Abstract
The aryl hydrocarbon receptor (AhR) repressor (AhRR) inhibits the AhR activity. AhRR acts by competing with AhR for heterodimer formation with the AhR nuclear translocator (Arnt) and preventing the AhR·Arnt complex from binding the xenobiotic-responsive elements. Here, we report that AhRR has three evolutionarily conserved SUMOylation consensus sequences within its C-terminal repression domain and that Lys-542, Lys-583, and Lys-660 at the SUMOylation sites are modified by SUMO-1 in vivo. Arginine mutation of the three lysines results in a significant reduction of transcriptional repression activity. SUMOylation of the three lysine residues is important for the interaction between AhRR and ANKRA2, HDAC4, and HDAC5, which are important corepressors for AhRR. Arnt, a heterodimer partner for AhRR, markedly enhanced the SUMOylation of AhRR. AhRR, but not AhR, also significantly enhanced the SUMOylation of Arnt. The SUMOylation of both AhRR and Arnt is important for the efficient transcriptional repression activity of the AhRR/Arnt heterodimer.
The aryl hydrocarbon receptor repressor (AhRR)2 is a member of the bHLH-PAS (basic helix-loop-helix and the Per-Arnt-Sim) protein superfamily and has a high similarity to AhR in the N-terminal bHLH-PAS A domain (1). The 5′-flanking promoter region of the mouse AhRR gene contains conserved xenobiotic-responsive element (XRE) sequences, and expression of the AhRR gene is induced by binding of the AhR/Arnt heterodimer to these XREs (2). AhRR forms a heterodimer with Arnt, another bHLH-PAS transcription factor, to inhibit AhR-dependent transactivation of the XRE-driven genes; thus, AhRR participates in a negative feedback loop in the AhR signaling pathway (1, 3, 4). Recently, we generated AhRR–/– mice, which show higher than wild type levels of ligand-induced expression of the AhR target gene, Cyp1a1 mRNA induction in some tissues (5). These mice also displayed a delayed response to skin carcinogenesis caused by benzo[a]pyrene (5). Recent work has also demonstrated that AhRR is a tumor suppressor gene (6); AhRR mRNA is consistently down-regulated in human malignant tissues from different anatomical origins; furthermore, ectopic expression of AhRR in tumor cells resulted in diminished cell growth (7) and reduced angiogenic potential. These observations provide new insight into the still largely unknown physiological functions of AhRR and form the basis for further studies of its mechanisms of action.
We reported previously that AhRR contains a transcriptional repression domain within its C-terminal region (8). Using the C-terminal region of AhRR as bait, we isolated ANKRA2 (ankyrin repeat, family A, 2) by CytoTrap yeast two-hybrid screening. AhRR was also shown to interact with HDAC4 and HDAC5, and these interactions are important for the transcriptional repression activity of AhRR (8).
Small ubiquitin-like modifiers (SUMO-1, SUMO-2, and SUMO-3) belong to a family of ubiquitin-like proteins, which are covalently attached to (or detached from) substrate proteins to regulate their functions. The post-translational modification of proteins by SUMO has been increasingly recognized as an important regulatory mechanism in a diverse range of cellular processes. SUMO precursors are processed by SUMO-specific proteases and activated by an E1 enzyme. The activated SUMO is transferred to the E2 conjugating enzyme Ubc9, which recognizes the SUMOylation consensus sequence ψKXE within target proteins. With the help of E3 ligases such as PIAS, the C-terminal glycine of SUMO is covalently linked to the ε-amino group of lysine in the SUMOylation consensus sequence of the target proteins (9, 10).
In contrast to ubiquitylation, SUMOylation has a wide range of substrate-specific functions and acts via multiple mechanisms, including alterations in the subcellular localization of target proteins, protein stability, protein-protein interactions, and protein-DNA binding activities. Many transcription factors are SUMOylated. In many cases, SUMO modification of transcription factors is associated with transcriptional repression through the suppression of transactivation activity or the enhancement of repression activity.
In this study, we report that three lysine residues within the C-terminal repression domain of AhRR can be modified by SUMO-1, resulting in enhancement of its transcriptional repression activity.
EXPERIMENTAL PROCEDURES
Plasmids—Constructions of pBOSGAL4DBD-AhRR, pBOSHA-AhRR, pBOSFLAG-ANKRA2, pG3TK-Luc, and plasmids encoding HDAC4-FLAG and HDAC5-FLAG was described previously (8). pSG5His-SUMO-1 was generated by fusing full-length human SUMO-1 cDNA with the His tag sequence and cloning into the expression vector pSG5 (Stratagene). pBOSHA-SUMO-1 and pBOSHA-SUMO-2 were generated by ligating the full-length human SUMO-1 or SUMO-2 cDNA with the SmaI site of pBOST7-HA. pBOS-Ubc9 was generated by inserting the EcoRI/SalI fragment of pBSK-mUbc9 into the pEFBOS vector (11) cleaved with EcoRI/SalI. pCMV3xFLAG-AhRR was constructed by ligating the blunt-ended EcoRI/SalI fragment from pBSK-mAhRR with the blunt-ended p3xFLAG-CMV-10 (Sigma). AhRR was cleaved from pBSK-AhRR (8) with EcoRI/XhoI, blunt-ended, and subsequently inserted into the EGFP-C3 vector, which was cut with SmaI to generate pEGFP-AhRR. pBOS-AhR, pBOS-Arnt, and pXRE4TK-Luc were described previously (1). To construct pBOSHA-AhR and pBOSHA-Arnt, pBSK-mAhR and pBSK-mArnt (1) were digested with HindIII/XbaI and NcoI/BamHI, respectively, and the blunt-ended fragments of mAhR and mArnt were ligated with the SmaI site of pBOST7-HA. pBOS3xFLAG-Arnt was described previously (12). Single, double, or triple amino acid mutations in AhRR (K542R, K583R, K660R) or Arnt (K245R) were generated using the QuikChange site-directed mutagenesis kit (Stratagene).
Cell Culture and Transfection—COS-7, Hepa-1c1c7 (Hepa-1), and HeLa cells were maintained, respectively, in high or low glucose Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (Sigma) and penicillin/streptomycin (Invitrogen) under 5.0% CO2 at 37 °C. Transfection was performed using Lipofectamine™ (Invitrogen).
Luciferase Assay—Hepa-1, COS-7, or HeLa cells (5.0 × 104 cells/well) were grown in 24-well dishes for 24 h and transfected with the expression plasmids indicated in the figure legends, such as pG3TK-Luc or pXRE4TK-Luc, and the expression plasmids for sea pansy luciferase as an internal control. Cells transfected with pXRE4TK-Luc were treated with 2 μm 3-MC or Me2SO for 12 h. Forty eight h after transfection, the cells were harvested, and luciferase was quantified using the Dual-Luciferase reporter assay system (Promega). Expressed firefly luciferase activity was normalized to the cotransfected sea pansy luciferase activity, which was used as a standard.
Co-immunoprecipitation and Immunoblot Analysis—The transfected COS-7 cells were lysed in radioimmune precipitation assay buffer for immunoprecipitation with the anti-HA antibody or in FLAG buffer for immunoprecipitation with the anti-FLAG antibody; buffer contained protease inhibitor mixture (Roche Applied Science) and 20 mm N-ethylmaleimide to preserve the SUMOylation. Whole cell lysates were used for immunoblot analysis either directly or after immunoprecipitation. Immunoprecipitation with anti-FLAG M2 agarose (Sigma) or anti-HA agarose (Sigma) was performed for 12 h, and the immunoprecipitates were washed according to the published procedure for immunoblot analysis. Immunoblot analysis was performed using anti-FLAG (Sigma), anti-HA (Sigma), anti-SUMO-1 (ALEXIS), anti-Arnt (13), and anti-AhRR antibodies (8).
Fluorescence Analysis—Hepa-1 cells were cultured to sub-confluency on coverslips and transfected with expression plasmids for EGFP-AhRR (wild type (WT) or 3KR), His-tagged SUMO-1, and Ubc9, with or without Arnt. After 48 h of transfection, cells were incubated with Hoechst DNA stain, and fluorescent images were observed using fluorescence microscopy (magnification ×1,000).
RESULTS
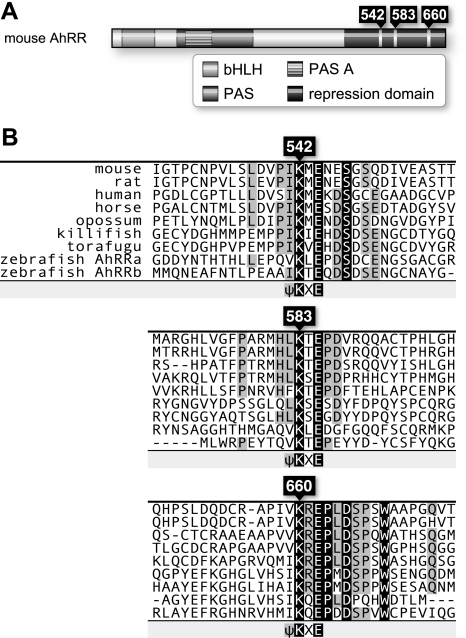
Three Evolutionarily Conserved SUMOylation Sites in AhRR—To gain insight into the function of AhRR, we inspected the amino acid sequence of mouse AhRR to identify structural motifs for potential covalent modification sites. We found three SUMOylation consensus sequences at amino acid positions 542, 583, and 660 (Fig. 1A). All of these sequences are located within the repression domain of AhRR (8). Three SUMOylation sites are well conserved across a broad range of vertebrate species, including mammals and fish (Fig. 1B). Other AhRR C-terminal regions have no obvious sequence conservation, suggesting that the putative SUMOylation motifs play an important role in AhRR transcriptional repression activity.
FIGURE 1.
Three conserved SUMOylation sequences in the C-terminal repression domain of AhRR. A, schematic representation of the full-length 701-amino acid mouse AhRR. The characterized domains represented are the basic helix-loop-helix (bHLH), Per-Arnt-Sim (PAS), and repression domains. Three putative SUMOylation sites are located within the repression domain, with the target lysine residues indicated. B, alignments of C-terminal regions of AhRR, including three SUMOylation sites found in mouse, rat, human, horse, opossum, killifish, torafugu, and zebrafish. Conserved amino acids are highlighted in black, and the conserved SUMOylation sites and lysine residues are indicated.
Lysines at Amino Acids 542, 583, and 660 of AhRR Are Modified by SUMO-1—To examine whether these potential SUMO modification sites are actually SUMOylated in vivo, we replaced the three conserved lysines with arginines to create the AhRR 3KR mutant. COS-7 cells were cotransfected with expression plasmids for 3xFLAG-AhRR WT or 3xFLAG-AhRR 3KR, along with His-SUMO-1 and Ubc9. In addition to a band for AhRR (∼80 kDa), Western blotting using anti-FLAG antibodies detected four bands with slower mobilities when 3xFLAG-AhRR WT was cotransfected, presumably corresponding to various SUMOylated AhRRs (Fig. 2A, left panel). These bands were also detected without adding Ubc9, although bands were weaker than those in the presence of added Ubc9 (data not shown). In contrast, when 3xFLAG-AhRR 3KR was cotransfected, no slower mobility bands were observed. When cell extracts were immunoprecipitated with anti-FLAG antibodies, followed by Western blotting using anti-SUMO-1 antibodies, four higher molecular weight bands with the same mobilities were detected (Fig. 2A, right panel). These results indicate that AhRR is actually SUMOylated in vivo, at Lys-542, Lys-583, and Lys-660.
FIGURE 2.
Lysines at 542, 583, and 660 amino acids of AhRR are modified by SUMO-1. A and F, COS-7 cells were cotransfected with indicated plasmids, and whole cell extracts (WCE) were prepared 48 h after transfection and immunoprecipitated (IP) with anti-FLAG antibodies. Immunoprecipitates were analyzed by immunoblotting (IB) with the indicated antibodies. Crude lysates were analyzed by immunoblotting to control protein expressions. B–E and G, COS-7 cells were cotransfected with the indicated expression plasmids. Cell extracts were prepared 48 h after transfection and analyzed by immunoblotting with the indicated antibodies as described above. Black and white arrowheads indicate SUMOylated or unmodified AhRR, respectively. In G, the right panel presents the SUMOylation sites and mobility of AhRR.
We further examined whether these lysines were also modified by SUMO-2, but when the SUMO-2 plasmid was cotransfected, no modified product of AhRR was observed (Fig. 2B). We detected a doublet band of SUMO-2 that was also observed in other reports overexpressing SUMO-2 (14, 15), although the reason is not clear. One possible explanation is that a higher molecular weight band corresponds to immature SUMO-2, which produced the mature SUMO-2 (lower band) by cleaving C-terminal 11 amino acids with SUMO-specific proteases (SENPs) (16). Likewise, addition of any of the PIAS family proteins, which are well characterized SUMO E3 ligases, did not result in increased SUMOylation of AhRR (data not shown). Taken together, these results indicate that AhRR is specifically modified by SUMO-1.
We next investigated whether the heterodimer partner Arnt affects the SUMOylation of AhRR. Arnt is also modified by SUMO-1 (17), and we were interested to know how the SUMOylation of each AhRR and Arnt affects that of the other. As shown in Fig. 2C, addition of Arnt significantly enhanced the SUMOylation of AhRR in a dose-dependent manner. Next, to examine whether SUMOylation of Arnt is necessary for enhancement of AhRR SUMOylation, we transfected cells with a plasmid expressing the Arnt K245R mutant plasmid; we observed no difference in AhRR SUMOylation regardless of whether the Arnt WT or K245R mutant was expressed (Fig. 2D). These results indicate that heterodimer formation between Arnt and AhRR, but not Arnt SUMOylation, enhances the SUMOylation of AhRR.
Then, we tested whether AhRR was SUMOylated by an endogenous SUMOylation system. When AhRR was expressed in COS-7 cells without exogenous SUMO-1 and Ubc9, four bands, corresponding to the SUMOylated AhRR bands, were also observed (Fig. 2E). Bands were of slightly lower mobility than the His-SUMO-1-modified AhRR because of the His tag (Fig. 2E, lanes 1 and 4). These bands were decreased in intensity when the dominant-negative SUMO-1 mutant with deletion of C-terminal GG residues was cotransfected and disappeared when AhRR 3KR was expressed (Fig. 2E, lanes 2 and 3). Cell extracts of AhRR-transfected COS-7 cells were immunoprecipitated with anti-FLAG antibodies, followed by Western blotting using anti-SUMO-1 antibodies, and four SUMOylated bands were detected (Fig. 2F). These results showed that AhRR was also SUMOylated by the endogenous SUMOylation system.
There are three putative SUMOylation sites in the AhRR amino acid sequence, and four SUMOylated bands were detected in the SUMOylation experiment. We were interested to investigate which sites are actually SUMOylated. We generated arginine mutations in the three lysine residues, either alone or in combination, to identify the SUMOylated bands of AhRR. As shown in Fig. 2G, SUMOylated AhRR bands with different mobilities were observed when AhRRs with different Lys-to-Arg mutations were expressed. From the electrophoretic mobilities and the sites of Lys-to-Arg replacement, we were able to attribute the bands to the specifically SUMOylated AhRR species shown in Fig. 2G. Taken together, these results showed that AhRR was mono-, di-, and tri-SUMOylated on Lys-542, Lys-583, and Lys-660, and all of these SUMOylations were significantly enhanced when Arnt was coexpressed with AhRR.
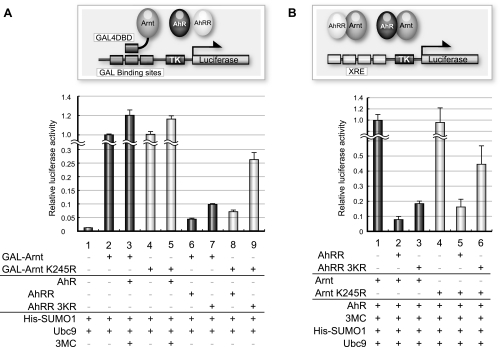
SUMO Modification of AhRR Is Important for Its Efficient Transcriptional Repression Activity—Because SUMOylation usually increases the activity of transcriptional repressors, we next examined the effects of SUMOylation on the function of AhRR as a transcriptional repressor. First, we performed a reporter assay using the luciferase gene under the control of the thymidine kinase promoter ligated with GAL4-binding sites (3xGAL-TK-Luc). Transcriptional repression activity of AhRR fused with the GAL4 DNA-binding domain (GAL4DBD) was assessed by measuring the luciferase gene expression driven by the thymidine kinase promoter. AhRR WT repressed the luciferase activity dose-dependently, whereas the 3KR mutant of AhRR significantly reduced the repressive activity compared with the WT (Fig. 3A). Expression level of AhRR WT and 3KR were normalized using Western blot analysis (data not shown). Next, we used a luciferase reporter gene driven by four tandemly repeated XREs to assess the repression activity of AhRR. AhRR WT repressed the luciferase expression driven by the XRE sequence to a greater degree than AhRR 3KR (Fig. 3B). For Fig. 3 (A and B), essentially the same results were also observed when SUMO-1 and Ubc9 were not exogenously expressed using HeLa cells (supplemental Fig. S1, A and B, lanes 1–3), indicating that endogenous SUMOylation activity is sufficient for AhRR repression activity. These results indicate that the SUMOylation of AhRR is important for its transcriptional repression activity.
FIGURE 3.
SUMO modification sites of AhRR and their effects on its transcriptional repression activity. A, shown is the transcriptional repression activity of AhRR WT and 3KR. Hepa-1 cells were transiently transfected with the expression plasmids for GAL4DBD-AhRR WT or 3KR, His-tagged SUMO-1, Ubc9, Arnt, and GAL-TK-Luc reporter gene containing three GAL-binding sites. Cell extracts were prepared 48 h after transfection and used for luciferase assays. The -fold repression is relative to the reporter gene alone. B, HeLa cells were transiently transfected with the expression plasmids for AhRR WT or 3KR, His-tagged SUMO-1, Ubc9, AhR, Arnt, and the 4xXRE-TK-Luc reporter gene containing four XREs. After 48 h of transfection, cells were treated with 2 μm of 3-MC or Me2SO; 24 h later, cell extracts were prepared and used for luciferase assays. C, Hepa-1 cells were transiently transfected with the expression plasmids for GAL4DBD-AhRR WT; 3KR or AhRR with Lys-to-Arg mutation(s) of each of the three lysines, either alone or in combination; His-tagged SUMO-1, Ubc9, Arnt, and GAL-TK-Luc reporter gene containing three GAL-binding sites. Cell extracts were prepared 48 h after transfection and used for luciferase assays. D, the subnuclear distribution of EGFP-AhRR is not changed in the SUMOylation mutant. Hepa-1 cells were transfected with expression plasmids of EGFP-AhRR WT or 3KR, His-tagged SUMO-1, and Ubc9, with or without Arnt. After 48 h of transfection, cells were incubated with Hoechst DNA stain (lower panels), and fluorescence images were taken (upper panels).
To determine the contribution of each of the three lysines to the repressive activity of AhRR, the GAL4 reporter assay was performed using the mutant AhRR-GAL4DBD fusion genes with the Lys-to-Arg mutations in all three lysine positions, either alone or in combination. As shown in Fig. 3C, the SUMOylation of each lysine residue contributes similarly to the repression activity of AhRR. These results indicate that SUMO-1 modification of Lys-542, Lys-583, and Lys-660 more or less contribute equally to the transcriptional repression activity of AhRR.
SUMOylation of AhRR Is Required for Interaction of AhRR with Corepressor Components—SUMO modification often regulates the subcellular localization of the target proteins. To examine whether SUMO modification affects nuclear localization of AhRR, we transfected Hepa-1 cells with EGFP-AhRR WT or EGFP-AhRR 3KR and observed their subcellular distributions using fluorescence microscopy. Both EGFP-AhRR WT and EGFP-AhRR 3KR gave a similar diffuse nuclear localization pattern (Fig. 3D, panels a and b). In contrast, coexpression of Arnt markedly changed the subcellular localization of both proteins to a speckled distribution pattern (Fig. 3D, panels c and d), with fewer than 40% of the transfected cells retaining a diffuse pattern. This speckled localization pattern is consistent with the Arnt localization pattern (17). The same results were also observed when SUMO-1 and Ubc9 were not exogenously expressed (data not shown). These results indicate that SUMO-1 modification of AhRR does not affect subcellular localization of AhRR, whereas Arnt alters the subnuclear localization of AhRR to the speckled pattern. SUMO modification is also known to play a key role in protein-protein interaction. Previously, we demonstrated that the C-terminal repression domain, which carries the three SUMO-1 modified lysines, interacts with ANKRA2, HDAC4, and HDAC5; these interactions are important for the repression activity of AhRR. Therefore, we examined whether the SUMOylation of AhRR affects the interaction with ANKRA2, HDAC4, and HDAC5. COS-7 cells were transfected with the HA-tagged AhRR WT or 3KR mutant, along with FLAG-tagged ANKRA2, HDAC4, or HDAC5. The cell extracts prepared from the transfected cells were immunoprecipitated with an anti-HA antibody. ANKRA2 was co-immunoprecipitated with AhRR WT, and HDAC4 was coprecipitated only when ANKRA2 was coexpressed (Fig. 4A, left and right panels, lanes 2 and 4), consistent with the previous report (8). However, when the HA-tagged mutant AhRR 3KR was transfected instead of AhRR WT, immunoprecipitates with an anti-HA antibody detected only a small amount of ANKRA2 and HDAC4 (Fig. 4, A, left and right panels, lane 3, and C), indicating that SUMOylation of AhRR is necessary for efficient interaction with ANKRA2 and HDAC4. As expected, HDAC5 was co-immunoprecipitated with AhRR WT even in the absence of ANKRA2 (Fig. 4B, left and right panels, lanes 2 and 4), but binding of HDAC5 with AhRR 3KR was much reduced (Fig. 4B, left and right panels, lane 3, and C). Taken together, these results show that the SUMOylation of AhRR is necessary for efficient interaction with ANKRA2, HDAC4, and HDAC5, resulting in its increased transcriptional repression activity.
FIGURE 4.
Interaction of SUMO-1-modified AhRR with ANKRA2, HDAC4, and HDAC5. A and B, COS-7 cells were cotransfected with expression plasmids for HA-tagged AhRR WT or 3KR, His-tagged SUMO-1, and Ubc9, with or without FLAG-tagged ANKRA2 and FLAG-tagged HDAC4 for A, or FLAG-tagged HDAC5 for B. Whole cell extracts (WCE) were prepared 48 h after transfection and immunoprecipitated (IP) with anti-HA antibodies. Immunoprecipitates were analyzed by immunoblot (IB) with the indicated antibodies (B, right panels). Crude lysates were analyzed by immunoblot to control for protein expression (B, left panels). C, the relative densities of the ANKRA2, HDAC4, and HDAC5 bands, which were co-immunoprecipitated with either AhRR WT or 3KR, were calculated with NIH Image software. Values are relative to AhRR WT.
AhRR, but Not AhR, Enhances SUMO-1 Modification of Arnt—Arnt forms a heterodimer with AhRR, colocalizes with AhRR in the nucleus, and enhances SUMOylation of AhRR (Figs. 2C and 3D). Arnt was previously reported to be SUMOylated, and SUMOylation of Arnt suppresses GAL4DBD-Arnt-mediated transactivation through dissociation from PML (17). Next, we examined whether AhRR or AhR affects the SUMO modification of Arnt. Plasmid 3xFLAG-tagged Arnt was transiently expressed in COS-7 cells, together with HA-tagged AhRR or HA-tagged AhR, and the SUMOylated Arnt was examined by Western blotting using an anti-Arnt antibody. Although SUMO-1-modified Arnt was barely detected in the absence of AhRR, SUMO-1-modified Arnt was significantly increased in a dose-dependent manner upon addition of AhRR (Fig. 5A, lanes 1–4). The same enhancing effect was observed when AhRR 3KR was cotransfected (Fig. 5A, lanes 5–7). Therefore, regardless of its SUMOylation, AhRR is necessary for enhancement of Arnt SUMOylation. AhR is also a partner molecule of Arnt for heterodimer formation. We were interested in examining how AhR affects the SUMO modification of Arnt. When HA-tagged AhR was coexpressed with Arnt, no enhancement in the SUMOylation of Arnt was observed, even when the ligands of AhR were added (Fig. 5A, lanes 8–13). Thus, AhRR and Arnt mutually enhance the SUMOylation of their partner molecules, which may result in enhancement of the repression activity of the AhRR/Arnt heterodimer.
FIGURE 5.
AhRR, but not AhR, and Arnt mutually stimulate one another's SUMO-1 modification. A, COS-7 cells were cotransfected with expression plasmids for HA-tagged AhRR WT or 3KR or HA-tagged AhR, 3xFLAG-tagged Arnt, His-tagged SUMO-1, and Ubc9. After 48 h of transfection, cells for lanes 11–13 were treated with 2 μm of 3-MC or Me2SO; 24 h later, whole cell extracts (WCE) were prepared and analyzed by immunoblot (IB) using the indicated antibodies. B and C, COS-7 cells were cotransfected with the indicated expression plasmids. Cell extracts were prepared 48 h after transfection and immunoprecipitated (IP) with the indicated antibodies. Immunoprecipitates were analyzed by immunoblot with the indicated antibodies. Crude lysates were analyzed by immunoblot to control for protein expression. Black and white arrowheads indicate SUMOylated or unmodified AhRR, respectively.
At the same time, these observations raised the question of whether these SUMOylations affect the interaction between AhRR and Arnt. To address this question, COS-7 cells were transfected with expression plasmids for 3xFLAG-tagged AhRR or 3xFLAG-tagged AhRR 3KR, together with HA-tagged Arnt WT or HA-tagged Arnt K245R. Cell extracts were prepared from the transfected cells and immunoprecipitated with anti-HA or anti-FLAG antibodies. No effect of SUMOylation was observed on the interaction between AhRR and Arnt (Fig. 5, B and C, lanes 1 and 2).
SUMOylation of Both AhRR and Arnt Is Important for Repressor Activity of the AhRR·Arnt Complex—We next assessed the potential effects of the SUMOylated Arnt on the repressor activity of the AhRR/Arnt heterodimer. Hepa-1 cells were transfected with expression plasmids for GAL4DBD-fused Arnt WT or K245R, together with His-SUMO-1 and Ubc9, and either of AhR, AhRR WT, or 3KR. GAL4DBD-Arnt WT showed an autonomous transactivation activity, and addition of AhR enhanced this transactivation (Fig. 6A, lanes 1–3). When non-SUMOylation mutant Arnt K245R was added, there was no difference in transactivation activity from Arnt WT, in agreement with the observation in Fig. 5A that Arnt was barely SUMOylated under these conditions (Fig. 6A, lanes 2–5). This result is also consistent with the previous report that the SUMOylation of Arnt did not affect the transactivation activity of AhR/Arnt or Hif-1α/Arnt (17). When we added AhRR WT, the luciferase expression was remarkably reduced, whereas the addition of AhRR 3KR inhibited the expression of luciferase activity to a lesser extent, with the residual luciferase activity significantly higher than with the wild type AhRR (Fig. 6A, lanes 6 and 7). This tendency was more remarkable when both non-SUMOylation mutants of AhRR and Arnt were used (Fig. 6A, lane 9). Next, to further assess the importance of SUMOylation of the AhRR/Arnt heterodimer for its repressor activity, we used a luciferase reporter gene driven by four tandemly repeated XREs. As shown in Fig. 6B, the result was almost the same as that as shown in Fig. 6A. These experiments were also performed without added SUMO-1 and Ubc9 using HeLa cells, which showed essentially the same results (supplemental Fig. S1B). Taken together, these results show that SUMOylation of both AhRR and Arnt is important for the efficient repressor activity of the AhRR/Arnt heterodimer (Fig. 7).
FIGURE 6.
SUMOylation of both AhRR and Arnt is important for the repression activity of the AhRR·Arnt complex. SUMOylation sites of both AhRR and Arnt are important for the transcriptional repression activity of the AhRR·Arnt complex. A, Hepa-1 cells were transiently transfected with the expression plasmids for GAL4DBD-Arnt WT or K245R, AhRR WT or 3KR or AhR, His-tagged SUMO-1, Ubc9, and the GAL-TK-Luc reporter gene containing three GAL-binding sites. After 48 h of transfection, cells for lanes 3 and 5 were treated with 2 μm of 3-MC or Me2SO; 24 h later, cell extracts were used for luciferase assays. Luciferase activities are shown as values relative to lane 2. B, COS-7 cells were transiently transfected with the expression plasmids for Arnt WT or K245R, AhRR WT, 3KR, or AhR, His-tagged SUMO-1, Ubc9, and 4xXRE-TK-Luc reporter gene containing four XRE sequences. After 48 h of transfection, cells were treated with 2 μm of 3-MC or Me2SO; 24 h later, cell extracts were prepared and used for luciferase assays. Luciferase activities are shown as values relative to lane 1.
FIGURE 7.
Proposed model for the transcriptional regulation mechanism of the AhR/Arnt activator complex and the AhRR/Arnt repressor complex. Unmodified Arnt forms a heterodimer with AhR and recruits coactivators such as CBP/p300 to form the transcriptional activator complex. Meanwhile, Arnt forms a heterodimer with AhRR, which significantly enhances SUMOylation of both proteins. SUMOylated AhRR recruits corepressors ANKRA2, HDAC4, and HDAC5 to form the transcriptional repressor complex.
DISCUSSION
When amino acid sequences of the C-terminal region of AhRR were compared among mammals and fish, three SUMOylation sites including minimal adjacent sequences were revealed to be distinctly conserved (Fig. 1B), suggesting that these sequences are involved in some fundamental biological processes. A newly reported composite motif named PDSM (phosphorylation-dependent SUMOylation motif), which contains the SUMO consensus sequence with an adjacent proline-directed phosphorylation site (ψKXEXXSP) (18–23), undergoes phosphorylation-dependent SUMOylation in many transcription factors, including heat shock proteins (HSPs), peroxisome proliferator-activated receptor γ, MEF2, and GATA-1. Accelerated SUMOylation of these factors by phosphorylation enhances the repression of transcriptional activity. The SUMOylation site at Lys-660 in mouse AhRR, which is well conserved across species, seems to resemble the SUMO consensus sequence accompanying the proline-directed phosphorylation (ψKXEXXXSP), but its functional analysis remains to be seen.
All of the conserved SUMOylation sites of AhRR were found to be SUMOylated more or less equally in our in vivo cell culture system. These SUMOylations in AhRR were significantly enhanced by the presence of Arnt, a partner molecule of the AhRR/Arnt heterodimer. Likewise, Arnt was reported previously to be SUMOylated at Lys-245 (17); this SUMOylation was also stimulated by AhRR, but not by AhR, another partner molecule of the transcription-active heterodimer AhR/Anrt. Thus, one of the partner molecules of the AhRR·Arnt repressor complex mutually enhances the SUMOylation of the other (Figs. 2C and 5A). The PIAS proteins are members of a well characterized SUMO E3 ligase family, consisting of PIAS1, PIASxα, PIASxβ, PIASγ, and PIAS3. However, none of the family members added into the cultured cells could enhance the SUMOylation of AhRR (data not shown). Unlike ubiquitin E3 ligases, apparently structurally unrelated proteins can serve as SUMO E3 ligases, providing a scaffold bridging between Ubc9 and SUMOylation substrates. Whereas the PIAS family apparently plays no role in AhRR modification, Arnt could significantly enhance SUMOylation of AhRR, and the reverse is true with respect to Arnt SUMOylation. Because both AhRR (data not shown) and Arnt (17) interact with Ubc9 and their partner molecules, SUMOylation substrates, it is reasonable to consider that AhRR and Arnt serve as the SUMO E3 ligase to each other. This could be substantiated experimentally by in vitro reconstitution of SUMOylation. Interestingly, it has recently been reported that AhR is able to act as an E3 ubiquitin ligase to estrogen receptors and androgen receptors in a ligand-dependent manner (24), indicating the potential role of bHLH-PAS proteins, including AhR, to be able to display an E3 ligase activity with a ubiquitin family protein, in addition to its transcriptional activity.
In the nucleus, unmodified Arnt interacts and colocalizes with PML, which enhances the transactivation activity of Arnt; in contrast, SUMOylated Arnt dissociates from PML, resulting in the suppression of Arnt transcription activity in the GAL4DBD-Arnt-mediated transactivation assay (17). In contrast to AhRR, SUMOylation of Arnt is enhanced by PIAS1, and a substantial amount of Arnt is present in the nucleus in SUMOylated form (17). When AhRR is synthesized and transported into the nucleus through the NLS signal, AhRR may form a heterodimer with Arnt or SUMOylated Arnt, resulting in the enhanced SUMOylation of AhRR in the heterodimer complex. Thus, the SUMOylated AhRR and Arnt complex becomes competent for recruitment of corepressor components such as ANKRA2, HDAC4, and HDAC5. It remains to be investigated whether this series of transcription suppressor complexes are formed directly on the XRE sequence of the target genes or in the nucleoplasm prior to DNA binding. Our previous data showed that treatment of ANKRA2 small interfering RNA resulted in reduction of the AhRR-mediated repression of Cyp1a1 mRNA expression under normal conditions (8). Because transcriptional inhibition by the AhRR·Arnt complex is reversed by trichostatin A, an inhibitor of histone deacetylase (8), not only competitive binding to the XRE sequence, but also the HDAC activity of the AhRR/Arnt heterodimer is involved in repression by the AhRR/Arnt heterodimer. In Figs. 3 and 6, AhRR 3KR also showed some repression activity even if all SUMOylation sites are mutated, which could be explained by the residual interaction of AhRR 3KR with ANKRA2, HDAC4, and HDAC5 or competitive binding mechanisms for the XRE sequence with the AhR/Arnt heterodimer (Figs. 3B and 6B).
In this study, we have revealed a novel mechanism of transcriptional repression by the AhRR/Arnt heterodimer involving SUMO-1 modification. Further studies are required to reveal the detailed molecular mechanisms by which AhRR and Arnt are SUMOylated and subsequently recruit the corepressor components, as well as the temporal relationship between these events and the switching on/off of the target genes.
Supplementary Material
Acknowledgments
We thank Y. Nemoto for clerical work.
This work was supported in part by a scientific research grant from the Ministry of Health, Labor, and Welfare of Japan and Solution Oriented Research for Science and Technology, Japan Science and Technology Agency.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: AhRR, aryl hydrocarbon receptor repressor; AhR, aryl hydrocarbon receptor; SUMO, small ubiquitin-like modifier; WT, wild type; EGFP, enhanced green fluorescent protein; XRE, xenobiotic-responsive element; HA, hemagglutinin; m, mouse; Arnt, AhR nuclear translocator; GAL4DBD, GAL4 DNA-binding domain; 3-MC, 3-methyl cholanthrene; PML, promyelocytic leukemia.
References
- 1.Mimura, J., Ema, M., Sogawa, K., and Fujii-Kuriyama, Y. (1999) Genes Dev. 13 20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., Mimura, J., Gradin, K., Kuroiwa, A., Watanabe, T., Matsuda, Y., Inazawa, J., Sogawa, K., and Fujii-Kuriyama, Y. (2001) J. Biol. Chem. 276 33101–33110 [DOI] [PubMed] [Google Scholar]
- 3.Karchner, S. I., Franks, D. G., Powell, W. H., and Hahn, M. E. (2002) J. Biol. Chem. 277 6949–6959 [DOI] [PubMed] [Google Scholar]
- 4.Nishihashi, H., Kanno, Y., Tomuro, K., Nakahama, T., and Inouye, Y. (2006) Biol. Pharm. Bull. 29 640–647 [DOI] [PubMed] [Google Scholar]
- 5.Hosoya, T., Harada, N., Mimura, J., Motohashi, H., Takahashi, S., Nakajima, O., Morita, M., Kawauchi, S., Yamamoto, M., and Fujii-Kuriyama, Y. (2008) Biochem. Biophys. Res. Commun. 365 562–567 [DOI] [PubMed] [Google Scholar]
- 6.Zudaire, E., Cuesta, N., Murty, V., Woodson, K., Adams, L., Gonzalez, N., Martinez, A., Narayan, G., Kirsch, I., Franklin, W., Hirsch, F., Birrer, M., and Cuttitta, F. (2008) J. Clin. Investig. 118 640–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanno, Y., Takane, Y., Izawa, T., Nakahama, T., and Inouye, Y. (2006) Biol. Pharm. Bull. 29 1254–1257 [DOI] [PubMed] [Google Scholar]
- 8.Oshima, M., Mimura, J., Yamamoto, M., and Fujii-Kuriyama, Y. (2007) Biochem. Biophys. Res. Commun. 364 276–282 [DOI] [PubMed] [Google Scholar]
- 9.Hay, R. T. (2005) Mol. Cell 18 1–12 [DOI] [PubMed] [Google Scholar]
- 10.Meulmeester, E., and Melchior, F. (2008) Nature 452 709–711 [DOI] [PubMed] [Google Scholar]
- 11.Mizushima, S., and Nagata, S. (1990) Nucleic Acids Res. 18 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sekine, H., Mimura, J., Yamamoto, M., and Fujii-Kuriyama, Y. (2006) J. Biol. Chem. 281 37507–37516 [DOI] [PubMed] [Google Scholar]
- 13.Sogawa, K., Nakano, R., Kobayashi, A., Kikuchi, Y., Ohe, N., Matsushita, N., and Fujii-Kuriyama, Y. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 1936–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong, L., and Yeh, E. T. (2006) J. Biol. Chem. 281 15869–15877 [DOI] [PubMed] [Google Scholar]
- 15.Motohashi, H., Katsuoka, F., Miyoshi, C., Uchimura, Y., Saitoh, H., Francastel, C., Engel, J. D., and Yamamoto, M. (2006) Mol. Cell. Biol. 26 4652–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, E. S. (2004) Annu. Rev. Biochem. 73 355–382 [DOI] [PubMed] [Google Scholar]
- 17.Tojo, M., Matsuzaki, K., Minami, T., Honda, Y., Yasuda, H., Chiba, T., Saya, H., Fujii-Kuriyama, Y., and Nakao, M. (2002) J. Biol. Chem. 277 46576–46585 [DOI] [PubMed] [Google Scholar]
- 18.Hietakangas, V., Anckar, J., Blomster, H. A., Fujimoto, M., Palvimo, J. J., Nakai, A., and Sistonen, L. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang, X. J., and Gregoire, S. (2006) Mol. Cell 23 779–786 [DOI] [PubMed] [Google Scholar]
- 20.Yamashita, D., Yamaguchi, T., Shimizu, M., Nakata, N., Hirose, F., and Osumi, T. (2004) Genes Cells 9 1017–1029 [DOI] [PubMed] [Google Scholar]
- 21.Gregoire, S., and Yang, X. J. (2005) Mol. Cell. Biol. 25 2273–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang, J., Gocke, C. B., and Yu, H. (2006) BMC Biochem. 7 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shalizi, A., Gaudilliere, B., Yuan, Z., Stegmuller, J., Shirogane, T., Ge, Q., Tan, Y., Schulman, B., Harper, J. W., and Bonni, A. (2006) Science 311 1012–1017 [DOI] [PubMed] [Google Scholar]
- 24.Ohtake, F., Baba, A., Takada, I., Okada, M., Iwasaki, K., Miki, H., Takahashi, S., Kouzmenko, A., Nohara, K., Chiba, T., Fujii-Kuriyama, Y., and Kato, S. (2007) Nature 446 562–566 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.