Abstract
Although the classical redox functions of co-enzyme NAD+ are firmly established in metabolism, there are numerous enzymes that catalyze cleavage of NAD+ to yield free ADP-ribose (ADPr) or related metabolites, whose functions remain largely unknown. Here we show that the Nudix (nucleoside diphosphate linked to another moiety X) hydrolase Ysa1 from Saccharomyces cerevisiae is a major regulator of cellular ADPr and O-acetyl-ADP-ribose (OAADPr). OAADPr is the direct product of NAD+-dependent protein deacetylases (sirtuins) and is readily converted to ADPr. Ysa1 cleaves ADPr/OAADPr into ribose phosphate/acetyl-ribose phosphate and AMP. In cells lacking Ysa1 (Δysa1), ADPr and OAADPr levels increased ∼50%, with a corresponding decrease in AMP. Strikingly, Δysa1 cells display higher resistance to exogenous reactive oxygen species (ROS) and 40% lower basal levels of endogenous ROS, compared with wild type. The biochemical basis for these differences in ROS-related phenotypes was investigated, and the results provide evidence that increased ADPr/OAADPr levels protect cells via the following two pathways: (i) lower ROS production through inhibition of complex I of the mitochondrial electron transport chain, and (ii) generation of higher levels of NADPH to suppress ROS damage. The latter occurs through diverting glucose into the pentose phosphate pathway by ADPr inhibition of glyceraldehyde-3-phosphate dehydrogenase, a central enzyme of glycolysis.
NAD+ is well known for its role as a hydride-transferring co-enzyme in many oxidation-reduction reactions of metabolism. However, NAD+ is also a substrate for NAD+ glycohydrolases, ADP-ribose transferases, poly(ADP-ribose) polymerases (PARPs),2 cyclic ADP-ribose synthases (1, 2), and sirtuins (3, 4), all of which cleave the glycosidic bond of NAD+ to produce nicotinamide and an ADP-ribosyl product. Notably, sirtuins catalyze NAD+-dependent lysine deacetylation to generate nicotinamide, deacetylated lysine, and OAADPr (5, 6). OAADPr has been proposed to act as a second messenger, signaling to other processes that NAD+-dependent protein deacetylation has occurred (7–9). The biological functions and in vivo metabolism of OAADPr and free ADPr are largely unknown.
Through a quantitative microinjection assay of starfish oocytes, both ADPr and OAADPr caused a delay/block in oocyte maturation, suggesting ADPr/OAADPr may have specific biological activity (10). In mammalian cells, intracellular ADPr/OAADPr can activate the TRPM2 (transient receptor melastatin-related ion channel 2) nonselective cationic channel (11–13). TRPM2 contains a conserved intracellular Nudix hydrolase domain (referred to as NudT9H) that directly binds ADPr/OAADPr, but it is incapable of cleaving the ligand because a major catalytic residue is missing (11, 14). Although still disputed, ADPr binding to NudT9H appears to be required for the well known oxidative stress activation of the channel (13, 15). Cell stress via puromycin treatment led to TRPM2-mediated cell death that was dependent on sirtuin deacetylases, presumably from the production of OAADPr (12).
Increasing evidence suggests that free ADPr may function as a cellular signal. ADPr can be produced from the coordinate actions of PARPs and poly(ADP-ribose) glycohydrolase (PARG), which cleave ADPr polymers to free ADPr (16, 17). Under massive genotoxic stress, hyper-stimulation of the NAD+-dependent PARPs depletes cellular NAD+, which is linked to catastrophic ATP loss and cell death (18, 19). The mechanism by which PARP1 hyperactivity in the nucleus impairs ATP production in mitochondria is unclear. The fact that PARP1 and poly(ADP-ribose) are localized in the nucleus adds a perplexing aspect. However, recent data suggest that PARP1-induced loss of ATP requires PARG (20). Under conditions of PARP1 hyperactivation, it has been suggested that the PARG-dependent production of ADPr can exit the nucleus and interfere with ATP production in mitochondria (21, 22). Thus ADPr could be the molecular signal released from the nucleus of cells undergoing massive poly(ADP-ribosyl)ation and rapidly triggers mitochondrial dysfunction.
In support for ADPr/OAADPr as potential signaling molecules, the existence of enzymes capable of metabolizing these compounds suggests that their cellular concentrations may be subject to tight regulation (23, 24). To understand the biological roles played by ADPr/OAADPr, it is essential to elucidate the degradation pathways that can modulate their levels. Previously we described the ability of several conserved members of the Nudix hydrolase family to hydrolyze in vitro the diphosphate linkage in ADPr/OAADPr, generating ribose phosphate or acetyl-ribose phosphate and AMP (10, 24). Here we examine the biochemical and cellular functions of the Nudix hydrolase Ysa1 (14) from Saccharomyces cerevisiae. We determined that Ysa1 is the major ADPr Nudix hydrolase and an important regulator of cellular ADPr/OAADPr levels. A Δysa1 strain displays increased resistance to both exogenously and endogenously generated ROS. Basal level of ROS decreased by 40% in the Ysa1 deletion strain. We provide biochemical evidence that increased ADPr/OAADPr levels protect cells via the following two pathways: (i) lower ROS production through the inhibition of complex I of the electron transport chain, and (ii) generation of higher NADPH levels to suppress ROS damage. The latter occurs by diverting glucose into the pentose phosphate pathway by ADPr inhibition of glycolysis.
EXPERIMENTAL PROCEDURES
Yeast Strains—Parental strains BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0), Δysa1 (Δysa1::Kanr), with BY4741 and BY4742 as parental strains, and TAP-Ysa1 strain with BY4742 as parental strain were purchased from Open Biosystems (Huntsville, AL). The TAP tag was inserted at the C-terminal end of the coding region of YSA1 gene, consisting of a calmodulin-binding peptide, a tobacco etch virus protease cleavage site, and two IgG binding domains of Staphylococcus aureus protein A (25).
Cultivation and Harvest Conditions—For the copper resistance spot test experiment, cells were spotted on copper containing YMD (2% (w/v) glucose, 0.67% (w/v) yeast nitrogen base without amino acids, supplemented with histidine (20 mg/liter), leucine (40 mg/liter), lysine (40 mg/liter), and uracil (20 mg/liter)), 2% (w/v) agar plates. For all other experiments, yeast strains were cultivated on 2% (w/v) agar plates or in liquid culture with rich YPD medium (1% (w/v) yeast extract, 2% (w/v) tryptone, 2% (w/v) glucose) at 30 °C. Liquid cultures were harvested by centrifugation at 1,600 × g for 20 min at 4 °C and rinsed once with ddH2O, and the pellet was stored at -20 °C.
Cell Lysis—Packed yeast cell volume was estimated based on cell concentration of 3 × 107 cell/ml with A600 of 1 (26), average cell size of 66 μm3 (27), and the harvested liquid culture size. The pellet was resuspended in 1–3 volumes of lysis buffer (either 20 mm Tris-HCl, pH 7.9, 10 mm MgCl2, 1 mm EDTA, 5% (v/v) glycerol, 1 mm dithiothreitol, 0.3 m ammonium sulfate, 1 mm phenylmethylsulfonyl fluoride for protein extraction; 10% (v/v) trifluoroacetic acid in water for LC-MS/MS analysis of ADPr/OAADPr; or as indicated below for determination of NAD+, NADH, AMP, ATP, and NADPH), resulting in a 2–4-fold dilution of the final cellular metabolite concentrations. This cell paste was mixed with an equal volume of glass beads and vortexed at maximum speed for 10 min at 4 °C. The resulting cell extract was spun at 15,800 × g for 30 min at 4 °C. The clear supernatant was collected and stored at -20 °C.
ADPr/OAADPr/NAADPr Metabolizing Assays and HPLC Analysis—To determine the degradation of ADPr/OAADPr/NAADPr within cell extracts, 1 mm of each small molecule was incubated with yeast cell extracts at 37 °C for 30 min in 50 mm Tris, pH 7.5, 10 mm MgCl2. The reactions were quenched with 1% (v/v) trifluoroacetic acid and loaded onto Shimadzu HPLC (LC 2010) and separated by C18 analytical reverse phase column (Grace Vydac, 201SP104) at a flow rate of 0.5 ml/min. The column was equilibrated with buffer A (0.05% (v/v) trifluoroacetic acid in water) for 5 min, and thereafter the reaction mixture was eluted in a 20-min gradient to 8% buffer B (0.02% (v/v) trifluoroacetic acid in acetonitrile), followed by a 20-min gradient to 40% B. The column was then rinsed with 100% buffer B for 10 min and re-equilibrated with buffer A for 10 min before the next injection. ADPr/OAADPr/NAADPr and their respective degradation products were monitored at 260 nm.
Halo Assay—The halo assay was performed following the protocol described in Guisbert et al. (28). Briefly, yeast cells were grown to saturation, and then 50 μl of the culture was rapidly mixed with 4 ml of 0.5% (w/v) agar at 50 °C and poured on top of standard YPD plates. Next, three dilutions of hydrogen peroxide (1.5, 2.9, and 4.4 m) were applied onto the plates as 5-μl drops. The plates were incubated at 30 °C for 1 day before being photographed.
H2O2 Survival Test—Yeast cells were grown to A600 of 0.6 before hydrogen peroxide was added to a final concentration of 3 mm. At each time point, the treatment was quenched by pelleting the cells and rinsing the cell pellets with ddH2O. Next, equal numbers of cells were plated onto standard YPD plates for recovery, and the plates were incubated at 30 °C for 2 days before the number of colonies were counted, using Epi Chem II Darkroom Imager (UVP).
Copper Treatment—For the spot test, yeast cells were grown overnight at 30 °C with constant shaking at 200 rpm. After rinsing with ddH2O, the cultures were serially diluted from A600 = 1–10-5. Each dilution (5 μl) was spotted on YMD plates, with the indicated concentrations of CuSO4, and incubated at 30 °C for 2 days. For the survival test, cells were grown to A600 of 0.6, and CuSO4 was added to the liquid culture to a final concentration of 9 mm. Control cultures without CuSO4 were also grown in parallel. After incubation at 30 °C for 24 h, cells were pelleted, rinsed with ddH2O, and normalized before being plated onto YPD agar plates for recovery. The plates were incubated at 30 °C for 2 days before being photographed.
Assessment of Cellular ROS and Mitochondrial Membrane Potential (ΔΨm)—Cellular ROS and mitochondrial membrane potential were measured using staining with dihydroethidium (DHE) or rhodamine 123 (Rho123), respectively. For ROS staining, cells were incubated in the presence of 5 μg/ml DHE for 15 min, and ROS oxidized DHE into ethidium. For mitochondrial membrane potential, cells were incubated with 2.5 μg/ml Rho123 for 35 min. In each experiment, 10,000 cells were analyzed per cell culture by FACSCalibur flow cytometer (BD Biosciences) equipped with multicolor analysis (λex 488 nm/λem 605 nm for ethidium; λex 507 nm/λem 529 nm for Rho123). Unstained cells served as controls.
Ysa1 Cellular Localization—Nuclei and mitochondria were isolated as described previously (29, 30). Anti-TAP antibody was purchased from Open Biosystems (Huntsville, AL). Anti-Isu1 antibody was a kind gift from the Craig laboratory (University of Wisconsin, Madison). Anti-H3 antibody was a kind gift from Catherine Fox (University of Wisconsin, Madison). Western blots were visualized using Pierce SuperSignal West Pico chemiluminescent substrate kit, according to the manufacturer's instructions.
Screening for ADPr/OAADPr Interacting Proteins—Protein (0.5 mg) from whole cell extract was mixed with 100 μl Cibacron blue resin (C1160, Sigma) pre-equilibrated in 20 mm Tris, pH 7, 1 mm 2-mercaptoethanol. The mixture was mixed at 60 rpm at 4 °C for 1 h. Then the resin was washed four times with 1 ml of wash buffer (20 mm Tris, pH 7.5, 1 mm 2-mercaptoethanol, 10 mm NaCl, 10 mm KCl, 1.5 mm MgCl2) and eluted with 200 μl of wash buffer containing the indicated concentrations of ADPr/OAADPr. The wash and elution samples (100 μl) were concentrated, resolved by SDS-PAGE, and stained with SYPRO Ruby (Molecular Probes) protein stain. Anti-Sir2 antibody was a kind gift from Catherine Fox (University of Wisconsin, Madison). Western blot of Sir2 was visualized as described in Ysa1 localization experiment.
In-gel Digestion and LC-MS/MS—SDS-PAGE bands were ingel digested with trypsin and analyzed using LC-MS/MS, as described (31).
Data Base Searching—Data were searched using Mascot and the Swiss Prot S. cerevisiae Database (Feb. 2006). The search parameters included peptide mass tolerance of 1.2 Da, fragment mass tolerance 0.6 Da, trypsin, allowing for five missed cleavages, and carbamidomethyl (C), deamidation (NQ), and oxidation (M) as variable modifications.
LC-MS/MS Measurement of ADPr/OAADPr Cellular Concentration—Cell extracts were analyzed by LC-MS/MS as described (32). Briefly, cell pellets were spiked with known amounts of isotopic standard 13C-OAADPr before being extracted to correct for sample losses during the processing. Extracts were then diluted with acetonitrile and directly analyzed using HILIC LC-MS/MS. Known concentrations of isotope standard 13C-OAADPr were used to generate a standard curve to quantify concentration of ADPr/OAADPr. Intracellular concentrations of ADPr and OAADPr were calculated from measured concentration, corrected for sample loss, and multiplied by dilution factors in the previously described cell lysis and LC-MS/MS sample preparation step.
ADPr Inhibition of GAPDH Activity—The assay mixture (0.3 ml) contained 100 mm potassium phosphate buffer, pH 7.6, 10 mm EDTA, 0.1 mm dithiothreitol, 1 mm glyceraldehyde 3-phosphate, 0.043 to 0.286 mm NAD+, 0.06 to 0.3 mm ADPr, and highly purified GAPDH from S. cerevisiae (Sigma, G5537, 70–140 units/mg, <0.01% contamination activity of 3-phosphoglyceric phosphokinase). The reaction was initiated by the addition of NAD+, and the activity was monitored by NAD+ reduction at 340 nm. Absorbance was recorded every 4 s during 10 min. Initial velocities were determined from the linear portion of the curves. The initial velocity data were fitted to the competitive inhibition equation using Kinetasyst (Intellikinetics, State College, PA). Data were displayed using KaleidaGraph (Synergy Software, Reading, PA).
Determination of Cellular NAD+, NADH, AMP, ATP, and NADPH—Cells extracts were generated as described above. To measure NAD+ and AMP concentrations, cells were lysed in 0.1 m HCl buffer; for ATP, cells were lysed in 5% (w/v) trichloroacetic acid; for NADH and NADPH, cells were lysed in 0.1 m NaOH. NAD+ was measured using the EnzyChrom™ NAD+/NADH assay kit from BioAssay Systems. NADH, AMP, and ATP were measured as described (33). NADPH concentrations were quantified using HPLC and NADPH standards following the method in (34). Intracellular concentrations of these metabolites were calculated from extract concentrations and multiplying by the appropriate dilution factors (described earlier).
RESULTS
Ysa1 Is the Major Cellular ADPr and OAADPr Nudix Hydrolase—To fully explore the biological functions of ADPr/OAADPr, it is critical to identify the enzymes that are capable of modulating their in vivo levels. Previously, we had demonstrated in vitro that several recombinant Nudix hydrolases, including Ysa1, were capable of hydrolyzing both ADPr and OAADPr with comparable activity (24). However, the biological importance of this specific activity had not been elucidated. Initially, we sought to establish whether endogenous Ysa1 is a significant contributor to the ADPr/OAADPr consuming activities in S. cerevisiae. Soluble protein extracts were generated from wild type and Δysa1 strains, and the ability of these extracts to cleave exogenously added ADPr and OAADPr to AMP was assessed using an HPLC-based assay. Wild type cell extract (Fig. 1A, top chromatogram) displayed a strong hydrolytic activity, readily converting ADPr (peak 1) to AMP (peak 2). In contrast, the Δysa1 cell extract (Fig. 1A, middle chromatogram) failed to generate significant AMP, comparable with the buffer control reaction (Fig. 1A, bottom chromatogram). Similarly, we examined the ability of wild type and Δysa1 strains to hydrolyze OAADPr, which exists as an ∼50:50 mixture of 2′ and 3′ isomers at neutral pH (5). As was the case with ADPr, OAADPr was efficiently cleaved to AMP in the wild type extracts, but not in the Δysa1 strain (Fig. 1B, compare top and middle chromatograms). Together, these data suggest that Ysa1 is the major ADPr/OAADPr Nudix hydrolase in S. cerevisiae.
FIGURE 1.
Ysa1 is the major ADPr/OAADPr-metabolizing enzyme in yeast. HPLCs (A260 nm versus time) of 1 mm ADPr/OAADPr/3′-NAADPr were incubated with cell lysates or buffer (50 mm Tris, pH 7.5, 10 mm MgCl2) controls at 37 °C for 30 min as follows: peak 1, ADPr; peak 2, AMP; peak 3, 3′-OAADPr; peak 4, 2′-OAADPr; peak 5, 3′-NAADPr. A, 1 mm ADPr with wild type cell extract (top panel), Δysa1 cell extract (middle panel), or buffer (bottom panel). B, 1 mm OAADPr with wild type cell extract (top panel), Δysa1 cell extract (middle panel), or buffer (bottom panel). C, 1 mm 3′-NAADPr with wild type cell extract (top panel), Δysa1 cell extract (middle panel), or buffer (bottom panel).
Although the results were consistent with Ysa1 directly cleaving OAADPr to AMP and acetylated ribose phosphate, we performed additional experiments to corroborate this conclusion. Because of the labile nature of the acetyl group on OAADPr, there is significant nonenzymatic hydrolysis to ADPr during the incubation time of the assay (Fig. 1B, peak 1, bottom chromatogram). Moreover, we previously noted an esterase activity in mammalian and yeast cell extracts that yields acetate and ADPr from OAADPr (24). This esterase activity was evident here, as greater levels of ADPr were observed in the Δysa1 strain than in the buffer control (Fig. 1B, compare middle and bottom chromatograms). To rule out the possibility that Ysa1 was hydrolyzing only ADPr and not both OAADPr and ADPr, we examined the ability of the two strains to cleave an analog of OAADPr, 3′-NAADPr (35). This OAADPr analog has nitrogen in place of the 3′-oxygen, forming an amide with the acetyl group, which is predicted to be resistant to both enzymatic and spontaneous hydrolysis. When 3′-NAADPr (Fig. 1C, peak 5) was incubated with cell extracts from the wild type and Δysa1 strains, indeed no hydrolysis at either the acetyl group or at the pyrophosphate linkage was observed in the Δysa1 cell extract (Fig. 1C, middle chromatogram), whereas the analog was converted to AMP (Fig. 1C, peak 2) in the wild type cell extract (Fig. 1C, top chromatogram). Thus, 3′-NAADPr is directly hydrolyzed to AMP by Ysa1. These data support the conclusion that Ysa1 is the major metabolizing activity capable of efficient hydrolysis of both OAADPr and ADPr.
Quantification of Cellular Metabolites—Based on assays using exogenously added ADPr/OAADPr, Ysa1 is the major metabolizing enzyme for these metabolites in yeast protein extracts. This observation led to the hypothesis that Ysa1 may control the cellular concentrations of these metabolites. To determine whether ADPr and OAADPr levels are affected in the Ysa1 deletion strain, we measured their concentrations by a recently developed LC/MS-MS method (32). Wild type and Δysa1 cells were lysed by bead beating in 10% (v/v) trifluoroacetic acid. NAD+, ADPr, and OAADPr are stable to this acid treatment; however, NADH is not. Because NADH is hydrolyzed to ADPr in the acidic cell lysis buffer (5, 32, 33), we independently measured NADH concentration by an alcohol dehydrogenase (ADH) assay (33), using acetaldehyde as substrate. The endogenous ADPr concentration was calculated by subtracting NADH concentrations from the total ADPr (ADPr + NADH hydrolysis). ADPr concentrations increased 50% in the Δysa1 cells as compared with wild type (Table 1), whereas OAADPr also increased 49% (32). To demonstrate that the increase in ADPr was the direct result of Ysa1 activity, we measured the product of the Nudix reaction, AMP. Indeed, the AMP levels (Table 1) decreased (22%) in the deletion strain, consistent with a loss in ADPr hydrolysis by Ysa1. Whole cell levels of NAD+ and ATP were not significantly different between the two strains (Table 1).
TABLE 1.
Ysa1 affects cellular levels of multiple NAD+ metabolites All metabolites and concentration measurements in this paper were performed with three biological replicates. Concentrations are reported ± S.E. p values were determined by unpaired t test.
| Wild type | Δysa1 | p value | |
|---|---|---|---|
| mm | mm | ||
| OAADPra | (0.57 ± 0.051) × 10–3 | (0.85 ± 0.14) × 10–3 | 0.0460 |
| ADPr + NADHb | 0.166 ± 0.016 | 0.216 ± 0.004 | 0.0406 |
| NADH | 0.061 ± 0.012 | 0.058 ± 0.019 | >0.1 |
| ADPrc | 0.105 ± 0.012 | 0.158 ± 0.011 | 0.0088 |
| NAD+ | 1.76 ± 0.08 | 1.71 ± 0.01 | >0.1 |
| AMP | 0.749 ± 0.067 | 0.587 ± 0.026 | 0.0431 |
| ATP | 0.904 ± 0.034 | 0.913 ± 0.051 | >0.1 |
These data are from Ref. 32; 10–3 was used to indicate that OAADPr concentration is in the micromolar range
Under our acidic extraction conditions, NADH hydrolyzes to ADPr (33). Thus, this reflects the sum of ADPr and NADH concentrations
ADPr concentrations were determined by subtracting NADH concentrations from the sum of ADPr and NADH concentrations
Δysa1 Cells Display Increased H2O2 Resistance—Having demonstrated that in vivo levels of ADPr/OAADPr and AMP are controlled by Ysa1, we next explored the biological consequences of ADPr/OAADPr hydrolysis through Ysa1. Given precedent for the generation of ADPr metabolites during oxidative stress and the implications linked to Nudix domain-containing proteins (11–13, 19, 21, 22), we first investigated the sensitivity of Δysa1 cells to the oxidant H2O2. Different concentrations of H2O2 were spotted onto plates covered with a lawn of yeast cells (Fig. 2A). Zones of growth inhibition (halos) surrounding the H2O2 spots appear within 24–48 h. As shown in Fig. 2A, the diameter of the halo on the Δysa1 plate was substantially smaller than those on the wild type plate. More strikingly, Δysa1 but not wild type cells exhibited re-growth inside the halos under H2O2 levels that prevented survival in wild type cells (Fig. 2A). Qualitatively, these results indicate increased resistance to H2O2 in the Δysa1 cells as compared with the wild type cells. To provide a more quantitative assessment, survival assays were performed in liquid cultures (Fig. 2B). Liquid cultures were treated with 3 mm H2O2 and incubated for varying time intervals before cells were washed and plated onto YPD agar, and survival rates were assessed. Colony-forming units were determined for each time point and normalized to the value at time 0. Throughout the entire survival curve, the Δysa1 cells displayed a dramatic recovery from H2O2 stress as compared with wild type cells. At 3 h of treatment, 1% wild type cells survived, whereas 40% of the Δysa1 cells survived (Fig. 2B). The survival data are consistent with the halo assay (Fig. 2A), indicating increased H2O2 resistance in the Δysa1 strain.
FIGURE 2.
Δysa1 cells show increased resistance to H2O2. A, halo assays show that Δysa1 cells display increased resistance to H2O2 stress versus wild type cells. Wild type (WT) and Δysa1 cells from liquid culture were spread onto YPD plates and then spotted with 5 μl of H2O2 (1.5 m, top panel; 2.9 m, middle panel; 4.4 m, bottom panel), and allowed to grow for 1 day before the plates were imaged to determine the amount of cell death, as visualized by the diameter of the “halo,” where cell death occurred (dark area) because of H2O2 treatment. The Δysa1 cells (right panels) display smaller halos for all H2O2 concentrations tested versus wild type cells (left panels). B, survival assays also show Δysa1 cells display increased resistance to H2O2 treatment compared with wild type cells. Graph shows percent cell survival versus time for wild type (diamonds) and Δysa1 (circles) cells, after treatment with 3 mm H2O2 for the specified time points.
Δysa1 Cells Exhibit Increased Copper Resistance—Although the Δysa1 strain displayed substantial resistance from exogenously added oxidative stress, it was unclear whether cell-derived ROS would result in a similar phenotypic difference between the two strains. It is well established that copper ion, a transition metal, generates endogenous ROS (36, 37), which will oxidatively damage biological macromolecules such as proteins and DNA. When sufficient irreversible damage accumulates, cell death results. To compare the effects of copper treatment, serial dilutions of the wild type and Δysa1 strains were spotted on CuSO4-containing YMD plates, and their growth was assessed (Fig. 3A). The growth assays indicated that the Δysa1 strain displayed increased copper resistance, consistent with the previous H2O2 treatment phenotype. To complement these observations, a survival test was performed (Fig. 3B). After treating liquid cultures with 9 mm CuSO4 for 26 h, an equal number of wild type and Δysa1 cells were plated onto YPD plates for 2 days. Although only 10% of the wild type cells survived, 30–40% of Δysa1 cells survived the CuSO4 treatment (Fig. 3B). Collectively, the H2O2 and CuSO4 studies indicate that loss of Ysa1 endows cells with a higher capacity to resist ROS stress, emanating from either endogenous or exogenous sources.
FIGURE 3.
Δysa1 cells show increased resistance to copper-induced stress. A, growth comparison of wild type and Δysa1 cells on YMD plates containing 0 mm CuSO4 (top panel), 0.3 mm CuSO4 (middle panel), and 1 mm CuSO4 (bottom panel). Serial dilutions of each culture were spotted onto YMD plates and grown at 30 °C for 2 days before imaging. B, survival test comparing wild type (WT) and Δysa1 cells under CuSO4 stress. Top panel, cells treated with 9 mm copper for 24 h and rescued on YPD plates for 2 days. Bottom panel, control cells without CuSO4 treatment. Comparison of the wild type (top left panel) and Δysa1 cells (top right panel) shows the Δysa1 cells have increased resistance to copper-induced stress.
Loss of Ysa1 Reduces Endogenously Produced ROS—Based on the differential sensitivity to ROS stress, we examined whether the increased ADPr/OAADPr levels observed in the Δysa1 cells under nonstressed conditions (Table 1) may alter basal levels of ROS. To test this hypothesis, wild type and Δysa1 cells were treated with the ROS indicator DHE, and fluorescence intensity was analyzed by flow cytometry. After crossing the cell membrane, DHE is oxidized by in vivo ROS to generate the fluorescent molecule ethidium. The Δysa1 cells displayed 40% lower ethidium staining as compared with wild type cells (Fig. 4A), indicating less endogenous ROS is present in the Δysa1 cells. Therefore, loss of Ysa1 and the subsequent increase in ADPr/OAADPr accompany a reduction in cellular ROS.
FIGURE 4.
Endogenous ROS levels and mitochondrial membrane potential ΔΨm decrease in Δysa1 cells and Ysa1 localizes to the mitochondria. A, Δysa1 cells display lower basal levels of ROS. Bar graph shows percent of cells positive for ROS/ethidium, wild type (WT, black bar), and Δysa1 (gray bar). Cells were stained with 5 μg/ml dihydroethidium in phosphate-buffered saline buffer for 15 min and analyzed via flow cytometry. Unstained cells were used as controls for background fluorescence. Results are averages (with standard errors) of three biological replicates. B, Δysa1 cells generate lower mitochondrial membrane potential compared with wild type cells. Cells were stained with rhodamine 123 (Rho123) and analyzed by flow cytometry. Rhodamine 123 signal, which reflects the mitochondrial membrane potential, is plotted as cell number versus averaged intensity signal for three wild type strain biological replicates (black) and three Δysa1 strains (gray) biological replicates. In the upper range (indicated by the black bar), 51% of wild type cells are accounted for where as only 33% Δ ysa1 cells are in the same range. C, Ysa1 (26 kDa) is localized to mitochondria. Western blots show that the TAP-Ysa1 construct (54 kDa) localizes to mitochondria. Cells expressing TAP-Ysa1 were lysed and separated into whole cell, cytoplasm, nuclei, and mitochondrial fractions. Ysa1 was detected using anti-TAP antibody, whereas H3 and Isu1 were used as marker proteins for nuclei and mitochondria, respectively. Top panel, TAP-Ysa1 was detected in whole cell extract and possibly in the nuclei. Bottom panel, strong signal for TAP-Ysa1 was detected in whole cell extract and mitochondria but not in the cytoplasmic fraction. Note, we also detect degraded forms (smaller bands of ∼34 and 26 kDa) of TAP-Ysa1 in both whole cell and mitochondrial samples. The vertical lines indicate where extraneous lanes were removed electronically from the blot; however, the image was not manipulated in any other manner.
Loss of Ysa1 Yields Lower Mitochondrial Membrane Potential (ΔΨm)—Given the lower levels of ROS produced by Δysa1 cells, we reasoned that increased ADPr/OAADPr levels might directly inhibit processes that generate ROS and/or promote pathways that suppress ROS cellular damage. First, we examined whether ADPr/OAADPr could inhibit mitochondrial electron transport. Previously, Zharova and Vinogradov (38) demonstrated that ADPr competitively inhibits the NADH dehydrogenase activity of complex I, preventing electrons from flowing into flavin. Electron leakage from flavin in complex I is generally considered a significant source of electrons producing ROS (39, 40). Interestingly, the inhibition by ADPr (Ki = 30–200 μm) was not affected by ΔΨm, and the rate of the energy-dependent NAD+ reduction by succinate (lower ΔΨm) was insensitive to ADPr (38). Together, these observations predict that cellular ADPr would lower endogenous ROS and would lower the mitochondrial membrane potential. We have demonstrated that indeed ROS is decreased in the Δysa1 strain (Fig. 4A). To test the hypothesis that loss of Ysa1 activity affects the mitochondrial membrane potential, we determined the relative ΔΨm between Δysa1 and wild type cells. Utilizing the fluorescent dye rhodamine 123 and flow cytometry (38, 41, 42), the differences in ΔΨm were measured. Shown in Fig. 4B is the overlay of three sets of experiments, comparing wild type and Δysa1 samples. Wild type cells showed significantly stronger rhodamine 123 staining, indicating a higher membrane potential (Fig. 4B). Over the higher fluorescent range where 51% wild type cells exist, only 33% Δysa1 cells fall in this range (Fig. 4B, bar). Thus, the observation that loss of Ysa1 leads to lower ROS and lower ΔΨm is entirely consistent with ADPr inhibition of complex I.
Ysa1 Localization in Mitochondria—From the above observation that Ysa1 affects mitochondrial electron transport chain activity and noting that Ysa1 (26 kDa) has an N-terminal mitochondrial localization signal predicted by PSORTII (43), we set out to determine the cellular localization of Ysa1. We used a genomically tagged Ysa1 strain with a TAP tag on the C terminus. Cells expressing the TAP-Ysa1 were fractionated on a Percoll density gradient into cytoplasm and nuclei or by differential centrifugation into cytoplasm and mitochondria. As shown in Fig. 4C, TAP-Ysa1 (54 kDa) is weakly detected in the nuclei fraction, whereas the positive control histone H3 is greatly enriched in the same fraction. In contrast, TAP-Ysa1 signal is greatly enhanced in the mitochondrial fraction compare with either cytoplasm or nuclei compartments (Fig. 4C). Mitochondrial matrix protein Isu1 is co-fractionated with TAP-Ysa1 in the mitochondrial fraction. Hence, these results support a mechanism in which Ysa1 can directly control ADPr/OAADPr levels in the mitochondria and modulate electron transport chain activity. Weak staining of TAP-Ysa1 in nuclei and cytoplasm may suggest that a small quantity of Ysa1 exists in those compartments.
ADPr/OAADPr Bind Glycolytic Enzymes—Next, we explored the possibility that increased ADPr/OAADPr levels may promote pathways that protect cells from ROS damage. It is well established that H2O2 and Cu2+ stress leads to increased cellular levels of ROS in yeast cells (37, 44, 45). To eliminate the damaging forms of oxygen rising from both reagents, cells reroute glucose metabolism from the glycolytic pathway to the NADPH-generating pentose phosphate pathway (46–48). The enzymes glutathione reductase and thioredoxin reductase both require NADPH to reduce oxidized glutathione and thioredoxin, which are essential cellular antioxidants (49, 50). Therefore, the re-routing of the metabolic flux serves as a rapid response mechanism to counteract the damaging effects of H2O2 and copper-induced oxidative stress. Given that ADPr/OAADPr is structurally similar to several adenosine nucleotide-containing co-enzymes and substrates (e.g. NAD(H) and ATP) utilized by glycolytic enzymes, we hypothesized that ADPr/OAADPr might interact with key glycolytic enzymes and inhibit their activity. We explored this possibility by developing an affinity-binding procedure to identify proteins that interact with ADPr/OAADPr. Briefly, we utilized Cibacron blue 3GA-agarose resin to adsorb cellular enzymes/proteins with an adenosyl nucleoside-binding site, and we followed this step with specific elution using ADPr/OAADPr. To first validate the procedure, we employed this method to bind and elute endogenous Sir2, a protein deacetylase that produces OAADPr as a product of NAD+-dependent deacetylation. Sir2 was retained on the resin and was specifically eluted with low levels (50–200 μm) of OAADPr (Fig. 5A). Having established a positive control, proteins eluted from the column by ADPr or OAADPr were identified by in-gel digest followed by LC/MS-MS (Fig. 5B). Elutions with ADPr or OAADPr yielded similar protein bands (Fig. 5B). Importantly, the experiment was performed >10 times, and in all cases, the same protein banding patterns were observed. Three major bands were identified as phosphoglycerate kinase (PGK) (Fig. 5B, protein band 1), ADH (Fig. 5B, protein band 2), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Fig. 5B, protein band 3). GAPDH and PGK are central glycolytic enzymes, and ADH is linked to glycolysis through its ability to re-oxidize NADH to NAD+, which is necessary to maintain glycolysis. Because GAPDH is reported to be down-regulated during the oxidative defense pathway (45, 47), we investigated whether yeast GAPDH activity might be inhibited directly by ADPr. We analyzed the steady-state inhibition of GAPDH by ADPr. Initial velocities for NADH formation at saturating glyceraldehyde 3-phosphate levels were determined at varied concentrations of NAD+ and ADPr as the inhibitor. The entire data set was fitted to the competitive inhibition equation, revealing an excellent fit. The average of three replicate experiments yielded a Ki value of 60.5 ± 15.4 μm for ADPr and a Km value of 17.5 ± 3.1 μm for NAD+. One representative data set is shown in Fig. 5C, where 1/rate (v) versus 1/[NAD+] results in a series of lines that intersect at the 1/v axis. These results are consistent with a previous study on rabbit muscle GAPDH inhibition by ADPr (Ki = 180 μm) (51). OAADPr inhibits the enzyme with similar IC50 values (data not shown). Thus, the collective results suggest that the higher ADPr/OAADPr concentrations in the Δysa1 strain facilitate the rerouting of metabolic flux from glycolysis to pentose phosphate pathway. If this were true, we would expect the Δysa1 cells to exhibit higher NADPH levels, which would offer protection from both exogenous and endogenous ROS.
FIGURE 5.
ADPr/OAADPr interact with glycolytic enzymes. A, positive control for affinity elution. Endogenous Sir2 interacts with OAADPr. Anti-Sir2 Western blot shows the elution profile of wild type yeast cell extract applied to Cibacron blue resin and eluted with the indicated concentrations of OAADPr. B, identification of novel ADPr/OAADPr-interacting proteins. SYPRO Ruby-stained SDS-polyacrylamide gel shows the Cibacron blue elution profile for a wild type cell extract eluted with either OAADPr or ADPr. Indicated bands were in-gel digested with trypsin and identified by LC-MS/MS as phosphoglycerate kinase (band 1), alcohol dehydrogenase (band 2), and glyceraldehyde-3-phosphate dehydrogenase (band 3). C, double-reciprocal plot of GAPDH inhibition by ADPr. Initial rates were determined from the rate of NAD+ reduction. Assays were performed in the presence of 1 mm glyceraldehyde 3-phosphate (GAP), and NAD+ concentrations ranged from 43 to 286 μm. ADPr exhibits competitive inhibition toward NAD+. The following ADPr concentrations were used: 60 (•), 100 (♦), 150 (▪), and 300 μm (▴). Data were fitted to competitive inhibition using Kinetasyst as described under “Experimental Procedures.” Shown here is one representative of three replicated experiments. Ki value of ADPr and Km value of NAD+ were 60.5 ± 15.4 and 17.5 ± 3.1 μm (average ± S.E.) respectively, averaged from three replicate experiments.
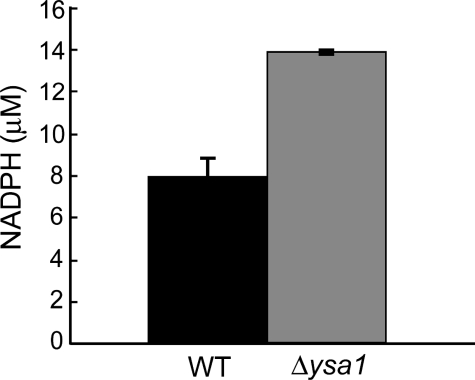
NADPH Is Increased in Δysa1 Cells—To provide evidence that increased levels of ADPr lead to higher NADPH levels, we performed an HPLC-based assay and determined cellular concentration of NADPH (34). As shown in Fig. 6, NADPH concentration increased from 8 ± 0.90 μm in wild type cells to 14 ± 0.10 μm in the Δysa1 cells, consistent with re-routing carbohydrate flux from glycolysis to the pentose phosphate pathway. This substantial increase (75%) in NADPH provides a pool of co-enzyme for ROS scavenging enzymes thioredoxin reductase and glutathione reductase in Δysa1 cells (50). Thus, increased cellular reducing power in the form of NADPH endows Δysa1 cells with stronger resistance to the toxic effects of ROS stress and the ability to maintain a lower basal level of cellular ROS.
FIGURE 6.
Δysa1 cells exhibit higher cellular NADPH concentrations. The cellular concentrations of NADPH in wild type (WT, black bar) and Δysa1 (gray bar) cells were determined by HPLC analysis to be 8 μm for wild type and 14 μm for Δysa1. Results are averages (with standard errors) of three biological replicates.
DISCUSSION
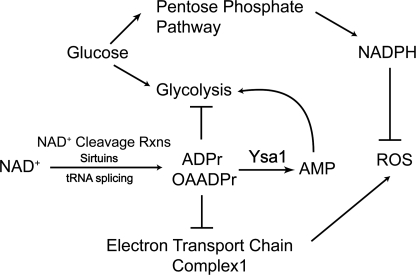
Here, we report the first in-depth biochemical and cellular investigation of a major ADPr/OAADPr metabolizing pathway involving the Ysa1 Nudix hydrolase. As summarized in the proposed model (Fig. 7), yeast lacking Ysa1 display increased accumulation of ADPr/OAADPr and a corresponding decrease in AMP. Strikingly, altered levels of ADPr/OAADPr and AMP are associated with higher resistance to oxidative stress and lower levels of basal ROS in Δysa1 cells. We explored the molecular basis for these phenotypes and provide biochemical evidence for two complementary pathways (Fig. 7). The results suggest that increased ADPr/OAADPr levels protect cells as follows: (i) via lower cellular ROS production through the inhibition of complex I of the electron transport chain, and (ii) via generation of higher NADPH levels to suppress ROS damage.
FIGURE 7.
Proposed functions for Ysa1 and ADPr/OAADPr in metabolic pathways and cellular redox. ADPr/OAADPr generated from NAD+ cleavage are directly hydrolyzed by Ysa1 into AMP. Accumulation of AMP activates glycolysis. Deletion of Ysa1 results in increased cellular ADPr/OAADPr and decreased AMP. As a consequence, ADPr/OAADPr inhibits glycolysis (e.g. GAPDH) and promotes NADPH production from the pentose phosphate pathway. Because NADPH is a major source of cellular reducing power for ROS-detoxifying enzymes, the increase in cellular NADPH strengthens the antioxidative stress response capability of Δysa1 cells. Additionally, increased ADPr/OAADPr in Δysa1 inhibits the mitochondrial electron transport chain through complex I, leading to lower in vivo ROS levels.
Mitochondria are a significant source of cellular ROS caused by electron leakage from complex I of the electron transport chain (39, 52). NADH binds to complex I, is oxidized to NAD+, and donates two electrons to flavin, which subsequently passes the electrons to ubiquinone through several Fe-S clusters. Classic electron transport chain inhibitors, including rotenone, amytal, and piericidin A, prevent electron transport from Fe-S centers to ubiquinone, lead to electron leakage to O2, and increased ROS production (53). However, prior reports suggested a distinct mechanism of complex I inhibition by ADPr (38), where ADPr prevents NADH from binding to the complex and electrons from reaching the flavin, which is a major source of electrons that initiate ROS (54). The inhibition potency of ADPr was not affected by ΔΨm, and the rate of the energy-dependent NAD+ reduction by succinate was insensitive to ADPr (38). These prior observations predict that cellular ADPr would lower the mitochondrial membrane potential. Indeed, we demonstrate that lower ROS and decreased ΔΨm strongly correlate with the elevated levels of ADPr in the Δysa1 cells (Fig. 4 and Table 1). ADPr inhibition of complex I is specific, as NAD+ does not significantly inhibit and ATP, ADP, or AMP only inhibit at concentrations of 10 mm (38, 40). Together, these results suggest that ADPr is a physiological modulator of ROS originating from mitochondrial electron transport.
In addition to suppressing the generation of ROS from mitochondria, ADPr in Δysa1 cells promotes increased levels of the cellular antioxidant NADPH (Fig. 6). One mechanism leading to increased NADPH levels is the inhibition of GAPDH. Using a novel affinity-based screening method to discover soluble proteins/enzymes that interact with ADPr/OAADPr, we identified GAPDH, PGK, and ADH. GAPDH and PGK catalyze sequential reactions in glycolysis, and ADH is an essential enzyme that recycles NAD from NADH to fuel glycolysis. We provide direct evidence that ADPr/OAADPr directly interacts with GAPDH, and ADPr was an effective inhibitor of the GAPDH reaction in vitro, yielding a Ki value of 60 μm. We postulated that inhibition might lead to a metabolic diversion of glucose equivalents into the pentose phosphate pathway (PPP), which would generate increased levels of NADPH. Indeed, the Δysa1 cells displayed a 75% increase in NADPH over wild type cells (Fig. 6). The idea that GAPDH is an important regulatory switch between glycolysis and PPP is supported by several studies. ROS-induced inactivation of GAPDH leads to increased levels of PPP metabolites and increased NADPH/NADP+, which is an essential cellular response to protect cells against ROS stress (44–47). NADPH is the co-enzyme employed by the oxidant defense pathways of thioredoxin reductase and glutathione reductase (49).
Here we provide both in vitro and in vivo evidence that micromolar levels of ADPr inhibit GAPDH activity, leading to increased production of NADPH through the PPP pathway (Fig. 7). As ROS have been proposed as the causative agent for the pathophysiology associated with several age-related diseases (55), accumulation of free ADPr/OAADPr might have an important regulatory role in metabolic switching to protect cells from ROS damage.
Using a TAP construct to elucidate the cellular localization of Ysa1, subcellular fractionation indicated strong enrichment in mitochondria (Fig. 4). Because weak staining was observed in nuclei and cytoplasmic preparations, a small fraction of total Ysa1 may also exist in these compartments. Enrichment in mitochondria is consistent with a predicted mitochondrial localization signal in the N terminus. Using a GFP-Ysa1 construct as part of a genome-wide effort to localize yeast proteins, Ysa1 was detected throughout the cell (56). Although it is unclear whether the green fluorescent protein tag interfered with its normal localization, the mitochondrial enrichment demonstrated here supports a model in which Ysa1 directly modulates mitochondrial levels of ADPr/OAADPr and affects activity of the electron transport chain. A whole cell change of ADPr/OAADPr levels by only 50% might be expected if the mitochondrial pool represents a smaller portion of the total cellular ADPr/OAADPr.
Our results also indicate a role for Ysa1 and ADPr/OAADPr metabolites in the cytosolic regulation of glycolysis. It is possible that mitochondrial Ysa1 controls a cytosolic pool of ADPr/OAADPr, if these molecules readily pass between the cytosol and mitochondria. There is indirect evidence that ADPr can be transported from nuclei to mitochondria (22) and that mitochondria-produced ADPr can be released into the cytosol (13). Alternatively, Ysa1 from nuclear and cytoplasmic compartments might directly modulate a pool of ADPr/OAADPr that is distinct from that generated in mitochondria. In support of this possibility, localization studies with green fluorescent protein-tagged Ysa1 suggested much broader localization (56).
There are a number of diverse sources of cellular ADPr and related metabolites. In all known cases, NAD+ serves as the donor substrate. Conserved among all living organisms, NAD+-dependent histone deacetylases produce OAADPr as a product (3, 5), which can be readily converted to ADPr by cellular esterases (24). Another source of ADPr in yeast is the dephosphorylation of ADP-ribose 1″-phosphate, a product of NAD+-dependent tRNA splicing (57, 58). Other potential sources of ADPr in yeast remain to be identified. This study is the first to quantify the cellular levels of ADPr and to demonstrate that Nudix hydrolases modulate both ADPr and OAADPr in yeast. In mammals, poly (ADP-ribose) degradation is predicted to yield substantial free ADPr (19). CD38, a cell surface NAD+ glycohydrolase, and mitochondrial NAD+ glycohydrolases are other relevant sources of free ADPr (2).
The idea that ADPr functions as an important signaling molecule is supported by accumulating evidence. For example, the mammalian TRPM2 ion channel is gated directly by both ADPr and OAADPr, which bind to an inactive Nudix domain found on the intracellular side of the channel (12, 13). In response to oxidative stress, TRPM2 is activated in a mechanism that requires PARP/PARG (12, 21, 59). Poly(ADP-ribose) is rapidly generated by PARP during ROS and genotoxic stress, leading to the rapid formation of free ADPr, catalyzed by PARG. Transient expression of NudT9, a highly active ADPr pyrophosphatase, abolishes the H2O2-induced Ca2+ influx through TRMP2, supporting a direct function of free ADPr (13).
Under massive genotoxic stress, PARP hyperactivation leads to catastrophic loss of NAD+ and ATP in mammalian tissue culture cells (18). Although the mechanism has not been elucidated, Cipriani et al. (21), suggested that ADPr might enter mitochondria and induce ATP depletion. It is unclear that these severe conditions reflect normal physiological control of ADPr levels. However, complete energy collapse would be consistent with overwhelming inhibition of glycolysis and mitochondrial function, as would be predicted from the proposed model (Fig. 7). Given that yeast cells grown in rich glucose medium are not dependent on mitochondria for production of ATP (46), we did not detect significant differences in ATP concentration between wild type and Δysa1. Also, we observed no significant change in NAD+ and NADH levels between wild type and Δysa1 strains, supporting a model in which a 50% increase of ADPr (to 158 μm) elicits an enhanced protective effect against ROS, but not to the extent that global metabolites ATP, NAD+, and NADH are affected. It is important to note that AMP levels show a corresponding decrease in Δysa1 cells versus wild type (Table 1). This drop in AMP might function synergistically with increased ADPr to down-regulate glycolysis (Fig. 7). AMP is an allosteric activator of glycolysis through a number of pathways (60). Whether the effects of low AMP with high ADPr/OAADPr combine to inhibit glycolysis and to promote the PPP will await further evaluation.
Although the yeast genome does not encode PARP or TRPM2, the proposed mechanisms (Fig. 7) for the biological functions of free ADPr/OAADPr and Nudix hydrolases are likely conserved among yeast and mammals. We propose that ADPr functions in fundamental metabolic processes, regulating the cellular responses to oxidative stress at two major levels of energy metabolism (Fig. 7). Mammalian genomes do encode two close homologs of yeast Ysa1, NudT5 (61) and NudT9 (62). Both NudT5 and NudT9 display robust in vitro activity toward ADPr (24). Interestingly, NudT9 is localized primarily to mitochondria, and NudT5 is largely cytoplasmic (62, 63). Future studies will be needed to examine whether NudT5 and NudT9 function as the mammalian counterparts to yeast Ysa1.
This study provides the first direct evidence that ADPr (and perhaps OAADPr) plays a central regulatory role in energy metabolism and the ability of cells to protect themselves from both endogenous and exogenous forms of oxidative stress. Ysa1 modulates the cellular levels of ADPr/OAADPr by cleaving these molecules to AMP and (acetylated) phosphoribose. Additional work is needed to identify the various pathways that contribute to the pools of ADPr/OAADPr, which are generated by NAD+-consuming systems. During normal growth, Ysa1 helps maintain a low basal level of ADPr/OAADPr. However, under cell stress, NAD+ cleavage to ADPr can provide a signal to slow down investment in energy-producing pathways and divert resources to provide a means to combat the effects of ROS (via the PPP). This stress-induced consumption of NAD+ is linked to the subsequent induction of nicotinamidase, a key player in the NAD+ salvage pathway from nicotinamide (64–68). In addition, Nudix hydrolases respond to environmental changes and stress. For example, expression of Ysa1 is differentially regulated under a variety of environmental stresses (69).
Because sirtuins are producers of ADPr/OAADPr and numerous studies have reported their beneficial effects on metabolism, stress response, and longevity (70, 71), it is tempting to suggest that many of these phenotypes result from the direct signaling functions of ADPr/OAADPr. However, overstimulation of ADPr-producing pathways might lead to cell death through a catastrophic cellular energy collapse, as reported for the PARP/PARG system (18, 21). Modest activation of the PARP/PARG pathway under mild oxidative stress facilitates DNA repair and cell survival. Thus ADPr/OAADPr levels provide a measured readout of cellular health, mediating the appropriate survival or self-destruct signal.
Acknowledgments
We thank Dr. Catherine Fox (University of Wisconsin, Madison) for help with the yeast strains, helpful suggestions, inspiring discussions, offering reagents, and sharing equipment. We thank Dr. Lindsay Comstock (Wake Forest University) for the synthesis of NAADPr. We greatly appreciate the gift of Isu1 antibody and mitochondrial isolation protocol from Dr. Amy J. Andrew (National Institutes of Health). We also thank Philipp Mueller (University of Wisconsin, Madison) and Zhonggang Hou (University of Wisconsin, Madison) for the gifts of H3 antibody, Sir2 antibody, and nuclei isolation protocol.
Footnotes
The abbreviations used are: PARP, poly(ADP-ribose) polymerase; ADPr, ADP-ribose; OAADPr, O-acetyl-ADP-ribose; Nudix, nucleoside diphosphate linked to another moiety X; ROS, reactive oxygen species; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; TRPM2, transient receptor melastin-related ion channel 2; PARG, poly(ADP-ribose) glycohydrolase; ADH, alcohol dehydrogenase; PGK, phosphoglycerate kinase; PPP, pentose phosphate pathway; HPLC, high pressure liquid chromatography; ddH2O, double distilled H2O; LC-MS/MS, liquid chromatography-tandem mass spectrometry; DHE, dihydroethidium.
References
- 1.Belenky, P., Bogan, K. L., and Brenner, C. (2007) Trends Biochem. Sci. 32, 12-19 [DOI] [PubMed] [Google Scholar]
- 2.Kim, H., Jacobson, E. L., and Jacobson, M. K. (1993) Science 261, 1330-1333 [DOI] [PubMed] [Google Scholar]
- 3.Imai, S., Armstrong, C. M., Kaeberlein, M., and Guarente, L. (2000) Nature 403, 795-800 [DOI] [PubMed] [Google Scholar]
- 4.Yang, T., and Sauve, A. A. (2006) AAPS J. 8, E632-E643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson, M. D., and Denu, J. M. (2002) J. Biol. Chem. 277, 18535-18544 [DOI] [PubMed] [Google Scholar]
- 6.Tanner, K. G., Landry, J., Sternglanz, R., and Denu, J. M. (2000) Proc. Natl. Acad. Sci. U. S. A. 97, 14178-14182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoff, K. G., and Wolberger, C. (2005) Nat. Struct. Mol. Biol. 12, 560-561 [DOI] [PubMed] [Google Scholar]
- 8.Liou, G. G., Tanny, J. C., Kruger, R. G., Walz, T., and Moazed, D. (2005) Cell 121, 515-527 [DOI] [PubMed] [Google Scholar]
- 9.Kustatscher, G., Hothorn, M., Pugieux, C., Scheffzek, K., and Ladurner, A. G. (2005) Nat. Struct. Mol. Biol. 12, 624-625 [DOI] [PubMed] [Google Scholar]
- 10.Borra, M. T., O'Neill, F. J., Jackson, M. D., Marshall, B., Verdin, E., Foltz, K. R., and Denu, J. M. (2002) J. Biol. Chem. 277, 12632-12641 [DOI] [PubMed] [Google Scholar]
- 11.Kuhn, F. J., and Luckhoff, A. (2004) J. Biol. Chem. 279, 46431-46437 [DOI] [PubMed] [Google Scholar]
- 12.Grubisha, O., Rafty, L. A., Takanishi, C. L., Xu, X., Tong, L., Perraud, A. L., Scharenberg, A. M., and Denu, J. M. (2006) J. Biol. Chem. 281, 14057-14065 [DOI] [PubMed] [Google Scholar]
- 13.Perraud, A. L., Takanishi, C. L., Shen, B., Kang, S., Smith, M. K., Schmitz, C., Knowles, H. M., Ferraris, D., Li, W., Zhang, J., Stoddard, B. L., and Scharenberg, A. M. (2005) J. Biol. Chem. 280, 6138-6148 [DOI] [PubMed] [Google Scholar]
- 14.Dunn, C. A., O'Handley, S. F., Frick, D. N., and Bessman, M. J. (1999) J. Biol. Chem. 274, 32318-32324 [DOI] [PubMed] [Google Scholar]
- 15.Perraud, A. L., Schmitz, C., and Scharenberg, A. M. (2003) Cell Calcium 33, 519-531 [DOI] [PubMed] [Google Scholar]
- 16.Miwa, M., and Sugimura, T. (1971) J. Biol. Chem. 246, 6362-6364 [PubMed] [Google Scholar]
- 17.Bonicalzi, M. E., Haince, J. F., Droit, A., and Poirier, G. G. (2005) Cell. Mol. Life Sci. 62, 739-750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berger, N. (1985) Radiat. Res. 101, 4-15 [PubMed] [Google Scholar]
- 19.Schreiber, V., Dantzer, F., Ame, J. C., and de Murcia, G. (2006) Nat. Rev. Mol. Cell Biol. 7, 517-528 [DOI] [PubMed] [Google Scholar]
- 20.Gao, H., Coyle, D. L., Meyer-Ficca, M. L., Meyer, R. G., Jacobson, E. L., Wang, Z. Q., and Jacobson, M. K. (2007) Exp. Cell Res. 313, 984-996 [DOI] [PubMed] [Google Scholar]
- 21.Cipriani, G., Rapizzi, E., Vannacci, A., Rizzuto, R., Moroni, F., and Chiarugi, A. (2005) J. Biol. Chem. 280, 17227-17234 [DOI] [PubMed] [Google Scholar]
- 22.Dumitriu, I. E., Voll, R. E., Kolowos, W., Gaipl, U. S., Heyder, P., Kalden, J. R., and Herrmann, M. (2004) Cell Death Differ. 11, 314-320 [DOI] [PubMed] [Google Scholar]
- 23.Ono, T., Kasamatsu, A., Oka, S., and Moss, J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103, 16687-16691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rafty, L. A., Schmidt, M. T., Perraud, A. L., Scharenberg, A. M., and Denu, J. M. (2002) J. Biol. Chem. 277, 47114-47122 [DOI] [PubMed] [Google Scholar]
- 25.Ghaemmaghami, S., Huh, W. K., Bower, K., Howson, R. W., Belle, A., Dephoure, N., O'Shea, E. K., and Weissman, J. S. (2003) Nature 425, 737-741 [DOI] [PubMed] [Google Scholar]
- 26.Ausubel, F. M. (ed) (2002) Short Protocols in Molecular Biology, Vol. 2, 5th Ed, pp. 9-13, John Wiley, Inc., New York [Google Scholar]
- 27.Roskams, J., and Rogers, L. (eds) (2002) Lab. Ref.: A Handbook of Recipes, Reagents, and Other Reference Tools for Use at the Bench (Section 7A), p. 196, Cold Spring Harbor, New York
- 28.Guisbert, K., Duncan, K., Li, H., and Guthrie, C. (2005) RNA (N. Y.) 11, 383-393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amati, B. B., and Gasser, S. M. (1988) Cell 54, 967-978 [DOI] [PubMed] [Google Scholar]
- 30.Andrew, A. J., Song, J. Y., Schilke, B., and Craig, E. A. (2008) Mol. Biol. Cell 19, 5259-5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hallows, W. C., Lee, S., and Denu, J. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103, 10230-10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, S., Tong, L., and Denu, J. M. (2008) Anal. Biochem. 383, 174-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bergmeyer, H. (ed) (1985) Methods of Enzymatic Analysis, 3rd Ed., Vol. 7, p. 253, VCH Publishers, Deerfield Beach, FL [Google Scholar]
- 34.Pollak, N., Niere, M., and Ziegler, M. (2007) J. Biol. Chem. 282, 33562-33571 [DOI] [PubMed] [Google Scholar]
- 35.Comstock, L. R., and Denu, J. M. (2007) Org. Biomol. Chem. 5, 3087-3091 [DOI] [PubMed] [Google Scholar]
- 36.Liang, Q., and Zhou, B. (2007) Mol. Biol. Cell 18, 4741-4749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valko, M., Morris, H., and Cronin, M. T. (2005) Curr. Med. Chem. 12, 1161-1208 [DOI] [PubMed] [Google Scholar]
- 38.Zharova, T. V., and Vinogradov, A. D. (1997) Biochim. Biophys. Acta 1320, 256-264 [DOI] [PubMed] [Google Scholar]
- 39.Adam-Vizi, V., and Chinopoulos, C. (2006) Trends Pharmacol. Sci. 27, 639-645 [DOI] [PubMed] [Google Scholar]
- 40.Schonfeld, P., and Wojtczak, L. (2008) Free Radic. Biol. Med. 45, 231-241 [DOI] [PubMed] [Google Scholar]
- 41.Du, L., Yu, Y., Chen, J., Liu, Y., Xia, Y., Chen, Q., and Liu, X. (2007) FEMS Yeast Res. 7, 860-865 [DOI] [PubMed] [Google Scholar]
- 42.Mathur, A., Hong, Y., Kemp, B. K., Barrientos, A. A., and Erusalimsky, J. D. (2000) Cardiovasc. Res. 46, 126-138 [DOI] [PubMed] [Google Scholar]
- 43.Horton, P., and Nakai, K. (1997) ISMB 5, 147-152 [PubMed] [Google Scholar]
- 44.Shanmuganathan, A., Avery, S. V., Willetts, S. A., and Houghton, J. E. (2004) FEBS Lett. 556, 253-259 [DOI] [PubMed] [Google Scholar]
- 45.Godon, C., Lagniel, G., Lee, J., Buhler, J. M., Kieffer, S., Perrot, M., Boucherie, H., Toledano, M. B., and Labarre, J. (1998) J. Biol. Chem. 273, 22480-22489 [DOI] [PubMed] [Google Scholar]
- 46.Shenton, D., and Grant, C. M. (2003) Biochem. J. 374, 513-519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ralser, M., Wamelink, M. M., Kowald, A., Gerisch, B., Heeren, G., Struys, E. A., Klipp, E., Jakobs, C., Breitenbach, M., Lehrach, H., and Krobitsch, S. (2007) J. Biol. 6, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Juhnke, H., Krems, B., Kotter, P., and Entian, K. D. (1996) Mol. Gen. Genet. 252, 456-464 [DOI] [PubMed] [Google Scholar]
- 49.Winyard, P. G., Moody, C. J., and Jacob, C. (2005) Trends Biochem. Sci. 30, 453-461 [DOI] [PubMed] [Google Scholar]
- 50.Jamieson, D. J. (1998) Yeast 14, 1511-1527 [DOI] [PubMed] [Google Scholar]
- 51.Eby, D., and Kirtley, M. E. (1971) Biochemistry 10, 2677-2682 [DOI] [PubMed] [Google Scholar]
- 52.Liu, Y., Fiskum, G., and Schubert, D. (2002) J. Neurochem. 80, 780-787 [DOI] [PubMed] [Google Scholar]
- 53.Nelson, D. L., and Cox, M. M. (2000) Lehninger Principles of Biochemistry, 3rd Ed, pp. 666-668 Worth Publishers, New York
- 54.Grivennikova, V. G., and Vinogradov, A. D. (2006) Biochim. Biophys. Acta 1757, 553-561 [DOI] [PubMed] [Google Scholar]
- 55.Galli, F., Piroddi, M., Annetti, C., Aisa, C., Floridi, E., and Floridi, A. (2005) Contrib. Nephrol. 149, 240-260 [DOI] [PubMed] [Google Scholar]
- 56.Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., and O'Shea, E. K. (2003) Nature 425, 686-691 [DOI] [PubMed] [Google Scholar]
- 57.Shull, N. P., Spinelli, S. L., and Phizicky, E. M. (2005) Nucleic Acids Res. 33, 650-660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zillmann, M., Gorovsky, M. A., and Phizicky, E. M. (1991) Mol. Cell. Biol. 11, 5410-5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buelow, B., Song, Y., and Scharenberg, A. M. (2008) J. Biol. Chem. 283, 24571-24583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanz, P. (2008) Curr. Protein Pept. Sci. 9, 478-492 [DOI] [PubMed] [Google Scholar]
- 61.Yang, H., Slupska, M. M., Wei, Y. F., Tai, J. H., Luther, W. M., Xia, Y. R., Shih, D. M., Chiang, J. H., Baikalov, C., Fitz-Gibbon, S., Phan, I. T., Conrad, A., and Miller, J. H. (2000) J. Biol. Chem. 275, 8844-8853 [DOI] [PubMed] [Google Scholar]
- 62.Perraud, A. L., Shen, B., Dunn, C. A., Rippe, K., Smith, M. K., Bessman, M. J., Stoddard, B. L., and Scharenberg, A. M. (2003) J. Biol. Chem. 278, 1794-1801 [DOI] [PubMed] [Google Scholar]
- 63.McLennan, A. G. (2006) Cell. Mol. Life Sci. 63, 123-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anderson, R. M., Bitterman, K. J., Wood, J. G., Medvedik, O., Cohen, H., Lin, S. S., Manchester, J. K., Gordon, J. I., and Sinclair, D. A. (2002) J. Biol. Chem. 277, 18881-18890 [DOI] [PubMed] [Google Scholar]
- 65.Anderson, R. M., Bitterman, K. J., Wood, J. G., Medvedik, O., and Sinclair, D. A. (2003) Nature 423, 181-185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Balan, V., Miller, G. S., Kaplun, L., Balan, K., Chong, Z. Z., Li, F., Kaplun, A., VanBerkum, M. F., Arking, R., Freeman, D. C., Maiese, K., and Tzivion, G. (2008) J. Biol. Chem. 283, 27810-27819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McClure, J. M., Gallo, C. M., Smith, D. L., Jr., Matecic, M., Hontz, R. D., Buck, S. W., Racette, F. G., and Smith, J. S. (2008) Genetics 180, 797-810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rongvaux, A., Andris, F., Van Gool, F., and Leo, O. (2003) BioEssays 25, 683-690 [DOI] [PubMed] [Google Scholar]
- 69.Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D., and Brown, P. O. (2000) Mol. Biol. Cell 11, 4241-4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guarente, L. (2007) Cold Spring Harbor Symp. Quant. Biol. 72, 483-488 [DOI] [PubMed] [Google Scholar]
- 71.Ghosh, H. S. (2008) Curr. Opin. Investig. Drugs 9, 1095-1102 [PubMed] [Google Scholar]