Abstract
System A transporters SNAT1 and SNAT2 mediate uptake of neutral α-amino acids (e.g. glutamine, alanine, and proline) and are expressed in central neurons. We tested the hypothesis that SNAT2 is required to support neurotransmitter glutamate synthesis by examining spontaneous excitatory activity after inducing or repressing SNAT2 expression for prolonged periods. We stimulated de novo synthesis of SNAT2 mRNA and increased SNAT2 mRNA stability and total SNAT2 protein and functional activity, whereas SNAT1 expression was unaffected. Increased endogenous SNAT2 expression did not affect spontaneous excitatory action-potential frequency over control. Long term glutamine exposure strongly repressed SNAT2 expression but increased excitatory action-potential frequency. Quantal size was not altered following SNAT2 induction or repression. These results suggest that spontaneous glutamatergic transmission in pyramidal neurons does not rely on SNAT2. To our surprise, repression of SNAT2 activity was not limited to System A substrates. Taurine, γ-aminobutyric acid, and β-alanine (substrates of the SLC6 γ-aminobutyric acid transporter family) repressed SNAT2 expression more potently (10×) than did System A substrates; however, the responses to System A substrates were more rapid. Since ATF4 (activating transcription factor 4) and CCAAT/enhancer-binding protein are known to bind to an amino acid response element within the SNAT2 promoter and mediate induction of SNAT2 in peripheral cell lines, we tested whether either factor was similarly induced by amino acid deprivation in neurons. We found that glutamine and taurine repressed the induction of both transcription factors. Our data revealed that SNAT2 expression is constitutively low in neurons under physiological conditions but potently induced, together with the taurine transporter TauT, in response to depletion of neutral amino acids.
Central neurons express the sodium-coupled neutral amino acid transporters SNAT1 and SNAT2 (also known as SLC38A1, GlnT, SAT1, or ATA1 and SLC38A2, SAT2, or ATA2, respectively) (1–10), two of the three SLC38 gene family members (reviewed in Ref. 11) that collectively account for the System A amino acid transport activity classically described in most cell types (12, 13). System A catalyzes the unidirectional uptake of small aliphatic neutral amino acids, especially alanine, cysteine, glutamine, glycine, methionine, proline, and serine. Among these, glutamine is around 10-fold more abundant than any other amino acid in extraneuronal space (14–16), which, combined with the high affinity of System A transporters for glutamine (Km = 0.2–0.5 mm), makes glutamine the predominant substrate for System A in neurons of the brain. Glutamine, in addition to glucose, is a major precursor for glutamate (17), and many neuronal cell bodies in the brain express phosphate-activated glutaminase (18, 19) and contain high levels of glutamate. Whereas System N transporters may serve the exodus of glutamine from the astrocyte (9, 20, 21), these observations present the possibility that SNAT1 and SNAT2 play a role in providing central neurons with the glutamine required for synthesis of transmitter glutamate (22), but the evidence is controversial (23).
Here we tested the hypothesis that SNAT2 provides neurons with glutamine required for neurotransmitter glutamate synthesis by measuring spontaneous glutamatergic activity in primary neuronal cultures in which we altered endogenous SNAT2 expression levels. Adaptive regulation of System A (up-regulation in amino acid-deprived (-AA)3 conditions and repression of System A activity when substrates are reintroduced) has been widely studied in peripheral cell types (12, 13). Such conditions have no effect on glutamine transport activities attributed to Systems L, N, or ASC (24). In nonneuronal cell types in which only one System A isoform is expressed, increased functional activity results from de novo synthesis of SNAT2 mRNA and protein in response to -AA treatment (25– 27). Adaptive regulation of System A was considered to be restricted to the effects of its own substrates, since the addition of System A substrates blocks the increase in transport activity observed in response to -AA treatment, whereas other amino acids (leucine, the classic System L inhibitor 2-aminobicyclo-(2,2,1)-heptane-2-carboxylic acid, anionic (glutamate), and cationic (arginine) amino acids) produce no such effect (24–31).
At the genomic level, an intronic amino acid response element (AARE) that is responsive to adaptive regulation and binds ATF4 (activating transcription factor 4, also known as CREB-2) and the CCAAT/enhancer-binding protein (C/EBPβ) has been identified in the SLC38A2 gene coding for SNAT2 (32, 33), and a role for SNAT2 as a nutrient amino acid sensor (exerting its effect upstream of the signaling mechanisms regulating the adaptive regulatory response) has been proposed (34, 35). The stimulation of System A by -AA treatment may alternatively be stress-induced and related to cell volume restoration through an expansion of the intracellular amino acid pool (36, 37) (but see Refs. 38 and 39). Adaptive regulation of System A has not previously been demonstrated in neurons, which express both System A isoforms, SNAT1 and SNAT2. The effect of incubation with System A substrates, particularly glutamine, on neuronal levels of glutamine transporters SNAT1 and SNAT2 has also not been examined.
EXPERIMENTAL PROCEDURES
Primary Neuronal Culture—Primary neuronal cultures were prepared as described (40, 41) with minor modifications. Cerebral cortices were removed rapidly from rat embryos at embryonic day 18 under stereomicroscopic observation, in ice-cold 1× PBS containing 6% glucose, and the meninges were discarded. Cortical tissue was placed in Neurobasal medium containing 0.02% bovine serum albumin, 0.1% papain, and 5 mm l-cysteine and incubated with gentle shaking at 37 °C for 20 min. The tissue pieces were then removed, rinsed three times with DMEM containing 10% fetal bovine serum and 2.5 μg/ml leupeptin, and dissociated by trituration using fire-polished Pasteur pipettes in complete Neurobasal medium supplemented with B27 (Invitrogen) and 0.5 mm Glutamax. Debris was allowed to settle for 10 min, and the cells in suspension were centrifuged for 10 min at 800 × g and resuspended in complete Neurobasal medium supplemented with 25 μm l-glutamate. Cells were counted in the presence of trypan blue and plated in 6-well clusters (0.75 × 106 cells/well) precoated with poly-d-lysine (25 μg/ml; Sigma). Cells were maintained in culture by refreshing the medium (50%) twice a week with Neurobasal medium supplemented with B27 and 0.5 mm Glutamax. The amino acid composition of Neurobasal medium is as follows 20 μm alanine, 5 μm asparagine, 10 μm cysteine, 500 μm glutamine, 400 μm glycine, 200 μm histidine, 200 μm methionine, 67 μm proline, 400 μm serine, 800 μm threonine. The overall concentration of SNAT2 substrates in neuronal culture medium is 2.6 mm. Other chemicals not present in Krebs-Ringer amino acid deprivation medium are as follows: 8 μm calcium pantothenate, 28 μm choline chloride, 8 μm folic acid, 40 μm inositol, 30 μm niacinamide, 20 μm pyridoxal, 1 μm riboflavin, 10 μm thiamine, 0.2 μm vitamin B12, 230 μm sodium pyruvate, 25,000 μm HEPES. The total osmolality of complete culture medium with B27 is ∼289 mosm, and that of Krebs-Ringer bicarbonate buffer used for -AA treatment is 300 mosm.
-AA of Cultured Neuronal Cells—Regulation studies were conducted in differentiating neuronal cultures (12 days in vitro), at the time of functional excitatory synapse maturation and major onset of regulated glutamate exocytosis via VGLUT1 (vesicular glutamate transporter isoform 1 (SLC17A7, BNPI)) (42). To treat the cells with -AA medium, the Neurobasal medium was removed and replaced with complete Neurobasal medium or with Krebs-Ringer bicarbonate buffer: 119 mm NaCl, 5.9 mm KCl, 1.2 mm MgSO4, 2.5 mm CaCl2, 5.6 mm glucose, 25 mm NaHCO3, pH 7.4, for various periods of time at 37 °C. Some experiments were also performed using Earle's balanced salt solution: 117.4 mm NaCl, 5.3 mm KCl, 0.81 mm MgSO4, 1.8 mm CaCl2, 5.55 mm glucose, 26.2 mm NaHCO3, pH 7.4. Various concentrations of unlabeled amino acids or drugs were also added at this time. For refeeding experiments, the Krebs-Ringer bicarbonate buffer was changed with fresh buffer containing the desired concentration of amino acid.
Immunocytochemistry—Primary neuronal cultures were fixed on ice in cold (-20 °C) methanol for 10 min, rinsed three times in PBS, blocked in 0.2% Triton, 6% normal goat serum in PBS at room temperature for 1 h, and incubated for 3 h in blocking buffer with a rabbit polyclonal antibody (1:1000) made against a glutathione S-transferase fusion of the N terminus of SNAT1 residues 1–63 (1) or SNAT2 residues 1–65 (2). Cells were then washed in PBS, incubated for 1 h in secondary antibodies coupled to Alexa-488 (highly cross-adsorbed anti-rabbit), diluted 1:200 in blocking buffer, washed, mounted with Prolong Gold antifade reagent (Invitrogen), and viewed. Epifluorescence microscopy was performed, and images were captured using an RT slider Spot camera under identical exposure conditions for comparative analysis.
Western Analysis—Western analysis was performed on postnuclear supernatant fractions containing protein inhibitors (5 μg/ml pepstatin, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 1 mm phenylmethylsulfonyl fluoride) following brief sonication and centrifugation at 2,000 × g. Western blotting was performed as described (43) using antibodies against SNAT1 (1:1000) and SNAT2 (1:1000) and visualized using peroxidase-conjugated anti-rabbit IgG secondary antibodies (Sigma) and SuperSignal West Pico chemiluminescent substrate (Pierce) followed by exposure to film.
Transport Assays in Cultured Neuronal Cells—We assayed System A functional activity (attributable to both SNAT1 and SNAT2) at the plasma membrane by measuring uptake of α-[1-14C]methylaminoisobutyric acid ([14C]MeAIB; PerkinElmer Life Sciences) in primary neocortical cell cultures. We assigned to System A activity the 50 μm [14C]MeAIB transport activity sensitive to inhibition by 4 mm alanine, MeAIB, or glutamine. To initiate uptake, cells were first rinsed with uptake medium (2 × 1 ml) comprising 125 mm NaCl, 4.8 mm KCl, 1.2 mm KH2PO4, 1.2 mm MgSO4, 1.2 mm CaCl2, 5.6 mm glucose, 25 mm HEPES (pH 7.4) and then incubated at 37 °C with uptake medium containing 1 μl/ml [14C]MeAIB (50.5 mCi/mmol) and 50 μm unlabeled MeAIB (in the absence or presence of inhibitor amino acids or drugs). At the end of the uptake period, we removed the uptake medium, rinsed the cells with ice-cold uptake medium (2.5 ml), and solubilized the cells in 1 ml of 1% SDS. From these samples, we removed 30-μl aliquots for protein determination using the Micro BCA kit (Pierce); to the remainder, we added 7.5 ml of EcoScint and measured 14C content by liquid scintillation counting.
Electrophysiological Recordings in Cultured Neuronal Cells—Whole cell patch clamp recordings were made using Axopatch 200B amplifiers. External solutions contained 140 mm NaCl, 5 mm KCl, 1.25 mm NaH2PO4, 10 mm HEPES, 2.5 mm CaCl2, 1.0 mm MgCl2, 10 mm glucose. Whole cell recording pipettes (3–5 μm) were pulled from borosilicate glass. The internal pipette solutions contained 120 mm potassium gluconate, 20 mm KCl, 4 mm NaCl, 10 mm HEPES, 0.1 mm EGTA, 4.0 mm Mg2ATP, 0.3 mm Tris2GTP, and 14 mm phosphocreatine (pH 7.25 with KOH). We recorded spontaneous firing under current clamp. The frequency and amplitude of miniature excitatory postsynaptic currents (mEPSC) recorded under voltage clamp (at a holding potential of -70 mV) were obtained using the Mini-Analysis software. Bicuculline (10 μm) was added to the external solution to eliminate GABA receptor-gated currents when measuring spontaneous excitatory activity. When mEPSCs were recorded, both bicuculline and tetrodotoxin (1 μm) were included in the external solution.
Real Time RT-PCR—Total RNA was isolated from 0.75 × 106 cells using commercial silica-based filter binding methods (RNeasy minikit; Qiagen). We reverse transcribed 1 μg of RNA using a mixture of oligo(dT) and random hexamers (iScript cDNA synthesis kit; Bio-Rad). Real time PCR was performed using the iCycler iQ detection system (Bio-Rad) according to the manufacturer's instructions. Reactions were performed in triplicate in a 25-μl total volume with 2 μl of template, 200 nm primers, and 12.5 μl of SYBRgreen PCR Master Mix (Applied Biosystems). The PCR cycle was as follows: 95 °C for 8 min and 40 cycles of 95 °C for 15 s and 60 °C for 45 s. The efficiency of the reaction for each target was first tested using 10-fold serial dilutions of plasmid containing the cognate gene (104 to 109 copies) to ensure >90% efficiency. Specific primer sets, designed to amplify ∼200-bp fragments, were for SNAT1 (5-cgttgtttgagctggccaagaagacg (forward) and 5′-gtattcttgtctccatcctggttcgtg (reverse)), SNAT2 (5′-gcatgaagaagaccgaaatgggaagg (forward) and 5′-cctggatgaaagtctgtttcgtacttc (reverse)), TauT (SLC6A6) (5′-tggcggtgataacttatatgacggtattg (forward) and 5′-gcagaggaggatgacaatgaccaag (reverse)), ATF4 (5′-cctgactctgctgcttatattactctaac (forward) and 5′-actccaggtgggtcataaggtttg (reverse)), C/EBPβ (5′-caacctggagacgcagcacaag (forward) and 5′-ctagcagtgacccgccgagg (reverse)), and β-actin (5′-taggcaccagggtgtgatggtggg (forward) and 5′-cgcagctgattgtagaaggtgtggtg (reverse)). 18 S primers (TaqMan Ribosomal RNA control reagents) were purchased from PerkinElmer Life Sciences. The PCR amplification of each product was further assessed using 10-fold dilutions of a rat brain cDNA library as a template and found to be linear over 5 orders of magnitude and >95% efficient. Duplicate reactions were set up in a 25-μl total volume with 5 pmol of each primer, 12.5 μl of 2× SYBRgreen Master Mix (Applied Biosystems), and 2 μl of template. The PCR cycle was 95 °C for 3 min, 42 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s, and a melt curve analysis was performed at the end of each experiment to verify that a single product per primer pair was amplified. The size of the amplified DNA fragment was verified by gel electrophoresis on a 3% agarose gel. Samples were compared using the relative CT method. The -fold increase or decrease was determined relative to a vehicle-treated control after normalizing to a housekeeping gene using 2-ΔΔCT, in which ΔCT represents (gene of interest CT) - (β-actin CT) and ΔΔCT is (ΔCT treated) - (ΔCT control). The β-actin-normalizing control was always included in the real time RT-PCR experiments.
Expression and Analysis of SNAT2 in Xenopus Oocytes—We performed laparotomy and ovariectomy on adult female Xenopus laevis frogs under 2-aminoethylbenzoate methanesulfonate anesthesia (0.1% in 1:1 water/ice, by immersion). Ovarian tissue was isolated and treated with collagenase A (Roche Applied Science), and defolliculate oocytes were isolated and stored at 17 °C in modified Barths medium, as described (44). SNAT2 was expressed in oocytes as described (2). We analyzed SNAT2 activity in oocytes by measuring amino acid-evoked currents and transport of 100 μm [14C]MeAIB as described (2, 6).
Chemicals—Cycloheximide (CHX) was dissolved in ethanol, and actinomycin D (ACD) was dissolved in DMSO. Solvents (0.05%) alone had no effect on SNAT2 expression in control or -AA conditions (data not shown). Amino acids were purchased from Sigma and dissolved in water or buffer, adjusting pH with NaOH when necessary. Radiochemicals were purchased from PerkinElmer Life Sciences. (S)-SNAP 5114 and NNC 711 were obtained from Tocris Bioscience.
Statistical Analyses—Data are expressed as mean ± S.E. from samples of size n. Statistical analyses (Student's t test, nonlinear regression or one-way analysis of variance followed by post hoc multiple comparisons using the Holm-Sidak method) were performed using Prism 3 (Graphpad) or SigmaStat 3.5 (Systat Software) with critical significance level p < 0.05. Nonlinear regression analysis used the least squares method, and errors presented are the S.E. of the estimates.
RESULTS
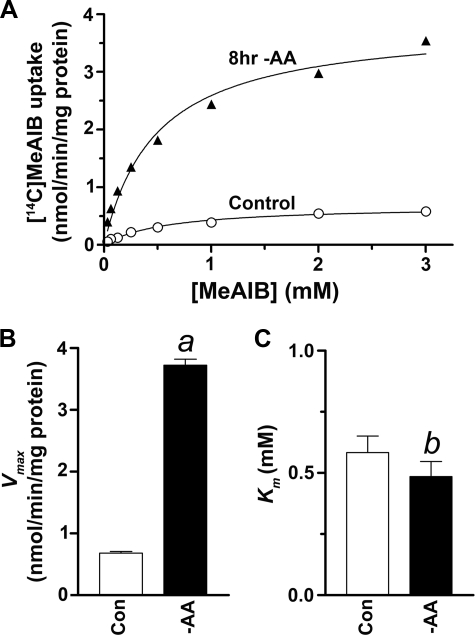
System A Undergoes Adaptive Regulation in Primary Neuronal Cultures—To assess System A transport activity at the plasma membrane, we assayed [14C]MeAIB uptake in rat neocortical neuron-enriched cultures at 12 days in vitro. Transport of 50 μm [14C]MeAIB was inhibited by ∼95% in the presence of saturating (10 mm) cold alanine. The remaining fraction was largely attributable to background labeling, so we took the alanine-sensitive [14C]MeAIB uptake as an index of System A activity. We found that System A transport activity was significantly up-regulated 5-fold following 8 h of incubation in Krebs-Ringer buffer containing 5.6 mm glucose (-AA) (Fig. 1). The increased System A transport activity was completely blocked in the presence of glutamine, alanine, or MeAIB (4 mm). We performed kinetic analyses of the uptake process and found that the increase in System A activity resulted from a 2-fold increase in Vmax following acute (1-h) -AA treatment (data not shown) and a 5-fold increase in Vmax following long term (8-h) -AA treatment, without effect on Km (Fig. 1, B and C), consistent with the effects of -AA treatment in peripheral cells (12). System A activity is therefore subject to adaptive regulation in primary neuronal cultures.
FIGURE 1.
The number of functional System A transporters on the plasma membrane (Vmax) is up-regulated by -AA treatment in neuron-enriched cultures. A, initial velocity (3 min) of MeAIB uptake as a function of concentration (0.1–3.0 mm) in control and following long term (8-h) -AA treatment. B and C, data in A were fitted by the Michaelis-Menten equation using nonlinear regression, revealing an increase in Vmax following 8-h -AA treatment (a, p < 0.001) without effect on Km (b, p = 0.83). Results shown are the averages of duplicate measurements in one representative experiment; we obtained similar results when the experiment was repeated in a second neuronal preparation.
Whereas a multiplicity of glutamine transport systems are expressed in rat brain primary cultures, namely Na+-independent System L (45), Na+-dependent SBAT1/B0AT2 (SLC6A15), (46, 47), d-aspartate sensitive transporter (48), and system A (7, 49), only that fraction of the [3H]glutamine uptake that is inhibited by MeAIB was increased by -AA treatment (data not shown). Therefore, System A is the only neuronal glutamine-transport system that undergoes adaptive regulation following amino acid deprivation.
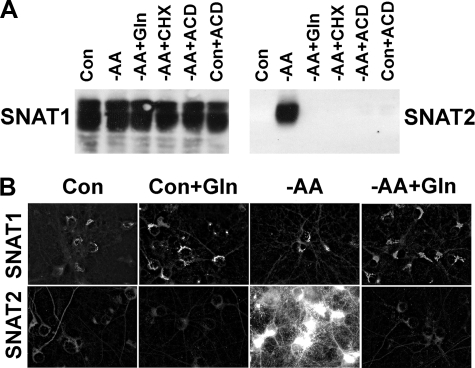
Adaptive Regulation of System A in Neurons Is Isoform-specific—Central nervous system neurons express two isoforms of System A transporter, so we examined the level of expression of SNAT1 and SNAT2 protein following -AA treatment. By Western analysis, SNAT1 immunoreactivity levels were relatively constant, whereas SNAT2 protein levels were selectively up-regulated in response to 8 h of -AA treatment (Fig. 2A). This up-regulation of SNAT2 was completely abolished in the presence of 4 mm glutamine. To determine whether the increase in System A activity following long term -AA treatment was due to changes in mRNA and protein synthesis, we examined the effects of CHX (75 μg/ml) or ACD (7.5 μg/ml). The increase in SNAT2 protein was blocked by both agents (Fig. 2A), suggesting that -AA treatment induces de novo synthesis of SNAT2 protein. These observations are consistent with the “derepression” of System A as previously described (12). Acute changes in System A activity (2-fold increase) following -AA treatment were not affected by CHX or ACD (data not shown). We also visualized subtype-specific regulation of System A by fluorescence microscopy with constant exposure times (Fig. 2B). SNAT2 immunoreactivity was prominently observed in most neurons in response to long term -AA treatment but not in similar cultures to which we added 4 mm glutamine and not in control neurons (amino acid-replete). We observed no change in fluorescence intensity for SNAT1 in cortical neurons following long term -AA treatment. Adaptive regulation of System A in neurons is therefore selective for SNAT2.
FIGURE 2.
Induction of SNAT2 but not SNAT1 protein synthesis in response to -AA treatment. A, Western analysis of SNAT1 and SNAT2 expression in control (Con) conditions (Neurobasal medium), -AA, and -AA in the presence of Gln (4 mm), CHX (75 μg/ml), or ACD (7.5 μg/ml). Control conditions with ACD are also shown. Blots were probed with anti-SNAT1 or -SNAT2 rabbit antibodies. B, selective up-regulation of SNAT2 by -AA treatment was blocked by glutamine. Note that SNAT2 induction occurred principally in neuronal somata and dendritic processes. Results shown are from a representative experiment that was repeated in an independent culture with similar results.
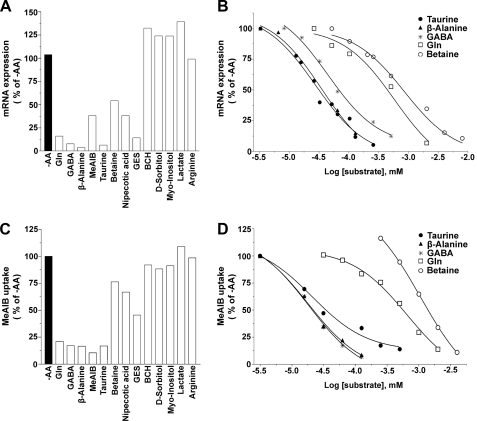
Down-regulation of SNAT2 in the Presence of SNAT2 Substrates—Since the up-regulation of System A following long term -AA treatment is due entirely to SNAT2, we used manipulation of incubation conditions to study the amino acid specificity of SNAT2 adaptive regulation in primary neurons. Glutamine at 4 mm completely reversed the long term (8-h -AA treatment) adaptive regulation of SNAT2 (Fig. 2), and at ∼0.5 mm, glutamine resulted in 50% repression (see Table 1 and Fig. 7). The IC50 values estimated for repression of SNAT2 expression by glutamine (Table 1) and also by alanine (data not shown) matched the half-maximal concentrations we have observed for SNAT2- and SNAT1-mediated transport of these substrates (i.e. K0.5 of 0.2–0.5 mm) (1–3, 50).
TABLE 1.
Potency of various compounds to repress SNAT2
|
Compound
|
IC50a
|
||
|---|---|---|---|
| RT-PCR | [14C]MeAIB | ||
| μm | |||
| Taurine | 19 | 18 | |
| β-Alanine | 28 | 14 | |
| GABA | 39 | 16 | |
| Glutamine | 496 | 570 | |
| Betaine | 819 | 1497 | |
Data are averages of two independent experiments
FIGURE 7.
Repression of SNAT2 by neutral amino acid osmolytes of the SLC6 GABA transporter subfamily. A, adaptive regulation was not restricted to System A substrates. Total cDNA was prepared from mRNA isolated from -AA-treated cultures incubated with various amino acids, amino acid analogues, and other compounds for 8 h. Real time RT-PCR of cDNA (1 μg) was used to assess the activity and relative potency of these compounds to repress SNAT2 mRNA induction. B, dose-response curves of neutral amino osmolytes of the SLC6 GABA transporter subfamily, compared with glutamine, added to cultures following -AA treatment. C, the reduction of SNAT2 functional activity by incubating cultures with these various compounds followed the same order as SNAT2 mRNA repression in A. D, dose-response curves of the effect of neutral amino acid osmolytes of SLC6 GABA transporter subfamily and glutamine on reducing SNAT2 functional activity in -AA conditions.
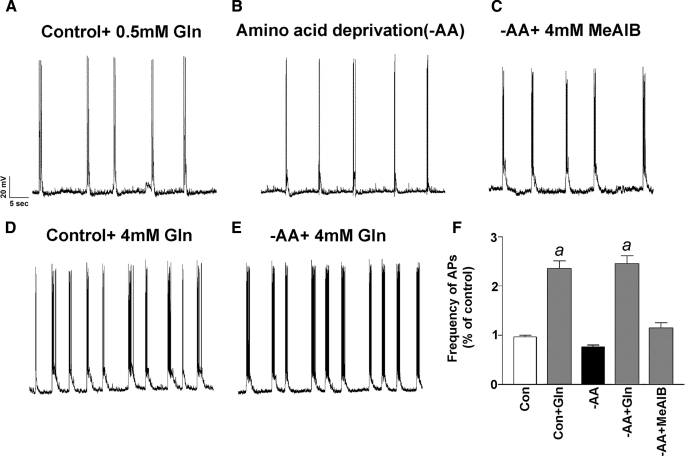
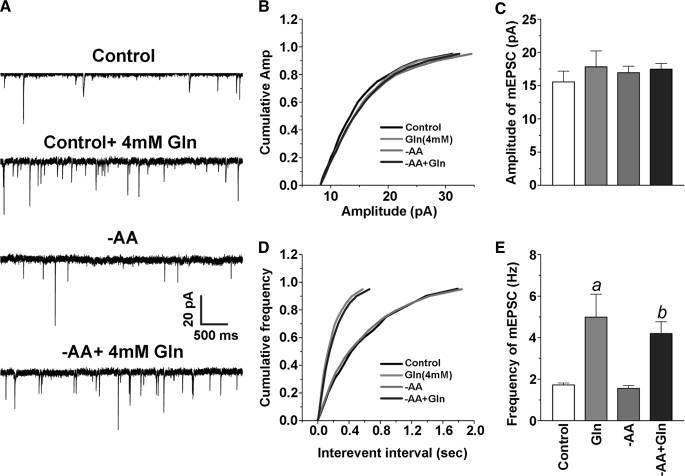
SNAT2 Expression Levels Do Not Affect Spontaneous Glutamatergic Activity—Having determined that we can manipulate SNAT2 expression levels by exploiting the adaptive regulation property of SNAT2, we tested whether SNAT2 plays an important role in providing excitatory neuron precursors for neurotransmitter glutamate by measuring spontaneous glutamatergic activity in neuronal cultures in the presence and absence of amino acids. Although low in abundance (and nondividing in Neurobasal medium), astrocytes were present in our cultures (GFAP staining) to provide endogenous glutamine to neurons for neurotransmission (data not shown). We used rat neocortical neuron-enriched cultures (12 days in vitro) during the major developmental period (7–18 days in vitro) in which functional maturation of glutamate vesicular storage and release in synapses occurs (42) in order to examine the relationship between SNAT2 expression and spontaneous glutamatergic activity. We first examined the spontaneous glutamatergic activity of neurons in control cultures and following adaptive regulation to assess active neuronal connectivity and integrity of our preparations and to correlate levels of SNAT2 and glutamine with excitatory transmission. We found no difference in the frequency of spontaneous action potentials or the frequency and magnitude of mEPSCs between control neurons and neurons deprived of amino acids (including glutamine) for 8 h (Fig. 3, A, B, and F). These results are consistent with a recent report indicating that glutamine supplied exogenously is not necessary for glutamatergic transmission in acute hippocampal slices (51). In contrast, incubation with 4 mm glutamine for 8 h resulted in a marked increase in spontaneous firing in both control and -AA conditions (Fig. 3, D–F). Since System A transport is rheogenic (2, 6, 52), currents can be detected by transient application of glutamine to neurons (5). The effect of MeAIB was less robust (Fig. 3, C and F), and this may relate to the fact that, whereas MeAIB is a specific substrate of System A, it is poorly transported, particularly true for SNAT1 (6). We found no relationship between SNAT2 levels and spontaneous action potentials. Glutamine also enhanced the frequency of mEPSCs (Fig. 3, A–C) and spontaneous action potential firing (Fig. 4, D and E); however, we found no difference in the amplitude of mEPSCs following -AA treatment or in the presence of 4 mm glutamine (Fig. 4, B and C).
FIGURE 3.
Altering endogenous SNAT2 levels does not affect spontaneous excitatory action potentials in neuronal cultured cells. A–E, typical traces recorded from neurons pretreated for 8 h with control + 0.5 mm Gln, control + 4 mm Gln, -AA, -AA + 4 mm Gln, and -AA + 4 mm MeAIB. Spontaneous action potentials were eliminated in the presence of DNQX (10 μm), an AMPA glutamate receptor antagonist. F, mean frequencies of spontaneous firing (normalized to the control) under various conditions (a, p < 0.05 versus -AA treatment). Con, control.
FIGURE 4.
Glutamine enhances mEPSCs. A, representative sweeps of mEPSCs recorded in cultured cortical neurons treated with vehicle, Gln (4 mm), -AA, and -AA + Gln (4 mm). The membrane potential was held at -70 mV. Bicuculline (10 μm) and tetrodotoxin (TTX) (0.5–1 μm) were included in the external solution. The synaptic events were analyzed using the MiniAnalysis program. B, cumulative probability of mEPSC amplitude recorded in neurons under different treatments. C, mean percentage changes in the amplitude of mEPSCs. D, cumulative probability of mEPSC frequency. E, mean percentage changes in the frequency of mEPSCs. a, p < 0.05 compared with the vehicle control; b, p < 0.05 compared with -AA treatment.
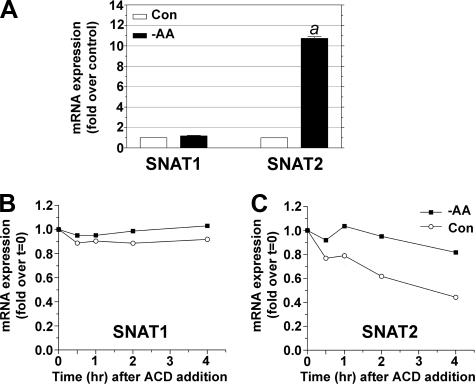
SNAT1 mRNA Is More Stable than Is SNAT2 mRNA—We assessed the molecular response of adaptive regulation of SNAT2 in neurons by real time RT-PCR. SNAT2 mRNA levels increased ∼10-fold following -AA treatment, whereas SNAT1 mRNA expression was unchanged (Fig. 5A). SNAT1 mRNA was more stable than SNAT2 mRNA under control conditions and was not affected by adaptive regulation (Fig. 5B). SNAT2 mRNA stability is increased significantly in -AA conditions (Fig. 5C). Noting also that SNAT2 protein stability is increased by -AA treatment in muscle cells (35), we speculate that SNAT2 protein levels should be low in the absence of de novo SNAT2 mRNA synthesis.
FIGURE 5.
SNAT2 mRNA stability is constitutively low but increased following -AA treatment. A, real time RT-PCR shows that SNAT2 mRNA was selectively and dramatically up-regulated compared with SNAT1 mRNA following -AA treatment. B, SNAT1 mRNA stability was relatively constant following the addition of ACD and unaffected by -AA treatment. C, SNAT2 mRNA stability was low in control conditions and greatly increased in -AA treatment. Starting levels are at t = 0 after the addition of ACD. Results in A are from three independent cultures. Results in B and C are averages of duplicates in two independent experiments. Con, control.
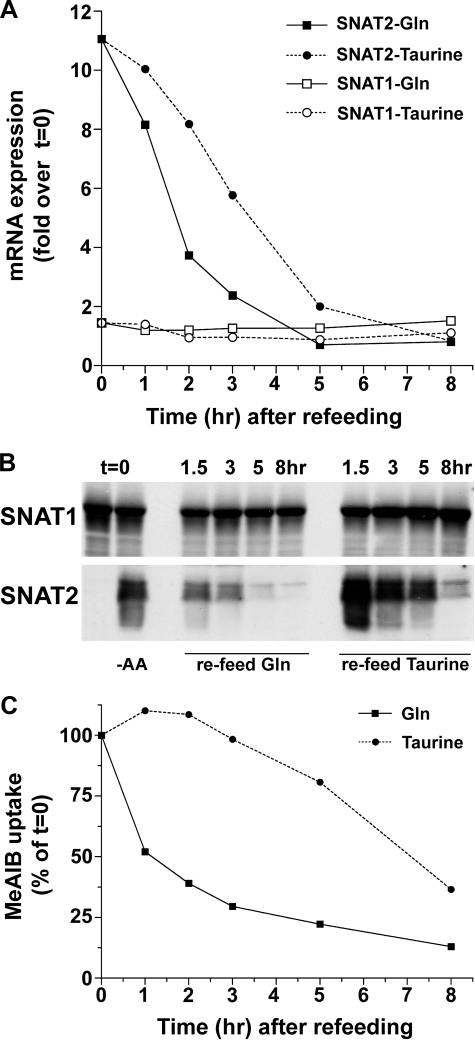
Rapid Down-regulation of SNAT2 in Response to Refeeding System A Substrates—We assessed SNAT2 stability following adaptive regulation induced by glutamine refeeding (4 mm). We used as a reference the non-System A substrate taurine, an important osmolyte in neurons. We observed more rapid disappearance of SNAT2 protein in response to glutamine than to taurine (Fig. 6). At 2 h of incubation, glutamine repressed SNAT2 mRNA expression more strongly than taurine, but at 5 h, both amino acids reduced SNAT2 mRNA to control levels (Fig. 6A). Taurine also down-regulated SNAT2 protein and activity; however, the effect was delayed compared with the effect of glutamine (Fig. 6, B and C). Our results therefore indicate that SNAT2 protein has a relatively short half-life in the presence of System A substrates.
FIGURE 6.
The decay in SNAT2 mRNA, total protein, and functional activity in response to glutamine refeeding of -AA-treated cultures is faster than by the non-System A substrate taurine. Cells were amino acid-deprived for 8 h, and then fresh -AA medium containing 4 mm glutamine or taurine was added. Cells were sampled for mRNA, protein, and functional analyses with time. A, real time RT-PCR analysis of SNAT1 and SNAT2 mRNA levels following glutamine or taurine refeeding of cultures pre-exposed to -AA conditions for 8 h. B, Western blot analysis; C,[14C]MeAIB uptake following 8-h -AA treatment and refeeding with glutamine or taurine. Results presented in A and C are averages of two independent experiments.
Reexamination of the Substrate Profile of System A—Since 8-h refeeding with taurine resulted in the disappearance of SNAT2 mRNA and protein expression (Fig. 6, A and B) and System A functional activity (Fig. 6C), we reexamined the gamut of amino acids capable of adaptively regulating System A in neurons. In addition to taurine, we tested the similar compounds β-alanine and β-aminoisobutyric acid (β-AIB), other known osmolytes (betaine, myo-inositol, and sorbitol), and GABA. When neuronal cultures were incubated with these compounds (at 1 mm in Krebs-Ringer bicarbonate buffer) (Fig. 7, A and C), we found that each of these amino acids (all substrates of the SLC6 GABA transporter subfamily) effectively repress SNAT2 (Fig. 7). In fact, GABA, β-alanine, β-AIB, and taurine are more potent than are the preferred System A substrates glutamine and alanine in blocking the adaptive increase in SNAT2 mRNA (Fig. 7, A and B). myo-Inositol and sorbitol do not repress SNAT2 expression, indicating that only α- and β-amino acids are effective.
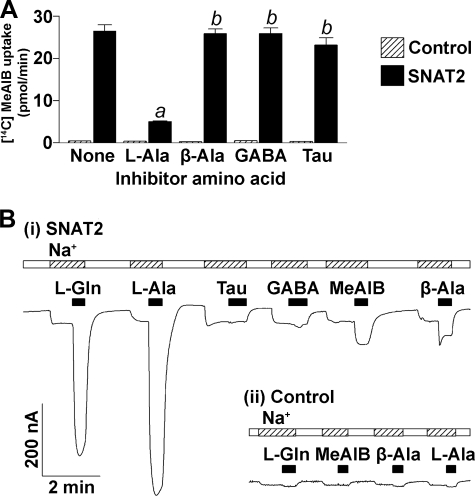
These findings prompted us to examine whether SNAT2 may be capable of also transporting taurine, GABA, or β-alanine, so we extended our previous analyses (2, 6) of the substrate profile of SNAT2 to these candidate substrates. Overexpression of SNAT2 in Xenopus oocytes stimulated the uptake of 100 μm [14C]MeAIB 60-fold over that observed in control oocytes (Fig. 8A). SNAT2-mediated [14C]MeAIB transport was inhibited 81 ± 1% by 1 mm l-alanine, whereas no inhibition was observed in the presence of β-alanine, GABA, or taurine (each at 1 mm). Whereas l-glutamine and l-alanine (10 mm) evoked large currents in oocytes expressing SNAT2 (Fig. 8B), taurine evoked no current and GABA, only a tiny current if at all. In oocytes expressing SNAT2, β-alanine evoked only very small currents that were (in three oocytes) 7.5 ± 1.6% of the current evoked by l-alanine. These data demonstrate that taurine and GABA are not SNAT2 substrates and suggest that no significant transport of β-alanine (at best a weak substrate of SNAT2) will proceed in the presence of physiological glutamine concentrations. We also found that GABA and β-alanine were not substrates for SNAT1 (data not shown).
FIGURE 8.
Amino acid transport and evoked currents in oocytes overexpressing SNAT2. A, uptake of 100 μm [14C]MeAIB was measured over 20 min (in 100 mm NaCl medium, pH 7.5) in control oocytes and oocytes expressing SNAT2; among SNAT2: a, p = 0.003; b, not significant (p > 0.23) compared with no inhibitor amino acid. B, amino acid-evoked currents in an oocyte expressing SNAT2 (i) and a control oocyte (ii). Oocytes were superfused in 100 mm choline chloride medium (pH 7.5) and then, for the periods shown by the hatched bars, in NaCl medium. Amino acids (10 mm) were superfused for the periods shown by the solid bars. (Tau and GABA evoked no currents in this control oocyte (data not shown).)
To determine whether the regulation of system A by nonsubstrates is unique to neurons or is observed in other peripheral cells, we examined the effect of -AA treatment in HeLa cells, a human epithelial carcinoma cell line. We found that [14C]MeAIB uptake increased 5-fold following 8 h of -AA treatment, which was blocked by 4 mm glutamine or MeAIB, as in neurons. However, taurine and GABA (4 mm) could not repress adaptive regulation of system A following -AA treatment in HeLa cells (data not shown).
Regulation of SNAT2 by Transporters in the SLC6 GABA Transporter Subfamily—Transporters in the GABA transporter subfamily (SLC6) include those serving for the transport of neurotransmitters (GABA), osmolytes (taurine, betaine, and GABA), and fuel (creatine) (53, 54). We found that GABA, β-alanine, and taurine inhibited the up-regulation of SNAT2 with roughly equivalent IC50 values (<50 μm), which were one-tenth the IC50 for glutamine (∼500 μm) (Fig. 7 and Table 1). We have ranked in Table 1 the potencies of a range of potential System A repressors at the mRNA level (from data in Fig. 7, A and B) and the functional level (from data in Fig. 7, C and D).
Since GABA is a major inhibitory transmitter in the brain, we investigated whether its ability to repress SNAT2 expression may be GABA receptor-mediated. We found that the 90% reduction in SNAT2 expression (at the level of RNA and [14C]MeAIB transport activity) induced by 200 μm GABA was unchanged in the presence of the GABAA receptor blockers bicuculline (100 μm) or picrotoxin (100 μm) or the GABAB-receptor blocker baclofen (100 μm) (data not shown). There exist in the rat four distinct proteins encoding GABA transporters, namely GAT-1 (GABA transporter 1; SLC6A1), GAT-2 (GABA transporter 2; SLC6A13), GAT-3 (GABA transporter 3; SLC6A11), and BGT-1 (betaine-sensitive GABA transporter (SLC6A12)) (53, 54). In our neuronal cultures, GABA transporter-selective compounds block only a portion of the total [3H]GABA transport; GAT-1 (NNC-711-sensitive) and GAT-2/3 (SNAP-5114 sensitive) each comprise at least 40%, and BGT-1 (betaine-sensitive) only a minor portion (10%) of the total [3H]GABA transport (data not shown). Interestingly, GAT-3 is abundantly expressed in pyramidal neurons during development in rat brain and accounts for a large fraction of GABA transport in vivo (55). GAT-1, the principle GABA reuptake transporter in GABAergic nerve endings (56), does not appear to be involved in GABA blockade of increased System A activity seen in adaptive regulation, because the GAT-1-specific blocker NNC 711 (100 μm) could not reverse the effect of GABA (200 μm). Instead, the GAT-2/3-specific blocker, SNAP-5114 (100 μm) suppresses by ∼50% the repression of SNAT2 mRNA by β-alanine and, to a lesser extent, that induced by GABA (data not shown). Whereas β-alanine is not a substrate for GAT-1, it is transported by GAT-2 and GAT-3 and weakly transported by BGT-1 (57–59). Hence, GABA and β-alanine are probably being taken up through the same GABA transporters, which may include GAT-3, and suppress adaptive regulation of SNAT2. The taurine transporter also efficiently transports β-alanine (60). Since taurine is the most potent repressor of SNAT2 mRNA (Fig. 5 and Table 1), we conjecture that the action of β-alanine to repress SNAT2 induction can be accounted for both by GABA transport and taurine transport systems in neurons. Betaine, a substrate of BGT-1 and a well recognized osmolyte in the kidney (61), also represses SNAT2 mRNA induction, but higher concentrations are required (Fig. 7). In further support of Na+-coupled and solute co-transport-mediated regulation of SNAT2 expression, we found that transporter-selective substrates (1 mm) such as nipecotic acid (a substrate for GAT-1 and GAT-3 (mouse GAT-4) (58)) and guanidinoethylsulfonate (a substrate of the taurine transporter (62)) are effective repressors of SNAT2 up-regulation at the mRNA level (Fig. 7A) and, less so, at the protein level (Fig. 7C).
Regulation of SNAT2 expression in neurons therefore comprises repression of SNAT2 mRNA synthesis by nonsubstrates, namely GABA, and several β-amino acids, such as taurine, β-alanine, and β-AIB, and acceleration of SNAT2 mRNA and protein degradation by System A substrates. Thus, SNAT2 expression is expected to be constitutively low under normal conditions but potently induced in response to depletion of neutral α- and β-amino acids.
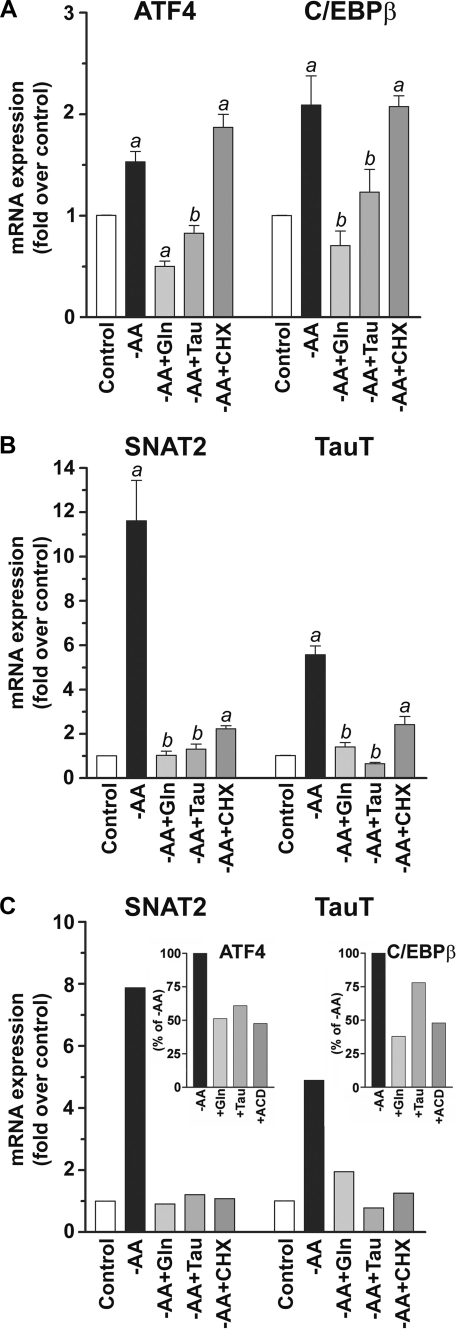
Mechanism of Repression of SNAT2 mRNA Induction by Glutamine and Taurine in Neurons—To determine whether Na+-coupled transport of solutes by neutral amino acid osmolyte transporters of the SLC6 GABA subfamily leads to repression of the SLC38A2 gene coding SNAT2 in neurons in the same way as does System A-mediated transport of glutamine, we examined the mechanism of SNAT2 mRNA repression. Previous studies have indicated that ATF4 mRNA transcription is increased by adaptive regulation. This transcription factor binds to the AARE of SNAT2 and is selectively repressed by System A substrates (32). We found that the induction of ATF4 mRNA (∼50%) was repressed by glutamine or taurine following -AA in neurons (Fig. 9A).
FIGURE 9.
Repression of the induction of transcription factors ATF4 and C/EBPβ and the transporters SNAT2 and TauT by glutamine and taurine. A, ATF4 and C/EBPβ mRNA expression increased following -AA treatment, but both were repressed in the presence of 4 mm glutamine or taurine. B, SNAT2 and TauT mRNA expression increased following -AA, but both were repressed in the presence of 4 mm glutamine or taurine. Results presented are the mean ± S.E. of five independent experiments. a, p < 0.01 compared with the vehicle control; b, not significant (p > 0.05) compared with the vehicle control. C, both glutamine and taurine repressed SNAT2 and TauT induction following -AA treatment using Earle's balanced salt solution. Insets, ATF4 and C/EBPβ mRNA induction was repressed by glutamine and taurine similarly as ACD (7.5 μg/ml). Data are from a representative experiment performed in triplicate.
To examine whether multiple pathways may converge upon the SNAT2 promoter during the adaptive response to -AA treatment in neurons, we examined the mRNA expression of other related transcription factors. The AARE sequence (5′-ATTGCATCA-3′) shows significant homology with the specific binding sequences of two transcription factor families, namely ATF4/CREB-2 and C/EBPβ (33). We therefore examined the expression of C/EBPβ following -AA treatment and its repression by glutamine and taurine. Like ATF4, we find an increase (∼2-fold) of C/EBPβ mRNA following -AA treatment, and this is repressed by glutamine and taurine (Fig. 9A). Induction of both ATF4 and C/EBPβ following -AA treatment is blocked by actinomycin D as expected (data not shown) but not by cycloheximide (Fig. 9A). Both transcription factors are increased by -AA, and the bZIP family of transcription factors may bind to genomic elements as heterodimers (63, 64). Thus, ATF4-C/EBPβ may both be required for SNAT2 gene induction in neurons. Blockade of the increased expression of either transcription factor (or both) may be sufficient to repress SNAT2 mRNA synthesis.
Adaptive Regulation of the Taurine Transporter TauT Is Repressed by Taurine and Glutamine—Since transported substrates of the amino acid osmolyte transporters within the SLC6 GABA transporter subfamily are potent repressors of SNAT2, we examined whether GABA transporters or the taurine transporter (TauT) also exhibit adaptive regulation following -AA treatment in neurons and assessed the effectiveness of System A substrates as repressors. Among the GABA transporters, only BGT-1 (also known as mGAT-2) mRNA levels were significantly up-regulated (∼2-fold) by -AA treatment in our cultures (data not shown); however, TauT mRNA levels increased ∼6-fold following prolonged -AA treatment (Fig. 9B). As expected, the inclusion of taurine in Krebs-Ringer bicarbonate buffer blocked the induction of TauT mRNA (Fig. 9B). Surprisingly, the inclusion of glutamine also blocked adaptive regulation of TauT (Fig. 9B). Induction of both SNAT2 and TauT mRNA is blocked by actinomycin D (data not shown) and cycloheximide (Fig. 9B). Similar results were also observed using Earle's balanced salt solution as the -AA treatment instead of Krebs-Ringer bicarbonate buffer (Fig. 9C). Collectively, our results indicate that these neutral α- and β-amino acids may serve a common role in central neurons that converge at the level of gene regulation of their respective neuronal transporters.
DISCUSSION
Adaptive Regulation of SNAT2 mRNA Expression by Amino Acid Deprivation in Neurons Is Not Selective for System A Substrates—We have demonstrated for the first time that SNAT2 is subject to adaptive regulation following total amino acid deprivation in neurons and that such adaptive regulation of SNAT2 is not limited to the effects of System A substrates. In fact, substrates of the SLC6 GABA transporter family, including GABA, β-alanine, β-AIB, and taurine, block SNAT2 mRNA induction ∼10-fold more potently than do System A substrates. We confirmed, in the oocyte overexpression system, that GABA and the β-amino acids described are not System A substrates. Not surprisingly, we uncovered an early report indicating that β-alanine represses the adaptive regulation of System A in Chinese hamster ovary cells (65), and an even earlier report showed that the hydrophobic amino acids tryptophan, phenylalanine, and tyrosine (but not leucine) could repress adaptive regulation of System A in an established line of rat hepatoma cells (66). Nevertheless, confusion as to the nature of control of SNAT2 expression is still apparent in the literature. β-Alanine and GABA are substrates of the taurine transporter TauT (67, 68) as well as of rat GAT-2/3 (57–59); orphan transporters whose substrates include GABA and taurine may also exist (69). In support of transport-mediated regulation of SNAT2, we found that transporter-selective substrates, such as guanidinoethylsulfonate (taurine transporter), nipecotic acid (GABA transporter), and MeAIB (System A), but not 2-aminobicyclo-(2,2,1)-heptane-2-carboxylic acid (System L), repressed adaptive regulation of SNAT2.
Our present findings support the concept that amino acid deprivation releases neuronal SNAT2 from its constitutive repression (i.e. “derepression”) at the genomic level. We found that this adaptive regulation is a property only of SNAT2 and not of SNAT1. Amino acid deprivation results in parallel increases in SNAT2 mRNA, plasma-membrane protein expression, and System A activity (Vmax effect). The presence of any System A substrate at saturating concentrations in the extracellular medium blocks this up-regulation of SNAT2. The mid-point of glutamine-inhibited SNAT2 up-regulation in neocortical neurons occurs at ∼0.5 mm, which lies within the range of K0.5 values reported for SNAT2 (2, 3, 50) and is close to the concentration of glutamine (∼0.4 mm) expected in extrasynaptic space (14–16). Glutamine is far more abundant there than is either taurine or GABA, but glutamine repressed SNAT2 far less potently. Taurine is present in extraneuronal space at concentrations of 35–50 μm (70). At these concentrations, taurine inhibits the induction of SNAT2 activity in our system, suggesting that taurine may negatively regulate expression of endogenous SNAT2 mRNA levels in the brain. We therefore expect that extracellular levels of glutamine and taurine (and also GABA during development) will determine neuronal SNAT2 expression levels.
Neurotransmitter Glutamate Regeneration Does Not Rely on SNAT2—Spontaneous activity of excitatory projections (which comprise the majority of cells in our culture) is unaffected by removal of exogenous amino acids for 8 h. Similar results were recently obtained using acute hippocampal slices, indicating that pyramidal neurons have the capacity to produce glutamate for long periods of time (51). Whereas resident astrocytes in culture (and slices) may export endogenous glutamine directly to pyramidal neurons via System N (SNAT3, SNAT5) (20, 21, 71), in our preparation, our data indicate that SNAT2 does not provide glutamine to neurons for glutamate neurotransmitter regeneration under these conditions, because inducing SNAT2 levels (>5-fold) or repressing SNAT2 levels to negligible levels had no effect on spontaneous action potential-derived excitatory postsynaptic potentials. Furthermore, any increase in cytoplasmic glutamate levels derived from glutamine imported by SNAT2 would have been expected to increase the quantal size of glutamate release (72, 73), but miniature quantal amplitude was not altered following SNAT2 induction or repression.
We have previously shown that SNAT1 and SNAT2 protein expression in neurons is restricted to somata and dendritic processes, and little, if any, co-localization with synaptophysin, a synaptic vesicle marker, is apparent (6, 8, 10). Although glutamine taken up in neuronal somata and dendrites may be converted to glutamate, the glutamate generated there is not readily available to synaptic vesicles in axon terminals of projection neurons to which the major vesicular glutamate transporter proteins VGLUT1 and VGLUT2 are restricted (43). Indeed, Broer and co-workers (74) have shown by NMR spectroscopy that System A-mediated glutamine transport supports a large pool of neuronal glutamate that turns over very slowly (presumably in cell soma and proximal dendritic regions), and instead, a separate histidine-sensitive glutamine transport system may be involved in providing glutamine for excitatory transmission. However, the presence of a glutamine transporter in nerve terminals that supports glutamate transmission has not been established. Our data do not rule out a role for System A glutamine transport in transmitter biosynthesis in nonprojection systems (excitatory or inhibitory), such as in interneurons (75, 76), various local feedback circuits, such as those described in motoneurons (77), or in smaller cells, such as glutamatergic sensory ganglia in which, for example, plasma membrane expression of SNAT1 is exceptionally high (6). Recent work hypothesizes that SNAT2 mediates replenishment of dendritic glutamate pools in pyramidal neurons for filling VGLUT3-containing vesicles (78). However, although scattered populations of VGLUT3-expressing neurons are found in the cerebral cortex (79, 80), SNAT2 is found in all pyramidal neurons (10). A role for System A in directly providing glutamine to inhibitory neurons for synaptic GABA release has recently been shown (75). SNAT1 is abundantly expressed in established GABAergic neurons (6, 8), as well as in excitatory neurons (1, 3, 5).
The selective regulation at the SLC38A2 gene locus encoding SNAT2 in neocortical neurons supports the notion that the multiple isoforms of System A serve unique roles in neuronal function. The presence of SNAT1 and/or SNAT2 protein on brain cells other than neurons, such as in leptomeninges, ependymal cells, and choroid plexus (6, 8), in astrocytes and oligodendrocytes (8, 81) as well as both excitatory and inhibitory neurons suggests roles outside of neurotransmitter glutamate regeneration. Interestingly, recent evidence strongly supports a role for SNAT1 in the brain to supply neutral amino acids for local protein synthesis related to synaptic remodeling and plasticity in pyramidal neuronal dendrites (82). Indeed, SNAT1 and SNAT2 expression is directly linked to amino acid delivery for protein synthesis in various peripheral tissues (83–85).
A Role for SNAT2 and TauT as Compatible Neutral Amino Acid Cytoprotectants in Neurons—The amino acid transporters of the SLC6 GABA transporter subfamily (53, 54, 62) are known to mediate substrate cotransport with Na+ and Cl- (Na/Cl/taurine, 2:1:1; Na/Cl/GABA, 3:2:1). SNAT2 displays stoichiometry of 1:1 (Na+/amino acid) (2). Such coupling with the Na+ electrochemical gradient permits these transporters to be concentrative. Efficient amino acid transport by members of the SLC6 GABA transporter subfamily or by System A (via SNAT2) may compensate for the loss of endogenous neutral amino acids in cell somata and proximal dendrites expected to occur during amino acid starvation. Amino acid transport by SNAT2 or by known osmolyte transporters (e.g. TauT, rGAT-2/3, and BGT-1) may be one mechanism by which excitatory and inhibitory neurons and astrocytes maintain cell volume within a very narrow range, by directing the transport of organic solutes. Na+-coupled solute transport could thus be utilized to achieve osmotic balance. The specificity of the SNAT2 adaptive regulatory process in other systems may therefore be determined by whichever transporters are expressed in any given cell type and by whichever transporter genes possess neutral AAREs.
Molecular studies indicate that SNAT2 plays a dominant role in the control of intracellular volume in fibroblasts following hyperosmotic stress (36, 37, 86, 87). Notably, intracellular volume is altered, and ultimately, cell shrinkage is a result that is common to both hyperosmotic conditions and adaptive regulation following amino acid depletion. The TauT, BGT-1, and SMIT (myo-inositol transporter (SLC5A3)) are each up-regulated by hypertonic stress (88, 89). myo-Inositol is in an organic osmolyte class (polyols) that differs from the methylamines (betaine) and the amino acids. We found that betaine is not as effective as is glutamine in repressing SNAT2 and that myo-inositol was ineffective. Thus, some degree of osmolyte (and amino acid) specificity in SNAT2 adaptive regulation argues against the involvement of membrane channels, such as the volume-sensitive organic osmolyte and anion channel (90, 91), in regulating SNAT2 expression. BGT-1 and SMIT are regulated by a tonicity-responsive element (Ton-E) in renal cell lines (92, 93). A different set of factors converges upon the SNAT2 gene locus to activate or repress transcription.
We have shown that both ATF4 and C/EBPβ mRNA expression increase following -AA treatment in neurons, and the induction of these transcription factors is blocked by the System A substrate glutamine. We have also shown that taurine is a potent repressor of SNAT2 mRNA induction following -AA treatment and that taurine also represses ATF4 and C/EBPβ expression. Thus, we postulate that both transcription factors are important in conferring specificity of transporter gene induction to this osmotic/nutritional stress in neurons. High affinity taurine transport is also adaptively regulated by substrate in vitro and in vivo (62, 94, 95). Surprisingly, we have shown that a System A substrate, such as glutamine, also represses TauT induction in neurons. Indeed, both SNAT2 and TauT are negatively regulated by System A substrates and taurine. Recently, Kilberg and co-workers (96) have reexamined the substrate specificity of SNAT2 adaptive regulation following complete amino acid deprivation and suggested that AARE-independent transcription of SNAT2 occurs in Krebs-Ringer bicarbonate buffer but not in Earle's balanced salt solution. We therefore examined the ability of glutamine and taurine to repress SNAT2, TauT, ATF4, and C/EBPβ expression using Earle's balanced salt solution and found essentially identical results as those presented with Krebs-Ringer bicarbonate buffer.
In conclusion, the coordinate pattern of regulation of the transport of neutral α- and β-amino acids in neurons responsive to total amino acid deprivation may serve a similar role to restore intracellular amino acids that are expected to be lost during -AA treatment. Taurine is classically regarded as a principal amino acid osmolyte in neurons (97), and it does not play a direct role in nutritive amino acid metabolism. Hence, its negative control over SNAT2 expression suggests that β-amino acids, such as taurine, and System A α-amino acid substrates, like glutamine, serve as compatible organic solutes in neuronal somata and dendrites in the brain. Interestingly, recent work indicates an adaptive increase in the level of expression of SNAT2 in the TauT knock-out mouse (98). Maintenance of cell volume via these relatively inert neutral amino acids instead of by inorganic ions, such as Na+, K+, Cl-, or Ca2+, is critical for neurons in the brain. Thus, the functionally interchangeable transport of α-and β-amino acids in the regulation of genes controlling the expression of their respective transporters suggests that these amino acids serve as compatible counteracting cyoprotectants in high osmolarity and other stresses.
This work was supported, in whole or in part, by National Institutes of Health Grants 1P29RR16816 (to H. V.) and NS36936 (to J. D. E.). This work was also supported by the University of Cincinnati (to B. M.).
Footnotes
The abbreviations used are: -AA, amino acid deprivation; AARE, amino acid response element; ACD, actinomycin D; β-AIB, β-aminoisobutyric acid; C/EBPβ, CCAAT/enhancer-binding protein; CHX, cycloheximide; GABA, γ-aminobutyric acid; MeAIB, α-methylaminoisobutyric acid; mEPSC, miniature excitatory postsynaptic current; PBS, phosphate-buffered saline; RT, reverse transcription.
References
- 1.Varoqui, H., Zhu, H., Yao, D., Ming, H., and Erickson, J. D. (2000) J. Biol. Chem. 275, 4049-4054 [DOI] [PubMed] [Google Scholar]
- 2.Yao, D., Mackenzie, B., Ming, H., Varoqui, H., Zhu, H., Hediger, M. A., and Erickson, J. D. (2000) J. Biol. Chem. 275, 22790-22797 [DOI] [PubMed] [Google Scholar]
- 3.Reimer, R. J., Chaudhry, F. A., Gray, A. T., and Edwards, R. H. (2000) Proc. Natl. Acad. Sci. U. S. A. 97, 7715-7720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu, S., Roderick, H. L., Camacho, P., and Jiang, J. X. (2001) J. Biol. Chem. 276, 24137-24144 [DOI] [PubMed] [Google Scholar]
- 5.Chaudhry, F. A., Schmitz, D., Reimer, R. J., Larsson, P., Gray, A. T., Nicoll, R., Kavanaugh, M., and Edwards, R. H. (2002) J. Neurosci. 22, 62-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackenzie, B., Schafer, M. K., Erickson, J. D., Hediger, M. A., Weihe, E., and Varoqui, H. (2003) J. Biol. Chem. 278, 23720-23730 [DOI] [PubMed] [Google Scholar]
- 7.Dolinska, M., Zablocka, B., Sonnewald, U., and Albrecht, J. (2004) Neurochem. Int. 44, 75-81 [DOI] [PubMed] [Google Scholar]
- 8.Melone, M., Quagliano, F., Barbaresi, P., Varoqui, H., Erickson, J. D., and Conti, F. (2004) Cereb. Cortex 14, 562-574 [DOI] [PubMed] [Google Scholar]
- 9.Cubelos, B., Gonzalez-Gonzalez, I. M., Gimenez, C., and Zafra, F. (2005) Glia 49, 230-244 [DOI] [PubMed] [Google Scholar]
- 10.Melone, M., Varoqui, H., Erickson, J. D., and Conti, F. (2006) Neuroscience 140, 281-292 [DOI] [PubMed] [Google Scholar]
- 11.Mackenzie, B., and Erickson, J. D. (2004) Pflugers Arch. Eur. J. Physiol. 447, 784-795 [DOI] [PubMed] [Google Scholar]
- 12.Christensen, H. N. (1990) Physiol. Rev. 70, 43-77 [DOI] [PubMed] [Google Scholar]
- 13.McGivan, J. D., and Pastor-Anglada, M. (1994) Biochem. J. 299, 321-334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gjessing, L. R., Gjesdahl, P., and Sjaastad, O. (1972) J. Neurochem. 19, 1807-1808 [DOI] [PubMed] [Google Scholar]
- 15.McGale, E. H., Pye, I. F., Stonier, C., Hutchinson, E. C., and Aber, G. M. (1977) J. Neurochem. 29, 291-297 [DOI] [PubMed] [Google Scholar]
- 16.Kanamori, K., and Ross, B. D. (2004) J. Neurochem. 90, 203-210 [DOI] [PubMed] [Google Scholar]
- 17.Hertz, L. (2004) Neurochem. Int. 45, 285-296 [DOI] [PubMed] [Google Scholar]
- 18.Conti, F., and Minelli, A. (1994) J. Histochem. Cytochem. 42, 717-726 [DOI] [PubMed] [Google Scholar]
- 19.Danbolt, N. C. (2001) Prog. Neurobiol. 65, 1-105 [DOI] [PubMed] [Google Scholar]
- 20.Chaudhry, F. A., Reimer, R. J., Krizaj, D., Barber, D., Storm-Mathisen, J., Copenhagen, D. R., and Edwards, R. H. (1999) Cell 99, 769-780 [DOI] [PubMed] [Google Scholar]
- 21.Deitmer, J. W., Broer, A., and Broer, S. (2003) J. Neurochem. 87, 127-135 [DOI] [PubMed] [Google Scholar]
- 22.Chaudhry, F. A., Reimer, R. J., and Edwards, R. H. (2002) J. Cell Biol. 157, 349-355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conti, F., and Melone, M. (2006) Neurochem. Int. 48, 459-464 [DOI] [PubMed] [Google Scholar]
- 24.Kilberg, M. S., Han, H. P., Barber, E. F., and Chiles, T. C. (1985) J. Cell. Physiol. 122, 290-298 [DOI] [PubMed] [Google Scholar]
- 25.Ling, R., Bridges, C. C., Sugawara, M., Fujita, T., Leibach, F. H., Prasad, P. D., and Ganapathy, V. (2001) Biochim. Biophys. Acta 1512, 15-21 [DOI] [PubMed] [Google Scholar]
- 26.Gazzola, R. F., Sala, R., Bussolati, O., Visigalli, R., Dall'Asta, V., Ganapathy, V., and Gazzola, G. C. (2001) FEBS Lett. 490, 11-14 [DOI] [PubMed] [Google Scholar]
- 27.Hyde, R., Christie, G. R., Litherland, G. J., Hajduch, E., Taylor, P. M., and Hundal, H. S. (2001) Biochem. J. 355, 563-568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Handlogten, M. E., and Kilberg, M. S. (1984) J. Biol. Chem. 259, 3519-3525 [PubMed] [Google Scholar]
- 29.Bracy, D. S., Handlogten, M. E., Barber, E. F., Han, H. P., and Kilberg, M. S. (1986) J. Biol. Chem. 261, 1514-1520 [PubMed] [Google Scholar]
- 30.Boerner, P., and Saier, M. H., Jr. (1985) J. Cell. Physiol. 122, 308-315 [DOI] [PubMed] [Google Scholar]
- 31.Dall'Asta, V., Franchi-Gazzola, R., Bussolati, O., Sala, R., Rotoli, B. M., Rossi, P. A., Uggeri, J., Belletti, S., Visigalli, R., and Gazzola, G. C. (1996) Biochem. Soc. Trans. 24, 864-869 [DOI] [PubMed] [Google Scholar]
- 32.Palii, S. S., Chen, H., and Kilberg, M. S. (2004) J. Biol. Chem. 279, 3463-3471 [DOI] [PubMed] [Google Scholar]
- 33.Jousse, C., Averous, J., Bruhat, A., Carraro, V., Mordier, S., and Fafournoux, P. (2004) Biochem. Biophys. Res. Commun. 313, 447-452 [DOI] [PubMed] [Google Scholar]
- 34.Hyde, R., Taylor, P. M., and Hundal, H. S. (2003) Biochem. J. 373, 1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hyde, R., Cwiklinski, E. L., MacAulay, K., Taylor, P. M., and Hundal, H. S. (2007) J. Biol. Chem. 282, 19788-19798 [DOI] [PubMed] [Google Scholar]
- 36.Franchi-Gazzola, R., Visigalli, R., Bussolati, O., Dall'Asta, V., and Gazzola, G. C. (1999) J. Biol. Chem. 274, 28922-28928 [DOI] [PubMed] [Google Scholar]
- 37.Franchi-Gazzola, R., Dall'Asta, V., Sala, R., Visigalli, R., Bevilacqua, E., Gaccioli, F., Gazzola, G. C., and Bussolati, O. (2006) Acta Physiol. (Oxf.) 187, 273-283 [DOI] [PubMed] [Google Scholar]
- 38.Lopez-Fontanals, M., Rodriguez-Mulero, S., Casado, F. J., Derijard, B., and Pastor-Anglada, M. (2003) J. Gen. Physiol. 122, 5-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pastor-Anglada, M., Derijard, B., and Casado, F. J. (2005) J. Gen. Physiol. 125, 41-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brewer, G. J., Torricelli, J. R., Evege, E. K., and Price, P. J. (1993) J. Neurosci. Res. 35, 567-576 [DOI] [PubMed] [Google Scholar]
- 41.De Gois, S., Schafer, M. K., Defamie, N., Chen, C., Ricci, A., Weihe, E., Varoqui, H., and Erickson, J. D. (2005) J. Neurosci. 25, 7121-7133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson, N. R., Kang, J., Hueske, E. V., Leung, T., Varoqui, H., Murnick, J. G., Erickson, J. D., and Liu, G. (2005) J. Neurosci. 25, 6221-6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varoqui, H., Schafer, M. K., Zhu, H., Weihe, E., and Erickson, J. D. (2002) J. Neurosci. 22, 142-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mackenzie, B. (1999) Biomembrane Transport, Academic Press, Inc., San Diego, CA
- 45.Thurlow, R. J., Hill, D. R., and Woodruff, G. N. (1996) Br. J. Pharmacol. 118, 449-456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takanaga, H., Mackenzie, B., Peng, J. B., and Hediger, M. A. (2005) Biochem. Biophys. Res. Commun. 337, 892-900 [DOI] [PubMed] [Google Scholar]
- 47.Broer, A., Tietze, N., Kowalczuk, S., Chubb, S., Munzinger, M., Bak, L. K., and Broer, S. (2006) Biochem. J. 393, 421-430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamarappoo, B. K., Raizada, M. K., and Kilberg, M. S. (1997) J. Neurochem. 68, 954-960 [DOI] [PubMed] [Google Scholar]
- 49.Su, T. Z., Campbell, G. W., and Oxender, D. L. (1997) Brain Res. 757, 69-78 [DOI] [PubMed] [Google Scholar]
- 50.Sugawara, M., Nakanishi, T., Fei, Y. J., Huang, W., Ganapathy, M. E., Leibach, F. H., and Ganapathy, V. (2000) J. Biol. Chem. 275, 16473-16477 [DOI] [PubMed] [Google Scholar]
- 51.Kam, K., and Nicoll, R. (2007) J. Neurosci. 27, 9192-9200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Albers, A., Broer, A., Wagner, C. A., Setiawan, I., Lang, P. A., Kranz, E. U., Lang, F., and Broer, S. (2001) Pflugers Arch. Eur. J. Physiol. 443, 92-101 [DOI] [PubMed] [Google Scholar]
- 53.Borden, L. A. (1996) Neurochem. Int. 29, 335-356 [DOI] [PubMed] [Google Scholar]
- 54.Nelson, N. (1998) J. Neurochem. 71, 1785-1803 [DOI] [PubMed] [Google Scholar]
- 55.Minelli, A., Barbaresi, P., and Conti, F. (2003) Brain Res. Dev. Brain Res. 142, 7-18 [DOI] [PubMed] [Google Scholar]
- 56.Minelli, A., Brecha, N. C., Karschin, C., DeBiasi, S., and Conti, F. (1995) J. Neurosci. 15, 7734-7746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lopez-Corcuera, B., Liu, Q. R., Mandiyan, S., Nelson, H., and Nelson, N. (1992) J. Biol. Chem. 267, 17491-17493 [PubMed] [Google Scholar]
- 58.Liu, Q. R., Lopez-Corcuera, B., Mandiyan, S., Nelson, H., and Nelson, N. (1993) J. Biol. Chem. 268, 2106-2112 [PubMed] [Google Scholar]
- 59.Liu, M., Russell, R. L., Beigelman, L., Handschumacher, R. E., and Pizzorno, G. (1999) Am. J. Physiol. 276, G206-G210 [DOI] [PubMed] [Google Scholar]
- 60.Liu, Q. R., Lopez-Corcuera, B., Nelson, H., Mandiyan, S., and Nelson, N. (1992) Proc. Natl. Acad. Sci. U. S. A. 89, 12145-12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia-Perez, A., and Burg, M. B. (1991) Physiol. Rev. 71, 1081-1115 [DOI] [PubMed] [Google Scholar]
- 62.Han, X., Patters, A. B., Jones, D. P., Zelikovic, I., and Chesney, R. W. (2006) Acta Physiol. (Oxf.) 187, 61-73 [DOI] [PubMed] [Google Scholar]
- 63.Fawcett, T. W., Martindale, J. L., Guyton, K. Z., Hai, T., and Holbrook, N. J. (1999) Biochem. J. 339, 135-141 [PMC free article] [PubMed] [Google Scholar]
- 64.Wolfgang, C. D., Chen, B. P., Martindale, J. L., Holbrook, N. J., and Hai, T. (1997) Mol. Cell. Biol. 17, 6700-6707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moffett, J., and Englesberg, E. (1984) Mol. Cell. Biol. 4, 799-808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heaton, J. H., and Gelehrter, T. D. (1977) J. Biol. Chem. 252, 2900-2907 [PubMed] [Google Scholar]
- 67.Fujita, T., Shimada, A., Wada, M., Miyakawa, S., and Yamamoto, A. (2006) Pharm. Res. 23, 689-696 [DOI] [PubMed] [Google Scholar]
- 68.Tomi, M., Tajima, A., Tachikawa, M., and Hosoya, K. (2008) Biochim. Biophys. Acta 1778, 2138-2142 [DOI] [PubMed] [Google Scholar]
- 69.Peterson, W. M., and Miller, S. S. (1995) J. Gen. Physiol. 106, 1089-1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lehmann, A., Carlstrom, C., Nagelhus, E. A., and Ottersen, O. P. (1991) J. Neurochem. 56, 690-697 [DOI] [PubMed] [Google Scholar]
- 71.Boulland, J. L., Osen, K. K., Levy, L. M., Danbolt, N. C., Edwards, R. H., Storm-Mathisen, J., and Chaudhry, F. A. (2002) Eur. J. Neurosci. 15, 1615-1631 [DOI] [PubMed] [Google Scholar]
- 72.Ishikawa, T., Sahara, Y., and Takahashi, T. (2002) Neuron 34, 613-621 [DOI] [PubMed] [Google Scholar]
- 73.Erickson, J. D., De Gois, S., Varoqui, H., Schafer, M. K., and Weihe, E. (2006) Neurochem. Int. 48, 643-649 [DOI] [PubMed] [Google Scholar]
- 74.Rae, C., Hare, N., Bubb, W. A., McEwan, S. R., Broer, A., McQuillan, J. A., Balcar, V. J., Conigrave, A. D., and Broer, S. (2003) J. Neurochem. 85, 503-514 [DOI] [PubMed] [Google Scholar]
- 75.Liang, S. L., Carlson, G. C., and Coulter, D. A. (2006) J. Neurosci. 26, 8537-8548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fricke, M. N., Jones-Davis, D. M., and Mathews, G. C. (2007) J. Neurochem. 102, 1895-1904 [DOI] [PubMed] [Google Scholar]
- 77.Herzog, E., Landry, M., Buhler, E., Bouali-Benazzouz, R., Legay, C., Henderson, C. E., Nagy, F., Dreyfus, P., Giros, B., and El Mestikawy, S. (2004) Eur. J. Neurosci. 20, 1752-1760 [DOI] [PubMed] [Google Scholar]
- 78.Jenstad, M., Quazi, A. Z., Zilberter, M., Haglerod, C., Berghuis, P., Saddique, N., Goiny, M., Buntup, D., Davanger, S., Haug, F. M., Barnes, C. A., McNaughton, B. L., Ottersen, O. P., Storm-Mathisen, J., Harkany, T., and Chaudhry, F. A. (2009) Cereb. Cortex, in press [DOI] [PubMed]
- 79.Herzog, E., Gilchrist, J., Gras, C., Muzerelle, A., Ravassard, P., Giros, B., Gaspar, P., and El Mestikawy, S. (2004) Neuroscience 123, 983-1002 [DOI] [PubMed] [Google Scholar]
- 80.Schafer, M. K., Varoqui, H., Defamie, N., Weihe, E., and Erickson, J. D. (2002) J. Biol. Chem. 277, 50734-50748 [DOI] [PubMed] [Google Scholar]
- 81.Maallem, S., Mutin, M., Gonzalez-Gonzalez, I. M., Zafra, F., and Tappaz, M. L. (2008) Neuroscience 153, 95-107 [DOI] [PubMed] [Google Scholar]
- 82.Burkhalter, J., Fiumelli, H., Erickson, J. D., and Martin, J. L. (2007) J. Biol. Chem. 282, 5152-5159 [DOI] [PubMed] [Google Scholar]
- 83.Evans, K., Nasim, Z., Brown, J., Butler, H., Kauser, S., Varoqui, H., Erickson, J. D., Herbert, T. P., and Bevington, A. (2007) J. Am. Soc. Nephrol. 18, 1426-1436 [DOI] [PubMed] [Google Scholar]
- 84.Novak, D., Quiggle, F., and Haafiz, A. (2006) Biochimie (Paris) 88, 39-44 [DOI] [PubMed] [Google Scholar]
- 85.Hyde, R., Hajduch, E., Powell, D. J., Taylor, P. M., and Hundal, H. S. (2005) FASEB J. 19, 461-463 [DOI] [PubMed] [Google Scholar]
- 86.Franchi-Gazzola, R., Gaccioli, F., Bevilacqua, E., Visigalli, R., Dall'Asta, V., Sala, R., Varoqui, H., Erickson, J. D., Gazzola, G. C., and Bussolati, O. (2004) Biochim. Biophys. Acta 1667, 157-166 [DOI] [PubMed] [Google Scholar]
- 87.Morimura, H., Shimada, S., Otori, Y., Saishin, Y., Yamauchi, A., Minami, Y., Inoue, K., Ishimoto, I., Tano, Y., and Tohyama, M. (1997) Brain Res. Mol. Brain Res. 44, 245-252 [DOI] [PubMed] [Google Scholar]
- 88.Burg, M. B., Kwon, E. D., and Kultz, D. (1997) Annu. Rev. Physiol. 59, 437-455 [DOI] [PubMed] [Google Scholar]
- 89.Olsen, M., Sarup, A., Larsson, O. M., and Schousboe, A. (2005) Neurochem. Res. 30, 855-865 [DOI] [PubMed] [Google Scholar]
- 90.Jackson, P. S., and Strange, K. (1993) Am. J. Physiol. 265, C1489-C1500 [DOI] [PubMed] [Google Scholar]
- 91.Roy, G., and Banderali, U. (1994) J. Exp. Zool. 268, 121-126 [DOI] [PubMed] [Google Scholar]
- 92.Rim, J. S., Atta, M. G., Dahl, S. C., Berry, G. T., Handler, J. S., and Kwon, H. M. (1998) J. Biol. Chem. 273, 20615-20621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takenaka, M., Preston, A. S., Kwon, H. M., and Handler, J. S. (1994) J. Biol. Chem. 269, 29379-29381 [PubMed] [Google Scholar]
- 94.Jones, D. P., Miller, L. A., and Chesney, R. W. (1990) Kidney Int. 38, 219-226 [DOI] [PubMed] [Google Scholar]
- 95.Jessen, H., and Jacobsen, C. (1997) Biochim. Biophys. Acta 1325, 309-317 [DOI] [PubMed] [Google Scholar]
- 96.Palii, S. S., Kays, C. E., Deval, C., Bruhat, A., Fafournoux, P., and Kilberg, M. S. (2009) Amino Acids, in press [DOI] [PMC free article] [PubMed]
- 97.Schousboe, A., Apreza, C. L., and Pasantes-Morales, H. (1992) Adv. Exp. Med. Biol. 315, 391-397 [DOI] [PubMed] [Google Scholar]
- 98.Ito, T., Kimura, Y., Uozumi, Y., Takai, M., Muraoka, S., Matsuda, T., Ueki, K., Yoshiyama, M., Ikawa, M., Okabe, M., Schaffer, S. W., Fujio, Y., and Azuma, J. (2008) J. Mol. Cell Cardiol. 44, 927-937 [DOI] [PubMed] [Google Scholar]