Abstract
The interaction between epithelial cells and the extracellular matrix is crucial for tissue architecture and function and is compromised during cancer progression. Dystroglycan is a membrane receptor that mediates interactions between cells and basement membranes in various epithelia. In many epithelium-derived cancers, β-dystroglycan is expressed, but α-dystroglycan is not detected. Here we report that α-dystroglycan is correctly expressed and trafficked to the cell membrane but lacks laminin binding as a result of the silencing of the like-acetylglucosaminyltransferase (LARGE) gene in a cohort of highly metastatic epithelial cell lines derived from breast, cervical, and lung cancers. Exogenous expression of LARGE in these cancer cells restores the normal glycosylation and laminin binding of α-dystroglycan, leading to enhanced cell adhesion and reduced cell migration in vitro. Our findings demonstrate that LARGE repression is responsible for the defects in dystroglycan-mediated cell adhesion that are observed in epithelium-derived cancer cells and point to a defect of dystroglycan glycosylation as a factor in cancer progression.
Normal epithelial cells are tightly associated with one another and with the underlying basement membrane to maintain tissue architecture and function. During cancer progression, primitive cancer cells escape from this control by modifying the binding affinities of their cell membrane receptors. Several receptors have been described as important for this process. Of these, the integrins are the best studied (1). The receptor dystroglycan has been reported to be required for the development and maintenance of epithelial tissues (2, 3). A direct requirement for dystroglycan in epithelia is further demonstrated by the profound effect that loss of dystroglycan expression has on cell polarity and laminin binding in cultured mammary epithelial cells (4, 5). However, dystroglycan is not only important in the establishment and maintenance of epithelial structure. Associations have also been made between the loss of α-dystroglycan immunoreactivity and cancer progression in tumors of epithelial origin, including breast, colon, cervix, and prostate cancers (4, 6–9). The dystroglycan loss of function could thus serve as an effective means by which cancerous cells modify their adhesion to the extracellular matrix (ECM).2
Dystroglycan is a ubiquitously expressed cell membrane protein that plays a key function in cellular integrity, linking the intracellular cytoskeleton to the extracellular matrix. The dystroglycan gene encodes a preprotein that is cleaved into two peptides (10). The C-terminal component, known as β-dystroglycan, is embedded within the cell membrane, whereas the N-terminal component, α-dystroglycan, is present within the extracellular periphery but remains associated with β-dystroglycan through non-covalent bonds. β-Dystroglycan binds to actin (11), dystrophin (11), utrophin (11), and Grb2 (12) through its C-terminal intracellular domain. α-Dystroglycan, on the other hand, binds to ECM proteins that contain laminin globular domains including laminins (13, 14), agrin (15), and perlecan (16), as well as to the transmembrane protein neurexin (17). α-Dystroglycan is extensively decorated by three different types of glycan modifications: mucin type O-glycosylation, O-mannosylation, and N-glycosylation. The state of α-dystroglycan glycosylation has been shown to be critical for the ability of the protein to bind to laminin globular domain-containing proteins of the ECM (18).
Previous studies of epithelium-derived cancers (4, 9) demonstrated that the loss of immunoreactivity of α-dystroglycan antibodies correlates with tumor grade and poor prognosis. This reduced detection of α-dystroglycan, however, is based on a loss of α-dystroglycan reactivity to antibodies (known as IIH6 and VIA4-1) that recognize the laminin-binding glyco-epitope of α-dystroglycan, i.e. the protein is only functional when it is glycosylated in such a way (henceforth, referred to as functional glycosylation). However, in most of the cancer samples that have been studied to date, β-dystroglycan is expressed at normal levels at the cell membrane. Thus, the aforementioned cancer-associated loss of α-dystroglycan expression may reflect a failure in the post-translational processing of dystroglycan rather than in the synthesis of α-dystroglycan itself.
A similar defect in dystroglycan has been reported in a group of congenital muscular dystrophies (19). This spectrum of human developmental syndromes involves the brain, eye, and skeletal muscle and shows a dramatic gradient of phenotypic severity that ranges from the most devastating in Walker-Warburg syndrome to the least severe in limb-girdle muscular dystrophy. Six distinct known and putative glycosyltransferases have been shown to underlie these syndromes: protein O-mannosyltransferase 1 (POMT1), protein O-mannosyltransferase 2 (POMT2), protein O-mannose β-1,2-acetylglucosaminyltransferase 1 (POMGnT1), like acetylglucosaminyltransferase (LARGE), Fukutin, and Fukutin-related protein (FKRP) (20–25). Indeed, all muscular dystrophy patients with mutations in any of these genes fail to express the functionally glycosylated α-dystroglycan epitope that is recognized by the IIH6 and VIA4-1 antibodies.
To investigate the molecular mechanism responsible for the loss of α-dystroglycan in epithelium-derived cancers and its role in metastatic progression, we examined the expression and glycosylation status of α-dystroglycan in a group of breast, cervical, and lung cancer cell lines. Here we report that although α-dystroglycan is expressed in the metastatic cell lines MDA-MB-231, HeLa, H1299, and H2030, it is not functionally glycosylated. In screening these cell lines for expression of the six known α-dystroglycan-modifying proteins, we observed that only one, LARGE, was extensively down-regulated. We also report that the ectopic restoration of LARGE expression in these cell lines led not only to the production of a functional dystroglycan but also to the reversion of certain characteristics associated with invasiveness, namely cell attachment to ECM proteins and cell migration.
EXPERIMENTAL PROCEDURES
Full experimental procedures and any associated references are available in the supplemental materials.
RESULTS
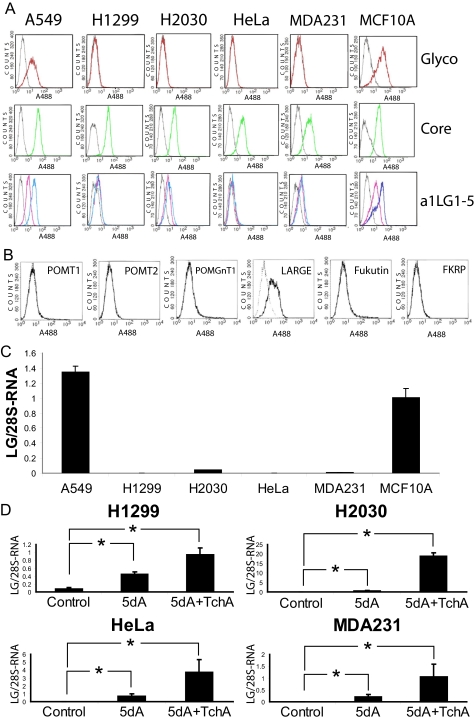
To characterize the mechanism that underlies the loss of α-dystroglycan detection in epithelium-derived cancers, we first studied the expression and glycosylation of α- and β-dystroglycan in a series of model human cancer cell lines derived from breast, lung, and cervical tissue. These include a breast cancer cell line (MDA-MB-231), a cervical cancer cell line (HeLa), and two lung cancer cell lines (H1299 and H2030), and results were compared with those from a control mammary epithelial cell line (MCF10A) and a low metastasis lung cancer cell line (A549). MDA-MB-231, H1299, H2030, and HeLa cells were originally isolated from human cancer biopsies (American Type Culture Collection) and have been proven to retain their invasive phenotype in in vitro as well as in mouse in vivo studies. The presence and glycosylation of dystroglycan in these cells was tested by FACS analysis, using one antibody that targets the α-dystroglycan core irrespective of its glycosylation status (GT20ADG). According to the FACS analysis, all of the cell lines of this cohort expressed similar quantities of core α-dystroglycan on the cell surface (Fig. 1A), demonstrating that the cancer-associated defect of dystroglycan was not due to an expression or trafficking defect. The expression of α-dystroglycan in these cells was confirmed by Western blotting (data not shown). We next tested the glycosylation status of dystroglycan by FACS analysis, using another antibody that targets the α-dystroglycan glyco-motif required for its binding to laminin (IIH6). The results (Fig. 1A) show that H1299, H2030, MDA231, and HeLa cells lacked functionally glycosylated dystroglycan.
FIGURE 1.
Dystroglycan glycosylation in control and metastatic cell lines derived from epithelia. A, cells in culture were detached with 10 mm EDTA and incubated with the antibody IIH6, which recognizes a glyco-epitope ofα-dystroglycan (Glyco-aDG), the antibody GT20ADG, which recognizes theα-dystroglycan core (Core-aDG), or a laminin-derived fusion protein (a1LG1–5; detected using an anti-Myc antibody). FACS analysis of laminin staining was performed in the presence of Ca2+/Mg2+ (blue trace) and in the presence of the cation chelator EDTA (pink trace). Fluorescent-labeled secondary antibodies were used for protein detection, and the analysis was carried out using a FACScan flow cytometer. Gray trace, secondary antibody only. These analyses showed that all the cell lines tested express both α-dystroglycan and β-dystroglycan at the cell surface but that the MDA-MB-231 (MDA231), H1299, H2030, and HeLa cell lines are negative for the α-dystroglycan glyco-epitope that is associated with the ability to bind laminin-111. B, MDA-MB-231 cells were infected with adenoviral vectors expressing the dystroglycan-modifying glycosyltransferases and tested by FACS analysis for reactivity to the IIH6 antibody. LARGE was the only gene that restored IIH6 staining. Solid line, primary and secondary antibody; broken line, secondary antibody only. C, total RNA was isolated from each of the cell lines studied, and cDNA was synthesized by random priming. For each cell line, LARGE and 28 S RNA (normalization control) were specifically amplified, in triplicate, in the presence of SYBR green. The expression of LARGE is shown as expression relative to that of the 28 S RNA in the same sample. Note that expression of the LARGE is virtually undetectable in the cell lines in which α-dystroglycan is hypoglycosylated, i.e. MDA-MB-231 (MDA231), H1299, H2030, and HeLa. D, effect of 5-deoxy-2′-azacytidine and TchA treatment on LARGE expression in cell lines expressing hypoglycosylatedα-dystroglycan. H1299, H2030, MDA-MB-231 (MDA231), and HeLa cells were subjected to treatment, for 96 h, with 5dA, either alone or with trichostatin A (5dA+TchA) added for the last 12 h of treatment. LARGE expression is presented as the ratio of LARGE mRNA versus 28 S RNA expression in the control cell line MCF10A. The standard error (error bars) was calculated using the Student's test (n = 4); *, p < 0.01.
To determine the consequences of this loss of functional glycosylation, we assessed the interaction of these cell lines with laminin-111, the main ECM ligand of dystroglycan in epithelia, by FACS analysis. This analysis was carried out using fusion protein a1LG1–5, which contains the dystroglycan-binding domains of laminin-111 (α1 G-like domains 1–5) fused to a Myc tag. α-Dystroglycan is known to bind to laminin-111 in a Ca2+-dependent fashion. Thus, to corroborate the specificity of the laminin-dystroglycan interaction, we incubated the cells with either Ca2+/Mg2+ or EDTA. We observed that all the cell lines in which α-dystroglycan was hypoglycosylated were defective for Ca2+-dependent laminin-111 binding at their cell surface (Fig. 1A).
To test whether any of the known dystroglycan modifiers (POMT1, POMT2, POMGnT1, Fukutin, FKRP, or LARGE) were defective in any of the cancer cell lines under study, we performed a complementation assay based on the FACS assays. To this end, we inserted the human cDNAs corresponding to each gene into adenoviral plasmids that also express green fluorescent protein (to control for infection). The only gene found to rescue MDA-MB-231 IIH6 staining on FACS scans was LARGE, indicating that a deficiency in this gene alone underlies the defect in dystroglycan glycosylation (Fig. 1B). This is not the result of coding sequence mutations, as sequence analysis revealed that LARGE and all the other known and putative glycosyltransferases that modify dystroglycan were found to be normal in MDA-MB-231 cells. Although several sequence variations relative to the deposited reference coding sequences were identified, subsequent screening of publicly available databases revealed these to be common polymorphisms (supplemental Table 1).
To determine the exact nature of the LARGE effect in the cancer cell lines, we evaluated the expression levels of dystroglycan and its modifying proteins by real-time PCR. We found that in each of the α-dystroglycan hypoglycosylated cell lines under study, LARGE was the only one for which we could not detect significant levels of mRNA (Fig. 1C and supplemental Fig. 1). Thus, hypoglycosylation of α-dystroglycan correlates with the loss of LARGE expression in these metastatic cell lines. We hypothesized that this down-regulation of LARGE is the mechanism behind the loss of dystroglycan receptor function.
To test whether LARGE silencing in these cell lines is the consequence of an epigenetic process, we treated cell cultures with the demethylating agent 5-aza-2′-deoxycytidine (5dA), alone and in combination with the histone deacetylase inhibitor trichostatin A (TchA). After the combined treatment, we found that LARGE transcription was significantly up-regulated in each of the four cell lines that exhibits α-dystroglycan hypoglycosylation (Fig. 1D). These results show that the LARGE gene is not deleted in these cells and that instead, its expression is reversibly silenced by an epigenetic mechanism. To determine whether the rescue of LARGE expression by this treatment is accompanied by recovery of proper α-dystroglycan glycosylation, we repeated the treatment in cell line H2030, which showed the highest degree of LARGE up-regulation. We used FACS analysis to test these cells for α-dystroglycan glycosylation. Approximately 50% of the H2030 cells were positive for the α-dystroglycan IIH6 glyco-epitope after the combined treatment, confirming that the epigenetic down-regulation of LARGE is functionally significant (data not shown).
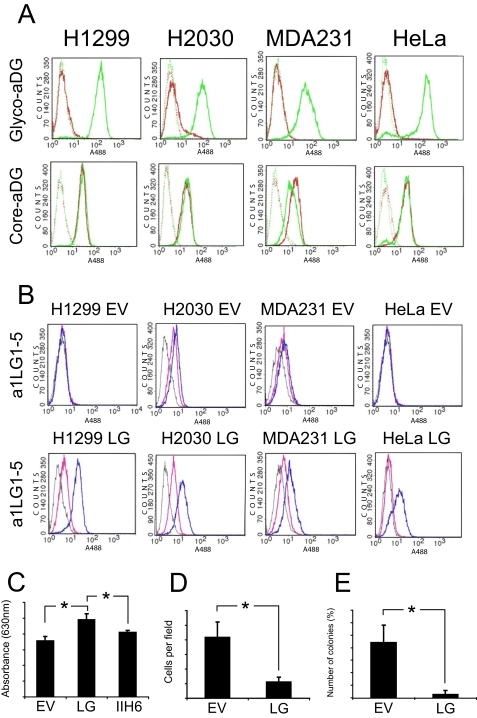
To investigate the functional role of LARGE in these epithelium-derived cancer cell lines, we modified each to stably express either LG or empty vector (EV, control). Each of the four LARGE-expressing modified cell lines showed high expression of the transgene, at both the transcriptional and the protein levels (data not shown). We used FACS analysis to evaluate whether recombinant LARGE expression restored normal α-dystroglycan glycosylation and functional laminin binding. We screened the modified cells for the glyco-epitope and found that α-dystroglycan was uniformly glycosylated in all cases (Fig. 2A), confirming the notion, first reflected by the complementation results shown in Fig. 1B, that LARGE down-regulation underlies cancer-related α-dystroglycan hypoglycosylation. Moreover, FACS analysis of laminin α1 binding showed that LARGE expression also restored the ability of the receptor to bind to its natural ECM ligand, laminin-111 (Fig. 2B).
FIGURE 2.
Effects of LARGE expression in cell lines expressing hypoglycosylated α-dystroglycan. A, after cancer cell lines with stable expression of LARGE were generated, the modified cells were tested by FACS using antibody IIH6 (Glyco-aDG) and antibody GT20ADG (Core-aDG). Each cell line expressing the empty virus (EV, red trace), was compared with its LARGE-expressing counterpart (LG, green trace). Solid line, primary and secondary antibody; broken line, secondary antibody only. In all cases, LARGE expression restored expression of the α-dystroglycan glyco-epitope. B, cancer cell lines that stably express LARGE were also tested by FACS analysis for the ability to bind the laminin α1-derived fusion protein a1LG1–5 in the presence of Ca2+/Mg2+ (blue trace) or EDTA (pink trace). Gray trace, secondary antibody only. LARGE expression was found to confer the ability to bind laminin α1 to the epithelium-derived cancer cells. C, assay for adhesion of cells to laminin-111. 96-well plates were coated with laminin-111, and then MDA231 EV (EV) and MDA231 LG (LG) cells were seeded at 1.5 × 105 cells per laminin-coated well. Cell attachment was measured by crystal violet staining 1 h later. Attachment of MDA231 LG cells was also measured in the presence of antibody IIH6 (Glyco-aDG). The standard error (error bars) was calculated using the Student's test (n = 4); *, p < 0.01. D, Transwell migration of MDA231 cells through Matrigel-coated 8-μm pore filters. MDA-MB-231 cells were incubated in the upper chambers, in culture medium without fetal bovine serum, for 24 h. During this time, they migrated toward the bottom chamber, which contained medium with 10% fetal bovine serum. Cells were counted from at least four random fields at ×20 magnification. The standard error was calculated using the Student's test (n = 6); *, p < 0.01. E, effect of LARGE expression on anchorage-independent growth of MDA-MB-231 cells. Cells were suspended in 0.3% agar medium and layered onto a 0.5% agar base layer (n = 3). After 28 days, colony number was assessed following crystal violet staining. The standard error was calculated using the Student's test (n = 3); *, p < 0.01.
We then tested whether restoring functional α-dystroglycan by expressing recombinant LARGE in these epithelium-derived cancer cells would affect phenotypes associated with invasive metastatic growth. We focused on the MDA231-LG cells that had been stably transfected with LARGE. In the case of the effects of LARGE expression on cell proliferation in monolayer cultures, there was no statistically significant difference in LARGE-expressing cells (data not shown). We tested the effect of LARGE expression on cell adherence to laminin-111-coated surfaces. We found that MDA231-LG cells bound to laminin-111 more effectively than did MDA231-EV cells (Fig. 2C) and that this binding could be blocked by competition with IIH6 antibody. Thus, the functional glyco-epitope of α-dystroglycan is directly involved in the binding of this protein to laminin.
To evaluate the effects of LARGE expression on the migration of MDA-MB-231 cells, we used a Transwell assay involving a reconstituted basement membrane (Matrigel) (Fig. 2D). We found that LARGE-mediated modification of α-dystroglycan in MDA231-LG cells consistently reduced the rates of cell migration through the Matrigel, which indicates that dystroglycan glycosylation by LARGE impairs the ability of the cell to escape from its laminin anchor and thus its ability to migrate. Finally, we assessed the effect of LARGE expression on anchorage-independent growth (in soft agar), which correlates well with tumorigenic potential in vivo (26). We found that LARGE reexpression inhibited colony formation (Fig. 2E), indicating that LARGE has an antitumorigenic function in these cells. Taken together, these data indicate that LARGE expression negatively influences cancer cell phenotypes associated with aggressive invasive and metastatic disease.
DISCUSSION
The ability of dystroglycan to bind to LG domain-containing ECM proteins and to mediate their polymerization is highly dependent on dystroglycan glycosylation status. The results presented here not only provide evidence for the importance of the functional glycosylation of dystroglycan in epithelial-derived cancers but also implicate epigenetic silencing of LARGE as the cause of the cancer-associated loss of functional dystroglycan glycosylation.
We have confirmed, by independent methods, thatα-dystroglycan is hypoglycosylated and non-functional in terms of laminin binding in metastatic cell lines. Although dystroglycan is present at the cell membrane in each case, defective glycosylation renders it unable to bind to the ECM protein laminin. These α-dystroglycan hypoglycosylated cells are not able to anchor to the basement membrane or organize ECM polymerization as effectively as normal epithelial cells, and their integration into the epithelium is compromised. These results demonstrate that LARGE is important for proper α-dystroglycan functional glycosylation and, thereby, for the ability of dystroglycan to function as an ECM protein receptor. The fact that dystroglycan is well synthesized and transported to the cell membrane makes it tempting to speculate that this cancer-associated form of dystroglycan may have a different function than the fully glycosylated one, shifting its affinity from laminin to a different ECM ligand.
The LARGE locus was first described as a region prone to loss of heterozygosity in cancer (27). We have shown that the mechanism governing LARGE inactivation in our cohort of epithelium-derived cell lines is reversible and that it is not a result of a chromosomal aberration. Instead, LARGE repression in our α-dystroglycan hypoglycosylated cell cohort supports the notion that LARGE is involved in cancer progression. Indeed, its expression can be partially restored by treatment with drugs that are known to force the re-expression of anti-oncogenes. Whether this recovery is a response to a general cellular program of gene silencing or is regulated by specific transcription factors is now under investigation. The rescue of functional α-dystroglycan glycosylation by LARGE in vitro and the fact that no mutation has been found in any of the dystroglycan-modifying genes in the MDA-MB-231 cell line support the notion that no other gene or mechanism is involved in this cancer-associated dystroglycan glycosylation defect. The reversibility of LARGE repression makes it tempting to speculate that LARGE is expressed during certain phases of cancer progression, for example, during the extravasation process, when it may be necessary for the cancer cell to interact with the vascular endothelium.
It is striking that six different known or putative glycosyltransferases (POMT1, POMT2, POMGnT1, LARGE, Fukutin, and FKRP) have only one identified target in brain and muscle, dystroglycan. In epithelium, dystroglycan also seems to be the main functional target of LARGE. The main features of the phenotype observed in our cell cohort after LARGE re-expression can be ascribed to α-dystroglycan based on the resemblance between a breast cancer cell line deficient for the α-dystroglycan protein (4) and the breast cancer cell line MDA-MB-231, which is deficient for LARGE. Also, dystroglycan knock-out mice recapitulate most of the features shown by the LARGE-deficient mouse (28–30). Although other targets of LARGE have not been identified yet, these findings indicate that α-dystroglycan is the major target of LARGE-dependent glycosylation and that this modification affects the maintenance of epithelia, as well as that of skeletal muscle and brain.
LARGE is a putative glycosyltransferase that contains two domains with homology to known catalytic sites, one related to β-1,3-N-acetylglucosaminyltransferase and another to bacterial glycosyltransferase (29). These two predicted catalytic domains of LARGE contain three DXD motifs that are typically conserved among glycosyltransferases that use nucleoside diphosphate sugars as donors. Because mutation of each DXD motif to NNN in human LARGE prevents the generation of functionally modified α-dystroglycan in Chinese hamster ovary transformant (31), it is very likely that LARGE actually acts as a glycosyltransferase, although the biochemical activity of LARGE has not yet been determined. This notion is supported by the fact that LARGE mutated patients and spontaneous myodystrophy mouse Largemyd present with similar clinical and biochemical phenotypes to patients with mutations in the known glycosyltransferases POMT1, POMT2, and POMGnT1.
Correct cell anchoring to the ECM is crucial to orchestrating and maintaining cellular structure and function. In the epithelium, these effects result from cooperation between the β1 integrins and dystroglycan complexes (1), which connect the ECM to intracellular actin and intermediate filaments and also elicit intracellular signal transduction and differentiation (1, 4, 32). In fact, the changes in integrins β1 and -4 and α2, -3, and -6 that have been reported in mammary tumors and cell lines lead to a loss of polarity and invasiveness, and it has been documented that integrin misplacement can lead to the loss of polarity in epithelial cells (33). Further studies will be needed to determine whether LARGE-mediated modification of α-dystroglycan has an effect on either integrin-mediated ligand binding or integrin-mediated signaling.
In conclusion, we have demonstrated in a cohort of highly metastatic epithelial cancer cell lines that epigenetic silencing of LARGE glycosyltransferase underlies the α-dystroglycan hypoglycosylation that renders this receptor unable to bind to its natural ligand, laminin-111. This repression can be partially rescued by treatment with 5dA and TchA. The consequences and significance of this repression are demonstrated by the rescue of α-dystroglycan receptor function by forced expression of LARGE. Ectopic expression of LARGE alters the cancer cell phenotype toward one that is significantly less invasive. This study thus demonstrates that LARGE is silenced in epithelium-derived cancer cells. The re-expression of the LARGE gene may thus represent a future avenue for the treatment of cancer.
Supplementary Material
Acknowledgments
We thank the members of the Campbell laboratory for comments on this work. We thank the staff of the Gene Transfer Vector Core Facility of the University of Iowa, supported by National Institutes of Health/NIDDK P30 DK 54759, for their assistance with viral vector production.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 CA130916 (to M. D. H.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, a supplemental figure, and a supplemental table.
This article was selected as a Paper of the Week.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: ECM, extracellular matrix; LARGE, like-acetylglucosaminyltransferase; LG, LARGE; EV, empty vector; TchA, trichostatin A; 5dA, 5-aza-2′-deoxycytidine; FACS, fluorescence-activated cell sorter; glyco-aDG, glyco-epitope of α-dystroglycan; POMT1, protein O-mannosyltransferase 1; POMT2, protein O-mannosyltransferase 2; POMGnT1, protein O-mannose β-1,2-acetylglucosaminyltransferase (POMGnT1); FKRP, Fukutin-related protein.
References
- 1.Muschler, J., Lochter, A., Roskelley, C. D., Yurchenco, P., and Bissell, M. J. (1999) Mol. Biol. Cell 10, 2817-2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durbeej, M., Larsson, E., Ibraghimov, B. O., Roberds, S. L., Campbell, K. P., and Ekblom, P. (1995) J. Cell Biol. 130, 79-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durbeej, M., and Campbell, K. P. (1999) J. Biol. Chem. 274, 26609-26616 [DOI] [PubMed] [Google Scholar]
- 4.Muschler, J., Levy, D., Boudreau, R. Henry, M., Campbell, K. P., and Bissell, M. J. (2002) Cancer Res. 62, 7102-7109 [PubMed] [Google Scholar]
- 5.Weir, M. L., Oppizzi, M. L., Henry, M. D., Onishi, A., Campbell, K. P., Bissell, M, J., and Muschler, J. L. (2006). J. Cell Sci. 119, 4047-4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henry, M. D., Cohen, M. B., and Campbell, K. P. (2001) Hum. Pathol. 32, 791-795 [DOI] [PubMed] [Google Scholar]
- 7.Sgambato, A., Migaldi, M., Montanari, M., Camerini, A., Brancaccio, A., Rossi, G., Cangiano, R., Losasso, C., Capelli, G., Trentini, G. P., and Cittadini, A. (2003) Am. J. Pathol. 162, 849-860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jing, J., Lien, C. F., Sharma, S., Rice, J., Brennan, P. A., and Górecki, D. C. (2004) Eur. J. Cancer 40, 2143-2151 [DOI] [PubMed] [Google Scholar]
- 9.Sgambato, A., Tarquini, E., Resci, F., De Paola, B., Faraglia, B., Camerini, A., Rettino, A., Migaldi, M., Cittadini, A., and Zannoni, G. F. (2006) Gynecol. Oncol. 103, 397-404 [DOI] [PubMed] [Google Scholar]
- 10.Ibraghimov-Beskrovnaya, O., Ervasti, J. M., Leveille, C. J., Slaughter, C. A., Sernett, S. W., and Campbell, K. P. (1992) Nature 355, 696-702 [DOI] [PubMed] [Google Scholar]
- 11.Ilsley, J. L., Sudol, M., and Winder, S. J. (2002) Cell. Signal. 14, 183-189 [DOI] [PubMed] [Google Scholar]
- 12.Yang, B., Jung, D., Motto, D., Meyer, J., Koretzky, G., and Campbell, K. P. (1995) J. Biol. Chem. 270, 11711-11714 [DOI] [PubMed] [Google Scholar]
- 13.Yamada, H., Denzer, A. J., Hori, H., Tanaka, T., Anderson, L. V. B., Fujita, S., Fukuta-Ohi, H., Shimizu, T., Ruegg, M. A., and Matsumura, K. (1996) J. Biol. Chem. 271, 23418-23423 [DOI] [PubMed] [Google Scholar]
- 14.Kanagawa, M., Saito, F., Kunz, S., Yoshida-Moriguchi, T., Barresi, R., Kobayashi, Y. M., Muschler, J., Dumanski, J. P., Michele, D. E., Oldstone, M. B., and Campbell, K. P. (2004) Cell 117, 953-964 [DOI] [PubMed] [Google Scholar]
- 15.Gee, S. H., Montanaro, F., Lindenbaum, M. H., and Carbonetto, S. (1994) Cell 77, 675-686 [DOI] [PubMed] [Google Scholar]
- 16.Talts, J. F., Andac, Z., Gohring, W., Brancaccio, A., and Timpl, R. (1999) EMBO J. 18, 863-870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugita, S., Saito, F., Tang, J., Satz, J., Campbell, K., and Südhof, T. C. (2001) J. Cell Biol. 154, 435-445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanagawa, M., Michele, D. E., Satz, J. S., Barresi, R., Kusano, H., Sasaki, T., Timpl, R., Henry, M. D., and Campbell, K. P. (2005) FEBS Lett. 579, 4792-4796 [DOI] [PubMed] [Google Scholar]
- 19.Michele, D. E., Barresi, R., Kanagawa, M., Saito, F., Cohn, R. D., Satz, J. S., Dollar, J., Nishino, I., Kelley, R. I., Somer, H., Straub, V., Mathews, K. D., Moore, S. A., and Campbell, K. P. (2002) Nature 418, 417-422 [DOI] [PubMed] [Google Scholar]
- 20.Beltrán-Valero de Bernabé, D., Currier, S., Steinbrecher, A., Celli, J., van Beusekom, E., van der Zwaag, B., Kayserili, H., Merlini, L., Chitayat, D., Dobyns, W. B., Cormand, B., Lehesjoki, A. E., Cruces, J., Voit, T., Walsh, C. A., van Bokhoven, H., and Brunner, H. G. (2002). Am. J. Hum. Genet. 71, 1033-1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Reeuwijk, J., Janssen, M., van den Elzen, C., Beltran-Valero de Bernabé, D., Sabatelli, P., Merlini, L., Boon, M., Scheffer, H., Brockington, M., Muntoni, F., Huynen, M. A., Verrips, A., Walsh, C. A., Barth, P. G., Brunner, H. G., and van Bokhoven, H. (2005). J. Med. Genet. 42, 907-912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida, A., Kobayashi, K., Manya, H., Taniguchi, K., Kano, H., Mizuno, M., Inazu, T., Mitsuhashi, H., Takahashi, S., Takeuchi, M., Herrmann, R., Straub, V., Talim, B., Voit, T., Topaloglu, H., Toda, T., and Endo, T. (2001) Dev. Cell 1, 717-724 [DOI] [PubMed] [Google Scholar]
- 23.Longman, C., Brockington, M., Torelli, S., Jimenez-Mallebrera, C., Kennedy, C., Khalil, N., Feng, L., Saran, R. K., Voit, T., Merlini, L., Sewry, C. A., Brown, S. C., and Muntoni, F. (2003) Hum. Mol. Genet. 12, 2853-2861 [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi, K., Nakahori, Y., Miyake, M., Matsumura, K., Kondo-Iida, E., Nomura, Y., Segawa, M., Yoshioka, M., Saito, K., Osawa, M., Hamano, K., Sakakihara, Y., Nonaka, I., Nakagome, Y., Kanazawa, I., Nakamura, Y., Tokunaga, K., and Toda, T. (1998) Nature 394, 388-392 [DOI] [PubMed] [Google Scholar]
- 25.Brockington, M., Blake, D. J., Prandini, P., Brown, S. C., Torelli, S., Benson, M. A., Ponting, C. P., Estournet, B., Romero, N. B., Mercuri, E., Voit, T., Sewry, C. A., Guicheney, P., and Muntoni, F. (2001) Am. J. Hum. Genet. 69, 1198-1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freedman, V. H., and Shin, S. (1974) Cell, 3, 355-360 [DOI] [PubMed] [Google Scholar]
- 27.Peyrard, M., Seroussi, E., Sandberg-Nordqvist, A., Xie, Y., Han, F., Fransson, I., Collins, J., Dunham, I., Kost-Alimova, M., Imreh, S., and Dumanski, J. P. (1999) Proc. Natl. Acad. Sci. U. S. A. 96, 598-603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satz, J. S., Barresi, R., Durbeej, M., Willer, T., Turner, A., Moore, S. A., and Campbell, K. P. (2008) Neurobiol. Dis. 28, 10567-10575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grewal, P. K., Holzfeind, P. J., Bittner, R. E., and Hewitt, J. E. (2001) Nat. Genet. 28, 151-154 [DOI] [PubMed] [Google Scholar]
- 30.Moore, S. A., Saito, F., Chen, J., Michele, D. E., Henry, M. D., Messing, A., Cohn, R. D., Ross-Barta, S. E., Westra, S., Williamson, R. A., Hoshi, T., and Campbell, K. P. (2002) Nature 418, 422-425 [DOI] [PubMed] [Google Scholar]
- 31.Brockington, M., Torelli, S., Prandini, P., Boito, C., Dolatshad, N. F., Longman, C., Brown, S. C., and Muntoni, F. (2005) Hum. Mol. Genet. 14, 657-665 [DOI] [PubMed] [Google Scholar]
- 32.Spence, H. J., Dhillon, A. S., James, M., and Winder, S. J. (2004) EMBO Rep. 5, 484-489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giancotti, F. G. (1996) J. Cell Sci. 109, 1165-1172 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.