Abstract
In this study, we sought to investigate the mechanism by which heterogeneous nuclear ribonucleoprotein (hnRNP) H and F regulate proteolipid protein (PLP)/DM20 alternative splicing. G-rich sequences in exon 3B, G1 and M2, are required for hnRNPH- and F-mediated regulation of the PLP/DM20 ratio and, when placed between competing 5′ splice sites in an α-globin minigene, direct hnRNPH/F-regulated alternative splicing. In contrast, the activity of the intronic splicing enhancer, which is necessary for PLP splicing, is only modestly reduced by removal of hnRNPH/F both in PLP and α-globin gene context. In vivo, hnRNPH reversed reduction of DM20 splicing induced by hnRNPH/F removal, whereas hnRNPF had little effect. Tethering of the MS2-hnRNPH fusion protein downstream of the DM20 5′ splice site increased DM20 splicing, whereas MS2-hnRNPF did not. Binding of U1 small nuclear ribonucleoprotein (U1snRNP) to DM20 is greatly impaired by mutation of G1 and M2 and depletion of hnRNPH and F. Reconstitution of hnRNPH/F-depleted extracts with either hnRNPH or F restored U1snRNP binding. We conclude that hnRNPH and F regulate DM20 splicing by recruiting U1snRNP and that hnRNPH plays a primary role in DM20 splice site selection in vivo. Decreased expression of hnRNPH/F in differentiated oligodendrocytes may regulate the PLP/DM20 ratio by reducing DM20 5′ splice site recognition by U1snRNP.
Alternative splicing of a single transcript is widely utilized to generate proteomic diversity in response to developmental, cell specific, and external signals (1). Typically, alternatively spliced sites are weak and the final splicing selection depends on the interplay of enhancers and silencers and the relative abundance and/or affinity of the RNA binding factors (2, 3). Early recognition of the 5′ splice sites is mediated by the U1snRNP2 through direct base pairing of the single-stranded 5′-end of U1snRNA with six conserved nucleotides at the 5′ splice site. Binding of U1snRNP and base pairing of the U1snRNA to the template is required for spliceosome assembly (4, 5).
A number of splicing factors that bind to either enhancers or silencers have been identified and shown to influence the efficiency of splice site recognition and spliceosome assembly (6). The hnRNPs are a large family of ubiquitously expressed RNA binding factors, which, in addition to regulating constitutive splicing, play an important role in alternative splicing (7, 8). hnRNPH and F are highly homologous proteins that bind to G-rich sequences present in exons, in introns, and in close proximity to the polyadenylation site (8–12). Depending on the gene context, these splicing factors have different affinity for their cognate sequences and appear to act in concert to regulate splicing of a number of genes (13, 14). Although they are most often inhibitors of alternatively spliced exons, they can also function as enhancers (14–18). G-rich sequences are highly conserved within introns in close proximity to splice sites (7, 10, 19–21). In genes containing short introns, these G-rich sequences allow splice site recognition through an intron-definition mechanism (22, 23). In an artificial α-globin gene construct containing duplicated 5′ splice sites flanked by identical G triplets, the G-rich sequences favor selection of the distal 5′ splice site through binding of U1snRNA to the G-rich sequence by direct base pairing (24).
The proteolipid protein (PLP) gene gives rise to two isoforms, PLP and DM20, through alternative splicing of competing 5′ splice sites resulting in either inclusion or exclusion of exon 3B. G-rich enhancers, named M2 and ISE, flank the DM20 and PLP 5′ splice sites, respectively (25–27). In addition, there are other G-rich sequences in exon 3B, and all are potential hnRNPH/F binding motifs (27, 28). The PLP/DM20 ratio increases in postnatal brain development and in differentiated oligodendrocytes, and this is temporally related to reduced expression of hnRNPH and F (27, 29). The DM20 isoform is preferentially expressed in embryonic brain and non-glial cells (30). Knockdown of hnRNPH and F synergistically increased the PLP/DM20 ratio, and this effect was mediated by M2 in concert with some or all of the G runs present in PLP exon 3B, whereas the ISE had little contribution to this regulation (27). hnRNPH and F have been shown to serve redundant functions in regulating alternative splicing and to cooperatively regulate other genes (17, 31, 32); however, to the best of our knowledge, a synergistic effect such as that observed with PLP alternative splicing was not previously reported.
In this study, we sought to investigate further the mechanism by which hnRNPH and F regulate the PLP/DM20 ratio. A proximal G-rich sequence (G1) and M2 are required for hnRNPH- and F-mediated regulation of the PLP/DM20 ratio. In contrast, the ISE enhancer's activity is only modestly reduced by removal of hnRNPH/F. In vivo, hnRNPH and F are not functionally redundant, and hnRNPH plays a primary role in regulating the PLP/DM20 ratio. In vitro, hnRNPH and hnRNPF were able to restore binding of U1snRNP, which was greatly reduced by removal of hnRNPH/F. The data show that both hnRNPH and F regulate DM20 splicing by recruiting U1snRNP and suggest that hnRNPH may have additional functions in regulating the splicing machinery.
EXPERIMENTAL PROCEDURES
Plasmids—The pcDNA-MS2-H and pcDNA-MS2-F plasmids expressing MS2 coat protein fused in-frame to Myc-hisA-tagged hnRNPH and F, respectively, were a generous gift of Dr. Mark McNally (33). The pFLAG-hnRNPH and pFLAG-hnRNPF plasmids were a generous gift of Dr. Mariano Garcia-Blanco (32). These plasmids were used in transfections into Oli-neu cells treated with siF/H, which targets a sequence common to mouse hnRNPH and F. The human hnRNPF has a two-nucleotide mismatch with the mouse sequence targeted by the siF/H, thus it is resistant to siRNAs-mediated degradation. The siF/H-targeted sequence is identical between the human and mouse hnRNPH transcripts, and the FLAG-hnRNPH transcript is targeted by siF/H and degraded (data not shown). To make FLAG-hnRNPH resistant to siF/H degradation, we changed two nucleotides (supplemental Table S1). The SRp40 was a kind gift of Dr. Stefan Stamm. The wild-type α-globin splicing construct was a kind gift of Dr. Andrew McCullough (23). Mutant PLP-neo (Figs. 1, 2A, and 5) and α-globin constructs (Figs. 2B and 3A) were generated by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). In the PLP-neo, the G runs were replaced by polyTs, as previously described (27). In the α-globin construct, the G runs were mutated as previously described (23) or replaced by G1M2 or ISE sequences from the PLP exon 3B as indicated in Fig. 3. The distance of the ISE and G1M2 sequences relative to the α-globin 5′ splice sites was the same as that in their endogenous position in the PLP gene. The SαGLO-ISE was generated by deleting the duplicated sequence containing the G1M2 in the α-glo-G1M2-ISE, thus leaving a single α-globin 5′ splice site and the downstream intron containing ISE (Fig. 2B). The SαGLO-ISEMT1, -MT2, and -MT3 were generated by site-directed mutagenesis of the SαGLO-ISE (Fig. 2B). The PLPneo-MS1 and PLPneo-NR were made by replacing twenty-two nucleotides spanning G1, M2, and the intervening five nucleotides with twenty-two nucleotides containing the MS2 protein binding motif or an unrelated sequence (Fig. 5). The PLP-neo-MS2 (data not shown) and -MS3 were made by replacing twenty-two nucleotides spanning M4–M6 and M3–M5 with the MS2 protein binding motif, respectively, in the PLP-neo-MS1 (Fig. 5). The Bcl-x minigene construct was a kind gift of Dr. Charles Chalfant (34).
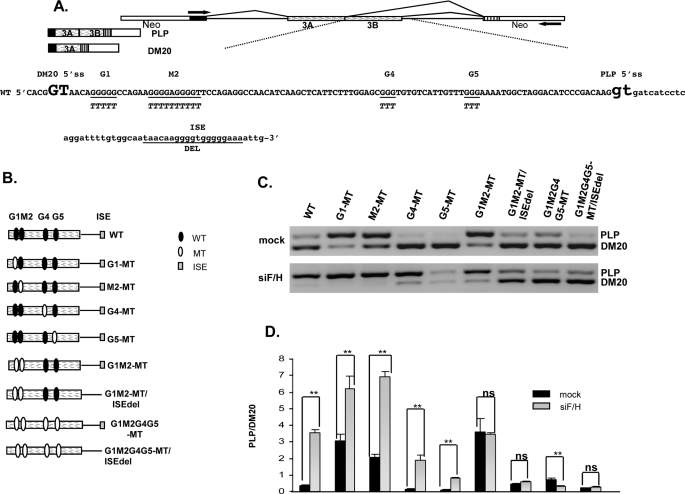
FIGURE 1.
G1 and M2 are required for hnRNPH- and F-mediated regulation of PLP/DM20 ratio. A, the PLP-neo splicing construct is shown. The arrows indicate the position of the PCR primers. The PLP and DM20 PCR products are shown. Partial sequences of PLP exon 3B (uppercase) and intron 3 (lowercase) in PLP-neo (WT) are shown, DM20 and PLP 5′ splice site are enlarged and in bold, G1, M2, G4, G5, and ISE are underlined, and the mutations are shown below the WT sequences. B, schematic representation of exon 3B and proximal intron 3 in all constructs is shown. C, representative RT-PCR analysis of WT-, G1-MT-, M2-MT-, G4-MT-, G5-MT-, G1M2-MT-, G1M2-MT/ISEdel-, G1M2G4G5-MT-, and G1M2G4G5-MT/ISEdel-derived PLP and DM20 products amplified from RNA isolated from Oli-neu cells treated with siF/H (n = 4) (30 PCR cycles). Mock are cells transfected with the same plasmids and treated with scrambled siRNA (30 PCR cycles). D, the bar graph shows the PLP/DM20 ratios ± S.E. (n = 4). *, p < 0.05 and **, p < 0.01, ns = non significant.
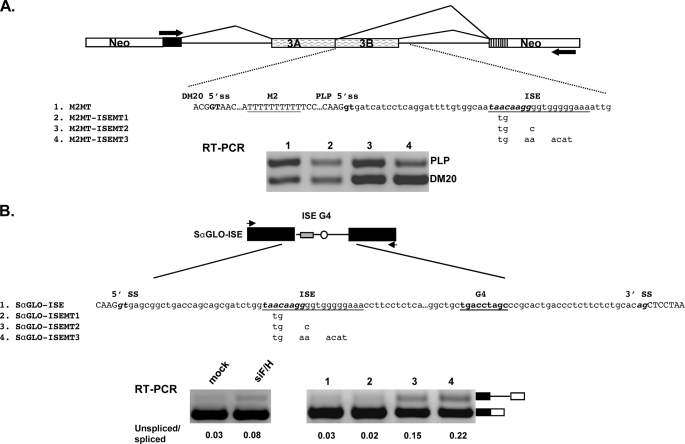
FIGURE 2.
Analysis of the ISE function in PLP and α-globin gene. A, the PLP-neo splicing construct is shown. The arrows indicate the position of the PCR primers. Partial sequences of PLP exon 3B (uppercase) and intron 3 (lowercase) in the PLP-neo are shown, DM20 and PLP 5′ splice sites are in bold, M2MT and ISE are underlined, and the putative SRp40 motif in the ISE is italicized and in bold. The mutations are shown below the ISE sequences. Representative RT-PCR analysis of M2MT-, M2MT-ISEMT1-, M2MT-ISEMT2-, and M2MT-ISEMT3-derived PLP and DM20 products amplified from RNA isolated from Oli-neu cells (n = 3) (30 PCR cycles). B, schematic of SαGLO-ISE construct is shown. The arrowheads indicate the position of primers used for PCR amplification. Partial sequences of the α-globin construct spanning the 5′ splice site, the intron, and the 3′ splice site. The ISE is underlined, and the putative SRp40 motif in the ISE is italicized and in bold. The nucleotides that replace the endogenous α-globin G4 sequence are shown in bold. The mutations are shown below the ISE. Representative RT-PCR analysis of SαGLO-ISE derived spliced and unspliced products amplified from RNA isolated from Oli-neu cells either treated with control siRNA (mock) or with siF/H (n = 2) (30 PCR cycles). Representative RT-PCR analysis of SαGLO-ISE-, SαGLO-ISEMT1-, SαGLO-ISEMT2-, and SαGLO-ISEMT3-derived spliced and unspliced products (n = 2). The ratio of unspliced/spliced products is shown.
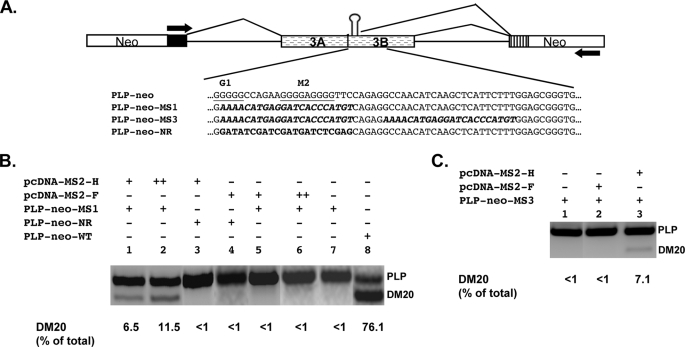
FIGURE 5.
hnRNPH directly promotes DM20 splicing in vivo. A, schematic of PLP-neo-WT and constructs in which sequences spanning G1 and M2 have been replaced with MS2 coat protein binding motif (PLP-neo-MS1) or an unrelated sequence (PLP-neo-NR). PLP-neo-MS3 was derived from PLP-neo-MS1 by replacing sequences spanning M3-M5 with the MS2 motif (see “Experimental Procedures”). The sequences are shown. The stem-loop represents the binding site for the MS2 coat protein. B, RT-PCR analysis of PLP and DM20 products derived from the minigenes. Oli-neu cells were transfected with 0.5 μg(+) and 1.0 μg(++) of the indicated plasmids and RNA was subjected to RT-PCR (30 PCR cycles). A representative experiment is shown (n = 2). The data are expressed as percent of the DM20 product versus the total PLP+DM20 product. C, RT-PCR analysis of PLP and DM20 products derived from PLP-neo-MS3. Oli-neu cells were transfected with 0.5 μg(+) of the indicated plasmids, and RNA was subjected to RT-PCR (30 PCR cycles). A representative experiment is shown (n = 2). The data are expressed as percent of the DM20 product versus the total PLP+DM20 product.
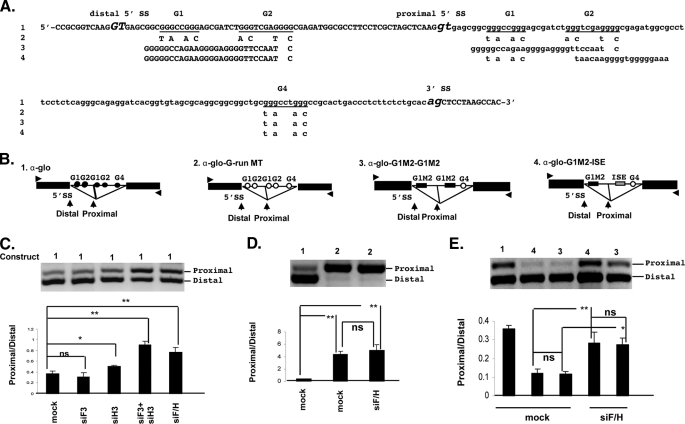
FIGURE 3.
hnRNPH and F regulate the proximal/distal 5′ splice site ratio of α-globin. A, partial sequences of the α-globin construct spanning the duplicated 5′ splice sites, the intron, and the 3′ acceptor splice site. The sequences introduced to generate the mutated constructs are shown below the wild-type sequences. B, schematic of all α-globin constructs is shown. The filled circles represent wild-type α-globin sequences and the open circles represent the mutated sequences. The arrowheads indicate the position of primers used for PCR amplification. C, representative RT-PCR analysis of proximal (P) and distal (D) α-globin products amplified from RNA isolated from Oli-neu cells treated with siH3, siF3, siH3+siF3, and siF/H (30 PCR cycles). Mock are cells treated with scrambled siRNA. The bar graph shows the P/D ratios ± S.E. (n = 4). *, p < 0.05 and **, p < 0.01. D, representative RT-PCR analysis of proximal (P) and distal (D) products derived from all G run mutated α-globin construct amplified from RNA isolated from Oli-neu cells treated with siF/H (30 PCR cycles). Mock are cells treated with control siRNA. The bar graph shows the P/D ratios ± S.E. (n = 4). **, p < 0.01. E, representative RT-PCR analysis of proximal (P) and distal (D) α-globin products derived from G1M2-ISE and G1M2-G1M2 α-globin constructs amplified from RNA isolated from Oli-neu cells treated with siF/H (30 PCR cycles). Mock are cells treated with scrambled siRNA. The wild-type α-globin is used as control. The bar graph shows the P/D ratios ± S.E. (n = 4). *, p < 0.05; **, p < 0.01; ns = non significant.
siRNAs, Cell Cultures, and Transfections—Pre-designed double-stranded siRNAs targeting hnRNPF (ID# 175722 (siF3)), hnRNPH (ID# 75775 (siH3)), the custom-made double-stranded siRNA, siF/H (27), and Silencer® Negative Control #1 siRNA were purchased from Ambion (Austin, TX). Oli-neu cells were cultured in SATO medium (Dulbecco's modified Eagle's medium containing 10 ng/ml biotin, 10 ng/ml apotransferrin, 100 μm putrescine, 2 μm progesterone, 2 μm sodium selenite, 2.5 μg/ml insulin) with 1% horse serum (27). Fibroblast L cells and the neuronal Neuro2A cells (ATCC) were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Cells were co-transfected with plasmid DNAs (0.5 μg, unless otherwise indicated) and siRNAs (80 nm) using the siPORT amine transfection reagent according to the Neofection protocol (Ambion) (27). Western blot analysis showed that expression of hnRNPF was not affected by treatment with siH3, and expression of hnRNPH was not affected by treatment with siF3, thus excluding compensation effects (data not shown). In overexpression studies, 0.5 μg of DNA of the splicing construct was co-transfected with 0.5, 1, and 1.5 μg of the expression plasmid DNA or 0.5 μg of the empty vector pcDNA3 using the siPORT amine transfection reagent.
RNA Extraction and RT-PCR—Total RNA was extracted from cultured cells using the RNeasy Mini Kit (Qiagen, Valencia, CA) and was treated with the DNA-free Kit (Ambion) according to the manufacturer's instructions. Reverse transcription was performed with 1 μg of total RNA using random hexamer primer mixture according to the manufacturer's instructions (BD Biosciences). The PCR products derived from the wild-type and mutated PLP-neo constructs were amplified by RT-PCR using a primer set previously described (Fig. 1) (27, 35). The proximal and distal splice products derived from the wild-type and mutated α-globin constructs were amplified by RT-PCR with a forward primer (5′-TGGTACCGAGCTCGGATCCGATGT-3′) and a reverse primer (5′-GATGGATATCTGCAGAATTCGGGA-3′) complementary to sequences in pcDNA3 vector (Figs. 2B and 3). The XL and XS products derived from the Bcl-x minigene construct were amplified by RT-PCR with primers described previously (36).
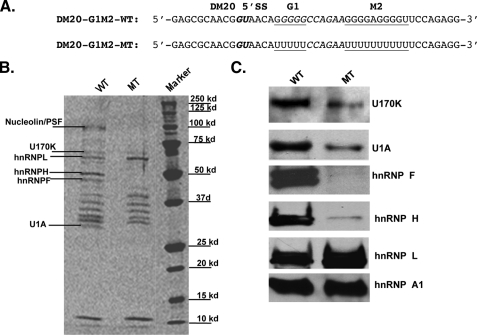
Nuclear Extracts, Recombinant Proteins, and RNA Affinity Precipitations—Nuclear extracts were prepared from Oli-neu differentiated in SATO medium containing 1 mm Bt2cAMP for 7 days or from Oli-neu treated with siF/H for 72 h in SATO medium without Bt2cAMP using the NEP kit (Pierce) according to the manufacturer's instructions (27). Poly(G) affinity column (Sigma)-mediated removal of hnRNPH and F from nuclear extracts was performed as described (37) Recombinant His-tagged hnRNPF and H proteins were generated in Escherichia coli and purified as described before (16). The wild-type RNA template spans 10 nucleotides upstream and 35 nucleotides downstream of the DM20 5′ splice site and contains the DM20 5′ splice site and the G-rich enhancers, G1 and M2 (DM20-G1M2-WT, Fig. 6). The mutated RNA template is identical to the wild-type except that G1 and M2 sequences were replaced by polyUs (DM20-G1M2-MT, Fig. 6). RNA templates were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). RNA affinity precipitations were performed with biotinylated RNA templates, as previously described (27) except that the complexes were not UV-cross-linked.
FIGURE 6.
Mutations of G1 and M2 interfere with binding of U1 70K and U1A in RNA affinity precipitates. A, RNA templates used in RNA affinity precipitations. The DM20 5′ splice site is in bold italics, and the natural G1 and M2 and the mutated poly-U sequences are underlined. B, representative sypro-ruby stained gel of RNA affinity precipitates with biotinylated RNA templates containing wild-type (WT) or poly-U mutated G1M2 (MT) and Oli-neu extracts (n = 3) (“Experimental Procedures”). M = protein markers. We have analyzed by liquid chromatography-MS/MS bands that are absent and one band that is more prominent in precipitates with the G1M2 mutated template. The protein's identity is shown. C, Western blot analysis of U1 70K, U1A, hnRNPA1, F, H, and L in the RNA affinity precipitates with Oli-neu extracts and either WT or MT template. The data were repeated in three separate experiments.
Protein Isolation and Identification by Liquid Chromatography-MS/MS and Western Blot Analysis—Proteins were eluted from the streptavidin beads in 100 μl of SDS-PAGE loading buffer at 95 °C for 5 min, and one-third of the mixtures were separated by 10% SDS-PAGE and either visualized by syproruby staining (Invitrogen) or blotted to nitrocellulose membrane. Selected bands were excised from the gel and analyzed by matrix-assisted laser desorption ionization-MS/MS and liquid chromatography-MS/MS (27) (Proteomics and Mass Spectrometry Core Facility, University of Kentucky). Nitrocellulose membranes were reacted with antibodies to: U1 70K (Aviva Systems Biology, San Diego, CA), U1A (Aviva Systems Biology), hnRNPH (Bethyl Laboratories) and hnRNPHF (rabbit polyclonal, generous gift of Dr. Douglas Black), A1 and L (Abcam), SRp40 (Santa Cruz Biotechnology), Myc tag (Invitrogen), and FLAG tag (Sigma) diluted 1:2000, horseradish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch Laboratories) diluted 1:2000, and developed with enhanced chemiluminescence (ECL, Amersham Biosciences). Blots were visualized with a Kodak 440CF Digital Image Station using one-dimensional analysis software.
RESULTS
The Synergistic Effect of hnRNPH/F Is Mediated by G1 and M2—We have shown previously that siRNA-mediated knock-down of hnRNPH and F synergistically increased the PLP/DM20 ratio in oligodendrocytes, and this effect was in large part dependent on M2 and G runs in exon 3B, named G1, G4, and G5.
To determine the G runs that are necessary for the hnRNPH/F synergism in addition to M2, we tested the impact of individual mutations of G1, G4, and G5 on the hnRNPH/F-mediated increase in PLP/DM20 ratio (Fig. 1, A and B). Oli-neu cells were transfected with the G1-MT, G4-MT, and G5-MT constructs and the plasmid-derived PLP and DM20 products were amplified by RT-PCR in RNA isolated from cells cultured for 72 h in growth medium. Although the PLP/DM20 ratio derived from G4-MT and G5-MT was 0.15 ± 0.02 and 0.1 ± 0.03, respectively, compared with 0.38 ± 0.02 with the WT construct, the PLP/DM20 ratio derived from G1-MT was 3.06 ± 0.4 (Fig. 1, C (upper panel) and D). The data suggest that G1 is an enhancer of the DM20 5′ splice site, whereas G4 and G5 appear to enhance PLP 5′ splice site selection, in keeping with the results obtained previously with M6-MT and M8-MT, mutations that encompass G4 and G5, respectively (27). G1-MT increased the PLP/DM20 ratio by 8-fold compared with ∼5-fold induced by M2-MT (3.06 ± 0.40 versus 2.05 ± 0.20) (Fig. 1, C (upper panel) and D (27)), suggesting that G1 is a stronger enhancer of DM20 5′ splice site than M2. For mutations of G1 and M2, G1M2-MT increased the PLP/DM20 ratio to 3.61 ± 0.8 (Fig. 1, C (upper panel) and D). Mutations of G1 and M2 combined with the ISE deletion, G1M2-MT/ISEdel, resulted in a PLP/DM20 ratio of 0.44 ± 0.06 (Fig. 1, C (upper panel) and D). This is higher than the wild-type ratio, but lower than the G1M2 mutation alone and reflects the loss of both the PLP 5′ splice site-enhancing ISE and the DM20 5′ splice site-enhancing G1 and M2 sequences. Likewise, the PLP/DM20 ratio derived from constructs in which G1M2, G4, and G5 are all mutated and the ISE is present, is 0.7 ± 0.1 and is higher than that derived from M2G1G4G5-MT/ISEdel (0.21 ± 0.01 (27)), reflecting the loss of the PLP-enhancing function of G4, G5, and the ISE (see next section) (Fig. 1, C (upper panel) and D). The data indicate that, in addition to M2, G1 is an enhancer of DM20 5′ splice site selection.
We next assessed whether siRNA-mediated knock-down of hnRNPH and F (siH/F) affected the PLP/DM20 ratio derived from G1-MT, G4-MT, G5-MT, and G1M2-MT in Oli-neu cells (Fig. 1C, lower panel). In keeping with published data, knockdown of hnRNPH and F caused a >9-fold increase in the PLP/DM20 ratio derived from the WT construct versus mock treated cells (3.57 ± 0.18 versus 0.38 ± 0.02). Knockdown of hnRNPH and F caused a >12-fold increase in PLP/DM20 ratio derived from G4-MT (1.9 ± 0.28 versus 0.15 ± 0.02) and an 8-fold increase from G5-MT (0.82 ± 0.05 versus 0.1 ± 0.03) compared with mock treated cells, indicating that these sequences are not required for hnRNPH/F-mediated regulation of PLP/DM20 ratio (Fig. 1, C (lower panel) and D). In contrast, knockdown of hnRNPH/F increased the PLP/DM20 ratio derived from G1-MT only by ∼2-fold compared with mock treated cells (6.18 ± 0.8 versus 3.06 ± 0.4) (Fig. 1, C (lower panel) and D). The PLP/DM20 ratio derived from M2-MT was increased >3-fold by knockdown of hnRNPH/F (6.91 ± 0.32 versus 2.05 ± 0.2) (Fig. 1, C (lower panel) and D) (27). The increase in the PLP/DM20 ratio derived from G1-MT and M2-MT is statistically significant. In contrast, removal of hnRNPH/F did not increase the PLP/DM20 ratio derived from G1M2-MT compared with mock treated cells (3.45 ± 0.12 versus 3.61 ± 0.8) and from G1M2MT/ISEdel compared with mock treated (0.58 ± 0.05 versus 0.44 ± 0.06) (Fig. 1, C (lower panel) and D). These data show that G1 and M2 are the primary cis-elements that mediate the hnRNPH/F synergistic regulation of the PLP/DM20 ratio.
The ISE Is Partially Regulated by hnRNPH and F—The presence of the ISE did not affect the hnRNPH/F-mediated regulation of the PLP/DM20 ratio in any of the mutants tested above, however, the PLP/DM20 ratio derived from M2G1G4G5-MT, in which all G runs are mutated and the ISE is present, was 2.3-fold lower in cells treated with siF/H compared with mock treated cells (0.3 versus 0.7) (Fig. 1, C and D). These data suggest that hnRNPH/F partially regulate the selection of PLP 5′ splice site through the ISE. Because removal of hnRNPH/F has a modest effect on the function of the ISE, we reasoned that other sequences, and consequently splicing factors, may contribute to the function of the ISE. The 5′ sequences adjacent to the G runs contain a putative SRp40 binding motif identified by ESEfinder (38, 39), thus we tested whether this motif is functionally important for the regulation of the PLP/DM20 ratio. We have introduced mutations that disrupt the SRp40 motif (Fig. 2A) either alone (ISEMT1, construct 2) or in combination with disruption of the first G run (ISEMT2, construct 3) or both G runs (ISEMT3, construct 4) in PLP-neo and in M2MT (construct 1). Because the PLP/DM20 ratio derived from M2MT is higher than that derived from PLP-neo, reduction in the PLP product is more easily quantified with this plasmid, and here we show these data. Oli-neu cells were transfected with the M2MT constructs, and the plasmid-derived PLP and DM20 products were amplified by RT-PCR in RNA isolated from cells cultured for 72 h in growth medium. We found that the PLP/DM20 ratio derived from ISEMT1 is ∼2-fold lower (1.04 ± 0.06 versus 1.84 ± 0.15), from ISEMT2 is ∼4-fold lower (0.56 ± 0.08 versus 1.84 ± 0.15), and from ISEMT3 is 6-fold lower (0.30 ± 0.06 versus 1.84 ± 0.15) than the PLP/DM20 ratio derived from the M2MT. Similar results were obtained with the PLP-neo mutated constructs (data not shown). These data suggest that the 5′ sequences of the ISE contribute to its enhancer's activity possibly through SRp40. To determine whether SRp40 regulates the PLP/DM20 ratio through the ISE motif, we have overexpressed SRp40 in Oli-neu transfected with PLP-neo and PLP-neo-ISE-MT2. The SRp40 increased the PLP/DM20 ratio derived from both constructs (supplemental Fig. S1), suggesting that the activity of the enhancer is not mediated by SRp40.
Next, we sought to investigate the function of the ISE in the selection of a 5′ splice site in the α-globin heterologous gene context. We made constructs in which the endogenous G1 and G2 triplets downstream of the α-globin 5′ splice site (23) were replaced by ISE (Fig. 2B, construct 1, SαGLO-ISE), ISEMT1 (Fig. 2B, construct 2, SαGLO-ISEMT1), ISEMT2 (Fig. 2B, construct 3, SαGLO-ISEMT3), and ISEMT3 (Fig. 2B, construct 4, SαGLO-ISEMT3). The plasmids were transfected into Oli-neu cells, and the α-globin spliced and unspliced products were amplified by RT-PCR in RNA isolated from cells cultured for 72 h in growth medium. The data are expressed as the ratio of unspliced versus spliced product (Fig. 2B). The α-globin was fully spliced with the SαGLO-ISE similarly to the endogenous G triplets (23), and a small increase in the unspliced product was induced by removal of hnRNPH/F (Fig. 2B), in keeping with results obtained in the PLP gene context. The amount of unspliced product was increased 5-fold with SαGLO-ISEMT2 and 7-fold with SαGLO-ISEMT3, whereas no increase was detected with SαGLO-ISEMT1 (Fig. 2B). The data indicate that the G runs of the ISE regulate 5′ splice site selection in the α-globin gene context. Mutations in these sequences have a greater effect on splicing efficiency than that induced by removal of hnRNPH/F, suggesting that other factors binding to these sequences may have a regulatory function. The sequences 5′ of the G runs are not sufficient to regulate the α-globin 5′ splice site. This finding is in contrast with the results obtained in the PLP gene context, suggesting that either these sequence are a weak enhancer or that they are part of a regulatory element that extends upstream in PLP intron 3 and is therefore absent in the α-globin construct. The latter interpretation is in keeping with the data showing that the SRp40 motif is not responsible for the function of the enhancer of the 5′ sequences (supplemental Fig. S1). Together, the data indicate that the function of the ISE is only partially regulated by hnRNPH/F in the PLP and α-globin context and is likely to be regulated by additional splicing factors that remain to be identified.
hnRNPH and F Cooperatively Regulate Selection of Competing 5′ Splice Sites in an α-Globin Minigene—We next asked whether the hnRNPH and F cooperation represents a more general mechanism that regulates competing 5′ splice sites flanked by G-rich sequences. To this end, we have used an α-globin construct with duplicated 5′ splice sites flanked by identical G runs (Fig. 3, A and B, construct 1), which closely resembles the configuration of PLP and DM20 5′ splice sites. The distal 5′ splice site is favored in HeLa cells (23). We have examined whether knockdown of hnRNPH, F, and H/F changes the ratio of spliced products derived from the α-globin construct. We determined the ratio of PCR products derived from proximal and distal 5′ splice sites in Oli-neu cells transfected with α-globin and treated with siH3, siF3, siF3+siH3, and siF/H. In cells treated with siH3, the ratio of proximal to distal product increased by 0.5-fold versus mock treated cells (0.51 ± 0.03 versus 0.37 ± 0.05), whereas in cells treated with either siH3+siF3 (0.91 ± 0.07) or siF/H (0.77 ± 0.09), the ratio increased 2-fold versus mock treated cells (Fig. 3C). In contrast, removal of hnRNPF did not significantly alter the ratio (0.31 ± 0.08 versus 0.37 ± 0.05) (Fig. 3C). These data show that selection of competing 5′ splice sites flanked by G-rich enhancers in a gene other than PLP is cooperatively regulated by hnRNPH and F, although not as efficiently as in the PLP gene. To demonstrate that the G runs are required for the hnRNPH/F regulation of α-globin gene splicing, we have mutated distal and proximal G runs by replacing 10 nucleotides spanning the G runs, as previously described (Fig. 3B, construct 2, and “Experimental Procedures”) (23). Mutation of all G runs led to a dramatic increase in the ratio of proximal to distal product 4.34 ± 0.55 versus 0.37 ± 0.02 for the α-globin (Fig. 3D) and abolished the regulation mediated by knockdown of hnRNPH/F (5.05 ± 0.89 versus 4.34 ± 0.55) (Fig. 3D). These data indicate that hnRNPH/F regulate competing 5′ splice sites through G-rich enhancers in the α-globin minigene and support the interpretation that the hnRNPH/F synergistic effect is not limited to PLP/DM20 splicing.
Next, we have tested whether the cell-specific context influences the hnRNPH/F-dependent regulation of the PLP/DM20 ratio. We knocked down hnRNPH, hnRNPF, and hnRNPH/F in the fibroblast L cells and in the neuronal cell line, Neuro2A cells transfected with PLP-neo (supplemental Fig. S2). Knockdown with either siF3 or siH3 was specific for the targeted protein and was not accompanied by a compensatory increase in the non-targeted hnRNP (data not shown), in keeping with published results (27). In both cell types, removal of hnRNPH, but not hnRNPF increased the PLP/DM20 ratio. Removal of both hnRNPH/F resulted in a greater increase in PLP/DM20 ratio. The data suggest that cell-specific factors do not contribute to the hnRNPH/F synergism.
G1M2 Regulate Selection of Competing 5′ Splice Sites in a Heterologous Gene—We next sought to determine whether G1M2 is able to regulate 5′ splice site selection in the heterologous α-globin gene context and mediate the hnRNPH/F synergistic regulation of proximal versus distal products. We generated an α-globin construct, in which the distal G runs are replaced with G1M2 and the proximal G runs by ISE, thus replicating the DM20 and PLP configuration (Fig. 3B, construct 4). In addition, we have made a construct in which G1M2 replaces both proximal and distal G groups, thus resembling the G runs configuration of the α-globin construct (Fig. 3B, construct 3). The ratio of the PCR products derived from α-glo-G1M2-G1M2 and α-glo-G1M2-ISE was 0.118 ± 0.012 and 0.122 ± 0.022, respectively, versus 0.372 ± 0.018 derived from α-globin (Fig. 3E). Knockdown of hnRNPH/F caused >2-fold increase in the ratio of the PCR products derived from α-glo-G1M2-G1M2 (0.275 ± 0.033) and α-glo-G1M2-ISE (0.283 ± 0.059) (Fig. 3E). These data support the interpretation that G1M2 enhances the distal 5′ splice site and mediates the hnRNPH/F regulation of proximal to distal splice site selection in a heterologous gene context. Notably, the ratio of splice products and the magnitude of increase in the ratio are lower in these constructs than in the PLP-neo, suggesting that other variables, such as the splice site strength and length of the intron, may influence G1M2 function and its ability to mediate hnRNPH and F regulation of competing 5′ splice sites.
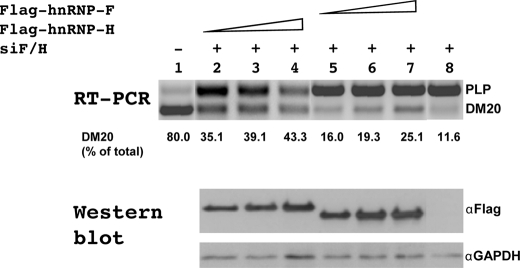
hnRNPH Is Primarily Responsible for the Regulation of DM20 Splicing in Vivo—To more directly investigate the function of hnRNPH and F in regulating DM20 splice site selection in vivo, we have taken two approaches. First, we have determined the effect of overexpressed FLAG-hnRNPH or FLAG-hnRNPF on the PLP/DM20 ratio derived from PLP-neo in Oli-neu cells treated with siF/H. FLAG-hnRNPH and FLAG-hnRNPF transcripts are resistant to siF/H-mediated degradation (see “Experimental Procedures” for details). The expression of each fusion protein was verified by Western blot with a FLAG antibody (Fig. 4). The plasmid-derived PLP and DM20 products were amplified by RT-PCR in RNA isolated from cells cultured for 72 h in growth medium, and the data are expressed as a percent of DM20 product over the total PLP+DM20 (Fig. 4). In Oli-neu cells treated with siF/H, the DM20 product is 11% (lane 8) versus 80% (lane 1) in cells treated with control siRNA. Overexpression of FLAG-hnRNPH induced a dose-dependent increase in the DM20 product (lanes 2–4) and increased the DM20 product up to 43%, whereas overexpression of FLAG-hnRNPF induced a smaller increase in the DM20 product (lanes 5–7), which was up to 25%. To demonstrate that the small increase in DM20 is not the result of a poorly functional hnRNPF protein, we have overexpressed FLAG-hnRNPF with a Bcl-x minigene into Oli-neu cells treated with siF/H. The XL and XS products are regulated by hnRNPH and F (Ref. 17 and supplemental Fig. S3). The plasmid-derived XL/XS ratio was 4-fold higher after treatment with siF/H than in untreated cells (8.47 versus 1.84, supplemental Fig. S3). Overexpressed FLAG-hnRNPF restored the XL/XS ratio to the control level (1.55 versus 1.89, supplemental Fig. S3). These data are consistent with the hnRNPF-dependent regulation of XL/XS ratio (17) and demonstrate that recombinant FLAG-hnRNPF is fully competent to regulate splicing. The results suggest that hnRNPH is a stronger regulator of DM20 splicing than hnRNPF.
FIGURE 4.
hnRNPH rescues DM20 splice site selection after siF/H treatment. Representative RT-PCR analysis of PLP-neo derived PLP and DM20 products amplified from RNA isolated from Oli-neu cells untreated (lane 1), treated with siF/H (lanes 2–8) and transfected with increasing amount (0.5, 1, and 1.5 μg) of pFLAG-hnRNPH DNA (lanes 2–4, respectively) or of pFLAG-hnRNPF DNA (lanes 5–7, respectively) or pcDNA (0.5 μg) (lane 8) (30 PCR cycles) (n = 2). The data are expressed as percent of DM20 product over the total PLP+DM20. The amount of FLAG-hnRNPH and FLAG-hnRNPF protein expressed is shown in the Western blot probed with FLAG antibody.
Next, we replaced the G1M2 sequences in PLP-neo with an MS2 coat protein binding motif (MS) or a non-related sequence (NR) (Fig. 5A) and expressed these genes in Oli-neu cells alone or with MS2-HMyc or MS2-FMyc fusion proteins, which will tether each protein to the same location in exon 3B. The PLP and DM20 splice products were determined by RT-PCR (Fig. 5B). The expression of MS2-HMyc and MS2-FMyc fusion proteins was assessed by Western blot of Oli-neu cell lysates with an antibody to the Myc tag (supplemental Fig. S4). Replacing G1M2 with the MS2 hairpin or with an unrelated sequence resulted in complete loss of the DM20 splice product, and only the PLP product was amplified by RT-PCR (Fig. 5B, lanes 7 and 3). Significantly, expression of MS2-HMyc caused a dose-dependent increase in the DM20 product from PLP-neo-MS1, whereas expression of MS2-FMyc had no effect (Fig. 5B, compare lanes 1 and 2 to lanes 5 and 6). We next asked whether enhancing tethering by increasing the number of MS motifs would result in greater DM20 splicing and potentially allow detection of an effect induced by MS2-FMyc. We co-transfected Oli-neu cells with a PLP-neo construct containing two MS motifs cloned one adjacent to the other (PLP-neo-MS3) with either MS2-HMyc or MS2-FMyc. The DM20 product derived from PLP-neo-MS3 was increased by MS2-HMyc to the same extent as the product derived from PLP-neo-MS1 (Fig. 5C, lane 3), the MS2-FMyc had no effect (Fig. 5C, lane 2). Similar results were obtained from PLP-neo-MS2, in which the two MS motifs are separated by a longer stretch of PLP sequences (data not shown). Neither MS2-HMyc nor MS2-FMyc increased the DM20 splice product derived from a PLP-neo construct in which G1M2 were replaced with unrelated sequences (Fig. 5B, lanes 3 and 4). The data indicate that hnRNPH directly regulates selection of the DM20 5′ splice site, while tethering of hnRNPF does not, suggesting that hnRNPF alone is not sufficient to direct selection of the 5′ splice site. Collectively, the results of these studies support the notion that hnRNPH plays a primary function in the regulation of DM20 splicing.
hnRNPH and F Recruit U1snRNP to the DM20 5′ Splice Site—To further elucidate the molecular basis of the functional difference between hnRNPH and F in regulating DM20 splicing; we have examined the biochemical interaction of hnRNPH and F with other components of the splicing machinery. We have determined whether G1M2 and hnRNPH and F regulate binding of U1snRNP to the DM20 template. We have performed RNA affinity precipitations with a biotinylated RNA template that contains the DM20 5′ splice site and exon 3B sequences encompassing G1 and M2 (DM20-G1M2, Fig. 6A and “Experimental Procedures”). Oli-neu nuclear extracts were incubated with either the DM20-G1M2 RNA template (WT) or an RNA template in which the G1 and M2 are replaced by polyUs (MT). The proteins bound to the templates were separated by SDS-PAGE and analyzed by liquid chromatography/MS/MS and by Western blot. Fifteen distinct bands were detected in the precipitates with the DM20-G1M2-WT template (Fig. 6B). Mutation of G1 and M2 resulted in disappearance or strong reduction of several specific protein bands: the ∼30-kDa band was identified as U1A, the 55- to 58-kDa bands were identified as hnRNPF and H, respectively, a 70-kDa band was identified as U1 70K, and a band of 100 kDa was identified as containing PTB-associated splicing factor and nucleolin (Fig. 6B). Western blot analysis of RNA precipitates with the G1- and M2-mutated template confirmed that U1 70K and U1A were significantly reduced, whereas hnRNPH and F were almost completely absent, compared with precipitates with the wild-type template (Fig. 6C). In contrast, Western analysis showed that binding of hnRNPL and hnRNPA1 was not affected by mutation of G1 and M2. The data suggest that the absence of G1 and M2 abolishes binding of hnRNPH and F, which in turn greatly reduces recruitment of components of the U1snRNP to the template.
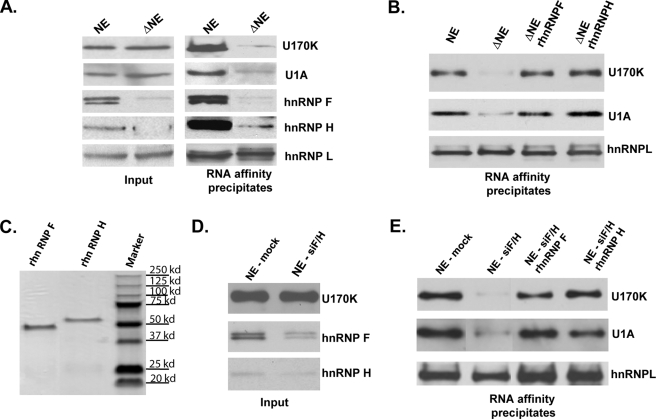
To demonstrate that removal of hnRNPH and F is responsible for loss of U1snRNP binding, RNA affinity precipitations were carried out with the DM20-G1M2-WT template and Oli-neu nuclear extracts that had been depleted of hnRNPH and F with poly-G affinity columns (37). As shown in Fig. 7A, hnRNPH and F were specifically and almost completely removed, whereas the U1 70K and U1A and hnRNPL were not affected. RNA affinity precipitations were carried out with the DM20-G1M2-WT template and either the natural or poly-G-depleted Oli-neu nuclear extracts and subjected to Western blot. Binding of U1 70K and U1A to the DM20-G1M2 template was almost completely abolished with the hnRNPH and F depleted extracts. The reduction of binding is specific, because binding of hnRNPL was not affected by depletion of hnRNPH and F (Fig. 7A). Similar results were obtained in RNA affinity precipitations with nuclear extracts prepared from Oli-neu cells treated with siF/H to remove endogenous hnRNPH and F (Fig. 7D). These results demonstrate that, in the absence of hnRNPH and F, U1snRNP does not bind to the DM20 template.
FIGURE 7.
Binding of U1 70K and U1A is reconstituted by recombinant hnRNPH and F. A, representative Western blot analysis of the input (one-tenth of nuclear extracts used in RNA affinity precipitations) (n = 3). hnRNPH and F are almost completely removed from Oli-neu extracts run through poly-G affinity column (ΔNE), whereas U1 70K and U1A as well as hnRNPL are present in similar amount as in the natural extracts (NE). Western blot analysis of RNA affinity precipitates with either natural Oli-neu nuclear extracts (NE) or poly-G depleted extracts (ΔNE) (n = 3). Binding of U1 70K and U1A to the template is almost completely abolished by depletion of hnRNPH and F. Binding of hnRNPHL is not affected by depletion of hnRNPH/F. B, representative Western blot analysis of RNA affinity precipitates with the natural nuclear extracts (NE), poly-G depleted extracts (ΔNE) and ΔNE supplemented with either recombinant hnRNPH or hnRNPF (n = 3). Binding of U1 70K and U1A to the template is reconstituted by hnRNPH and F. C, Coomassie Blue-stained SDS gel showing the homogeneous preparation of recombinant hnRNPH and hnRNPF. D, representative Western blot analysis of the input (one-tenth of nuclear extracts used in RNA affinity precipitations) (n = 2). hnRNPH and F are almost completely removed from Oli-neu extracts prepared from cells treated with siF/H (NE-siF/H), whereas U1 70K is present in similar amount as in the extract prepared from mock treated cells (NE-mock). E, representative Western blot analysis of RNA affinity precipitates with NE, siF/H-NE, and siF/H NE supplemented with either recombinant hnRNPH or hnRNPF (n = 2). Binding of U1 70K and U1A to the RNA template is reconstituted by either hnRNPH or F.
Next, we sought to determine whether hnRNPH and hnRNPF differ in their ability to recruit U170K and U1A to DM20-G1M2-WT. To this end, we have added either bacterially expressed purified His-hnRNPH or His-hnRNPF (Fig. 7C) to Oli-neu nuclear extracts that were depleted of hnRNPH/F with polyG (Fig. 7B) or by siF/H treatment in vivo (Fig. 7E) and examined U1 70K and U1A binding to the DM20-G1M2 template. Reconstitution of the extract with either hnRNPH or hnRNPF restored binding of U170K and U1A to the RNA template (Fig. 7, B and E), indicating that both are effective in recruiting U1snRNP to the DM20. Addition of His-hnRNPH and F together did not increase recruitment of U1 70K and U1A to the DM20 template compared with the individual protein (data not shown). These data show that hnRNPH and F are equally effective in recruiting U1snRNP, suggesting a redundant function of hnRNPH and F in stabilizing the assembly of U1snRNP.
DISCUSSION
In this work, we have characterized the molecular mechanisms underlying the regulation of PLP exon 3B inclusion/exclusion mediated by hnRNPH/F and G-rich enhancers in exon 3B. Notably, exon 3B is almost 100% conserved at the nucleotide level across species (40), suggesting a strong role of these sequences in RNA-processing regulation. Exon 3B harbors five distinct G runs of which G1 and M2 are clustered in close proximity to the DM20 5′ splice site and act as enhancers of the DM20 splice site. G1 is a stronger enhancer than M2, possibly because it contains a longer stretch of five uninterrupted Gs. In contrast, G4 and G5, although they represent consensus binding motifs for hnRNPH family of splicing factors (28), do not participate in the hnRNPH/F mediated regulation of PLP/DM20 ratio. The clustering of enhancers close to the DM20 5′ splice site supports the notion that an active mechanism, centered on these G-rich sequences, has evolved to select DM20 over the PLP 5′ splice site in cells in which the DM20 isoform is preferentially expressed such as oligodendrocyte progenitor cells in the embryonic brain, Schwann cells and non-glial cells (41). This regulation is mediated by hnRNPH and F through G1M2 enhancers and is not dependent on cell type, consistent with a proposed function in the selection of DM20 in immature glial cells and in non-glial cells.
G1 and M2 form a functional unit and mediate hnRNPH and F regulation of competing 5′ splice sites in the PLP gene and in a heterologous α-globin, suggesting that their function is in large part independent from the gene context. In addition, hnRNPH and F cooperatively regulate the distal of two α-globin 5′ splice sites through the natural G triplets present in this gene, indicating that the synergism of hnRNPH and F may represent a more general mechanism by which competing 5′ splice sites flanked by G-rich enhancers are regulated. However, removal of hnRNPH/F induces a relatively small change in the α-globin distal to proximal transcript ratio compared with the effect on the PLP/DM20 ratio, suggesting that other factors, such as splice site strength or intron length, may play a role. Previously, α-globin G runs were shown to directly recruit U1snRNP through base-pairing of G-rich sequences with U1snRNA nucleotides not involved in 5′ splice site interaction and to help position the U1snRNA on to the 5′ splice site (24). In the PLP gene, hnRNPH and F regulate the PLP/DM20 ratio by recruiting and/or stabilizing the U1snRNP binding likely through protein-protein interactions. Whether direct base pairing also occurs between the G1M2 and U1snRNA, similarly to the G runs of the α-globin, and whether this mechanism contributes to selection of DM20 5′ splice site remain to be determined.
Unlike other genes in which hnRNPH and F have redundant functions in vivo (17, 31, 32), we demonstrate that hnRNPH and F are not functionally redundant in regulating DM20 splicing. Expression of hnRNPH alone is able to regulate selection of the DM20 splice site after removal of hnRNPH/F in vivo, whereas hnRNPF only weakly regulates DM20 splicing, although the transfected hnRNPH and F proteins are expressed in an equivalent amount. Furthermore, tethering of hnRNPF directly downstream of the DM20 5′ splice site, even in the presence of two binding motifs, did not have an effect on DM20 splicing, whereas tethering of hnRNPH induced a dose-dependent increase in DM20 splicing. These data suggest that hnRNPF alone is not sufficient to directly regulate the DM20 splicing.
The ability of FLAG-hnRNPF to exert a modest regulation on the DM20 splicing in the PLP gene context may depend on the presence of G1M2 and flanking sequences, which would allow binding of other factors to these sequences. A number of proteins, including hnRNPA1 and L, bind to the DM20-G1M2 template (Fig. 6) and to M2 (27) and may interact with hnRNPH and F and participate in the regulation of splicing. Nucleolin and PTB-associated splicing factor bind to DM20-G1M2 and depend on G1M2. Both nucleolin and PTB-associated splicing factor are ubiquitously expressed polyfunctional proteins, which participate in several aspects of RNA metabolism and processing (42, 43). Interestingly, PTB-associated splicing factor forms a complex with the non-snRNP-associated U1A (44), and this complex may interact with hnRNPH/F.
In this study, we show that G1M2 and hnRNPH/F are critical for the recruitment of components of U1snRNP to the DM20 template, suggesting that this represents a major mechanism, which regulates the increase in PLP/DM20 ratio in differentiated oligodendrocytes where the abundance of hnRNPH and F is reduced (27). Although the effect of hnRNPH/F on U1snRNP recruitment is clearly demonstrated by our studies, the mechanism whereby hnRNPH and F recruit U1snRNP remains to be elucidated. It is tempting to speculate that hnRNPH/F may either antagonize a repressor or associate with an activator leading to recruitment of U1snRNP to the DM20 5′ splice site. Some of the splicing factors identified in the proteomics analysis and shown to be differentially affected by mutations in G1M2 and loss of hnRNPH/F binding are potential candidates for such interactions. Importantly, hnRNPH and F are equally effective in recruiting U1snRNP, thus excluding the possibility that differences in assembly of the E complex account for the functional role of hnRNPH. It is tempting to speculate that hnRNPH plays a unique role in later steps of spliceosome assembly, such as formation of ATP-dependent splicing complexes, because it was shown to occur in the HIV splicing (28) or that binding of hnRNPH modifies the RNA structure, especially through a predicted quadruplex G run structure formed by G1 and M2 (45, 46), leading to greater accessibility to other splicing factors or that hnRNPH selectively inhibits a repressor of the DM20 splice site.
The ISE downstream of the PLP 5′ splice site regulates the increase in PLP/DM20 in the developing brain in vivo, and absence of the ISE leads to predominance of DM20 product, demonstrating its antagonistic effect on the G1M2 (29). Unlike G1M2, ISE is only partially and weakly regulated by hnRNPH/F both in the PLP and the α-globin gene contexts. Importantly, mutations of the G runs of the ISE have a stronger effect on the PLP/DM20 ratio than removal of hnRNPH/F, suggesting that other splicing factors play a critical role in the ISE function. hnRNPA1 and L bind to the G runs of the ISE (27), thus these splicing factors may contribute to the function of the ISE. Furthermore, sequences 5′ of the G runs in the PLP gene context have an enhancing function on PLP 5′ splice site selection, however, a putative SRp40 binding motif does not appear to mediate this enhancer's function, supporting the notion that other splicing factors may participate in the regulation. These factors remain to be identified.
In summary, we have shown that hnRNPH and F regulate the PLP/DM20 ratio through G1M2 and recruit U1snRNP to the DM20 5′ splice site. Because both hnRNPH and F are equally effective in recruiting U1snRNP, the functional difference between hnRNPH and hnRNPF in vivo may be dependent on other steps of splicing. On the basis of the data, we propose that reduced abundance of hnRNPH/F in differentiated oligodendrocytes regulates the PLP/DM20 ratio by decreasing the overall efficiency of DM20 5′ splice site recognition by U1snRNP.
Supplementary Material
Acknowledgments
We thank Dr. Martha Peterson for critical reading of the manuscript, Dr. D. Black for the hnRNPF antibody and the recombinant hnRNPH and hnRNPF constructs, Dr. A. McCullough for the wild-type α-globin construct, Dr. M. McNally for pcDNA-MS2-H and pcDNA-MS2-F constructs, Dr. M. Garcia-Blanco for FLAG-hnRNPH and FLAG-hnRNPF, Dr. Stefan Stamm for SRp40, and Dr. Charles Chalfant for Bcl-x. We also thank Giulio Disanto for performing RT-PCR analyses of the PLP-neo mutant constructs.
This work was supported by National Institutes of Health Grant RO1NS053905 from NINDS. This work was also supported by the European Leukodystrophy Association. The mass spectrometric analysis was performed at the University of Kentucky, Center for Structural Biology Protein Core Facility. This core facility is supported in part by funds from NCRR/NIH Grant P20 RR020171.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
Footnotes
The abbreviations used are: U1snRNP, U1 small nuclear ribonucleoprotein; U1snRNA, U1 small nuclear RNA; hnRNP, heterogeneous nuclear ribonucleoprotein; PLP, proteolipid protein; siRNA, small interference RNA; RT, reverse transcription; MS, MS2 coat protein binding motif; NR, non-related sequence; WT, wild type; MT, mutated; Bt2cAMP, dibutyryl cyclic AMP; ISE, intronic splicing enhancer.
References
- 1.Modrek, B., and Lee, C. J. (2003) Nat. Genet. 34 177-180 [DOI] [PubMed] [Google Scholar]
- 2.Black, D. L. (2003) Annu. Rev. Biochem. 72 291-336 [DOI] [PubMed] [Google Scholar]
- 3.Hertel, K. J. (2008) J. Biol. Chem. 283 1211-1215 [DOI] [PubMed] [Google Scholar]
- 4.Green, M. R. (1991) Annu. Rev. Cell Biol. 7 559-599 [DOI] [PubMed] [Google Scholar]
- 5.Seraphin, B., and Rosbash, M. (1989) Cell 59 349-358 [DOI] [PubMed] [Google Scholar]
- 6.Matlin, A. J., Clark, F., and Smith, C. W. (2005) Nat. Rev. Mol. Cell Biol. 6 386-398 [DOI] [PubMed] [Google Scholar]
- 7.Han, K., Yeo, G., An, P., Burge, C. B., and Grabowski, P. J. (2005) PLoS Biol. 3 e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krecic, A. M., and Swanson, M. S. (1999) Curr. Opin. Cell Biol. 11 363-371 [DOI] [PubMed] [Google Scholar]
- 9.Caputi, M., and Zahler, A. M. (2001) J. Biol. Chem. 276 43850-43859 [DOI] [PubMed] [Google Scholar]
- 10.Marcucci, R., Baralle, F. E., and Romano, M. (2007) Nucleic Acids Res. 35 132-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matunis, M. J., Xing, J., and Dreyfuss, G. (1994) Nucleic Acids Res. 22 1059-1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veraldi, K. L., Arhin, G. K., Martincic, K., Chung-Ganster, L. H., Wilusz, J., and Milcarek, C. (2001) Mol. Cell. Biol. 21 1228-1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alkan, S. A., Martincic, K., and Milcarek, C. (2006) Biochem. J. 393 361-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caputi, M., and Zahler, A. M. (2002) EMBO J. 21 845-855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, C. D., Kobayashi, R., and Helfman, D. M. (1999) Genes Dev. 13 593-606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou, M. Y., Rooke, N., Turck, C. W., and Black, D. L. (1999) Mol. Cell. Biol. 19 69-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garneau, D., Revil, T., Fisette, J. F., and Chabot, B. (2005) J. Biol. Chem. 280 22641-22650 [DOI] [PubMed] [Google Scholar]
- 18.Min, H., Chan, R. C., and Black, D. L. (1995) Genes Dev. 9 2659-2671 [DOI] [PubMed] [Google Scholar]
- 19.Nussinov, R. (1988) J. Theor. Biol. 133 73-84 [DOI] [PubMed] [Google Scholar]
- 20.Nussinov, R. (1989) J. Biomol. Struct. Dyn. 6 985-1000 [DOI] [PubMed] [Google Scholar]
- 21.Sirand-Pugnet, P., Durosay, P., Brody, E., and Marie, J. (1995) Nucleic Acids Res. 23 3501-3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlo, T., Sterner, D. A., and Berget, S. M. (1996) RNA 2 342-353 [PMC free article] [PubMed] [Google Scholar]
- 23.McCullough, A. J., and Berget, S. M. (1997) Mol. Cell. Biol. 17 4562-4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCullough, A. J., and Berget, S. M. (2000) Mol. Cell. Biol. 20 9225-9235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobson, G. M., Huang, Z., Sperle, K., Stabley, D. L., Marks, H. G., and Cambi, F. (2002) Ann. Neurol. 52 477-488 [DOI] [PubMed] [Google Scholar]
- 26.Nave, K. A., Lai, C., Bloom, F. E., and Milner, R. J. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 5665-5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, E., Dimova, N., and Cambi, F. (2007) Nucleic Acids Res. 35 4164-4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaub, M. C., Lopez, S. R., and Caputi, M. (2007) J. Biol. Chem. 282 13617-13626 [DOI] [PubMed] [Google Scholar]
- 29.Wang, E., Dimova, N., Sperle, K., Huang, Z., Lock, L., McCulloch, M. C., Edgar, J. M., Hobson, G. M., and Cambi, F. (2008) Exp. Neurol. 214 322-330 [DOI] [PubMed] [Google Scholar]
- 30.LeVine, S. M., Wong, D., and Macklin, W. B. (1990) Dev. Neurosci. 12 235-250 [DOI] [PubMed] [Google Scholar]
- 31.Crawford, J. B., and Patton, J. G. (2006) Mol. Cell. Biol. 26 8791-8802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mauger, D. M., Lin, C., and Garcia-Blanco, M. A. (2008) Mol. Cell. Biol. 28 5403-5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McNally, L. M., Yee, L., and McNally, M. T. (2006) J. Biol. Chem. 281 2478-2488 [DOI] [PubMed] [Google Scholar]
- 34.Massiello, A., Salas, A., Pinkerman, R. L., Roddy, P., Roesser, J. R., and Chalfant, C. E. (2004) J. Biol. Chem. 279 15799-15804 [DOI] [PubMed] [Google Scholar]
- 35.Wang, E., Huang, Z., Hobson, G. M., Dimova, N., Sperle, K., McCullough, A., and Cambi, F. (2006) J. Cell. Biochem. 97 999-1016 [DOI] [PubMed] [Google Scholar]
- 36.Paronetto, M. P., Achsel, T., Massiello, A., Chalfant, C. E., and Sette, C. (2007) J. Cell Biol. 176 929-939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carlo, T., Sierra, R., and Berget, S. M. (2000) Mol. Cell. Biol. 20 3988-3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cartegni, L., Wang, J., Zhu, Z., Zhang, M. Q., and Krainer, A. R. (2003) Nucleic Acids Res. 31 3568-3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, P. J., Zhang, C., Wang, J., Chew, S. L., Zhang, M. Q., and Krainer, A. R. (2006) Hum. Mol. Genet. 15 2490-2508 [DOI] [PubMed] [Google Scholar]
- 40.Kurihara, T., Sakuma, M., and Gojobori, T. (1997) Biochem. Biophys. Res. Commun. 237 559-561 [DOI] [PubMed] [Google Scholar]
- 41.Campagnoni, A. T., and Macklin, W. B. (1988) Mol. Neurobiol. 2 41-89 [DOI] [PubMed] [Google Scholar]
- 42.Mongelard, F., and Bouvet, P. (2007) Trends Cell Biol. 17 80-86 [DOI] [PubMed] [Google Scholar]
- 43.Shav-Tal, Y., and Zipori, D. (2002) FEBS Lett. 531 109-114 [DOI] [PubMed] [Google Scholar]
- 44.Lutz, C. S., Cooke, C., O'Connor, J. P., Kobayashi, R., and Alwine, J. C. (1998) RNA 4 1493-1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kikin, O., D'Antonio, L., and Bagga, P. S. (2006) Nucleic Acids Res. 34 W676-W682, Web Server issue [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kikin, O., Zappala, Z., D'Antonio, L., and Bagga, P. S. (2008) Nucleic Acids Res. 36 D141-D148, Database issue [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.