Abstract
The current studies show FGF15 signaling decreases hepatic forkhead transcription factor 1 (FoxO1) activity through phosphatidylinositol (PI) 3-kinase-dependent phosphorylation. The bile acid receptor FXR (farnesoid X receptor) activates expression of fibroblast growth factor (FGF) 15 in the intestine, which acts through hepatic FGFR4 to suppress cholesterol-7α hydroxylase (CYP7A1) and limit bile acid production. Because FoxO1 activity and CYP7A1 gene expression are both increased by fasting, we hypothesized CYP7A1 might be a FoxO1 target gene. Consistent with recently reported results, we show CYP7A1 is a direct target of FoxO1. Additionally, we show that the PI 3-kinase pathway is key for both the induction of CYP7A1 by fasting and the suppression by FGF15. FGFR4 is the major hepatic FGF receptor isoform and is responsible for the hepatic effects of FGF15. We also show that expression of FGFR4 in liver was decreased by fasting, increased by insulin, and reduced by streptozotocin-induced diabetes, implicating FGFR4 as a primary target of insulin regulation. Because insulin and FGF both target the PI 3-kinase pathway, these observations suggest FoxO1 is a key node in the convergence of FGF and insulin signaling pathways and functions as a key integrator for the regulation of glucose and bile acid metabolism.
Hepatic cholesterol is converted to bile acids, secreted into the gallbladder, and during a meal is released into the small intestine to enhance digestion and absorption of dietary lipids and fat-soluble vitamins. The majority of the bile acid pool (95%) is recycled back to the liver, whereas the remaining 5% is eliminated through fecal excretion (1, 2). This is an important route for the elimination of excess cholesterol and underscores the importance that bile acids play in regulating mammalian cholesterol metabolism. The initial and rate-controlling step in the classic pathway for cholesterol conversion into bile acids is catalyzed by cholesterol 7α-hydroxylase (CYP7A1).2 CYP7A1 regulation is primarily transcriptional, and expression of its gene is dynamically regulated by hormones and metabolites (2–6). Importantly, bile acids themselves regulate CYP7A1 gene expression through a multicomponent negative feedback pathway.
One of the molecular pathways for bile acid regulation is initiated by the activation of the farnesoid X receptor (FXR) responding directly to bile acid agonists (7). Ligand-activated FXR directly binds to a site in the promoter for the small heterodimer partner (SHP) gene and induces expression of SHP mRNA (8, 9). The translated SHP protein lacks the signature nuclear receptor zinc finger DNA binding domain but uses its conserved dimerization motif to form protein-protein contacts with DNA bound activators, usually other nuclear receptors, to inhibit or interfere with their activation potential (10, 11).
The first identified target for SHP repression was the CYP7A1 promoter, and SHP was proposed to interfere with activation by the DNA-bound monomeric liver receptor homologue 1 (LRH-1) nuclear receptor (8, 9). However, hepatic nuclear factor-4 (HNF-4), another nuclear receptor that stimulates CYP7A1 (12, 13), is also a target for SHP repression as well (14). Bile acids activate the JNK pathway, which may also play an important role in inhibition of CYP7A1 gene expression by a SHP-independent mechanism (15). Other kinase signaling pathways have been implicated in regulating CYP7A1, but mechanistic information is incomplete (16–18).
More recently, FXR has been shown to activate expression of both the human fibroblast growth factor (FGF) 19 in primary cultures of human hepatocytes (19) and its mouse orthologue, FGF15, in the intestine in response to bile acids (20). FGF15 along with FGF21 and FGF23 compose a subfamily of FGFs that are regulated by nuclear receptors and function as metabolic hormones. Although FGF15 regulates bile acid metabolism, FGF21 has an important role in glucose metabolism, peroxisome proliferator-activated receptor-α-dependent activation of fatty acid oxidation, ketogenesis, and growth hormone function (21–24), whereas FGF23 regulates phosphate metabolism (25). Most FGFs require strong binding to cell surface heparin sulfate proteoglycans to stabilize binding to their cognate FGF receptor. However, these three metabolic FGF hormones do not bind tightly to heparin sulfate; rather, they utilize one of two distinct but related cell surface co-receptors called klotho or β-klotho, apparently as co-receptors (26–29).
FGF15 produced in the distal small intestine signals through FGFR4 in hepatocytes to inhibit expression of the liver CYP7A1 gene, providing an intriguing example of tissue communication in metabolic regulation (24). Another report demonstrated that serum levels of FGF19 in humans are increased by bile acid feeding and decreased by a bile acid sequestrant (30). Thus, it is likely that FGF-dependent bile acid regulation is conserved between rodents and humans.
FXR and bile acids have been implicated as integrated regulators of bile acid and glucose metabolism (31, 32). In support of this connection, we found that CYP7A1 was induced by fasting and during streptozotocin induced diabetes (33), two stressful metabolic conditions where insulin signaling and glucose metabolism are compromised. More recently, other reports have also revealed an important role for bile acids in glucose metabolism (34, 35).
Insulin has long been known to inhibit CYP7A1 (6, 36), but the mechanism has not been fully revealed. Insulin binding to its cell surface receptor initiates a signaling cascade through the PI 3-kinase pathway, resulting in phosphorylation and activation of the serine/threonine protein kinase Akt. Akt in turn phosphorylates forkhead transcription factor 1 (FoxO1) on three key residues, Thr-24, Ser-253, and Ser-316, converting FoxO1 from a predominantly nuclear to a predominantly cytoplasmic location, rendering it unable to activate its nuclear target genes (37–42).
This redistribution and inhibition of FoxO1 occurs in response to insulin and other nutrient and growth signals that activate the PI 3-kinase pathway in many tissues and cell types including liver (42–45). Overall, nuclear localization and activation of FoxO1 is associated with stressful conditions, which in the liver occurs during fasting (43). Key hepatic FoxO1 target genes include phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (46–48) that encode key gluconeogenic enzymes, which are turned on to produce glucose when serum levels are low and insulin signaling is compromised.
Signaling through both insulin and FGF target the PI 3-kinase pathway (38, 40, 42, 49), and both inhibit CYP7A1. Because FoxO1 is negatively regulated by PI 3-kinase signaling and both FoxO1 and CYP7A1 are induced by fasting, we hypothesized that FoxO1 might be involved in the negative regulation of CYP7A1. Here, we show that FoxO1 directly regulates CYP7A1 gene expression and that, similar to insulin, FGF15/19 signaling leads to a PI 3-kinase-dependent phosphorylation and inhibition of FoxO1 in mice and in cultures of primary hepatocytes. Thus, FoxO1 is at a critical junction in hepatic physiology where it links bile acids with glucose metabolism.
EXPERIMENTAL PROCEDURES
Plasmids—The rat pGL3R7α-342 has been described previously (a gift from Dr. G. Gil, Virginia Commonwealth University) (50). Point mutations were introduced into putative FoxO1 binding sites designated FoxO1/1 or FoxO1/2 by QuikChange site-directed mutagenesis (Stratagene) to generate pGL3R7α-342/M1, pGL3R7α-342/M2, pGL3R7α-342/M1 and -2, and pGL3R7α-228/M1. The following primers were used: M1, 5′-CTAGTAGGAGGACAAATAGTGGGTGCTTTGGTCACTCAAGTTCA-3′; M2, 5′-TGACAGATGTGCTCATCTGGGTACTTCTTTTTCTACACACAG-3′.
pGL3R7α-228 was constructed by PCR-based amplification using the following primers: forward primer, 5′-ATGTTATGTCAGCACATGAGG-3′; reverse primer, 5′-AAAAGCAGGAAAATTTCCAAAGG-3′. The PCR product was digested with SstI and HindIII, which were added to the forward and reverse primers, respectively, for cloning purpose and inserted into the SstI and HindIII sites of pGL3-basic. The human CYP7A1 promoter construct ph-371/+24-Luc (36) was obtained from Dr. J. Chiang (Northeastern Ohio University).
Cytomegalovirus-SHP was a gift from Dr. D. Mangelsdorf (University of Texas Southwestern Medical Center). FLAG-FoxO1/WT, FLAG-FoxO1/TSS, and TK-IRS3 were gifts from Dr. T. Unterman (University of Illinois College of Medicine). pCMV6-Akt-WT and pCMV6-Akt-K179M were provided by T. Franke (Columbia University) through Dr. D. Fruman (University of California, Irvine, CA).
Cell Culture and Transient Transfection Assay—HepG2 cells were maintained in minimum essential medium supplemented with 10% fetal bovine serum at 37 °C and 5% CO2. Transient transfection was performed by the calcium phosphate coprecipitation method as described (33). Values represent the mean of duplicates ± S.D. Each experiment was repeated at least three times.
Animal Studies—All animal experiments were approved by the Institutional Animal Care and Use Committee at UC Irvine (protocol 97–1545). FoxO1flox/flox mice expressing Cre recombinase under the α1-antitrypsin promoter (51) were obtained from Dr. D. Accili (Columbia University). These mice were crossed, generating liver-specific FoxO1 knock-out mice (confirmed by PCR of genomic DNA as described (51)) and their littermates, FoxO1flox/flox, which were used as controls. These mice are in the FVB/N strain. Fasting experiments were performed with 8–9-week-old male mice, as described below.
In other studies 4-week-old B6129 male mice were obtained from Taconic and maintained on a 12-h light/dark cycle with free access to food and water. The mice were allowed to adapt to new environments for at least 1 week before experiments. At 8 weeks of age mice were fasted for 24 h, and all mice were sacrificed at 8:00 a.m. (end of the dark cycle). Livers and ileums were removed and frozen in liquid nitrogen and stored at –80 °C until RNA was isolated.
For the induction of diabetes, 4-week-old 129SVE male mice (Taconic) were treated with streptozotocin by intraperitoneal injections (100 μg/g of body weight) daily for 3 days to induce type I diabetes, as described previously (33). Mice were sacrificed, and livers were processed as described above.
Overexpression of FGF15 or FoxO1 Δ256 in mice was achieved through adenoviral delivery. For this, recombinant adenoviruses expressing either GFP, FGF15 (a gift from Dr. S. Kliewer, University of Texas Southwestern Medical Center), or FoxO1 Δ256 (a gift from D. Accili, Columbia University) were first propagated in 293 cells and purified by CsCl gradient centrifugation. A total of 1 × 109 plaque-forming units of each adenovirus was administered into 8-week-old 129SVE male mice (Taconic) by intravenous injection (4 animals in each group). 7 days after adenovirus inoculation mice were sacrificed for RNA and protein analysis. Where indicated, mice were fasted before sacrifice as described above.
Primary Mouse Hepatocyte Preparation—Primary mouse hepatocytes were isolated from 7–9-week-old C57BL/6 male mice (The Jackson Laboratory) by liver collagenase perfusion as described previously (52) with minor modifications. Briefly, mice were anesthetized using a ketamine/xylazine mixture. The liver was perfused with Earle's balanced salt solution (EBSS) (Invitrogen) supplemented with 0.5 mm EGTA through the cannulated portal vein at a flow rate of 4 ml/min for 15 min followed by perfusion with EBSS supplemented with 0.3 mg/ml collagenase (Wako Chemicals) and 4.8 mm CaCl2 for 20 min. The liver was dissected from the mouse, and dissociated cells were dispersed gently in Williams' E medium supplemented 10% fetal bovine serum. Cells were filtered through a 100-μm nylon cell strainer (BD Falcon), and hepatocytes were separated by a density gradient centrifugation using 45% Percoll (Sigma) solution. Hepatocyte viability was monitored by trypan blue exclusion, and more than 90% of cells were consistently viable. The isolated cells were plated in collagen-coated 6-well dishes at a density of 6 × 105/well. After a 4-h attachment, cells were overlaid with Matrigel (Collaborative Biomedical Products, Bedford, MA) and maintained in serum-free Williams' E medium supplemented with 10 nm dexamethasone, 2 mm glutamine, 100 units/ml penicillin, 100 units/ml streptomycin. 48 h after isolation, cells were treated with 100 nm human insulin (Sigma) for 24 h or 80 ng/ml FGF19 (R&D Systems) for 6 h and harvested for RNA analysis. Insulin or FGF19 additions were staggered so that cells were harvested at the same time. For the dose-response experiment, cells were treated with 0, 40, or 80 ng/ml FGF19 for 6 h, and RNA was isolated. Cells were treated with 100 nm wortmannin (Calbiochem) for 30 min before the treatment with FGF19. For Western analysis, cells were treated with 30 nm insulin or 80 ng/ml FGF19 for 30 min, and total cell lysates were prepared. Cells were treated with 100 nm wortmannin (Calbiochem) or 20 μm SP600125 (Calbiochem) 2.5 μm PIK 90 (Axon Medchem BV, The Netherlands) for 30 min before the treatment with FGF19 or insulin where indicated.
RNA Isolation and Analysis—Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. For quantitative real time RT-PCR (q-PCR), RNA was reverse-transcribed using the iScript cDNA synthesis kit (BioRad) at 42 °C for 30 min. cDNA was amplified and quantified using the iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer's instructions in the iQ5 real-time PCR detection system (Bio-Rad) under the following conditions: initial denaturation at 95 °C for 5 min and 40 cycles of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s. q-PCR was performed in triplicate for each sample and repeated at least three times. Ct values were used to calculate the relative expression level normalized to the expression of the housekeeping ribosomal protein L32. The results were expressed as the mean ± S.D. A melting curve analysis was performed for each sample after PCR amplification to verify that the amplicon is homogeneous in the absence of primer dimmers and DNA contamination. The sequences of primers used in q-PCR are available upon request. RNA from individual animals (4–6 animals in each group) was analyzed separately, and a Student t test was used for comparative statistical analysis as indicated in the individual figure legends.
Glucose Production Assay—Primary mouse hepatocytes were isolated from 8-week-old C57BL/6 male mice after overnight fasting, as described above. After a 4-h attachment in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), primary cells were cultured overnight in DMEM with 5% FBS. The medium was then replaced with serum- and glucose-free Dulbecco's modified Eagle's medium (pH 7.4) supplemented with 2 mm sodium lactate without phenol red. Where indicated, 20, 40, or 80 ng/ml FGF19 was added to the medium. After a 3-h incubation, the medium was collected, and glucose concentrations were measured using the Amplex red glucose assay kit (Invitrogen).
Electrophoretic Mobility Shift Assays—Nuclear extracts were prepared from 293T cells transfected with a vector expressing FLAG-FoxO1, and 5 μg of nuclear protein was used in electrophoretic mobility shift assays as described previously (33). For supershift experiments anti-FLAG M2 mouse monoclonal (Sigma) was incubated with the nuclear extract for 20 min on ice before incubation with the labeled probes. Competition experiments were performed in binding reactions where a 100-fold molar excess of unlabeled probes was incubated with nuclear extracts for 20 min before incubation with the labeled probes. The sequences of one strand of the complementary oligonucleotide probes were as follows: wild type FoxO1/1, 5′-AGGACAAATAGTGTTTGCTTTGGTCACTCA-3′; mutant FoxO1/1, 5′-AGGACAAATAGTGggTGCTTTGGTCACTCA-3′; wild type FoxO1/2, 5′-TGTGCTCATCTGTTTACTTCTTTTTC-3′; mutant FoxO1/2, 5′-TGTGCTCATCTGggTACTTCTTTTTC-3′; insulin-like growth factor binding protein-1/insulin response element, 5′-CACTAGCAAAACAAACTTATTTTGAACAC-3′.
Protein Isolation and Blotting—Total cell lysates from primary mouse hepatocytes or mouse livers infected with either adenovirus expressing GFP or FGF15 were fractionated on 8% SDS-PAGE, transferred to nitrocellulose membranes, and analyzed first by Ponceau S staining to confirm that equal amounts of total protein were both loaded and transferred in each lane. Then the blot was incubated with an indicated antibody followed by a secondary antibody conjugated to horseradish peroxidase. Reactivity was then detected with the ECL kit (Pierce). The primary antibodies were obtained as follows: anti-hemagglutinin (HA) (clone 12CA5) from Roche Applied Science, total FoxO1, phospho-FoxO1 (Thr-24) and phospho-Akt (Ser473) from Cell Signaling Technology, total Akt from Santa Cruz Biotechnology, anti-FLAG M2 mouse monoclonal from Sigma, mouse FGFR4 from R&D systems (AF2265), and mouse β-actin from Sigma (A1978). Antibody against hepatic nuclear factor 4 was from Dr. F. Sladek (University of California, Riverside, CA).
Adenovirus Infection of Primary Mouse Hepatocytes—Primary mouse hepatocytes were seeded in 100-mm dishes at a density of 4 × 106 cells/dish. 16 h after incubation in Matrigel, cells were infected with a recombinant adenovirus expressing FoxO1-WT or FoxO1-Δ256, a dominant negative FoxO1 lacking the carboxyl-terminal domain (from Dr. D. Accili, Columbia University (40, 48)) or GFP at a multiplicity of infection of ∼25 and incubated for 24 h in serum-free Williams' E medium. Hepatocytes were collected for chromatin immunoprecipitation assays or RNA analysis for q-PCR as described above and in the figure legends. Total cell lysates were also prepared for Western blotting to confirm the expression of FoxO1.
Chromatin Immunoprecipitation Assays—Chromatin immunoprecipitation assays were performed essentially as described previously (5) with minor modifications. Primary mouse hepatocytes were fixed in 1% formaldehyde for 10 min and lysed. Immune complexes were prepared using anti-HA (clone 12CA5) (Roche Applied Science) or the FoxO1antibody (Cell Signaling Technology catalog no. 9454). The following primers were used to amplify the specific promoters: mCYP7A1 forward primer (–367), 5′-TGGAAAGCTTCTGCCTGTTT-3′; mCYP7A1 reverse primer (–208), 5′-CGAAGGTCTGTCCCTCATGT-3′; mYY1 exon 4 forward, 5′-GCTGCACAAAGATGTTCAGGGATAA-3′; mYY1 exon 4 reverse, 5′-CTGAAAGGGCTTTTCTCCAGTATG-3′.
RESULTS
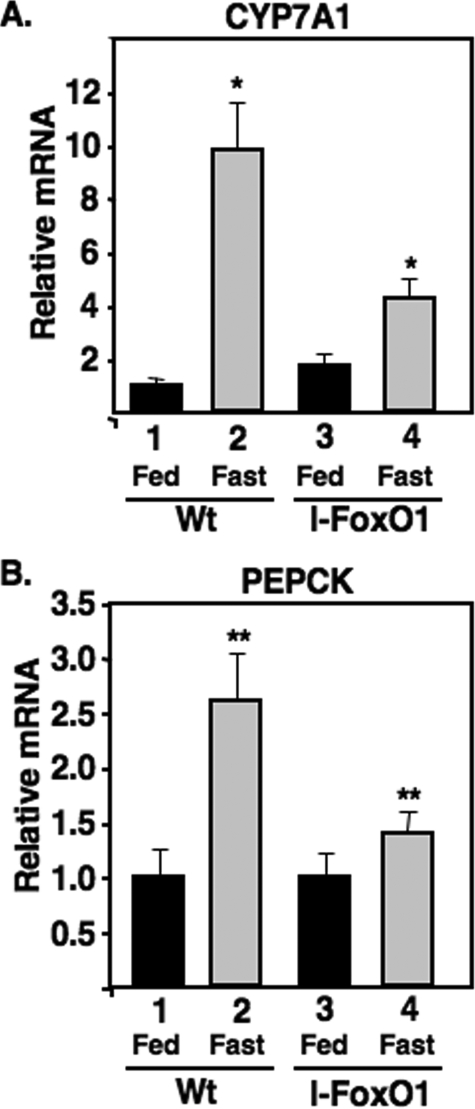
Fasting Induction of CYP7A1 Requires Hepatic FoxO1—FoxO1 is an important transcriptional regulator for hepatic gluconeogenesis during fasting, and because CYP7A1 is also induced by fasting, we hypothesized that FoxO1 might also regulate CYP7A1 gene expression. In support of this possibility, when FoxO1 was expressed at elevated levels in a transgenic mouse model, CYP7A1 was induced (53). To test the role of FoxO1 in CYP7A1 more directly, we evaluated CYP7A1 induction in l-FoxO1 mice where the FoxO1 gene was deleted specifically in liver (51). As shown in Fig. 1, the induction of CYP7A1 gene expression by fasting was severely blunted in the l-FoxO1 mice relative to their litter mate controls.
FIGURE 1.
CYP7A1 induction by fasting is blunted in liver-specific FoxO1 knock-out mice. A and B, total RNA was isolated from fed or fasted (24 h) I-FoxO1 and littermate control mice. mRNA levels were analyzed by q-PCR as described under “Experimental Procedures.” Results are expressed as -fold change relative to those of the fed littermate control. The mean values obtained from triplicates in each group as shown with error bars. The p values were obtained by comparing the mean of mRNA levels of littermate control to that of I-FoxO1 under fasted conditions. p < 0.005 (*) and p < 0.05 (**) for CYP7A1 and PEPCK, respectively.
FoxO1 Activation of the CYP7A1 Promoter—Analysis of the proximal region of the murine and human CYP7A1 promoters revealed two conserved putative FoxO1 binding sites, and electrophoretic mobility shift assays and transient DNA transfections showed that FoxO1 bound specifically to both murine sites and that one site at –281 was crucial for FoxO1 transactivation (supplemental Figs. 1–3). These results are consistent with another recent report showing that FoxO1 activates the rat CYP7A1 promoter (54). Our studies also revealed that the crucial FoxO1 site is conserved in the human promoter, which also bound FoxO1 in an electrophoretic mobility shift assay and was activated by FoxO1 in transfection assays (supplemental Fig. 4).
We also analyzed the binding of FoxO1 to the genomic mouse CYP7A1 promoter in hepatocytes using the chromatin immunoprecipitation technique. For this analysis primary mouse hepatocytes were infected with adenoviruses expressing either an HA epitope-tagged version of FoxO1 or a control GFP-expressing virus, and an antibody to the HA epitope was used to precipitate FoxO1-bound chromatin followed by q-PCR with primers for the CYP7A1 promoter or a control nonspecific region of the genome from the YY1 locus (Fig. 2A). The results show that the CYP7A1 promoter was specifically enriched by the precipitation with the HA antibody.
FIGURE 2.
FoxO1 binds to and is required for activation of CYP7A1 gene promoter. A, chromatin was prepared from primary mouse hepatocytes infected with an adenovirus expressing GFP (lanes 1 and 4) or HA-tagged FoxO1 (lanes 2 and 3 and lanes 5 and 6). Chromatin immunoprecipitation assays were performed with an anti-HA (lanes 1, 3, 4, and 6) or mouse IgG (lanes 2 and 5), and immunoprecipitates were analyzed by q-PCR. Results are expressed as -fold change in comparing the level of DNA amplification specifically precipitated by the HA antibody from the chromatin prepared from cells infected with FoxO1 relative to that infected with GFP. The level of DNA amplification precipitated by a normal mouse IgG fraction with chromatin from the FoxO1-infected cells (lanes 2 and 5) and the level of recruitment of FoxO1 to a non-relevant region of the genome at the YY1 locus (lane 6) are shown as negative controls. The data represent the mean of triplicates for two individual experiments and include error bars. Total FoxO1 expression was monitored by immunoblotting total chromatin-associated proteins with the HA epitope antibody from GFP- or HA-FoxO1-infected cells as indicated (inset). B, mouse primary hepatocytes were cultured in the absence of insulin and infected with adenovirus constructs expressing GFP or FoxO1-Δ256, a dominant negative version of FoxO1, as noted. Cells were harvested 24 h after infection, and total RNA was analyzed by q-PCR for CYP7A1 and FoxO1 and normalized to ribosomal protein L32 RNA levels as described under “Experimental Procedures.” In the comparisons marked by an asterisk, p < 0.0007 (*).
A Dominant Negative FoxO1 Construct Inhibits CYP7A1 Expression in Mouse Primary Hepatocytes—To address the role of FoxO1 in activation of CYP7A1 by an independent method, we infected primary hepatocytes with an adenovirus expressing FoxO1-Δ256 (40, 48), a dominant negative version of FoxO1 lacking the carboxyl-terminal domain, and this treatment significantly reduced CYP7A1 mRNA levels (Fig. 2B).
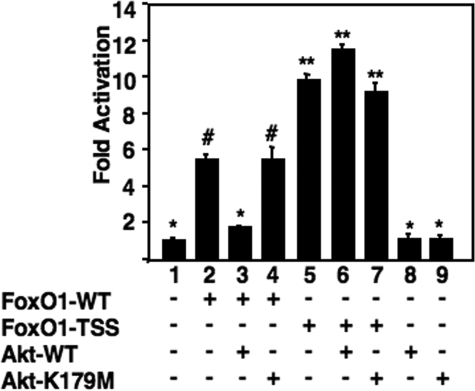
FoxO1 Transactivation of the CYP7A1 Promoter Is Inhibited by Akt—Insulin signaling through the PI 3-kinase pathway activates Akt, which in turn phosphorylates FoxO1, leading to its nuclear exclusion and inhibition of target gene activation (37, 38, 40, 42). To determine whether this signaling pathway might also play a role in regulation of CYP7A1, we transfected an Akt expression construct and showed that it prevented the FoxO1-dependent stimulation of the CYP7A1 reporter promoter (Fig. 3). In contrast, a kinase-defective version of Akt (Akt-K179M) (55) was unable to reverse the FoxO1 stimulation. Similarly, when a constitutively active version of FoxO1, where all three Akt phosphorylation sites are changed to alanines (FoxO1-TSS) (56), was transfected along with the wild type Akt construct, robust FoxO1 stimulation was still observed (Fig. 3).
FIGURE 3.
FoxO1 transactivation of the CYP7A1 promoter is inhibited by Akt. HepG2 cells were transfected with expression vectors for FoxO1-WT (0.5 μg), FoxO1-TSS (0.5 μg), Akt-WT (0.5 μg), or Akt-K179M (0.5 μg) along with pGL3R7α-342 (2 μg) as indicated. Results are expressed as corrected luciferase light units divided by the internal control signal for β-galactosidase activity. -Fold activation was calculated as in supplemental Fig. 2. All samples showing statistically significant differences when compared together (p < 0.01) are indicated by different symbols.
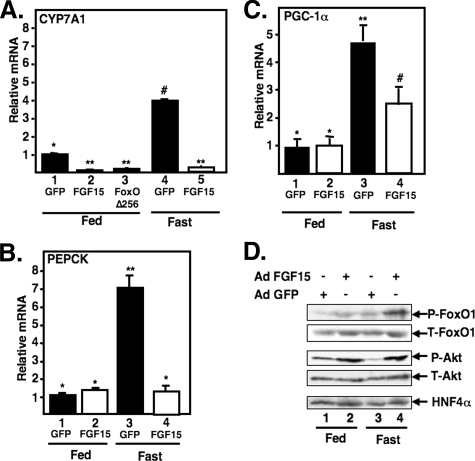
FGF15 Inhibits Expression of CYP7A1 and Other FoxO1 Target Genes through Phosphorylation of FoxO1 in Mice—Like insulin, FGF signaling feeds into the PI 3-kinase pathway (49). Because insulin signaling decreases FoxO1 activity, we hypothesized that the recently described FGF15-dependent inhibition of CYP7A1 might decrease FoxO1 in a similar manner. To test this hypothesis and to determine whether FGF15 might play a more global role in FoxO1-dependent gene expression, we evaluated the effects of FGF15 overexpression on FoxO1 phosphorylation and expression of CYP7A1 and other FoxO1 target genes in mice. Recombinant adenoviruses expressing GFP or FGF15 were introduced into chow-fed wild type mice. After 6 days, half of the animals in each infected group were fasted for 24 h, and all animals were sacrificed the following morning. Consistent with our earlier observations (Fig. 1) (33), expression of CYP7A1, PEPCK, and the transcriptional co-activator PGC-1α were induced by fasting in the control animals infected with the GFP expression adenovirus (Fig. 4A). In the animals expressing viral-encoded FGF15, CYP7A1 expression was significantly repressed in both chow-fed and fasted mice. This provides further evidence of a role for FoxO1 in CYP7A1 expression; infection of chow-fed mice with an adenovirus expressing a dominant negative version of FoxO1 (Δ256). Interestingly, expression of PEPCK was not affected by FGF15 in the chow-fed mice, but the fasting-dependent induction was completely prevented. PGC-1α mRNA expression is also activated by FoxO1 (57), and FGF15 also blunted its induction by fasting (Fig. 4C).
FIGURE 4.
FGF15 inhibits CYP7A1 and induces phosphorylation of FoxO1 in livers of mice. Mice were inoculated with the specified adenovirus (Ad) constructs and treated as described under “Experimental Procedures.” Total RNA was isolated and analyzed for CYP7A1 (A), PEPCK (B), or PGC-1α (C) gene expression. mRNA levels were analyzed from pooled RNA in each group (4–6 animals in each group) by q-PCR as described under “Experimental Procedures.” Results are expressed as -fold change relative to those of chow-fed mice infected with Ad-GFP, FGF15, or FoxO1 Δ256 as indicated. The mean values obtained from triplicates in each group are shown with error bars. Where pairwise comparisons provide p < 0.01, samples are labeled with a different symbol. D, protein extracts were prepared from pieces of the same mouse livers described in A–C. Pooled protein samples from animals in each group were analyzed by 8% SDS-PAGE and Western blotting with the indicated antibodies. P-FoxO1, phospho-FoxO1; T-FoxO1, total FoxO1; P-Akt, phospho-Akt, T-Akt, total Akt.
Immunoblotting analyses of hepatic protein extracts from these same animals demonstrated that the phosphorylation of both FoxO1 and Akt was increased in the Ad-FGF15-treated animals in both fasting and chow-fed animals (Fig. 4D). These results are consistent with FGF15 stimulating hepatic signaling through the PI 3-kinase pathway, resulting in inactivation of FoxO1 and repression of CYP7A1 and other FoxO1 target genes.
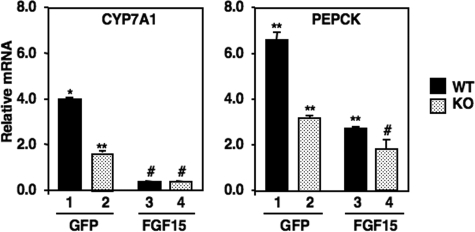
FGF15 Overexpression Decreases Expression of CYP7A1 and PEPCK in l-FoxO1 Mice—The data so far presented indicate that FoxO1 activates the CYP7A1 promoter, and FoxO1 is regulated by FGF15 signaling. To determine whether FoxO1 represents the only pathway for FGF15 inhibition, we evaluated the effects of overexpressing FGF15 on the fasting-dependent induction of CYP7A1 and PEPCK in WT and l-FoxO1 mice (Fig. 5). As in Fig. 1, the fasting-dependent induction of both CYP7A1 and PEPCK was blunted in the l-FoxO1 animals. The lower level of fasting induction for PEPCK was only partially blunted by Ad-FGF15 infection; however, CYP7A1 expression was reduced to the same level in both WT and l-FoxO1 samples. These results support a role for FoxO1 in FGF15 signaling, but they also indicate that PI 3-kinase regulation of FoxO1 is not the only pathway by which FGF15 inhibits CYP7A1.
FIGURE 5.
FGF15 adenovirus effects on CYP7A1 and PEPCK gene expression in WT and l-FoxO1 mice. Wild type or l-FoxO1 littermate control floxed mice were inoculated with the specified adenovirus constructs and fed a chow diet or fasted as described under “Experimental Procedures.” Total RNA was isolated and analyzed for CYP7A1 (left) or PEPCK (right) gene expression as noted. The data are plotted as relative expression compared with ad libitum-fed control samples (set at 1.0). Where pairwise comparisons provide p < 0.01, samples are labeled with a different symbol. KO, knock out.
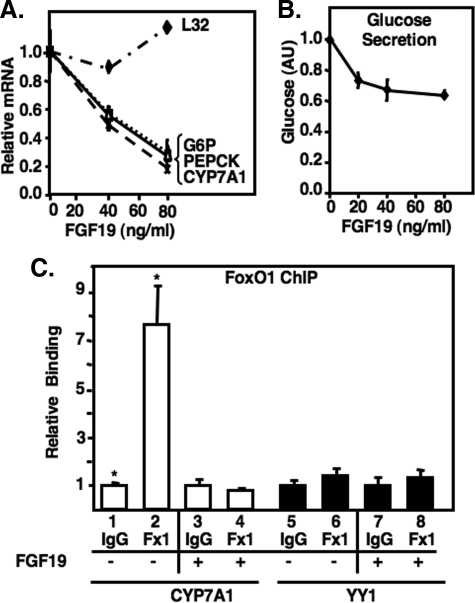
FGF19 Inhibits Expression of CYP7A1 and Other FoxO1 Target Genes in Primary Hepatocytes—The negative effects of FGF15 signaling on FoxO1 target gene expression were also evaluated in primary mouse hepatocytes treated with recombinant FGF19, the human orthologue of mouse FGF15. The results in Fig. 6 show a dose-dependent inhibition of FoxO1 target genes CYP7A1, PEPCK, and glucose-6-phosphatase in response to recombinant FGF19 treatment (Fig. 6A) as well as a direct loss of FoxO1 binding to the CYP7A1 promoter in hepatocytes as assayed by chromatin immunoprecipitation assay (ChIP, Fig. 6C). Along with the decrease in gluconeogenic gene expression, glucose production by FGF19 from primary hepatocytes was also inhibited by 40% (Fig. 6B).
FIGURE 6.
FGF19 inhibits CYP7A1 and other FoxO1 target genes in primary hepatocytes. A, primary mouse hepatocytes were treated with 0, 40 ng/ml, or 80 ng/ml FGF19 for 6 h, and total RNA was isolated and analyzed for expression of the indicted genes by q-PCR as described under “Experimental Procedures.” Data were analyzed as described in the legend to Fig. 4. L32 mRNA levels are shown as a negative control. B, primary mouse hepatocytes were cultured in serum- and glucose-free medium with or without 20, 40, or 80 ng/ml of FGF19 as indicated. After a 3-h incubation, the medium was assayed for glucose production as described under “Experimental Procedures.” p = 0.006, comparing 0 and 80 ng/ml FGF19. C, chromatin was prepared from primary mouse hepatocytes treated with 80 ng/ml FGF19 for 6 h, as described under “Experimental Procedures.” Chromatin immunoprecipitation (ChIP) assays were performed with an anti-FoxO1 or control mouse IgG fraction, and immunoprecipitates were analyzed by q-PCR. Results are expressed as -fold change in comparing the level of DNA amplified from chromatin specifically precipitated by the FoxO1 antibody with that precipitated by a control mouse IgG. The recruitment of FoxO1 to a non-relevant region of the genome in the YY1 locus is shown as a negative control. Data represent the mean of triplicates and include error bars.*, p = 0.02.
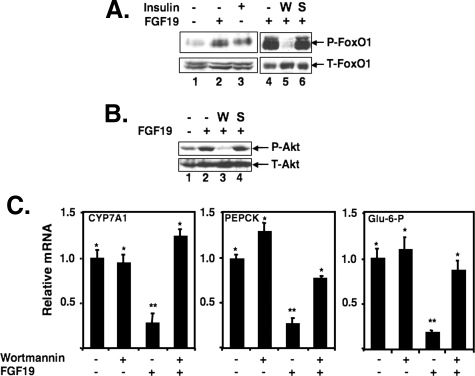
The Effects of FGF19 on FoxO1 Target Genes Require the PI 3-Kinase Pathway—To determine whether FGF19-dependent phosphorylation of FoxO1 and inhibition of target gene expression was mediated through the PI 3-kinase pathway, primary mouse hepatocytes were treated with wortmannin, a PI 3-kinase pathway inhibitor (58), or SP600125, a JNK pathway inhibitor (59), before the treatment with FGF19. Wortmannin almost completely blocked phosphorylation by FGF19, whereas SP600125 had no effect (Fig. 7A). Similar results were observed for Akt (Fig. 7B). Additionally, wortmannin also prevented the FGF19-dependent down-regulation of FoxO1-dependent target gene expression (Fig. 7C), providing further evidence that FGF19 inhibition of FoxO1 target gene expression requires signaling through the PI 3-kinase pathway.
FIGURE 7.
FGF19 induces phosphorylation of FoxO1 in primary mouse hepatocytes. A, primary mouse hepatocytes were treated with 80 ng/ml FGF19 or 30 nm insulin for 30 min. Where indicated, primary mouse hepatocytes were treated with 100 nm wortmannin (W) or 20 μm SP600125 (S) for 30 min before treatment with FGF19. Total cell lysates were analyzed by 8% SDS-PAGE and Western blotting with an antibody against total-FoxO1 (T-FoxO1) or phospho-FoxO1 (P-FoxO1). B, primary mouse hepatocytes were treated with 80 ng/ml FGF19 for 30 min. Total cell lysates were analyzed as in A using an antibody against total-Akt (T-Akt) or phospho-Akt (P-Akt). C, total RNA was isolated from primary mouse hepatocytes treated with either wortmannin (100 nm) or FGF19 (80 ng/ml) alone or both, as indicated. Wortmannin was added at 30 min before the treatment of FGF19 for 6 h. mRNA levels were analyzed by q-PCR as described in the legend to Fig. 4.
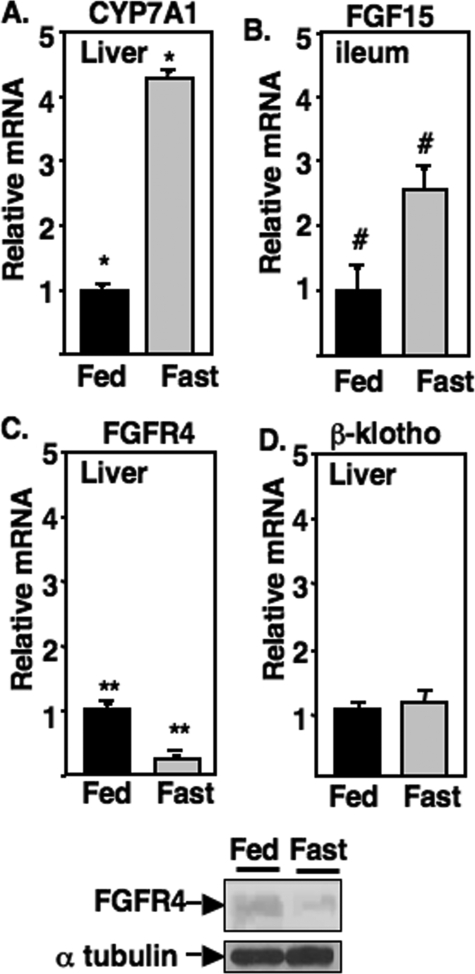
Fasting Reduces FGFR4 mRNA and Protein Levels in Mouse Liver—Decreased phosphorylation of FoxO1 through insulin-dependent PI 3-kinase signaling provides an explanation for the increase in CYP7A1 mRNA levels during fasting. Thus, we wondered whether the bile acid feedback regulation of hepatic CYP7A1 through intestine-derived FGF15 might also target FoxO1 through PI 3-kinase signaling. In fact, a recent report demonstrated that FGF signaling through PI 3-kinase regulates the related FoxG1 protein in the vertebrate forebrain (60). If the FGF19/15 signaling pathway plays a role in the regulation of CYP7A1 during fasting, then either intestinal FGF15 production or hepatic expression of FGFR4 should be decreased by fasting. As shown in Fig. 8, along with induction of CYP7A1, intestinal FGF15 mRNA was significantly induced (2.5-fold), whereas hepatic levels of FGFR4 mRNA and protein were significantly reduced by fasting, suggesting that reduced hepatic FGF15 signaling during fasting results from a decrease in hepatic FGFR4. There was no change in expression of the hepatic FGFR4 co-receptor β-klotho by fasting (Fig. 8D).
FIGURE 8.
Fasting reduces FGFR4 mRNA and protein levels in mouse liver. Total RNA was isolated from liver and ileum of mice that were fed ad libitum a normal chow diet (Fed) or fasted for 24 h (Fast). mRNA levels were analyzed by q-PCR described under “Experimental Procedures.” Results are expressed as -fold change relative to those of the normal, and the mean values obtained from triplicates of pooled RNA from six animals in each group are shown with error bars. At the bottom, FGFR4 protein expression in fed and fasted mice was analyzed by immunoblotting with α-tubulin as control. *, p = 0.000005; **, p = 0.00008; #, p = 0.0077.
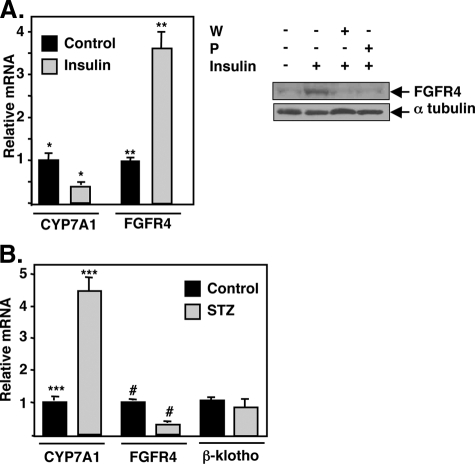
FGFR4 Is Regulated by Insulin—The decrease in FGFR4 expression by fasting suggests FGFR4 might be a direct target of insulin signaling. Consistent with this idea, hepatic FGFR4 mRNA and protein expression were induced by insulin treatment of primary hepatocytes, and two different PI 3-kinase inhibitors prevented the protein induction (Fig. 9A). Additionally, the mRNA was repressed in livers of mice that were made diabetic by injection with streptozotocin (STZ, Fig. 9B). Once again, expression of the co-receptor β-klotho was not altered by streptozotocin.
FIGURE 9.
Insulin regulates FGFR4 mRNA and protein expression. A, primary cultures of mouse hepatocytes were treated with 100 nm insulin, total RNA or protein was prepared as described under “Experimental Procedures,” and specific mRNAs were analyzed by q-PCR and normalized to ribosomal protein L32 mRNA. FGFR4 protein levels were measured by immunoblotting with total protein extracts from control, insulin-treated, or insulin-treated plus wortmannin (W) or PIK 90 (P) as described under “Experimental Procedures.” The same blot was stripped and re-probed with an antibody against α-tubulin. B, mice were injected with streptozotocin (STZ) to induce Type I diabetes as described under “Experimental Procedures.” Serum glucose levels were 141.50 ± 13.89 mg/dl for the control (N) and 433.25 ± 16.47 mg/dl for the streptozotocin-treated groups (p < 0.0001). Total RNA was prepared, and expression of CYP7A1, FGFR4, and β-klotho mRNAs were analyzed by q-PCR and normalized to ribosomal protein L32 mRNA. *, p = 0.00002; **, p = 0.0001; ***, p = 0.0003; #, p = 0.00009.
DISCUSSION
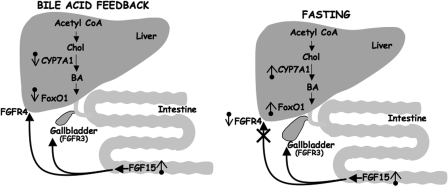
Our previous work demonstrated that CYP7A1 expression was induced by fasting (33), a condition known to result in elevated nuclear FoxO1. Inagaki et al. (20) reported that intestinal production of FGF15 plays a key role in bile acid feedback of hepatic CYP7A1, and when we (Figs. 1, 2, 3 and 6) and others (54, 61) determined that CYP7A1 was a direct target of FoxO1 activation, we hypothesized that FGF15 might regulate hepatic FoxO1 activity (Fig. 10, left). Consistent with this prediction, our current results show that hepatic FoxO1 is inhibited by FGF15 signaling through the PI 3-kinase pathway.
FIGURE 10.
Model for FGF signaling in intestinal/hepatic communication and metabolic regulation. Chol, cholesterol; BA, bile acids.
In addition to intestinal FGF15, hepatic expression of SHP is also important for bile acid feedback of CYP7A1 (20), and SHP was first shown to inhibit activation of CYP7A1 through interacting with the key CYP7A1 promoter factor liver receptor homologue 1 (8, 9). SHP also interacts with FoxO1 and inhibits its transcriptional activity as well (62). It is also noteworthy that efficient activation of the CYP7A1 promoter by FoxO1 also requires the liver receptor homologue 1 DNA binding site,3 suggesting the proteins work in concert to stimulate CYP7A1 expression. Taken together, the available evidence supports key roles for both SHP and FoxO1 in efficient repression of CYP7A1 by FGF15.
In Fig. 5, the fasting induction of CYP7A1 was still efficiently repressed in the l-Foxo1 mouse when FGF15 was overexpressed by adenovirus infection. The magnitude of the inhibition for PEPCK was reduced, suggesting FoxO1 is at least partly responsible for transmitting the FGF15 signal. However, when taken together with all of our other data, these results indicate that FGF15 likely inhibits CYP7A1 through more than one signaling pathway. This is not unexpected, as there are already documented redundant pathways for regulation of CYP7A1 (63). Additionally, FGF receptor signaling is known to feed into ERK (extracellular signal-regulated kinase)-, p38-, and JNK-signaling pathways (64) in addition to PI 3-kinase, and there is evidence for JNK-mediated inhibition of CYP7A1 (63).
FoxO1, liver receptor homologue 1, and hepatic nuclear factor 4 are all key activators of the CYP7A1 promoter, and all are targets for coactivation by PGC-1α. The PGC-1α gene promoter is also activated by FoxO1 (57), and the co-activator function of PGC-1α protein is inhibited directly by PI 3-kinase-dependent phosphorylation (65). Our results show FGF15 also inhibits the fasting-dependent increase in PGC-1α mRNA (Fig. 4). Thus, in addition to targeting the direct action of FoxO1, FGF15 and insulin both inhibit CYP7A1 expression through repressing PGC-1α, which would have pleiotropic effects on the CYP7A1 promoter.
Liver mRNA and protein for FGFR4, the isoform responsible for the hepatic effects of FGF15 (20), was repressed by fasting (Fig. 8), indicating that hepatic FGF15 signaling is attenuated during nutrient deprivation because of the decrease in FGFR4 (Fig. 10, right). Yu et al. (66) also showed that CYP7A1 mRNA levels are elevated in FGFR4 knock-out mice. Thus, there is an increase in CYP7A1 expression in two different in vivo situations where FGFR4 mRNA levels are reduced.
Our experiments reveal that FGFR4 is regulated by insulin, and because insulin and FGF both feed into the PI 3-kinase pathway (our results and Ref. 60), growth factor signaling through FGFR4 may play an important role in hepatic insulin action. In support of this idea, FGFR4 knock-out mice have impaired glucose tolerance (67), and transgenic expression of FGF19 reverses diabetes in the leptin-deficient ob/ob mouse (68).
Even though FGFR4 levels decline, the major reason that both CYP7A1 and PEPCK are induced during fasting is probably due to attenuated insulin signaling and the resulting increase in nuclear FoxO1. However, the decrease in FGFR4 signaling probably also plays a supportive role. Thus, it was interesting that when FGF15 was provided to mice through an adenovirus construct (Fig. 4), it was sufficient to activate PI 3-kinase signaling and inhibit the fasting-dependent activation of both CYP7A1 and PEPCK. This suggests that either the lower level of FGFR4 or another FGFR isoform is capable of responding to high levels of FGF15 supplied by the adenovirus vector. Additionally, recombinant FGF19 decreased FoxO1-dependent gene activation and glucose production in primary hepatocytes (Figs. 6 and 7). These observations suggest that exogenous delivery of FGF15/19 might be therapeutically beneficial to activate hepatic PI 3-kinase signaling and decrease hepatic glucose production under diabetic conditions where insulin signaling is compromised.
The expansion and contraction of the gall bladder to either fill with or secrete bile acids is regulated by FGF15 and cholecystokinin. During the early phase of a meal, cholecystokinin is secreted from the proximal intestine to enhance gall bladder contraction, delivering bile acids to the intestine to aid in digestion of fats and fat-soluble vitamins (69). As the bile acids transit the intestine, they increase production of FGF15 from the distal intestine to regulate bile acid production in the liver. Additionally, intestinal FGF15 stimulates gall bladder filling with bile, which is enhanced during fasting (70). Interestingly, the fasting-dependent increase in intestinal FGF15 mRNA in our experiments (Fig. 7) provides a possible mechanism for this key nutritional response (Fig. 9, right). The mouse gallbladder expresses only minor amounts of FGFR4 but relatively higher levels of FGFR3, which is also a target for FGF15 signaling and probably functions to relay the FGF15 signal for gallbladder filling (70). It is likely that induction of hepatic CYP7A1 at a time when gall bladder filling is stimulated is important to provide the correct balance of bile acids to mix with available cholesterol and phospholipids for optimal bile consistency.
Hepatic expression of FGF21, which is highly related to FGF15/19, is induced during fasting and activates fatty acid oxidation and ketogenesis in the liver through a peroxisome proliferator-activated receptor-α-dependent process (22, 23). However, evidence suggests that the hepatic effects of FGF21 may be indirect, as FGF21 does not efficiently signal through FGFR4, which as mentioned above is the major hepatic FGFR isoform (27). Our results showing that FGFR4 declines with fasting (Fig. 8) are consistent with this conclusion. It is likely that the hepatic effects of FGF21 are through FGF21 acting in adipocytes where FGFR1 is the predominant receptor isoform and potently responds to FGF21 (21, 27). One of the significant fasting-dependent adipocyte responses to FGF21 has been proposed to be an increase in lipolysis, resulting in elevated free fatty acids, which are absorbed by the liver where they can function as ligand agonists for hepatic peroxisome proliferator-activated receptor α. However, a recent report suggested the FGF21 decreases lipolysis (71), indicating there is still much to learn in this interesting area.
Taken together with these additional observations, our results provide evidence for an intriguing pattern of nutritional and tissue-specific regulation of FGFR isoforms 1, 3, and 4 that explains the specific metabolic specificity of FGF15/19 and -21. Differential expression of the FGFR co-receptors klotho and β-klotho could provide additional tissue specificity in signaling through these hormonal FGFs (26, 27, 29). However, changes in expression of β-klotho mRNA, the co-receptor responsible for FGFR4 signaling in the liver, were not observed in our studies (Figs. 8 and 9).
Our results also suggest some of the positive effects attributable to bile acids and FXR on hepatic insulin signaling are likely due to their activation of FGF15 expression in the intestine, which in turn stimulates hepatic PI 3-kinase activity through FGFR4. This conclusion is supported by recent studies that show FXR agonist treatment increased hepatic PI 3-kinase signaling and lowered glucose levels in wild type and a genetic model of diabetic mice (34, 35).
The critical FoxO1 binding site identified here is conserved in the human CYP7A1 promoter, which was also activated by FoxO1 (supplemental Fig. 4). Additionally, dietary manipulation of bile acids modulate the circulating levels of FGF19 in humans, indicating that the FGF signaling pathway plays a significant role in regulation of human bile acid metabolism (30). Thus, the mechanism described here is likely conserved in humans.
Finally, because FoxO1 activation is associated with cell cycle arrest and growth inhibition in other physiologic contexts (72), it is likely that the recently described stimulation of liver regeneration by bile acids (73) is also mediated at least in part through FGF15 inhibition of hepatic FoxO1 action.
Supplementary Material
Acknowledgments
We thank Dr. D. Accili for the FoxO1 floxed and α1-antitrypsin Cre recombinase mouse lines. We also thank Drs. D. Accili, J. Chiang, T. Franke, D. Fruman, G. Gil, D. Mangelsdorf, and T. Unterman for plasmids, R. Gerard (University of Texas Southwestern Medical Center) for the FGF15 adenovirus expression construct, and F. Sladek for hepatic nuclear factor 4 antibody. We are also grateful to Drs. G. Gil and D. Fruman for comments on the manuscript and T. Unterman and S. Kliewer for helpful suggestions and communicating unpublished data.
This work was supported, in whole or in part, by National Institutes of Health Grant DK71021 (to T. O.). This work was also supported by American Heart Association Scientist Development Grant 0730189N (to D.-J. S.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
Footnotes
The abbreviations used are: CYP7A1, cholesterol-7α hydroxylase; PEPCK, phosphoenolpyruvate carboxykinase; FGF, fibroblast growth factor; FGFR, FGF receptor; FoxO1, forkhead transcription factor 1; FXR, farnesoid X receptor; PGC-1, peroxisome proliferator-activated receptor-γ coactivator 1; SHP, small heterodimer partner; HA, hemagglutinin; JNK, c-Jun N-terminal kinase; PI, phosphatidylinositol; WT, wild type; GFP, green fluorescent protein; q-PCR, quantitative real time RT-PCR.
D. J. Shin, unpublished observations.
References
- 1.Chiang, J. Y. (2002) Endocr. Rev. 23 443–463 [DOI] [PubMed] [Google Scholar]
- 2.Russell, D. W. (2003) Annu. Rev. Biochem. 72 137–174 [DOI] [PubMed] [Google Scholar]
- 3.Jelinek, D. F., Andersson, S., Slaughter, C. A., and Russell, D. W. (1990) J. Biol. Chem. 265 8190–8197 [PMC free article] [PubMed] [Google Scholar]
- 4.Ness, G. C., Pendleton, L. C., and Zhao, Z. (1994) Biochim. Biophys. Acta 1214 229–233 [DOI] [PubMed] [Google Scholar]
- 5.Shin, D. J., Plateroti, M., Samarut, J., and Osborne, T. F. (2006) Nucleic Acids Res. 34 3853–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Twisk, J., Hoekman, M. F., Lehmann, E. M., Meijer, P., Mager, W. H., and Princen, H. M. (1995) Hepatology 21 501–510 [PubMed] [Google Scholar]
- 7.Makishima, M., Okamoto, A. Y., Repa, J. J., Tu, H., Learned, R. M., Luk, A., Hull, M. V., Lustig, K. D., Mangelsdorf, D. J., and Shan, B. (1999) Science 284 1362–1365 [DOI] [PubMed] [Google Scholar]
- 8.Goodwin, B., Jones, S. A., Price, R. R., Watson, M. A., McKee, D. D., Moore, L. B., Galardi, C., Wilson, J. G., Lewis, M. C., Roth, M. E., Maloney, P. R., Willson, T. M., and Kliewer, S. A. (2000) Mol. Cell 6 517–526 [DOI] [PubMed] [Google Scholar]
- 9.Lu, T. T., Makishima, M., Repa, J. J., Schoonjans, K., Kerr, T. A., Auwerx, J., and Mangelsdorf, D. J. (2000) Mol. Cell 6 507–515 [DOI] [PubMed] [Google Scholar]
- 10.Seol, W., Choi, H.-S., and Moore, D. D. (1996) Science 272 1336–1339 [DOI] [PubMed] [Google Scholar]
- 11.Fang, S., Miao, J., Xiang, L., Ponugoti, B., Treuter, E., and Kemper, J. K. (2007) Mol. Cell. Biol. 27 1407–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper, A. D., Chen, J., Botelho-Yetkinler, M. J., Cao, Y., Taniguchi, T., and Levy-Wilson, B. (1997) J. Biol. Chem. 272 3444–3452 [PubMed] [Google Scholar]
- 13.De Fabiani, E., Mitro, N., Anzulovich, A. C., Pinelli, A., Galli, G., and Crestani, M. (2001) J. Biol. Chem. 276 30708–30716 [DOI] [PubMed] [Google Scholar]
- 14.Lee, Y.-K., Dowhan, D. H., Hadzopoulou-Cladaras, M., and Moore, D. D. (2000) Mol. Cell. Biol. 20 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta, S., Stravitz, R. T., Dent, P., and Hylemon, P. B. (2001) J. Biol. Chem. 276 15816–15822 [DOI] [PubMed] [Google Scholar]
- 16.Han, S. I., Studer, E., Gupta, S., Fang, Y., Qiao, L., Li, W., Grant, S., Hylemon, P. B., and Dent, P. (2004) Hepatology 39 456–463 [DOI] [PubMed] [Google Scholar]
- 17.Qiao, D., Chen, W., Stratagoules, E. D., and Martinez, J. D. (2000) J. Biol. Chem. 275 15090–15098 [DOI] [PubMed] [Google Scholar]
- 18.Stravitz, R. T., Rao, Y. P., Vlahcevic, Z. R., Gurley, E. C., Jarvis, W. D., and Hylemon, P. B. (1996) Am. J. Physiol. 271 G293–G303 [DOI] [PubMed] [Google Scholar]
- 19.Holt, J. A., Luo, G., Billin, A. N., Bisi, J., McNeill, Y. Y., Kozarsky, K. F., Donahee, M., Y., W. D., Mansfield, T. A., Kliewer, S. A., Goodwin, B., and Jones, S. A. (2003) Genes Dev. 17 1581–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inagaki, T., Choi, M., Moschetta, A., Peng, L., Cummins, C. L., McDonald, J. G., Luo, G., Jones, S. A., Goodwin, B., Richardson, J. A., Gerard, R. D., Repa, J. J., Mangelsdorf, D. J., and Kliewer, S. A. (2005) Cell Metab. 2 217–225 [DOI] [PubMed] [Google Scholar]
- 21.Kharitonenkov, A., Shiyanova, T. L., Koester, A., Ford, A. M., Micanovic, R., Galbreath, E. J., Sandusky, G. E., Hammond, L. J., Moyers, J. S., Owens, R. A., Gromada, J., Brozinick, J. T., Hawkins, E. D., Wroblewski, V. J., Li, D. S., Mehrbod, F., Jaskunas, S. R., and Shanafelt, A. B. (2005) J. Clin. Investig. 115 1627–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inagaki, T., Dutchak, P., Zhao, G., Ding, X., Gautron, L., Parameswara, V., Li, Y., Goetz, R., Mohammadi, M., Esser, V., Elmquist, J. K., Gerard, R. D., Burgess, S. C., Hammer, R. E., Mangelsdorf, D. J., and Kliewer, S. A. (2007) Cell Metab. 5 415–425 [DOI] [PubMed] [Google Scholar]
- 23.Badman, M. K., Pissios, P., Kennedy, A. R., Koukos, G., Flier, J. S., and Maratos-Flier, E. (2007) Cell Metab. 5 426–437 [DOI] [PubMed] [Google Scholar]
- 24.Inagaki, T., Lin, V. Y., Goetz, R., Mohammadi, M., Mangelsdorf, D. J., and Kliewer, S. A. (2008) Cell Metab. 8 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White, K. E., Evans, W. E., O'Riordan, J. L. H., Speer, M. C., Econs, M. J., Lorenz-Depiereux, B., Grabowski, M., Meltinger, T., and Strom, T. M. (2000) Nat. Genet. 26 345–34811062477 [Google Scholar]
- 26.Goetz, R., Beenken, A., Ibrahimi, O. A., Kalinina, J., Olsen, S. K., Eliseenkova, A. V., Xu, C., Neubert, T. A., Zhang, F., Linhardt, R. J., Yu, X., White, K. E., Inagaki, T., Kliewer, S. A., Yamamoto, M., Kurosu, H., Ogawa, Y., Kuro-o, M., Lanske, B., Razzaque, M. S., and Mohammadi, M. (2007) Mol. Cell. Biol. 27 3417–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurosu, H., Choi, M., Ogawa, Y., Dickson, A. S., Goetz, R., Eliseenkova, A. V., Mohammadi, M., Rosenblatt, K. P., Kliewer, S. A., and Kuro, O. M. (2007) J. Biol. Chem. 282 26687–26695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurosu, H., Ogawa, Y., Miyoshi, M., Yamamoto, M., Nandi, A., Rosenblatt, K. P., Baum, M. G., Schiavi, S., Hu, M. C., Moe, O. W., and Kuro-o, M. (2006) J. Biol. Chem. 281 6120–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogawa, Y., Kurosu, H., Yamamoto, M., Nandi, A., Rosenblatt, K. P., Goetz, R., Eliseenkova, A. V., Mohammadi, M., and Kuro-o, M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7432–7437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundasen, T., Galman, C., Angelin, B., and Rudling, M. (2006) J. Intern. Med. 260 530–536 [DOI] [PubMed] [Google Scholar]
- 31.Houten, S. M., Watanabe, M., and Auwerx, J. (2006) EMBO J. 25 1419–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, F. Y., Lee, H., Hubbert, M. L., Edwards, P. A., and Zhang, Y. (2006) Trends Biochem. Sci. 31 572–580 [DOI] [PubMed] [Google Scholar]
- 33.Shin, D.-J., Campos, J. A., Gil, G., and Osborne, T. F. (2003) J. Biol. Chem. 278 50047–50052 [DOI] [PubMed] [Google Scholar]
- 34.Ma, K., Saha, P. K., Chan, L., and Moore, D. D. (2006) J. Clin. Investig. 116 1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, Y., Lee, F. Y., Barrera, G., Lee, H., Vales, C., Gonzalez, F. J., Willson, T. M., and Edwards, P. A. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 1006–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, D. P., Stroup, D., Marrapodi, M., Crestani, M., Galli, G., and Chiang, J. Y. (1996) J. Lipid Res. 37 1831–1841 [PubMed] [Google Scholar]
- 37.Accili, D., and Arden, K. C. (2004) Cell 117 421–426 [DOI] [PubMed] [Google Scholar]
- 38.Brunet, A., Bonni, A., Zigmond, M. J., Lin, M. Z., Juo, P., Hu, L. S., Anderson, M. J., Arden, K. C., Blenis, J., and Greenberg, M. E. (1999) Cell 96 857–868 [DOI] [PubMed] [Google Scholar]
- 39.Hadari, Y. R., Gotoh, N., Kouhara, H., Lax, I., and Schlessinger, J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 8578–8583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakae, J., Park, B. C., and Accili, D. (1999) J. Biol. Chem. 274 15982–15985 [DOI] [PubMed] [Google Scholar]
- 41.Ong, S. H., Hadari, Y. R., Gotoh, N., Guy, G. R., Schlessinger, J., and Lax, I. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 6074–6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rena, G., Woods, Y. L., Prescott, A. R., Peggie, M., Unterman, T. G., Williams, M. R., and Cohen, P. (2002) EMBO J. 21 2263–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barthel, A., Schmoll, D., and Unterman, T. G. (2005) Trends Endocrinol. Metab. 16 183–189 [DOI] [PubMed] [Google Scholar]
- 44.Brunet, A., Park, J., Tran, H., Hu, L. S., Hemmings, B. A., and Greenberg, M. E. (2001) Mol. Cell. Biol. 21 952–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woods, Y. L., Rena, G., Morrice, N., Barthel, A., Becker, W., Guo, S., Unterman, T. G., and Cohen, P. (2001) Biochem. J. 355 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altomonte, J., Richter, A., Harbaran, S., Suriawinata, J., Nakae, J., Thung, S. N., Meseck, M., Accili, D., and Dong, H. (2003) Am. J. Physiol. 285 E718–E728 [DOI] [PubMed] [Google Scholar]
- 47.Hall, R. K., Yamasaki, T., Kucera, T., Waltner-Law, M., O'Brien, R., and Granner, D. K. (2000) J. Biol. Chem. 275 30169–30175 [DOI] [PubMed] [Google Scholar]
- 48.Nakae, J., Kitamura, T., Silver, D. L., and Accili, D. (2001) J. Clin. Investig. 108 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamothe, B., Yamada, M., Schaeper, U., Birchmeier, W., Lax, I., and Schlessinger, J. (2004) Mol. Cell. Biol. 24 5657–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.del Castillo-Olivares, A., and Gil, G. (2000) Nucleic Acids Res. 28 3587–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsumoto, M., Pocai, A., Rossetti, L., Depinho, R. A., and Accili, D. (2007) Cell Metab. 6 208–216 [DOI] [PubMed] [Google Scholar]
- 52.LeCluyse, E. L., Bullock, P. L., Parkinson, A., and Hochman, J. H. (1996) Pharm. Biotechnol. 8 121–159 [DOI] [PubMed] [Google Scholar]
- 53.Zhang, W., Patil, S., Chauhan, B., Guo, S., Powell, D. R., Le, J., Klotsas, A., Matika, R., Xiao, X., Franks, R., Heidenreich, K. A., Sajan, M. P., Farese, R. V., Stolz, D. B., Tso, P., Koo, S. H., Montminy, M., and Unterman, T. G. (2006) J. Biol. Chem. 281 10105–10117 [DOI] [PubMed] [Google Scholar]
- 54.Li, T., Kong, X., Owsley, E., Ellis, E., Strom, S., and Chiang, J. Y. (2006) J. Biol. Chem. 281 28745–28754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Franke, T. F., Yang, S. I., Chan, T. O., Datta, K., Kazlauskas, A., Morrison, D. K., Kaplan, D. R., and Tsichlis, P. N. (1995) Cell 81 727–736 [DOI] [PubMed] [Google Scholar]
- 56.Guo, S., Rena, G., Cichy, S., He, X., Cohen, P., and Unterman, T. (1999) J. Biol. Chem. 274 17184–17192 [DOI] [PubMed] [Google Scholar]
- 57.Daitoku, H., Yamagata, K., Matsuzaki, H., Hatta, M., and Fukamizu, A. (2003) Diabetes 52 642–649 [DOI] [PubMed] [Google Scholar]
- 58.Arcaro, A., and Wymann, M. P. (1993) Biochem. J. 296 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bennett, B. L., Sasaki, D. T., Murray, B. W., O'Leary, E. C., Sakata, S. T., Xu, W., Leisten, J. C., Motiwala, A., Pierce, S., Satoh, Y., Bhagwat, S. S., Manning, A. M., and Anderson, D. W. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 13681–13686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Regad, T., Roth, M., Bredenkamp, N., Illing, N., and Papalopulu, N. (2007) Nat. Cell Biol. 9 531–540 [DOI] [PubMed] [Google Scholar]
- 61.Li, T., Ma, H., and Chiang, J. Y. (2008) J. Lipid Res. 49 1981–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamagata, K., Daitoku, H., Shimamoto, Y., Matsuzaki, H., Hirota, K., Ishida, J., and Fukamizu, A. (2004) J. Biol. Chem. 279 23158–23165 [DOI] [PubMed] [Google Scholar]
- 63.Wang, L., Lee, Y. K., Bundman, D., Han, Y., Thevananther, S., Kim, C. S., Chua, S. S., Wei, P., Heyman, R. A., Karin, M., and Moore, D. D. (2002) Dev. Cell 2 721–731 [DOI] [PubMed] [Google Scholar]
- 64.Eswarakumar, V. P., Lax, I., and Schlessinger, J. (2005) Cytokine Growth Factor Rev. 16 139–149 [DOI] [PubMed] [Google Scholar]
- 65.Li, X., Monks, B., Ge, Q., and Birnbaum, M. J. (2007) Nature 447 1012–1016 [DOI] [PubMed] [Google Scholar]
- 66.Yu, C., Wang, F., Kan, M., Jin, C., Jones, R. B., Weinstein, M., Deng, C. X., and McKeehan, W. L. (2000) J. Biol. Chem. 275 15482–15489 [DOI] [PubMed] [Google Scholar]
- 67.Huang, X., Yang, C., Luo, Y., Jin, C., Wang, F., and McKeehan, W. L. (2007) Diabetes 56 2501–2510 [DOI] [PubMed] [Google Scholar]
- 68.Fu, L., John, L. M., Adams, S. H., Yu, X. X., Tomlinson, E., Renz, M., Williams, P. M., Soriano, R., Corpuz, R., Moffat, B., Vandlen, R., Simmons, L., Foster, J., Stephan, J. P., Tsai, S. P., and Stewart, T. A. (2004) Endocrinology 145 2594–2603 [DOI] [PubMed] [Google Scholar]
- 69.Chandra, R., and Liddle, R. A. (2007) Curr. Opin. Endocrinol. Diabetes Obes. 14 63–67 [DOI] [PubMed] [Google Scholar]
- 70.Choi, M., Moschetta, A., Bookout, A. L., Peng, L., Umetani, M., Holmstrom, S. R., Suino-Powell, K., Xu, H. E., Richardson, J. A., Gerard, R. D., Mangelsdorf, D. J., and Kliewer, S. A. (2006) Nat. Med. 12 1253–1255 [DOI] [PubMed] [Google Scholar]
- 71.Arner, P., Pettersson, A., Mitchell, P. J., Dunbar, J. D., Kharitonenkov, A., and Ryden, M. (2008) FEBS Lett. 582 1725–1730 [DOI] [PubMed] [Google Scholar]
- 72.Arden, K. C. (2006) Exp. Gerontol. 41 709–717 [DOI] [PubMed] [Google Scholar]
- 73.Huang, W., Ma, K., Zhang, J., Qatanani, M., Cuvillier, J., Liu, J., Dong, B., Huang, X., and Moore, D. D. (2006) Science 312 233–236 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.