene 15, 1043–1049 [DOI] [PubMed] [Google Scholar]
- 18.Wohlgemuth, S., Kiel, C., Kramer, A., Serrano, L., Wittinghofer, F., and Herrmann, C. (2005) J. Mol. Biol. 348, 741–758 [DOI] [PubMed] [Google Scholar]
- 19.Glading, A., Han, J., Stockton, R. A., and Ginsberg, M. H. (2007) J. Cell Biol. 179, 247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beraud-Dufour, S., Gautier, R., Albiges-Rizo, C., Chardin, P., and Faurobert, E. (2007) FEBS J. 274, 5518–5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lafuente, E. M., van Puijenbroek, A. A., Krause, M., Carman, C. V., Freeman, G. J., Berezovskaya, A., Constantine, E., Springer, T. A., Gertler, F. B., and Boussiotis, V. A. (2004) Dev. Cell 7, 585–595 [DOI] [PubMed] [Google Scholar]
- 22.Han, J., Lim, C. J., Watanabe, N., Soriani, A., Ratnikov, B., Calderwood, D. A., Puzon-McLaughlin, W., Lafuente, E. M., Boussiotis, V. A., Shattil, S. J., and Ginsberg, M. H. (2006) Curr. Biol. 16, 1796–1806 [DOI] [PubMed] [Google Scholar]
- 23.Menasche, G., Kliche, S., Chen, E. J., Stradal, T. E., Schraven, B., and Koretzky, G. (2007) Mol. Cell. Biol. 27, 4070–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katagiri, K., Maeda, A., Shimonaka, M., and Kinashi, T. (2003) Nat. Immunol. 4, 741–748 [DOI] [PubMed] [Google Scholar]
- 25.Katagiri, K., Ohnishi, N., Kabashima, K., Iyoda, T., Takeda, N., Shinkai, Y., Inaba, K., and Kinashi, T. (2004) Nat. Immunol. 5, 1045–1051 [DOI] [PubMed] [Google Scholar]
- 26.Katagiri, K., Imamura, M., and Kinashi, T. (2006) Nat. Immunol. 7, 919–928 [DOI] [PubMed] [Google Scholar]
- 27.Miertzschke, M., Stanley, P., Bunney, T. D., Rodrigues-Lima, F., Hogg, N., and Katan, M. (2007) J. Biol. Chem. 282, 30629–30642 [DOI] [PubMed] [Google Scholar]
- 28.Medeiros, R. B., Dickey, D. M., Chung, H., Quale, A. C., Nagarajan, L. R., Billadeau, D. D., and Shimizu, Y. (2005) Immunity 23, 213–226 [DOI] [PubMed] [Google Scholar]
- 29.Jeong, H. W., Li, Z., Brown, M. D., and Sacks, D. B. (2007) J. Biol. Chem. 282, 20752–20762 [DOI] [PubMed] [Google Scholar]
- 30.Arthur, W. T., Quilliam, L. A., and Cooper, J. A. (2004) J. Cell Biol. 167, 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerard, A., Mertens, A. E., van der Kammen, R. A., and Collard, J. G. (2007) J. Cell Biol. 176, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwamborn, J. C., and Puschel, A. W. (2004) Nat. Neurosci. 7, 923–929 [DOI] [PubMed] [Google Scholar]
- 33.Sato, T., Irie, K., Okamoto, R., Ooshio, T., Fujita, N., and Takai, Y. (2005) Cancer Sci. 96, 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambert, J. M., Lambert, Q. T., Reuther, G. W., Malliri, A., Siderovski, D. P., Sondek, J., Collard, J. G., and Der, C. J. (2002) Nat. Cell Biol. 4, 621–625 [DOI] [PubMed] [Google Scholar]
- 35.Zaldua, N., Gastineau, M., Hoshino, M., Lezoualc'h, F., and Zugaza, J. L. (2007) FEBS Lett. 581, 5814–5818 [DOI] [PubMed] [Google Scholar]
- 36.Krugmann, S., Andrews, S., Stephens, L., and Hawkins, P. T. (2006) J. Cell Sci. 119, 425–432 [DOI] [PubMed] [Google Scholar]
- 37.Yamada, T., Sakisaka, T., Hisata, S., Baba, T., and Takai, Y. (2005) J. Biol. Chem. 280, 33026–33034 [DOI] [PubMed] [Google Scholar]
- 38.Sakurai, A., Fukuhara, S., Yamagishi, A., Sako, K., Kamioka, Y., Masuda, M., Nakaoka, Y., and Mochizuki, N. (2006) Mol. Biol. Cell 17, 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dube, N., Kooistra, M. R., Pannekoek, W. J., Vliem, M. J., Oorschot, V., Klumperman, J., Rehmann, H., and Bos, J. L. (2008) Cell. Signal. 20, 1608–1615 [DOI] [PubMed] [Google Scholar]
- 40.Liao, Y., Satoh, T., Gao, X., Jin, T. G., Hu, C. D., and Kataoka, T. (2001) J. Biol. Chem. 276, 28478–28483 [DOI] [PubMed] [Google Scholar]
- 41.Rebhun, J. F., Castro, A. F., and Quilliam, L. A. (2000) J. Biol. Chem. 275, 34901–34908 [DOI] [PubMed] [Google Scholar]
- 42.Yoshikawa, Y., Satoh, T., Tamura, T., Wei, P., Bilasy, S. E., Edamatsu, H., Aiba, A., Katagiri, K., Kinashi, T., Nakao, K., and Kataoka, T. (2007) Mol. Biol. Cell 18, 2949–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, Y., Asuri, S., Rebhun, J. F., Castro, A. F., Paranavitana, N. C., and Quilliam, L. A. (2006) J. Biol. Chem. 281, 2506–2514 [DOI] [PubMed] [Google Scholar]
- 44.Song, C., Hu, C. D., Masago, M., Kariyai, K., Yamawaki-Kataoka, Y., Shibatohge, M., Wu, D., Satoh, T., and Kataoka, T. (2001) J. Biol. Chem. 276, 2752–2757 [DOI] [PubMed] [Google Scholar]
- 45.Song, C., Satoh, T., Edamatsu, H., Wu, D., Tadano, M., Gao, X., and Kataoka, T. (2002) Oncogene 21, 8105–8113 [DOI] [PubMed] [Google Scholar]
- 46.Bai, Y., Edamatsu, H., Maeda, S., Saito, H., Suzuki, N., Satoh, T., and Kataoka, T. (2004) Cancer Res. 64, 8808–8810 [DOI] [PubMed] [Google Scholar]
- 47.Ikuta, S., Edamatsu, H., Li, M., Hu, L., and Kataoka, T. (2008) Cancer Res. 68, 64–72 [DOI] [PubMed] [Google Scholar]
- 48.Oestreich, E. A., Wang, H., Malik, S., Kaproth-Joslin, K. A., Blaxall, B. C., Kelley, G. G., Dirksen, R. T., and Smrcka, A. V. (2007) J. Biol. Chem. 282, 5488–5495 [DOI] [PubMed] [Google Scholar]
- 49.Schmidt, M., Evellin, S., Weernink, P. A., von Dorp, F., Rehmann, H., Lomasney, J. W., and Jakobs, K. H. (2001) Nat. Cell Biol. 3, 1020–1024 [DOI] [PubMed] [Google Scholar]
- 50.Jin, T. G., Satoh, T., Liao, Y., Song, C., Gao, X., Kariya, K., Hu, C. D., and Kataoka, T. (2001) J. Biol. Chem. 276, 30301–30307 [DOI] [PubMed] [Google Scholar]