Abstract
The receptor tyrosine kinase c-Kit plays a critical role in hematopoiesis, and gain-of-function mutations of the receptor are frequently seen in several malignancies, including acute myeloid leukemia, gastrointestinal stromal tumors, and testicular carcinoma. The most common mutation of c-Kit in these disorders is a substitution of the aspartic acid residue in position 816 to a valine (D816V), leading to constitutive activation of the receptor. In this study, we aimed to investigate the role of Src family kinases in c-Kit/D816V signaling. Src family kinases are necessary for the phosphorylation of wild-type c-Kit as well as of activation of downstream signaling pathways including receptor ubiquitination and the Ras/Mek/Erk pathway. Our data demonstrate that, unlike wild-type c-Kit, the phosphorylation of c-Kit/D816V is not dependent on Src family kinases. In addition, we found that neither receptor ubiquitination nor Erk activation by c-Kit/D816V required activation of Src family kinases. In vitro kinase assay using synthetic peptides revealed that c-Kit/D816V had an altered substrate specificity resembling Src and Abl tyrosine kinases. We further present evidence that, in contrast to wild-type c-Kit, Src family kinases are dispensable for c-Kit/D816V cell survival, proliferation, and colony formation. Taken together, we demonstrate that the signal transduction pathways mediated by c-Kit/D816V are markedly different from those activated by wild-type c-Kit and that altered substrate specificity of c-Kit circumvents a need for Src family kinases in signaling of growth and survival, thereby contributing to the transforming potential of c-Kit/D816V.
The receptor for stem cell factor (SCF),2 c-Kit, is a type III receptor tyrosine kinase that belongs to the same subfamily as the platelet-derived growth factor receptors, the Flt3 receptor, and the macrophage colony-stimulating factor receptor (1). The c-Kit gene is identical to the white spotting locus (W) in the mouse. c-Kit is expressed in the hematopoietic system, in the gastrointestinal system, in melanocytes, and in germ cells, and therefore loss-of-function mutations in c-Kit lead to defects in hematopoiesis, melanogenesis, and gametogenesis. Stimulation of the c-Kit receptor with its ligand, SCF, leads to receptor dimerization and activation of its intrinsic tyrosine kinase activity. Specific tyrosine residues are autophosphorylated, which results in the activation of downstream signaling pathways, including the Ras/Erk pathway and the PI3-kinase pathway (for review, see Ref. 2).
One of the crucial steps in oncogenic transformation of cells is the gain of independence of external growth stimuli. This can be achieved in several different ways, including mutations that render receptor tyrosine kinases constitutively active in the absence of ligand stimulation. In the case of c-Kit, these mutations most commonly occur either in exon 11 (encoding the juxtamembrane region) and are found predominantly in gastrointestinal stromal tumors or in exon 17 (encoding the activation loop of the kinase domain). A frequently occurring type of mutation in exon 17 in c-Kit is at codon 816. This type of mutation has been found in several human malignancies including acute myeloid leukemia, mastocytosis, germ cell tumors of the seminoma or dysgerminoma types, sinonasal natural killer/T-cell lymphomas, and in intracranial teratomas (3–10). These mutations at codon 816 lead to conversion of an aspartic acid residue to a valine, a tyrosine, a phenylalanine, an asparagine, or a histidine residue. The recently elucidated crystal structure of the c-Kit kinase domain has helped define the mechanism of activation by this type of mutation (11). In the unstimulated wild-type c-Kit, the juxtamembrane region inserts directly into the clefts between the amino- and carboxyl-terminal lobes of the kinase domain, disrupting the c-Kit control helix, and physically blocking the conserved kinase DFG motif from attaining a productive conformation. The activation loop folds back over the substrate binding groove and interacts with the active center of the kinase as a pseudosubstrate. It is not fully known how mutation of aspartic acid 816 leads to activation of c-Kit. It has been suggested either that the mutation inverts the conformation of the protein backbone so that the side chain of arginine 815 is being flipped from its position in the autoinhibited or that the effect of aspartic acid 816 mutations may be derived from its ability to stabilize the small positively charge α-helical dipole through its negative charge of its side chain. Asp-816 mutations in c-Kit promote receptor autophosphorylation and thereby constitutively activate downstream signaling pathways independent of SCF binding and therefore contribute to cell transformation (12). Imatinib (Gleevec) is a well known inhibitor of c-Kit juxtamembrane mutations and has been used in the treatment of gastrointestinal stromal tumors with activating mutations in the juxtamembrane region of c-Kit. In contrast, cells expressing c-Kit/D816V are resistant to Imatinib, whereas the Abl/Src dual inhibitor Dasatinib also inhibits the D816V mutant of c-Kit (13).
Negative regulation of c-Kit signaling has been shown to occur mainly through ubiquitin-mediated internalization and degradation of the receptor (14, 15). Ubiquitination is mediated by ubiquitin E3 ligases that attach ubiquitin to their target proteins, resulting in either monoubiquitination or polyubiquitination. Key components in this machinery are the Cbl family of ubiquitin E3 ligases, represented by Cbl, Cbl-b, and Cbl-c (16). Signaling through receptor tyrosine kinases must be tightly regulated, and inhibition of Cbl activation and receptor ubiquitination can lead to cell transformation (17). It has been shown that both direct and indirect binding of Cbl to wild-type c-Kit can induce Cbl activation and receptor ubiquitination followed by receptor internalization and degradation (15, 18). In contrast, the mechanisms behind negative regulation of the oncogenic c-Kit/D816V are so far unknown.
Extracellular signal-regulated kinase (Erk) proteins are the evolutionary conserved products of the two genes, Erk1 and Erk2, and are central proteins in the Ras/Erk signaling pathway. Erk1 and Erk2 are activated by dual phosphorylation on their regulatory tyrosine and threonine residues (19). The serine/threonine kinase Akt, also known as protein kinase B, is activated downstream of PI3-kinase and plays a central role in signaling induced by growth factors, cytokines, and other cellular stimuli (20). The Ras/Erk pathway and the PI3-kinase pathway are key signaling pathways involved in the regulation of cell proliferation, survival, and differentiation induced by c-Kit. A key player in the relay of signals from c-Kit into the cells is Src. Binding of Src to Tyr-568 in c-Kit leads to its activation and subsequent phosphorylation of Shc and activation of the Ras/Erk pathway (21). PI3-kinase is activated by c-Kit through two alternate routes, either through direct binding to Tyr-721 in c-Kit (22) or through indirect binding to the scaffolding protein Gab2, which associates to c-Kit via the adapter protein Grb2. Activation of PI3-kinase is dependent on Src-mediated phosphorylation of Gab2 (23). Other investigators have shown that neither Erk nor Akt is constitutively activated in cells expressing c-Kit/D816V (24).
In this report, we demonstrate that in c-Kit/D816V-expressing Ba/F3 cells, a low constitutive activation of both Erk and Akt exists and that this activation can be further augmented by SCF stimulation. We also present data showing that c-Kit/D816V evades the need of Src family kinases for receptor ubiquitination and Erk activation by having an altered substrate specificity resembling Src family kinases. We conclude that Src family kinases play different roles in wild-type c-Kit and c-Kit/D816V-induced cell survival and growth.
EXPERIMENTAL PROCEDURES
Cytokines, Antibodies, and Peptides—Recombinant human SCF was purchased from Prospec Tany (Rehovot, Israel). The rabbit antiserum KitC1, recognizing the carboxyl-terminal tail of human c-Kit, was purified as described (25). Antibodies recognizing pTyr-568, pTyr-703, pTyr-Y721, and pTyr-936 in human c-Kit were generated by immunizing rabbits with synthetic phosphopeptides corresponding to the phosphorylation sites and extensive affinity purification (26). β-actin antibody was from Sigma. The following antibodies were used in the study: Cbl, phospho-Erk (Santa Cruz, Biotechnology, Santa Cruz, CA), phospho-Akt (Cell Signaling Technology, Danvers, MA), 4G10 (Upstate Biotechnology, Charlottesville, VA), ubiquitin antibody P4D1 (Covance, Denver, PA), PE-labeled c-Kit antibody 104D2 (Biolegend, San Diego, CA), and Shc (BD Biosciences). Src optimal peptide (AEEEIYGEFEAKKKK) and Abl optimal peptide (EAIYAAPFAKKK) were synthesized according to (27). All peptides were synthesized by JPT Peptide Technology (Berlin, Germany), except the Src optimal peptide that was kindly synthesized by Ulla Engström at the Ludwig Institute for Cancer Research, Uppsala, Sweden.
Kits and Reagents—The QuikChange mutagenesis kit was from Stratagene (La Jolla, CA). Src family kinase inhibitor SU6656 and ATP were from Sigma. Lipofectamine 2000 was from Invitrogen. Chemiluminescent horseradish peroxidase substrate was from Millipore (Billerica, MA). The annexin V-PE apoptosis detection kit was from BD Biosciences. [γ-32P]ATP was from GE Healthcare.
Cell Culture—The EcoPack virus packaging cell line was grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Ba/F3 cells were grown in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin, and 10 ng/ml recombinant murine IL-3. To establish Ba/F3 cell lines expressing c-Kit, EcoPack cells were transfected with either wild-type or mutant c-Kit constructs in the pMSCVpuro vector. Supernatants were collected and used to infect Ba/F3 cells followed by a 2-week selection process in puromycin. Expression levels of c-Kit were confirmed by flow cytometry and immunoblotting. c-Kit-expressing Ba/F3 cells were grown in the same medium as Ba/F3 cells supplemented with 1.2 μg/ml puromycin.
Cell Stimulation, Immunoprecipitation, and Western Blotting—Ba/F3 cells were starved for 5 h in medium without serum and without IL-3 followed by SCF stimulation (100 ng/ml) for the indicated periods of time. Cell lysis, immunoprecipitation, and Western blotting were performed as described (23).
Kinase Activity Assay—Ba/F3 cells were starved for 5 h in medium without serum and without IL-3 and subsequently stimulated with SCF (100 ng/ml) for 5 min. Thereafter cells were washed once in ice-cold phosphate-buffered saline, lysed, immunoprecipitated with the KitC1 antibody, and processed for in vitro kinase assay as described (28) with the exception that Src optimal and Abl optimal peptide, respectively, was used. To determine the Km of the Src optimal peptide, a kinase reaction was performed varying the peptide concentration (5, 10, 20, 40, and 80 μm) on immunoprecipitates of c-Kit or Lyn, respectively.
Cell Survival and Proliferation Assay—Ba/F3 cells expressing wild-type c-Kit or various mutants of c-Kit were washed three times with phosphate-buffered saline, resuspended in Ba/F3 complete medium without IL-3, and seeded in 6-well plates with either 100 ng/ml SCF or 10 ng/ml IL-3 or without any cytokine as control. After a 48-h incubation, cells were stained with the annexin V-PE apoptosis detection kit (BD Biosciences). The living cells, apoptotic cells, and dead cells were counted using flow cytometry. For the cell proliferation assay, living cells were counted under the microscope.
Colony Formation in Semisolid Medium—Ba/F3 cells were washed three times with phosphate-buffered saline to wash away IL-3 and mixed with methylcellulose hematopoietic colony assay medium (MethoCult M3231, Stem Cell Technologies) at the concentration of 30,000 cells/ml, in the presence or absence of 100 ng/ml SCF, and incubated at 37 °C in a humidified atmosphere for 5 days.
RESULTS
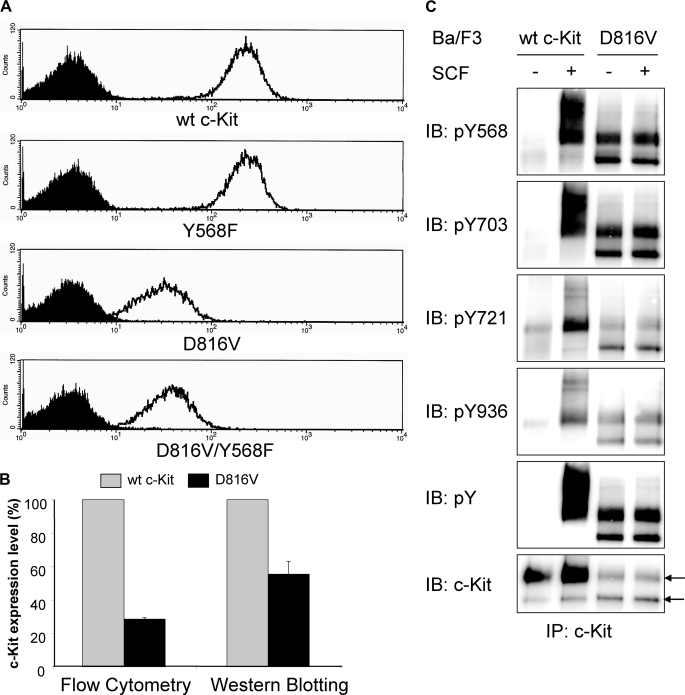
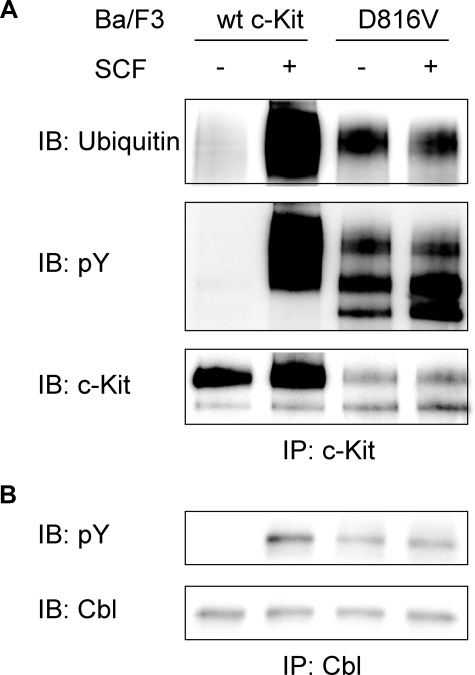
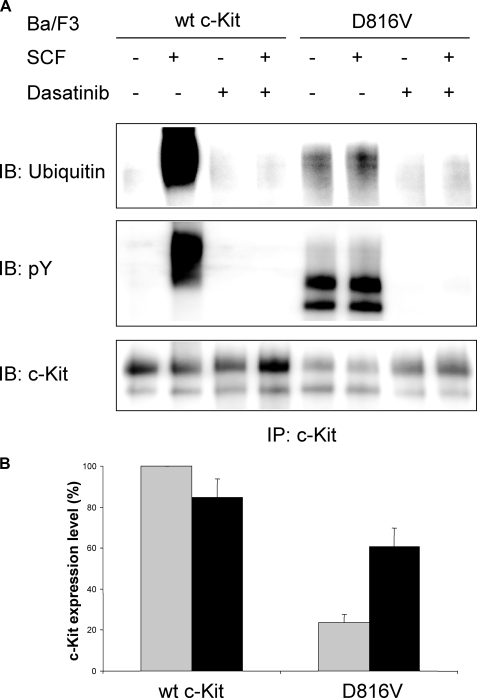
Constitutive Phosphorylation and Ubiquitination of c-Kit/D816V Leads to Lower Cell Surface Expression of c-Kit as Compared with Wild-type Receptor—It has been shown that inhibition of negative regulation of receptor tyrosine kinases due to impaired Cbl activation and subsequent receptor ubiquitination contributes to transformation in a number of mutated, oncogenic receptor tyrosine kinases (reviewed in Ref. 29). In this study, we sought to investigate the ubiquitination status of the oncogenic D816V mutant of c-Kit to examine whether diminished ubiquitination of this mutant contributes to its oncogenic phenotype. Both wild-type c-Kit and the D816V mutant of c-Kit were stably transfected into the hematopoietic cell line Ba/F3 followed by analysis of receptor surface expression. The D816V mutant of c-Kit showed much lower surface expression than wild-type receptor when examined by flow cytometry and Western blotting (Fig. 1, A and B). Next we wanted to investigate the phosphorylation of individual tyrosine residues on the D816V c-Kit mutant to see whether the phosphorylation sites would be different from wild-type c-Kit. In SCF-induced c-Kit signaling, phosphorylated Tyr-568 is responsible for activation of Src kinases, which in turn phosphorylate and activate both the ubiquitin E3 ligase Cbl and the Ras/Erk signaling pathway. Phosphorylated Tyr-703 and Tyr-936 are known to associate with the adapter protein Grb2, which indirectly recruits both Cbl as well as the scaffolding protein Gab2 to c-Kit (18, 23). Another important phosphorylation site is Tyr-721, the binding site for PI3-kinase, which in previous studies was shown to be of great importance for c-Kit/D816V-induced cell transformation (24, 30). Using phosphospecific antibodies against pTyr-568, pTyr-703, pTyr-721, and pTyr-936 in c-Kit, we investigated the ligand-induced phosphorylation of wild-type receptor as compared with D816V mutant. We found that Tyr-568, Tyr-703, Tyr-721, and Tyr-936 were all constitutively phosphorylated in c-Kit/D816V (Fig. 1C). Thus, the binding sites for signal transduction molecules downstream of c-Kit were all phosphorylated in c-Kit/D816V and potentially involved in signal transduction downstream of c-Kit/D816V. Because cell surface expression of c-Kit/D816V was significantly lower than wild-type c-Kit (Fig. 1A), we were interested in investigating whether this was due to differences in Cbl activation and ubiquitination. We could show that cells expressing c-Kit/D816V exhibited constitutive Cbl phosphorylation as well as constitutive receptor ubiquitination (Fig. 2, A and B). To investigate whether the constitutive ubiquitination and subsequent rapid receptor turnover contributed to the lower levels of cell surface expression of c-Kit/D816V as compared with wild-type receptor, we used the tyrosine kinase inhibitor Dasatinib to block the constitutive activation of c-Kit/D816V. Dasatinib treatment blocked receptor phosphorylation and receptor ubiquitination and, furthermore, increased the cell surface expression of c-Kit/D816V dramatically (p < 0.05 by Mann-Whitney test; Fig. 3, A and B). Taken together, our results suggest that the constitutive activation of c-Kit/D816V through ubiquitin-mediated degradation contributes to its low cell surface expression.
FIGURE 1.
Expression and activation of wild-type (wt) c-Kit and c-Kit/D816V in Ba/F3 cells. A, Ba/F3 cells expressing wild-type c-Kit, c-Kit/Y568F, c-Kit/D816V, or c-Kit/D816V/Y568F were stained with PE-conjugated anti-c-Kit antibody or isotype control followed by examination of receptor cell surface expression using flow cytometry (open curve, anti-c-Kit; filled curve, isotype control). B, expression of c-Kit was quantified based on both flow cytometry and Western blotting. Error bars indicate standard deviation. C, Ba/F3 cells expressing wild-type c-Kit or c-Kit/D816V were starved, SCF-stimulated (100 ng/ml) and subjected to immunoprecipitation (IP) using a c-Kit antibody. Phosphospecific antibodies toward pTyr-568 (pY568), pTyr-703 (pY703), pTyr-721 (pY721), and pTyr-936 (pY936) of c-Kit were used to detect the activation of individual tyrosine residues. As controls, phosphotyrosine (pY) and c-Kit expression levels were assessed. Arrows indicate the position of the 130-kDa immature band of c-Kit and the 145-kDa mature band of c-Kit, respectively. IB, immunoblot.
FIGURE 2.
Constitutive activation of Cbl and ubiquitination of c-Kit in Ba/F3 cells expressing c-Kit/D816V. Ba/F3 cells expressing c-Kit/D816V were starved and stimulated with SCF (100 ng/ml). A, lysates were subjected to immunoprecipitation (IP) using a c-Kit antibody followed by Western blot (IB) analysis using antibodies against ubiquitin, phosphotyrosine(pY), and c-Kit. wt, wild type. B, lysates were subjected to immunoprecipitation using a Cbl antibody followed by Western blot analysis using antibodies against phosphotyrosine and Cbl. Ba/F3 cells expressing wild-type c-Kit were used as control.
FIGURE 3.
Dasatinib inhibits the constitutive ubiquitination of c-Kit/D816V and increases the cell surface expression of the receptor. Ba/F3 cells expressing wild-type (wt) c-Kit or c-Kit/D816V mutant were incubated with or without Dasatinib (2 nm) for 24 h. A, cells were starved and stimulated with SCF (100 ng/ml) and subjected to immunoprecipitation using a c-Kit antibody. Levels of ubiquitination, tyrosine phosphorylation, and c-Kit expression were analyzed by Western blot (IB). pY, phosphotyrosine. B, cells were stained with PE-conjugated anti-c-Kit antibody or isotype control followed by examination of expression using flow cytometry. Cell surface expression levels of c-Kit were quantified, and the expression level of wild-type c-Kit without incubation with Dasatinib was considered as 100% and compared with other cells (gray column: no incubation; black column: incubation with Dasatinib). The data shown are the average of three independent experiments with error bars defining the standard deviation.
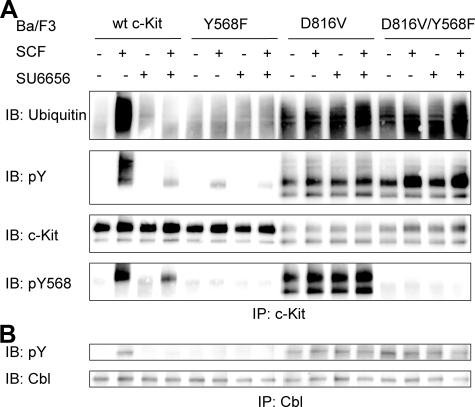
Src Family Kinases Are Not Required for the Phosphorylation and Ubiquitination of c-Kit/D816V—Src family kinases have been shown to play an important role in phosphorylation and ubiquitination induced by wild-type c-Kit (15, 31). To investigate the role of Src family kinases in the phosphorylation and ubiquitination of c-Kit/D816V, we generated mutants of the Src binding site Tyr-568 in both wild-type c-Kit and c-Kit/D816V background, which were transduced into Ba/F3 cells. Using Y568F mutants and the Src-selective inhibitor SU6656, we could examine the involvement of Src kinases in c-Kit/D816V-mediated phosphorylation. Both Y568F mutant and Src inhibitor SU6656 blocked the phosphorylation and ubiquitination of wild-type c-Kit. To our surprise, neither the Y568F mutant nor SU6656 blocked the phosphorylation and ubiquitination of c-Kit/D816V (Fig. 4A), suggesting differences in downstream signaling pathways between wild-type c-Kit and c-Kit/D816V. Because the ubiquitin E3 ligase Cbl is responsible for SCF-stimulated ubiquitination of wild-type c-Kit, we examined the phosphorylation of Cbl in c-Kit/D816V. In agreement with the effect of Y568F and SU6656 on receptor ubiquitination, in contrast to the phosphorylation mediated by wild-type c-Kit, neither Y568F nor SU6656 could block the phosphorylation of Cbl induced by c-Kit/D816V, (Fig. 4B).
FIGURE 4.
Src family kinases are not required for the phosphorylation and ubiquitination of c-Kit/D816V. Ba/F3 cells expressing wild-type (wt) c-Kit, c-Kit/Y568F, c-Kit/D816V or c-Kit/D816V/Y568F were starved. A, thereafter cells were preincubated with the Src inhibitor SU6656 (2 μm) for 30 min before SCF stimulation (100 ng/ml) followed by immunoprecipitation (IP) using a c-Kit antibody. After SDS-PAGE and electrotransfer, membranes were probed with ubiquitin antibody and reprobed with phosphotyrosine (pY) and c-Kit antibodies. IB, immunoblot; pY568, pTyr-568. B, cells were preincubated with the Src inhibitor SU6656 (2 μm) for 30 min before SCF stimulation (100 ng/ml) and immunoprecipitation using a Cbl antibody. After SDS-PAGE and electrotransfer, membranes were probed with phosphotyrosine and Cbl antibodies to show the activation of Cbl.
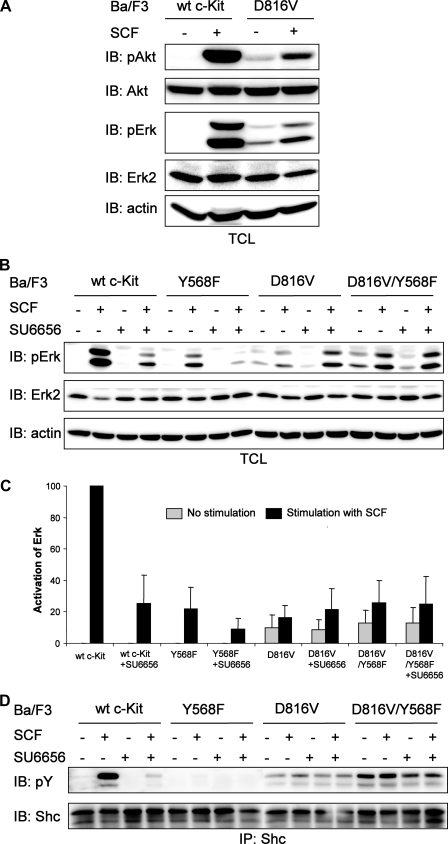
c-Kit/D816V-induced Activation of Erk Is Not Dependent on Src Family Kinases—Next we examined the c-Kit/D816V-induced activation of Erk and Akt in Ba/F3 cells. c-Kit/D816V could induce weak constitutive activation of both Erk and Akt, and this activation was further increased by SCF stimulation (Fig. 5A). In wild-type c-Kit signaling, phosphorylation of Tyr-568 leads to activation of Src family kinases, which in turn phosphorylates Shc and finally activates the Ras/Erk signaling pathway (21). Because we found Src family kinases to be dispensable for c-Kit/D816V-induced receptor phosphorylation and ubiquitination, we wanted to investigate how c-Kit/D816V activates the Ras/Erk pathway. Ba/F3 cells expressing wild-type c-Kit or c-Kit/D816V, with or without the Src binding mutation Y568F, were preincubated with the Src kinase inhibitor SU6656 followed by SCF stimulation and analyzed for Erk phosphorylation. We could show that Erk phosphorylation in cells expressing wild-type c-Kit was blocked by introduction of either the Y568F mutation or the Src-selective inhibitor SU6656. In the case of c-Kit/D816V, Erk phosphorylation was not dependent on Src family kinases (Fig. 5, B and C). Furthermore, in contrast to wild-type c-Kit signaling, the c-Kit/D816V-induced activation of the adaptor protein Shc was not dependent on Src kinase activity (Fig. 5D).
FIGURE 5.
Inhibition of Src family kinases has different effects on Erk activation in Ba/F3 cells expressing wild-type (wt) c-Kit and c-Kit/D816V. A, Ba/F3 cells expressing wild-type c-Kit or c-Kit/D816V were starved and thereafter stimulated with SCF (100 ng/ml). Whole cell lysates were investigated by Western blot (IB) analysis using antibodies against phospho-Erk (pErk) and phospho-Akt (pAkt). Filters were reprobed with Erk2, Akt, and β-actin antibodies to verify equal loading. B, Ba/F3 cells expressing wild-type c-Kit, c-Kit/Y568F, c-Kit/D816V, or c-Kit/D816V/Y568F were starved followed by preincubation with the Src inhibitor SU6656 (2 μm) for 30 min before SCF stimulation (100 ng/ml). Total cell lysates (TCL) were subjected to Western blot analysis using an antibody against phospho-Erk. Erk2 and β-actin were used as loading control. C, quantitation of data obtained in panel B. The level of Erk phosphorylation in Ba/F3 cells expressing wild-type c-Kit was set to 100. The data represent the average of triplicate determinations with the error bars showing standard deviation. D, cell lysates were immunoprecipitated (IP) using a Shc antibody, and samples were analyzed by Western blot using antibodies against phosphotyrosine and Shc. pY, phosphotyrosine.
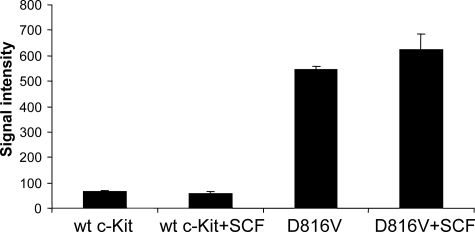
c-Kit/D816V Gains Src- and Abl-like Activity—The above results suggest that signaling through c-Kit/D816V is able to circumvent the requirement of Src family kinases. To further address the question of whether c-Kit/D816V gains a Src-like kinase activity, we performed in vitro kinase assays using either the so-called Src optimal peptide or the Abl optimal peptide as substrates. These peptides are highly selective for their respective kinase while being poor substrates for receptor tyrosine kinases (27). In contrast to wild-type c-Kit, the c-Kit/D816V mutant readily phosphorylated both peptides, and the activity could be further increased by SCF stimulation (Fig. 6; Abl kinase data not shown). Kinetic analysis of D816V c-Kit was performed and compared with Lyn, which is the most abundant Src family kinase in Ba/F3 cells. The Km for D816V c-Kit with respect to the Src optimal peptide was estimated to ∼20 μm, whereas Lyn showed a Km of ∼80 μm (data not shown). This was in the same range as has been described for Src (33 μm (27). This suggests that c-Kit/D816V circumvents a need for Src family kinases by mimicking Src in its kinase specificity.
FIGURE 6.
c-Kit/D816V gains Src activity. Ba/F3 cells expressing wild-type (wt) or c-Kit/D816V were starved, stimulated with SCF (100 ng/ml), and subsequently immunoprecipitated using a c-Kit antibody. The immunoprecipitates were used in a kinase assay together with Src optimal peptide as substrate. The phosphorylation of the substrate by wild-type c-Kit and c-Kit/D816V was detected by Fuji FLA 3000, and signal intensity was quantified by the Multigauge software. Error bars indicate standard deviation.
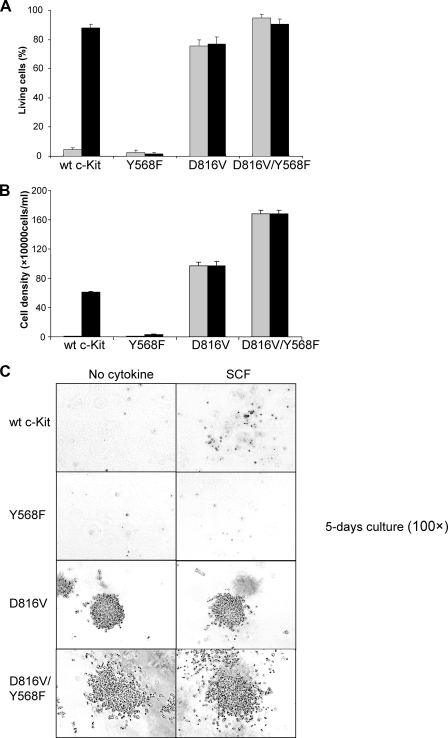
Phosphorylation of Tyr-568 Is Dispensable for c-Kit/D816V-induced Cell Survival—To specify the role of Tyr-568 in cell signaling, we next examined cell survival in Ba/F3 cells expressing either wild-type c-Kit or c-Kit/D816V. Cells were kept in medium in the presence or absence of SCF. Living cells, apoptotic cells, and dead cells were identified using the annexin-PE detection kit staining and flow cytometry analysis. As expected, Ba/F3 cells expressing wild-type c-Kit survived in the presence of SCF, whereas this effect was abolished in the Y568F mutant. In contrast, Ba/F3 cells expressing c-Kit/D816V survived in the absence of ligand and, furthermore, ligand stimulation did not further enhance cell survival. Additionally, in contrast to wild-type c-Kit, introduction of the Y568F mutation did not affect cell survival (Fig. 7A).
FIGURE 7.
Phosphorylation of Tyr-568 is not required in c-Kit/D816V-induced cell survival, proliferation, and growth in methylcellulose. Ba/F3 cells expressing wild-type (wt) c-Kit, c-Kit/Y568F, c-Kit/D816V, or c-Kit/D816V/Y568F were washed and thereafter seeded in 6-well plates in 100,000 cells/ml with SCF (100 ng/ml, black column) or without cytokine (gray column) as control followed by a 48-h incubation. A, cells were stained with the annexin V-PE apoptosis detection kit and subsequently subjected to flow cytometry to examine the proportion of living cells, apoptotic cells, and dead cells. Error bars indicate standard deviation. B, living cells were counted using trypan blue exclusion. Error bars indicate standard deviation. C, colony formation in methylcellulose.
Tyr-568 Inhibits Cell Proliferation Induced by the c-Kit/D816V Mutant—We thereafter wanted to explore the role of Tyr-568 in c-Kit-induced cell proliferation. Ba/F3 cells expressing wild-type c-Kit or c-Kit/D816V, with or without the Y568F mutation, were incubated with SCF for 48 h and subsequently scored for trypan blue exclusion. Ba/F3 cells expressing wild-type c-Kit proliferated upon SCF stimulation, and in agreement with the results from the survival assay, no living cells were detected if Tyr-568 was mutated. Ba/F3 cells expressing c-Kit/D816V could proliferate without SCF, and the addition of ligand did not further accelerate cell proliferation (Fig. 7B). The cells expressing Y568F mutation in addition to D816V had higher proliferation as compared with cells that only expressed c-Kit/D816V. Our data indicate a role of Tyr-568 in the inhibition of c-Kit/D816V-induced cell proliferation.
c-Kit/D816V Induces Colon Formation in Methylcellulose Independent of Activation of Src—Because recruitment of Src to Tyr-568 of c-Kit is important for wild-type c-Kit-mediated proliferation and survival, but not for c-Kit/D816V, we wanted to investigate whether the growth in semisolid medium was also independent of Src recruitment to Tyr-568 of c-Kit. These data clearly show that although colony formation by wild-type c-Kit-expressing Ba/F3 cells even in the presence of ligand was very weak, the c-Kit/D816V induced colony formation (Fig. 7C). Introduction of the Y568F mutation in c-Kit/D816V led to formation of even bigger colonies as compared with c-Kit/D816V. The cells in the colonies also appeared to have a greater tendency to scatter.
DISCUSSION
The mechanisms of signal transduction by the wild-type c-Kit are quite well characterized. Ligand binding to the receptor causes dimerization and activation of its intrinsic kinase activity, leading to phosphorylation of downstream signal transduction molecules, involving the Ras/Erk pathway and the PI3-kinase pathway (for review, see Ref. 2). In contrast, the mechanism by which the oncogenic mutant D816V of c-Kit signals is only partially known. In this study, we have analyzed the downstream signaling events in D816V/c-Kit and compared it with wild-type c-Kit signaling. Mutations analogous to D816V have been found in the closely related receptor tyrosine kinase Flt3, at aspartic acid 835, which is mutated in a subset of acute myeloid leukemia patients (32). The analogous residue in the c-Met receptor (D1264N) has been found to be mutated in papillary renal carcinoma, where it stabilized the active form of the receptor. D1264N displays altered kinase specificity in that CrkII, a known Abl kinase substrate, is phosphorylated by D1264N but not by wild-type c-Met (33). Interestingly, another oncogenic mutant of c-Met, M1268T, elevates c-Src phosphorylation, and its oncogenic activity was found to be inhibited by dominant negative forms of c-Src (34). In this study, we show that the D816V mutant of c-Kit gains both Src-like and Abl-like kinase activity. This is not due to increased binding of Src to c-Kit because the Y568F mutant of D816V shows the same ability to phosphorylate the Src optimal peptide (data not shown). It has previously been shown that the D816V mutant alters the specificity of c-Kit to resemble the Abl kinase (35), but that study failed to detect Src-like kinase activity in the D816V mutant. The reason for the discrepancy between the two studies is unclear. One possibility could be that the peptide sequence used in the study by Piao et al. (35) slightly differs from the one described by Songyang et al. (27), which is identical to the peptide that was used in this study.
A lowered degree of cell surface expression of oncogenic receptor mutants has been described in numerous cases of receptor tyrosine kinases. The mechanism is not fully understood. It has been shown that the D816V mutant of c-Kit drives transformation through intracellularly receptors trapped to the Golgi (36). Constitutive phosphorylation of Cbl and subsequent ubiquitin-mediated degradation could also contribute to the low cell surface expression, as proposed in this study.
Several phosphorylation sites in c-Kit are multifunctional docking sites for signal transduction molecules. One such site is Tyr-568 in the juxtamembrane region of c-Kit, which has been shown to associate with Src family kinases, CHK, the protein tyrosine phosphatase SHP2, Cbl, and the adapter protein APS (15, 21, 37–39). Thus, mutation of Tyr-568 will affect both signal transduction molecules involved in positive signaling (Src, SHP2) as well as in negative signaling (CHK, Cbl, and APS). One of the proteins known to be targeted for ubiquitination and degradation by the D816V mutant is the protein tyrosine phosphatase SHP1 (35). SHP1 is a well known negative regulator of c-Kit signaling (40). The observed increased survival and proliferation of the D816V/Y568F mutant as compared with the D816V mutant could also very well be ascribed to such effects. Other investigators have found the cytoplasmic tyrosine kinase Fes to be preferentially activated by the D816V mutant but not by wild-type c-Kit (41).
Although several signal transduction pathways were constitutively activated in D816V/c-Kit-expressing Ba/F3 cells, the addition of ligand further increased the phosphorylation of both Erk and Akt (Fig. 5, A and B). This is puzzling because the receptor is already constitutively active in the absence of ligand. However, in Jurkat cells transfected with D816V/c-Kit, SCF triggered a more potent chemotactic response than in cells transfected with wild-type c-Kit (42). This suggests that ligand does something qualitatively different from the oncogenic mutants. Furthermore, in the case of the receptor tyrosine kinase Met, the oncogenic mutant D1228N was dependent on the presence of ligand for full transformation of NIH3T3 cells (43). One explanation might be that ligand induces internalization of the receptors to intracellular compartments where it interacts with specific signal transduction molecules. It is clear from several studies that the subcellular localization of oncogenic receptor tyrosine kinases is critical for their signaling capabilities; e.g. trapping of the oncogenic Flt3-ITD (internal tandem duplication) mutant to the endoplasmic reticulum leads to selective phosphorylation of STAT5, whereas wild-type Flt3 does not phosphorylate STAT5 (44).
To summarize, our data show that the D816V mutant is not only an activating mutation that mimics the ligand-bound receptor but that it also has a qualitatively different substrate specificity that leads to activation of signal transduction molecules and downstream biological responses that are quite distinct from those induced by the wild-type receptor. The mechanism by which this mutant causes altered kinase specificity is not fully understood. However, this opens for the possibility that the D816V phosphorylates additional substrate molecules that are not phosphorylated by wild-type c-Kit that could provide novel targets for selective therapy against the oncogenic versions of c-Kit. Future work is aiming at identifying such signal transduction molecules.
Acknowledgments
We thank Susanne Bengtsson for expert technical assistance and Dr. Rasheed Khan for help with statistical analysis.
This work was supported by grants from the Swedish Cancer Society, the Swedish Research Council, the Gunnar Nilsson Cancer Foundation, the AlfredÖsterlund Foundation, the Malmö University Hospital Foundation, the Malmö University Hospital Cancer Foundation, the Royal Physiographical Society, and the Wallenberg Foundation.
Footnotes
The abbreviations used are: SCF, stem cell factor; PI3-kinase, phosphoinositide 3-kinase; Flt3, Fms-like tyrosine kinase 3; M-CSF, macrophage colony stimulating factor; IL-3, interleukin 3; Erk, extracellular signal-regulated kinase; PE, phycoerythrin.
References
- 1.Lennartsson, J., and Rönnstrand, L. (2006) Curr. Cancer Drug Targets 6, 65–75 [DOI] [PubMed] [Google Scholar]
- 2.Rönnstrand, L. (2004) CMLS Cell. Mol. Life Sci. 61, 2535–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ning, Z. Q., Li, J., and Arceci, R. J. (2001) Leuk. Lymphoma 41, 513–522 [DOI] [PubMed] [Google Scholar]
- 4.Beghini, A., Peterlongo, P., Ripamonti, C. B., Larizza, L., Cairoli, R., Morra, E., and Mecucci, C. (2000) Blood 95, 726–727 [PubMed] [Google Scholar]
- 5.Tian, Q., Frierson, H. F., Jr., Krystal, G. W., and Moskaluk, C. A. (1999) Am. J. Pathol. 154, 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagata, H., Worobec, A. S., Oh, C. K., Chowdhury, B. A., Tannenbaum, S., Suzuki, Y., and Metcalfe, D. D. (1995) Proc. Natl. Acad. Sci. U. S. A. 92, 10560–10564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Longley, B. J., Tyrrell, L., Lu, S. Z., Ma, Y. S., Langley, K., Ding, T. G., Duffy, T., Jacobs, P., Tang, L. H., and Modlin, I. (1996) Nat. Genet. 12, 312–314 [DOI] [PubMed] [Google Scholar]
- 8.Pauls, K., Wardelmann, E., Merkelbach-Bruse, S., Büttner, R., and Zhou, H. (2004) Virchows Arch. 445, 651–654 [DOI] [PubMed] [Google Scholar]
- 9.Sakuma, Y., Sakurai, S., Oguni, S., Satoh, M., Hironaka, M., and Saito, K. (2004) Cancer Sci. 95, 716–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hongyo, T., Li, T., Syaifudin, M., Baskar, R., Ikeda, H., Kanakura, Y., Aozasa, K., and Nomura, T. (2000) Cancer Res. 60, 2345–2347 [PubMed] [Google Scholar]
- 11.Mol, C. D., Dougan, D. R., Schneider, T. R., Skene, R. J., Kraus, M. L., Scheibe, D. N., Snell, G. P., Zou, H., Sang, B. C., and Wilson, K. P. (2004) J. Biol. Chem. 279, 31655–31663 [DOI] [PubMed] [Google Scholar]
- 12.Orfao, A., Garcia-Montero, A. C., Sanchez, L., and Escribano, L. (2007) Br. J. Haematol. 138, 12–30 [DOI] [PubMed] [Google Scholar]
- 13.Schittenhelm, M. M., Shiraga, S., Schroeder, A., Corbin, A. S., Griffith, D., Lee, F. Y., Bokemeyer, C., Deininger, M. W., Druker, B. J., and Heinrich, M. C. (2006) Cancer Res. 66, 473–481 [DOI] [PubMed] [Google Scholar]
- 14.Zeng, S., Xu, Z., Lipkowitz, S., and Longley, J. B. (2005) Blood 105, 226–232 [DOI] [PubMed] [Google Scholar]
- 15.Masson, K., Heiss, E., Band, H., and Rönnstrand, L. (2006) Biochem. J. 399, 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thien, C. B., and Langdon, W. Y. (2005) Growth Factors 23, 161–167 [DOI] [PubMed] [Google Scholar]
- 17.Marmor, M. D., and Yarden, Y. (2004) Oncogene 23, 2057–2070 [DOI] [PubMed] [Google Scholar]
- 18.Sun, J., Pedersen, M., Bengtsson, S., and Rönnstrand, L. (2007) Exp. Cell Res. 313, 3935–3942 [DOI] [PubMed] [Google Scholar]
- 19.Shaul, Y. D., and Seger, R. (2007) Biochim. Biophys. Acta 1773, 1213–1226 [DOI] [PubMed] [Google Scholar]
- 20.Hennessy, B. T., Smith, D. L., Ram, P. T., Lu, Y., and Mills, G. B. (2005) Nat. Rev. Drug Discov. 4, 988–1004 [DOI] [PubMed] [Google Scholar]
- 21.Lennartsson, J., Blume-Jensen, P., Hermanson, M., Pontén, E., Carlberg, M., and Rönnstrand, L. (1999) Oncogene 18, 5546–5553 [DOI] [PubMed] [Google Scholar]
- 22.Serve, H., Hsu, Y. C., and Besmer, P. (1994) J. Biol. Chem. 269, 6026–6030 [PubMed] [Google Scholar]
- 23.Sun, J., Pedersen, M., and Rönnstrand, L. (2008) J. Biol. Chem. 283, 27444–27451 [DOI] [PubMed] [Google Scholar]
- 24.Chian, R., Young, S., Danilkovitch-Miagkova, A., Rönnstrand, L., Leonard, E., Ferrao, P., Ashman, L., and Linnekin, D. (2001) Blood 98, 1365–1373 [DOI] [PubMed] [Google Scholar]
- 25.Blume-Jensen, P., Siegbahn, A., Stabel, S., Heldin, C. H., and Rönnstrand, L. (1993) EMBO J. 12, 4199–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voytyuk, O., Lennartsson, J., Mogi, A., Caruana, G., Courtneidge, S., Ashman, L. K., and Rönnstrand, L. (2003) J. Biol. Chem. 278, 9159–9166 [DOI] [PubMed] [Google Scholar]
- 27.Songyang, Z., Carraway, K. L. I., Eck, M. J., Harrison, S. C., Feldman, R. A., Mohammadi, M., Schlessinger, J., Hubbard, S. R., Smith, D. P., Eng, C., Lorenzo, M. J., Ponder, B. A. J., Mayer, B. J., and Cantley, L. C. (1995) Nature 373, 536–539 [DOI] [PubMed] [Google Scholar]
- 28.Hansen, K., Johnell, M., Siegbahn, A., Rorsman, C., Engström, U., Wernstedt, C., Heldin, C. H., and Rönnstrand, L. (1996) EMBO J. 15, 5299–5313 [PMC free article] [PubMed] [Google Scholar]
- 29.Peschard, P., and Park, M. (2003) Cancer Cell 3, 519–523 [DOI] [PubMed] [Google Scholar]
- 30.Hashimoto, K., Matsumura, I., Tsujimura, T., Kim, D. K., Ogihara, H., Ikeda, H., Ueda, S., Mizuki, M., Sugahara, H., Shibayama, H., Kitamura, Y., and Kanakura, Y. (2003) Blood 101, 1094–1102 [DOI] [PubMed] [Google Scholar]
- 31.Shivakrupa, R., and Linnekin, D. (2005) Cell. Signal. 17, 103–109 [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto, Y., Kiyoi, H., Nakano, Y., Suzuki, R., Kodera, Y., Miyawaki, S., Asou, N., Kuriyama, K., Yagasaki, F., Shimazaki, C., Akiyama, H., Saito, K., Nishimura, M., Motoji, T., Shinagawa, K., Takeshita, A., Saito, H., Ueda, R., Ohno, R., and Naoe, T. (2001) Blood 97, 2434–2439 [DOI] [PubMed] [Google Scholar]
- 33.Miller, M., Ginalski, K., Lesyng, B., Nakaigawa, N., Schmidt, L., and Zbar, B. (2001) Proteins 44, 32–43 [DOI] [PubMed] [Google Scholar]
- 34.Nakaigawa, N., Weirich, G., Schmidt, L., and Zbar, B. (2000) Oncogene 19, 2996–3002 [DOI] [PubMed] [Google Scholar]
- 35.Piao, X., Paulson, R., van der Geer, P., Pawson, T., and Bernstein, A. (1996) Proc. Natl. Acad. Sci. U. S. A. 93, 14665–14669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang, Z., Kreisel, F., Cain, J., Colson, A., and Tomasson, M. H. (2007) Mol. Cell. Biol. 27, 267–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price, D. J., Rivnay, B., Fu, Y., Jiang, S., Avraham, S., and Avraham, H. (1997) J. Biol. Chem. 272, 5915–5920 [DOI] [PubMed] [Google Scholar]
- 38.Kozlowski, M., Larose, L., Lee, F., Le, D. M., Rottapel, R., and Siminovitch, K. A. (1998) Mol. Cell. Biol. 18, 2089–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wollberg, P., Lennartsson, J., Gottfridsson, E., Yoshimura, A., and Rönnstrand, L. (2003) Biochem. J. 370, 1033–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paulson, R. F., Vesely, S., Siminovitch, K. A., and Bernstein, A. (1996) Nat. Genet. 13, 309–315 [DOI] [PubMed] [Google Scholar]
- 41.Voisset, E., Lopez, S., Dubreuil, P., and De Sepulveda, P. (2007) Blood 110, 2593–2599 [DOI] [PubMed] [Google Scholar]
- 42.Taylor, M. L., Dastych, J., Sehgal, D., Sundström, M., Nilsson, G., Akin, C., Mage, R. G., and Metcalfe, D. D. (2001) Blood 98, 1195–1199 [DOI] [PubMed] [Google Scholar]
- 43.Michieli, P., Basilico, C., Pennacchietti, S., Maffe, A., Tamagnone, L., Giordano, S., Bardelli, A., and Comoglio, P. M. (1999) Oncogene 18, 5221–5231 [DOI] [PubMed] [Google Scholar]
- 44.Schmidt-Arras, D., Böhmer, S. A., Koch, S., Müller, J., Blei, L., Cornils, H., Bauer, R., Korasikha, S., Thiede, C., and Böhmer, F. D. (2009) Blood, in press [DOI] [PubMed]