Abstract
ATP synthase uses a unique rotational mechanism to convert chemical energy into mechanical energy and back into chemical energy. The helix-turn-helix motif, termed “DELSEED-loop,” in the C-terminal domain of the β subunit was suggested to be involved in coupling between catalysis and rotation. Here, the role of the DELSEED-loop was investigated by functional analysis of mutants of Bacillus PS3 ATP synthase that had 3–7 amino acids within the loop deleted. All mutants were able to catalyze ATP hydrolysis, some at rates several times higher than the wild-type enzyme. In most cases ATP hydrolysis in membrane vesicles generated a transmembrane proton gradient, indicating that hydrolysis occurred via the normal rotational mechanism. Except for two mutants that showed low activity and low abundance in the membrane preparations, the deletion mutants were able to catalyze ATP synthesis. In general, the mutants seemed less well coupled than the wild-type enzyme, to a varying degree. Arrhenius analysis demonstrated that in the mutants fewer bonds had to be rearranged during the rate-limiting catalytic step; the extent of this effect was dependent on the size of the deletion. The results support the idea of a significant involvement of the DELSEED-loop in mechanochemical coupling in ATP synthase. In addition, for two deletion mutants it was possible to prepare an α3β3γ subcomplex and measure nucleotide binding to the catalytic sites. Interestingly, both mutants showed a severely reduced affinity for MgATP at the high affinity site.
F1F0-ATP synthase catalyzes the final step of oxidative phosphorylation and photophosphorylation, the synthesis of ATP from ADP and inorganic phosphate. F1F0-ATP synthase consists of the membrane-embedded F0 subcomplex, with, in most bacteria, a subunit composition of ab2c10, and the peripheral F1 subcomplex, with a subunit composition of α3β3γδε. The energy necessary for ATP synthesis is derived from an electrochemical transmembrane proton (or, in some organisms, a sodium ion) gradient. Proton flow down the gradient through F0 is coupled to ATP synthesis on F1 by a unique rotary mechanism. The protons flow through (half) channels at the interface of the a and c subunits, which drives rotation of the ring of c subunits. The c10 ring, together with F1 subunits γ and ε, forms the rotor. Rotation of γ leads to conformational changes in the catalytic nucleotide binding sites on the β subunits, where ADP and Pi are bound. The conformational changes result in the formation and release of ATP. Thus, ATP synthase converts electrochemical energy, the proton gradient, into mechanical energy in the form of subunit rotation and back into chemical energy as ATP. In bacteria, under certain physiological conditions, the process runs in reverse. ATP is hydrolyzed to generate a transmembrane proton gradient, which the bacterium requires for such functions as nutrient import and locomotion (for reviews, see Refs. 1–6).
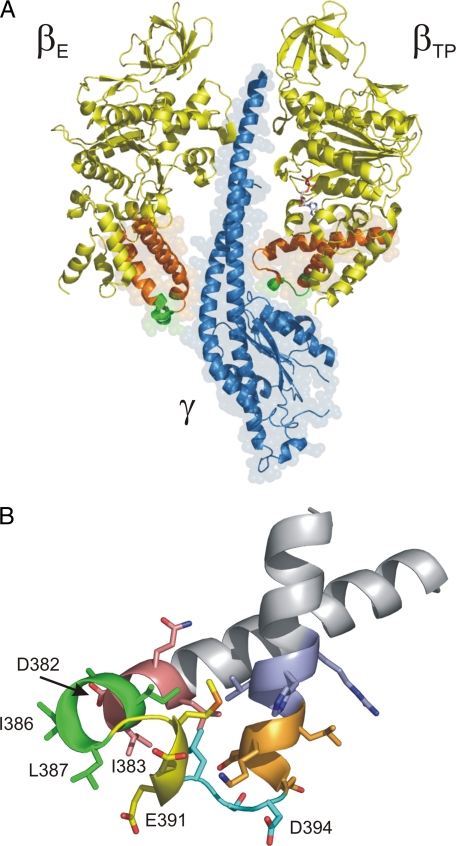
F1 (or F1-ATPase) has three catalytic nucleotide binding sites located on the β subunits at the interface to the adjacent α subunit. The catalytic sites have pronounced differences in their nucleotide binding affinity. During rotational catalysis, the sites switch their affinities in a synchronized manner; the position of γ determines which catalytic site is the high affinity site (Kd1 in the nanomolar range), which site is the medium affinity site (Kd2 ≈ 1 μm), and which site is the low affinity site (Kd3 ≈ 30–100 μm; see Refs. 7 and 8). In the original crystal structure of bovine mitochondrial F1 (9), one of the three catalytic sites, was filled with the ATP analog AMP-PNP,2 a second was filled with ADP (plus azide) (see Ref. 10), and the third site was empty. Hence, the β subunits are referred to as βTP, βDP, and βE. The occupied β subunits, βTP and βDP, were in a closed conformation, and the empty βE subunit was in an open conformation. The main difference between these two conformations is found in the C-terminal domain. Here, the “DELSEED-loop,” a helix-turn-helix structure containing the conserved DELSEED motif, is in an “up” position when the catalytic site on the respective β subunit is filled with nucleotide and in a “down” position when the site is empty (Fig. 1A). When all three catalytic sites are occupied by nucleotide, the previously open βE subunit assumes an intermediate, half-closed (βHC) conformation. It cannot close completely because of steric clashes with γ (11).
FIGURE 1.
The βDELSEED-loop. A, interaction of the βTP and βE subunits with theγ subunit.β subunits are shown in yellow andγ in blue. The DELSEED-loop (shown in orange, with the DELSEED motif itself in green)of βTP interacts with the C-terminal helixγ and the short helix that runs nearly perpendicular to the rotation axis. The DELSEED-loop of βE makes contact with the convex portion of γ, formed mainly by the N-terminal helix. A nucleotide molecule (shown in stick representation) occupies the catalytic site of βTP, and the subunit is in the closed conformation. The catalytic site on βE is empty, and the subunit is in the open conformation. This figure is based on Protein Data Bank file 1e79 (32). B, deletions in the βDELSEED-loop. The loop was “mutated” in silico to represent the PS3 ATP synthase. The 3–4-residue segments that are removed in the deletion mutants are color-coded as follows: 380LQDI383, pink; 384IAIL387, green; 388GMDE391, yellow; 392LSD394, cyan; 395EDKL398, orange; 399VVHR402, blue. Residues that are the most involved in contacts with γ are labeled. All figures were generated using the program PyMOL (DeLano Scientific, San Carlos, CA).
The DELSEED-loop of each of the three β subunits makes contact with the γ subunit. In some cases, these contacts consist of hydrogen bonds or salt bridges between the negatively charged residues of the DELSEED motif and positively charged residues on γ. The interactions of the DELSEED-loop with γ, its movement during catalysis, the conservation of the DELSEED motif (see Table 1), and a number of mutagenesis experiments led to the assumption that the DELSEED-loop might play an essential role in coupling between catalysis and rotation of γ (12–14). Thus, the finding that an AALSAAA mutant in the α3β3γ complex of ATP synthase from the thermophilic Bacillus PS3, where several hydrogen bonds/salt bridges to γ are removed simultaneously, could drive rotation of γ with the same torque as the wild-type enzyme (14) came as a surprise. On the other hand, it seems possible that it is the bulk of the DELSEED-loop, more so than individual interactions, that drives rotation of γ. According to a model favored by several authors (6, 15, 16) (see also Refs. 17–19), binding of ATP (or, more precisely, MgATP) to the low affinity catalytic site on βE and the subsequent closure of this site, accompanied by its conversion into the high affinity site, are responsible for driving the large (80–90°) rotation substep during ATP hydrolysis, with the DELSEED-loop acting as a “pushrod.” A recent molecular dynamics (20) study supports this model and implicates mainly the region around several hydrophobic residues upstream of the DELSEED motif (specifically βI386 and βL387)3 as being responsible for making contact with γ during the large rotation substep.
TABLE 1.
Conservation of residues in the DELSEED-loop Amino acids found in selected species in the turn region of the DELSEED-loop. Listed are all positions subjected to deletions in the present study. Residue numbers refer to the PS3 enzyme. Consensus annotation: p, polar residue; s, small residue; h, hydrophobic residue; –, negatively charged residue; +, positively charged residue.
In the present study, we investigated the function of the DELSEED-loop using an approach less focused on individual residues, by deleting stretches of 3–7 amino acids between positions β380 and β402 of ATP synthase from the thermophilic Bacillus PS3. We analyzed the functional properties of the deletion mutants after expression in Escherichia coli. The mutants showed ATPase activities, which were in some cases surprisingly high, severalfold higher than the activity of the wild-type control. On the other hand, in all cases where ATP synthesis could be measured, the rates where below or equal to those of the wild-type enzyme. In Arrhenius plots, the hydrolysis rates of the mutants were less temperature-dependent than those of wild-type ATP synthase. In those cases where nucleotide binding to the catalytic sites could be tested, the deletion mutants had a much reduced affinity for MgATP at high affinity site 1. The functional role of the DELSEED-loop will be discussed in light of the new information.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids—Plasmid pTR19-ASDS, which carries the genes for the F1F0-ATP synthase from thermophilic Bacillus PS3 (21), was used to generate mutations in the different positions of the C-terminal domain of the β subunit: Δ380LQDI383, Δ384IAIL387, Δ388GMDE391, Δ392LSD394, Δ395-EDKL398, Δ399VVHR402, Δ381QDIIAIL387, Δ388GMDELSD394, Δ390DELSDED396, and Δ392LSDEDKL396. The mutagenic oligonucleotides were designed in such a way that, in addition to the desired mutation, a restriction site would be eliminated or generated to facilitate screening. Deletions were introduced by polymerase chain reaction using the QuikChange II XL mutagenesis kit (Stratagene). Wild-type and mutated plasmids were transformed into E. coli strain DK8, which does not express E. coli ATP synthase (22).
The first seven deletion mutations were also introduced in plasmid pNM1. Plasmid pNM1 is a derivative of plasmid pKAGB1 (23). pKAGB1 is used to express a Cys- and Trp-less form of the α3β3γ subcomplex of PS3. pNM1 contains an additional mutation to generate an α3(βY341W)3γ subcomplex, which allows monitoring of nucleotide binding to the three catalytic sites. For expression, pNM1 and the derived deletion mutants were transformed into E. coli strain JM103 Δ(uncB-uncD).
Isolation of Inverted Membrane Vesicles and Determination of F1F0 Content in E. coli Membranes—E. coli strain DK8 harboring wild-type or mutated pTR19-ASDS plasmids was aerobically cultivated at 37 °C for 18 h in 2× YT medium (24) containing 100 μg/ml ampicillin. Inverted membrane vesicles from E. coli cells expressing thermophilic F1F0 were prepared as described (21, 25). The amount of wild-type F1F0 in E. coli membrane preparations was determined by SDS-PAGE as visualized by staining with Coomassie Brilliant Blue (21). The relative amount of mutant F1F0 in the membranes was estimated via Western blots, using an anti-β antibody (Agrisera, Vännäs, Sweden) or an anti-α/anti-β antibody (kindly provided by Dr. Bill Brusilow, Wayne State University). The staining intensity was quantified using a Photodyne imaging system and ImageJ acquisition software (National Institutes of Health).
Isolation of the α3β3γ Subcomplex—After expression in E. coli, the α3β3γ subcomplex of PS3 was prepared essentially as described (23). α3β3γ was stored as ammonium sulfate precipitate at 4 °C.
Functional Analysis of Mutant Strains and Enzymes—Growth of strains expressing wild-type or mutant PS3 ATP synthase on succinate plates and growth in limiting glucose was determined as described previously (26).
ATPase activities were assayed in a buffer containing 50 mm Tris/H2SO4, 10 mm ATP, and 4 mm MgSO4, pH 8.0. For the α3β3γ subcomplex, the assay was performed at 60 °C, for membrane vesicles at 42 °C. The reaction was started by the addition of 10–20 μg/ml α3β3γ or 50–150 μg/ml membrane vesicles and stopped after 1, 2, or 10 min (depending on the activity) by the addition of sodium dodecyl sulfate (final concentration 5% (w/v)). The released Pi was measured as described (27). 1 unit of enzymatic activity corresponds to 1 μmol ATP hydrolyzed (equivalent to 1 μmol of Pi produced) or synthesized per min. Turnover numbers (kcat) in membrane vesicles were calculated based on the F1F0 content measured as described above. The temperature dependence of ATPase activities of membrane vesicles was measured in the buffer given above. The temperature was varied between 20 and 50 °C. The reaction mixture was preincubated for 10 min at the desired temperature. Activation energies, and entropic and enthalpic components were calculated from Arrhenius plots as described (28).
ATP synthesis activity in vitro was measured as follows. Inverted membrane vesicles were suspended in a solution containing 20 mm HEPES/KOH, 100 mm KCl, 5 mm MgCl2, 1 mm ADP, 5 mm KPi, 10% glycerol, pH 7.5, and incubated at 42 °C. The reaction was initiated by the addition of 2 mm NADH. After 10, 40, and 70 s, aliquots of the reaction mixture (each containing 20 μg of membrane protein) were transferred into boiling buffer of 100 mm Tris/H2SO4, 4 mm EDTA, pH 7.75, for heat denaturation. The samples were incubated at 100 °C for 2 min, cooled on ice, and centrifuged for 1 min at 1000 × g. The amount of ATP was determined by the luciferin/luciferase method (CLS II ATP bioluminescence kit, Roche Applied Science). Light emission was measured at 562 nm in a Fluorolog 3 spectrofluorometer (HORIBA Jobin Yvon, Edison, NY).
NADH- and ATP-driven H+-pumping in membrane vesicles was measured via fluorescence quenching of ACMA at 42 °C. To a buffer of 10 mm HEPES/KOH, 100 mm KCl, and 5 mm MgCl2, pH 7.5, 0.5 mg/ml membrane vesicles and 0.3 μg/ml ACMA were added. Proton pumping was initiated by adding NADH or ATP to a final concentration of 1 mm and terminated by adding CCCP (final concentration 1 μm). The excitation wavelength was 410 nm and the emission wavelength 480 nm.
Binding of MgATP to the catalytic sites of the purified α3β3γ subcomplex was measured using the fluorescence of the inserted Trp residue, βW341 (8). Before use, the α3β3γ ammonium sulfate precipitate was pelleted by centrifugation and redissolved in a buffer containing 50 mm Tris/HCl, 10 mm CDTA, pH 8.0. After 1 h of incubation at 23 °C, the α3β3γ subcomplex was passed through two subsequent centrifuge columns containing Sephadex G-50, equilibrated with 50 mm Tris/HCl and 0.1 mm EDTA, pH 8.0. After this treatment, the enzyme subcomplex is essentially nucleotide-free (29). Fluorescence titrations were performed in a buffer containing 50 mm Tris/H2SO4, 2.5 mm MgSO4, pH 8.0, with ATP added in the appropriate concentrations. Each protein sample was used to acquire maximally two data points. Kd values were determined by fitting of theoretical curves to the experimental data points by nonlinear least-squares analysis, assuming a model with three different independent sites (8).
Protein concentrations of membrane vesicles were determined by the Lowry method (30) and those of purified α3β3γ subcomplex by the Bradford method (31). Both assays used bovine serum albumin as standard.
Modeling—Homology modeling including energy minimization refinement was performed using the program PRIME (Schroedinger Inc.). Templates were the structures of bovine mitochondrial β subunits in closed (βTP and βDP), half-closed (βHC), and empty (βE) conformations, taken from Protein Data Bank files 1h8e (11) and 1e79 (32). The γ subunit was included during the energy minimization step.
RESULTS
Overview of Deletion Mutants in the DELSEED-loop—Fig. 1B shows the helix-turn-helix structure known as the DELSEED-loop in the C-terminal domain of the β subunit of ATP synthase. As can be seen from Table 1, not only the DELSEED motif itself but the whole tip of the loop is strongly conserved. To assess the function of the loop, we designed deletion mutants that had 3–4 contiguous residues removed. The exact length of the deletion (3 or 4) was selected to make a “reconnection” of both ends of the protein chain as easy as possible as judged by eye. Originally, all mutations were made in E. coli ATP synthase; however, none of the deletion mutants assembled properly. Thus, the mutations were repeated in ATP synthase of the thermophilic bacterium Bacillus PS3 expressed in E. coli. In this system, all of the mutant enzymes with deletions of 3–4 residues removed could be obtained in membrane-bound form. The deletions were: Δ380LQDI383, Δ384IAIL387, Δ388GMDE391, Δ392LSD394, Δ395EDKL398, and Δ399VVHR402 (color-coded in Fig. 1B). It should be noted that DELSEED is actually 390DELSDED396 in the thermophilic enzyme. In a second mutagenesis round, several longer deletions of 7 residues were generated, mostly by combining two consecutive shorter deletions, Δ381QDIIAIL387, Δ388GMDELSD394, and Δ392LSD-EDKL396, plus Δ390DELSDED396 where the DELSEED motif itself is removed. With the exception of the Δ381QDIIAIL387 deletion mutant, in all other cases the amount of F1F0 found in membrane vesicles was not sufficient for inclusion in this study (<2% of wild type). In the case of the Δ390DELSDED396 deletion mutant, this finding reflects an earlier result (14) where no α3β3γ subcomplex of thermophilic F1 containing this deletion could be obtained.
Oxidative Phosphorylation in Vivo—As described previously (21), wild-type PS3 F1F0 expressed in E. coli (strain pTR19-ASDS/DK8) allowed growth on plates containing succinate as the sole carbon source, demonstrating that the PS3 enzyme is able to catalyze ATP synthesis in vivo. The growth of strain pTR19-ASDS/DK8 was about 75% as strong as that of pBWU13.4/DK8, which expresses wild-type E. coli ATP synthase. In contrast, none of the deletion mutants showed detectable growth on succinate plates after overnight incubation at 37 °C. Prolonged incubation (3 days) resulted in the growth of some of the mutants, especially Δ388GMDE391 and Δ392LSD394. However, as there was also growth, albeit very weak, even on the negative control plate containing strain pUC118/DK8 (presumably due to remnants of the LB medium used to streak out the bacteria), this line of experiments was aborted.
In growth yield assays in limiting glucose, strain pTR19-ASDS/DK8 expressing wild-type PS3 F1F0 grew to a turbidity (measured as absorbance at 595 nm) of 62% of that of the control strain pBWU13.4/DK8 expressing wild-type E. coli ATP synthase. The negative (unc-) control, strain pUC118/DK8, reached 38% of the value for strain pBWU13.4/DK8 and 61% of the value for strain pTR19-ASDS/DK8. Three of the deletion mutants, Δ380LQDI383, Δ388GMDE391, and Δ392LSD394, showed growth yields significantly higher than the negative control but clearly below the value for the wild-type PS3 enzyme (Table 2). Deletion mutants Δ395EDKL398 and Δ399VVHR402 exhibited growth yields similar to those of the negative control, whereas Δ384IAIL387 and Δ381QDIIAIL387 grew less well. The latter result may be indicative of a highly uncoupled enzyme or of severe proton leaks through the membranes due to incorrectly or incompletely inserted proteins.
TABLE 2.
Oxidative phosphorylation in vivo and ATP synthesis activities in vitro Oxidative phosphorylation in vivo and ATP synthesis activities of membrane preparations of deletion mutants and controls were measured as described under “Experimental Procedures.” The positive control was strain pTR19-ASDS/DK8, which expresses Bacillus PS3 ATP synthase in E. coli; this strain served as background for the deletions. The negative control was strain pUC118/DK8, which expresses neither PS3 ATP synthase nor the endogenous E. coli enzyme. The amount of F1F0 in the membrane preparations was measured by quantitative immunoblot analysis as described under “Experimental Procedures.” Turnover rates were calculated using a molecular mass of 531 kDa for the holoenzyme, taking into account the differing amounts of ATP synthase in the individual membrane preparations. ND, no activity detectable beyond background.
| Strain/mutation | Growth yields in limiting glucose | Amount of F1F0 on membranes | NADH-driven ATP synthesis activity | Turnover rate (kcat) |
|---|---|---|---|---|
| % wild type | % total protein | milliunits/mg | s–1 | |
| Wild type | 100 | 20 | 71 | 3.2 |
| pUC118/DK8 (unc–) | 61 | 0 | —a | |
| Δ380LQDI383 | 69 | 20 | 42 | 1.9 |
| Δ384IAIL387 | 47 | 10 | 33 | 3.0 |
| Δ388GMDE391 | 73 | 15 | 51 | 3.1 |
| Δ392LSD394 | 80 | 20 | 48 | 2.2 |
| Δ395EDKL398 | 58 | 2.0 | ND | |
| Δ399VVHR402 | 61 | 2.6 | ND | |
| Δ381QDIIAIL387 | 45 | 2.4 | 8 | 3.0 |
Strain pUC118/DK8 showed a small amount of ATP production, probably due to adenylate kinase activity of the membranes (52). All given values are corrected for this activity
ATP Synthesis Activity of Membrane Preparations—In addition to the in vivo growth assays to monitor oxidative phosphorylation, we measured NADH-driven ATP synthesis in vitro.At 42 °C, the wild-type PS3 enzyme in E. coli membrane vesicles showed an ATP synthesis activity of 71 milliunits/mg membrane protein. Except for Δ395EDKL398 and Δ399VVHR402, the other deletion mutants also exhibited some ATP synthesis activity. As well as being a direct consequence of a mutation, lack of activity can be due to lack of expression or to oligomeric instability of the enzyme; thus, it was necessary to quantify the amount of enzyme on the membranes. The amount of wild-type PS3 F1F0 in E. coli membrane preparations was found to be ∼20% of the total membrane protein (this study and Ref. 21). Membrane preparations containing PS3 ATP synthase with the deletions Δ380LQDI383, Δ384IAIL387, Δ388GMDE391, Δ392LSD394, Δ395EDKL398, Δ399VVHR402, and Δ381QDIIAIL387 had between 10 and 100% of the amount of the wild-type enzyme (Table 2). The lack of activity of the Δ395EDKL398 and Δ399VVHR402 mutants could reflect the low amount of enzyme in the membrane preparations. On the other hand, the Δ381QDIIAIL387 mutant ATP synthase, present in similarly low quantity, showed significant synthesis activity. After correction for the different amounts of enzyme, the turnover rates of the deletion mutants (except for Δ395EDKL398 and Δ399VVHR402) were between 60 and 100% of that of the wild-type enzyme.
ATPase Activity of Membrane Preparations—As seen in Table 3, all deletion mutants showed significant ATPase activity. After correction for the different amount of enzyme on the membranes, the turnover rates (at 42 °C) reached 10–15% of the wild type for Δ395EDKL398 and Δ399VVHR402 to astonishingly high values of 350 and 500% of wild type in Δ381QDIIAIL387 and Δ384IAIL387, respectively. Inclusion of the uncoupler CCCP (1 μm) in the assay medium, to avoid a buildup of back-pressure from the proton gradient, increased the activity of wild-type F1F0 by 1.8-fold. In contrast, for all investigated deletion mutants (Δ380LQDI383, Δ384IAIL387, Δ388GMDE391, and Δ392LSD394) the activity increased only slightly (1.1–1.3-fold) in the presence of CCCP. Just taken by itself, this finding could be attributed to the failure to build up a significant proton gradient or to the inability of the proton gradient to exert sufficient back-pressure in this specific mutant, i.e. uncoupling. To obtain a more quantitative description of coupling efficiency, we calculated the ratio of the rates for ATP synthesis and hydrolysis (Table 3). Coupling appears to be impaired in all mutants except Δ388GMDE391, which has a synthesis/hydrolysis ratio close to that of the wild type.
TABLE 3.
ATPase activities of membrane vesicles of deletion mutants ATPase activities of membrane preparations of deletion mutants and controls were measured as described under “Experimental Procedures.” Except where indicated, the assay medium did not contain CCCP. The positive control was strain pTR19-ASDS/DK8, which expresses Bacillus PS3 ATP synthase in E. coli; this strain served as background for the deletions. The negative control was strain pUC118/DK8, which expresses neither PS3 ATP synthase nor the endogenous E. coli enzyme. The values for the amount of F1F0 in the membrane preparations were taken from Table 2. Turnover rates were calculated using a molecular mass of 531 kDa for the holoenzyme, taking into account the differing amounts of ATP synthase in the individual membrane preparations. The synthesis/hydrolysis ratio was obtained by dividing ATP synthesis activities from Table 2 by the ATPase activities in this table.
| Strain/mutation | Amount of F1F0 on membranes | Membrane ATPase activity | Turnover rate kcat | Synthesis/hydrolysis |
|---|---|---|---|---|
| % | units/mg | s–1 | ||
| Wild type | 20 | 2.5 | 111 | 0.028 |
| Wild type (+CCCP) | 20 | 4.4 | 195 | |
| pUC118/DK8 (unc–) | 0 | 0.011 | ||
| Δ380LQDI383 | 20 | 3.4 | 150 | 0.012 |
| Δ384IAIL387 | 10 | 6.3 | 558 | 0.005 |
| Δ388GMDE391 | 15 | 1.6 | 94 | 0.032 |
| Δ392LSD394 | 20 | 5.7 | 252 | 0.008 |
| Δ395EDKL398 | 2.0 | 0.037 | 16 | 0 |
| Δ399VVHR402 | 2.6 | 0.043 | 15 | 0 |
| Δ381QDIIAIL387 | 2.4 | 1.1 | 406 | 0.007 |
NADH- and ATP-induced Proton Pumping—NADH-induced ACMA quenching was not reduced in membrane vesicles containing deletion mutants. The quenching reached values around 80–85% just as with wild-type PS3 enzyme, indicating that the mutations do not prevent the buildup of a considerable proton gradient. Thus, the deletions did not cause stability problems which increased the “leakiness” of the membranes. It should be noted, however, that reduced stability does not always manifest itself in enhanced proton leak rates. With the possible exception of Δ395EDKL398 and Δ399VVHR402, for the other deletion mutants this result was expected based on their normal or close-to-normal ATP synthesis rates.
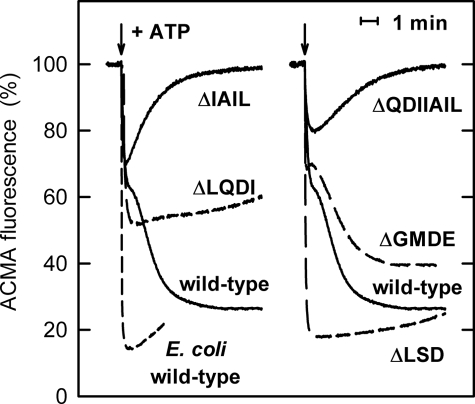
Upon ATP hydrolysis, wild-type PS3 ATP synthase in E. coli membrane vesicles forms a proton gradient that is only slightly smaller than that produced by the “native” E. coli enzyme. ATP-induced ACMA quenching reached values of ∼75% with the PS3 enzyme and 85% with wild-type E. coli ATP synthase. In the former case, but not in the latter, quenching seemed to occur in two phases, a fast phase, which was completed after 10–15 s and was associated with 35–40% quenching, followed by a slow phase of >100 s (Fig. 2). Similar behavior had been observed previously (see e.g. Fig. 3A in Ref. 21 and Fig. 2C in Ref. 33) but was not addressed. As the assay contains an excess of Mg2+ over ATP, it might be speculated that during the slow phase only a fraction of the enzyme molecules is active, whereas the majority is in an MgADP-inhibited form. This view is supported by the finding that the addition of external 1 mm Pi, which has been shown to suppress MgADP inhibition (34), reduces the second phase or even abolishes it completely (data not shown).
FIGURE 2.
ATP-driven H+-pumping. The quenching of ACMA fluorescence upon the addition of ATP (indicated by an arrow) is shown. “Wild-type” refers to membrane preparations of strain pTR19-ASDS/DK8, which expresses wild-type ATP synthase from the thermophilic Bacillus PS3 in E. coli membranes. This strain served as the background of the deletion mutants. For comparison, the fluorescence trace obtained with membranes carrying native E. coli ATP synthase is shown also. In all cases, after addition of 1 μm CCCP to dissipate the proton gradient, the ACMA fluorescence returned to within 3% of its original value.
Of the deletion mutants, Δ395EDKL398 and Δ399VVHR402 did not produce measurable ATP-induced ACMA quenching; the membrane ATPase activity was apparently not high enough to overcome the natural proton leak rate of the membranes. In contrast, all other deletion mutants could use ATP hydrolysis to form a proton gradient, demonstrating that the ATPase activity was not completely uncoupled. Like wild-type PS3 ATP synthase, the Δ388GMDE391 mutant showed biphasic kinetics, reaching a total value of about 60% quenching (Fig. 2). Of the other deletion mutants, Δ380LQDI383 and Δ392LSD394 did not show an obvious second quenching phase. Both reached their respective final quenching values of ∼45 and ∼ 80% nearly instantaneously. With the Δ384IAIL387 and Δ381QDIIAIL387 deletion mutants, only the initial fast phase resulted in quenching. In the slower phase, ATP-driven H+-pumping appeared to be reduced to such an extent that it could no longer compensate for the natural leak rate of the membranes. It should be noted that in all cases after the addition of 1 μm CCCP to dissipate the proton gradient, ACMA fluorescence returned to within 3% of its original value.
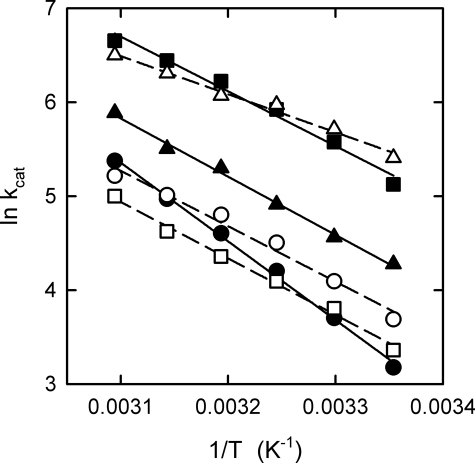
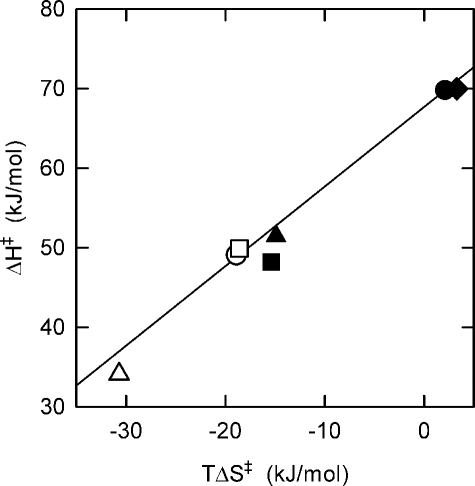
Arrhenius Analysis—Turnover rates at saturating ATP concentrations were measured as a function of temperature; because of their low membrane ATPase activities, the Δ395EDKL398 and Δ399VVHR402 deletion mutants were not included in this analysis. The results for the other deletion mutants and the wild-type enzyme are shown in Fig. 3 in the form of Arrhenius plots (ln kcat versus 1/T). From the slope of the regression lines, the activation energy (Ea) and changes of enthalpy (ΔH‡), entropy (ΔS‡), and Gibbs free energy of activation (ΔG‡) for the rate-limiting step of the overall reaction can be calculated (see Table 4 for values at 50 °C). First, it should be noted that the kcat for wild-type ATP synthase steadily increases, as expected, by a factor of >2 for each 10 °C increase in temperature over the entire observation range. This is in contrast to a recent report (35) using the α3β3γ subcomplex of PS3 ATP synthase, which found, surprisingly, a virtually temperature-independent steady-state ATPase activity for the range between 25 and 45 °C. The reason for this difference in behavior is not that, as in the case described here working with the membrane-embedded holoenzyme, a proton gradient is built up, which is obviously not possible using an α3β3γ subcomplex. The addition of CCCP to our experimental setup, to dissipate the proton gradient, did not alter the results significantly (Table 4). As to the deletion mutants, their ΔG‡ values are close to the wild-type value. In contrast, there are profound differences in Ea and ΔH‡, and consequently, because ΔG‡ = ΔH‡ - TΔS‡, in TΔS‡. The Ea and ΔH‡ values for the mutants with deletions of 3–4 amino acids were all rather similar, ∼70% of the respective wild-type value. The Ea and ΔH‡ values for the 7-amino acid deletion mutant, Δ381QDIIAIL387, was even lower, ∼50% of the wild-type values. Fig. 4 summarizes the data in the form of a free energy diagram, plotting ΔH‡ as a function of TΔS‡. The results indicate that in the rate-limiting step of the overall reaction in the mutants less interactions have to be formed or broken than in the wild-type enzyme. In the enzyme with the longer deletion of 7 residues the number of remaining energetically relevant interactions was even smaller than in the mutants with the shorter deletions of 3–4 amino acid residues.
FIGURE 3.
Arrhenius analysis of the ATPase activity of the deletion mutants. ATPase activities were measured at different temperatures (25–50 °C) and were calculated as turnover rates (kcat, in s-1). From the negative slope of the regression lines, the activation energy, Ea, of the rate-limiting step of the overall reaction can be determined (see Table 3). Filled circles, wild-type PS3 ATP synthase; open circles, Δ380LQDI383 deletion mutant; filled squares, Δ384IAIL387; open squares, Δ388GMDE391; filled triangles, Δ392LSD394; open triangles, Δ381QDIIAIL387.
TABLE 4.
Arrhenius analysis of ATPase activities of deletion mutants ATPase activities were measured at temperatures between 25 and 50 °C as described under “Experimental Procedures.” Except where indicated, the assay medium did not contain CCCP. From the resulting Arrhenius plots (In kcat versus 1/T), the activation energy, Ea, and changes of enthalpy, ΔH‡, entropy, ΔS‡ (expressed as TΔS‡), and Gibbs free energy of activation, ΔG‡, for the rate-limiting step of the overall reaction at 50 °C were calculated. ΔΔ values give the difference of the respective parameter between wild-type and mutant enzymes (or, for wild-type enzyme, between the activities in the absence and presence of CCCP).
| Strain/mutation | Ea | ΔH‡ | ΔΔH‡ | TΔS‡ | Δ(TΔS‡) | ΔG‡ | ΔΔG‡ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kJ/mol | |||||||||||||
| Wild type | 72.5 | 69.8 | 2.2 | 67.6 | |||||||||
| Wild type (+CCCP) | 72.7 | 70.0 | 0.2 | 3.4 | 1.2 | 66.6 | –1.0 | ||||||
| Δ380LQDI383 | 51.8 | 49.1 | –20.7 | –18.8 | –21.0 | 67.9 | 0.3 | ||||||
| Δ384IAIL387 | 50.9 | 48.2 | –21.6 | –15.3 | –17.5 | 63.5 | –4.1 | ||||||
| Δ388GMDE391 | 52.6 | 49.9 | –19.9 | –18.5 | –20.7 | 68.4 | 0.8 | ||||||
| Δ392LSD394 | 54.2 | 51.5 | –18.3 | –14.8 | –17.0 | 66.3 | –1.3 | ||||||
| Δ381QDIIAIL387 | 36.8 | 34.1 | –35.7 | –30.6 | –32.8 | 64.7 | –2.9 | ||||||
FIGURE 4.
Free energy plot of ATPase activity. ΔH‡ and TΔS‡ values were taken from Table 3. Except when indicated otherwise, ATPase activities were determined in the absence of the uncoupler CCCP. Filled circle, wild-type PS3 ATP synthase; open circle, Δ380LQDI383 deletion mutant; filled square, Δ384IAIL387; open square, Δ388GMDE391; filled triangle, Δ392LSD394; open triangle, Δ381QDIIAIL387; filled diamond, wild-type PS3 ATP synthase measured in the presence of CCCP. The line is an isobar for ΔG‡ = 67.6 kJ/mol, corresponding to the value for the wild-type enzyme (determined in the absence of CCCP).
MgATP Binding to the Catalytic Sites in the α3β3γ Subcomplex—To measure nucleotide binding to the catalytic sites of the deletion mutants, we inserted the same set of depletions into strain pNM1/JM103Δ(uncB-uncD), which expresses an α3(βY341W)3γ subcomplex of PS3 ATP synthase. Unfortunately, only two of the deletion mutants, Δ388GMDE391 and Δ392LSD394, could be obtained as an α3β3γ subcomplex in sufficient yield and purity to allow fluorescence-based nucleotide binding studies. As expected from the membrane vesicle experiments, both mutant subcomplexes are active ATPases. The following activities were measured at 60 °C: α3(βY341W)3γ, 31.4 units/mg; Δ388GMDE391, 7.9 units/mg; Δ392LSD394, 11.3 units/mg. Assuming a molecular mass of 352 kDa for the subcomplex, this corresponds to turnover rates of 184 s-1, 46s-1, and 66 s-1, respectively. Interestingly, the activities of both mutant subcomplexes were substantially below those for the parental α3(βY341W)3γ, whereas as F1F0 holoenzyme in membranes the same mutants had approximately the same (Δ388GMDE391) or even a higher activity (Δ392LSD394) than the wild-type control (Table 3). Part, although probably not all, of this discrepancy can be related to the higher assay temperature for the α3β3γ subcomplexes (60 versus 42 °C for the membrane-bound ATPase), considering the stronger temperature dependence of the activity of the wild-type enzyme as compared with that of the mutants (Fig. 3).
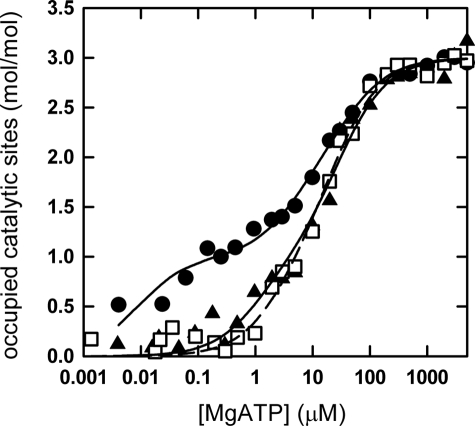
MgATP binding was measured using the fluorescence of the inserted Trp βW341 as signal. The results are shown in Fig. 5, and Kd values are given in Table 5. The α3(βY341W)3γ subcomplex exhibited a binding pattern resembling that determined for E. coli F1 (7, 8, 36) and F1F0 (37), with three sites of different binding affinity for the Mg2+-nucleotide. The Kd2 value for binding to the medium affinity catalytic site was somewhat higher than determined previously for the α3β3γ subcomplex of PS3 (38–40), which might be because of differences in the buffer composition. The deletion mutants showed a pronounced change in the binding behavior. The binding affinity of the medium affinity site 2 decreased to the level of the low affinity site 3. More importantly, the affinity of the high affinity site 1 decreased by at least 2 orders of magnitude. Thus, the two deletion mutants no longer have a true “high affinity” site.
FIGURE 5.
MgATP binding to the catalytic sites. MgATP binding to the three catalytic sites of the α3β3γ subcomplex of PS3 ATP synthase was measured in a buffer containing 50 mm Tris/H2SO4, 2.5 mm MgSO4, pH 8.0, at 23 °C (see “Experimental Procedures”). α3(βY341W)3γ, filled dots; Δ388GMDE391 in α3(βY341W)3γ, open squares; Δ392LSD394 inα3(βY341W)3γ, filled triangles. The lines are fitted binding curves based on the Kd values given in Table 5.
TABLE 5.
MgATP binding to the catalytic sites MgATP binding to the three catalytic sites was measured at 23 °C as described under “Experimental Procedures.”
| Enzyme/mutation | Kd1 | Kd2 | Kd3 |
|---|---|---|---|
| μm | |||
| PS3 α3(βY341W)3γ | 0.01 | 5.0 | 38 |
| E. coli βY331W F1a | 0.02 | 1.4 | 28 |
| E. coli βY331W F1,ε-depletedb | 0.12 | 2.8 | 23 |
| Δ388GMDE391 in PS3 α3(βY341W)3γ | 3 | 23 | 23 |
| Δ392LSD394 in PS3 α3(βY341W)3γ | 1 | 29 | 29 |
For comparison, the values for E. coli F1 (α3β3γδε), taken from Ref. 8, are listed. E. coli residue βY331 corresponds to PS3 residue βY341; thus, the Trp residue used to monitor nucleotide binding is inserted at the equivalent position in both enzymes
Values for ε-depleted E. coli F1 (α3β3γδ), taken from Ref. 36, are listed. The presence of ε increases the affinity at site 2 and, especially, at site 1 in the E. coli enzyme. It is not known whether ε has a similar effect in the PS3 enzyme. ε is obviously absent in the α3β3γ subcomplex assayed here
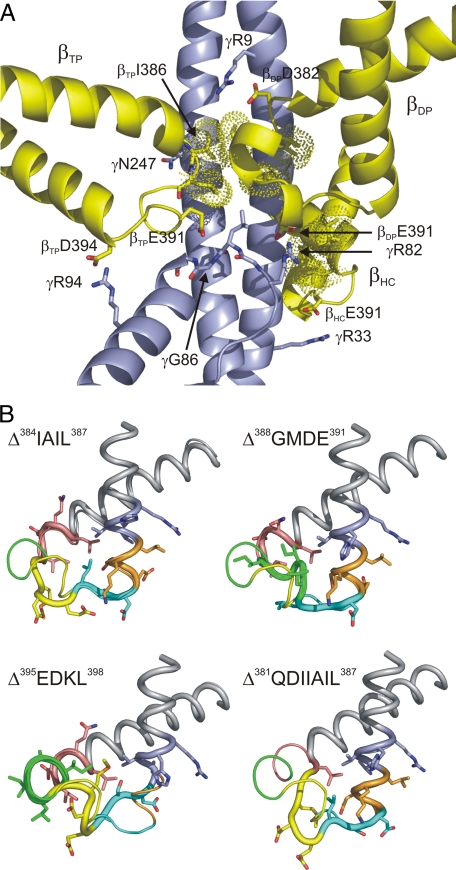
Modeling the Deletion Mutants—To understand the functional consequences of the deletions in terms of the enzyme mechanism on a residue level, we modeled the deletions into the available structures of the β subunit (βTP, βDP, βHC, and βE). In the wild-type enzyme, in all three β subunits the region of the DELSEED-loop investigated here (residues 380–402 in the PS3 enzyme) makes contact with γ via hydrogen bonds/salt bridges as well as van der Waals and hydrophobic interactions (Fig. 6A). In all three β subunits, βE391 is involved in these contacts. In βTP, the carboxylate groups of βD390 and βE391 can hydrogen bond to the peptide bond nitrogen of γG86, whereas the carboxylate group of βD394 can form a hydrogen bond/salt bridge with γR94 (Interestingly, the βD394/γR94 pair in the Bacillus enzyme is replaced by a Glu/Lys pair in ATP synthases from E. coli and bovine mitochondria; both amino acid pairs can cover approximately the same distance). Both γG86 and γR94 are located in a short helix that runs nearly perpendicular to the long N- and C-terminal helices of γ. Furthermore, a hydrogen bond is possible between the peptide bond oxygen of βI386 and the side chain of γN247 in the C-terminal helix. In βDP, the carboxylate group of βE391 can interact with the guanidino group of γR82 and the peptide bond nitrogen of γL84. In addition, a hydrogen bond/salt bridge can be formed between the carboxylate function of βD382 and the guanidino group of γR9 in the N-terminal helix. In the third β subunit, at least in its half-closed form (11) (i.e. when a nucleotide is bound to the catalytic site), the side chains of βE391 and γR33 interact. In all three β subunits, residues βI386 and βL387 are involved in van der Waals and hydrophobic interactions, and residue βI383 makes contact with γ in βDP and βHC.
FIGURE 6.
The βDELSEED-loop. A, interactions between the βDELSEED-loop and γ. The DELSEED-loops of the three β subunits are shown in yellow, and γ is shown in blue. Amino acid residues involved in hydrogen bonds/salt bridges are shown in stick representation and labeled using PS3 residue numbers. Dots indicate the van der Waals surface of the side chains of residues βI383, βI386, and βL387 involved in contacts with γ. This figure is based on Protein Data Bank file 1h8e (11), modified to represent the Bacillus PS3 ATP synthase using PRIME. B, models of the βDELSEED-loop of the deletion mutants. The main chain of the respective deletion mutant is shown as the thicker line in cartoon representation and the main chain of the wild-type enzyme as the thinner line. Side chains (in stick representation) are given only for the deletion mutants. Color-coding is as described in the legend for Fig. 1B.
Modeling of the deletion mutants indicated that the deletions leave the overall shape of the DELSEED-loop mostly intact (see Fig. 6B for selected cases). Even in the case of the 7-residue deletion, in all three β subunits the loop still extends far enough to make contact with γ. This is achieved mainly by “unraveling” of the helices flanking the turn or the short helical segment within the turn. Instead of a helical conformation, the respective amino acid stretches assume a straighter, more linear conformation, which can cover a longer distance. As of the residues investigated here, βE391 is involved in the largest number of β/γ hydrogen bonds/salt bridges, it is not surprising that this number is most reduced in the Δ388GMDE391 deletion mutant, from 7 to 8 in the wild-type enzyme to about 3 in the mutant. Δ392LSD394 and Δ381QDIIAIL387 have ∼4 residual β/γ hydrogen bonds/salt bridges. In Δ392LSD394, the deletion is just downstream of βE391 and changes the conformation of the glutamate side chain. Δ381QDIIAIL387 experiences the most pronounced conformational rearrangements overall of all the mutants investigated here, due to the size of the deletion. As most hydrophobic interactions involve residues βI386, βL387, and, to a lesser degree, βI383, these interactions, as expected, are the most reduced in mutants where these residues are deleted (Δ384IAIL387 and especially Δ381QDIIAIL387). On the other hand, according to the models, in the Δ395EDKL398 and Δ399VVHR402 deletion mutants both β/γ hydrogen bonds/salt bridges and van der Waals/hydrophobic interactions are nearly completely preserved.
DISCUSSION
The goal of the present study was to gain a better understanding of the role of the βDELSEED-loop in the coupling of catalysis and subunit rotation of ATP synthase. Thus far, mutational analysis of this region has focused on the negatively charged loop residues, such as βD382 (41), and those of the 390DELSEED396 motif itself, especially βE391 (12, 14, 42). To use a less restrictive approach, we constructed deletions of 3, 4, or 7 residues in the turn region of the loop of ATP synthase from the thermophilic bacterium Bacillus PS3. All deletions of 3–4 residues plus one of 7 residues could be expressed in E. coli and were incorporated into the membrane in sufficient amounts to allow functional analysis. The results show that the βDELSEED-loop plays an important role in the catalytic mechanism. It is not possible, however, to single out individual β/γ interactions that might be essential for function. In contrast, there appears to be a multitude of relevant contacts, each of which makes a rather limited contribution. Even the Δ381QDIIAIL387 mutant, where the majority of residues involved in contacts to γ is missing (such as βD382, βI383, βI386, and βL387, all of which are strictly conserved), is still functional to such an extent that one has to assume that it operates by the normal rotational mechanism.
All deletion mutants that we analyzed displayed the ability to hydrolyze ATP, and all except Δ395EDKL398 and Δ399VVHR402 showed ATP synthesis activity in vitro. Δ395EDKL398 and Δ399VVHR402 were also the only two mutants in which ATP hydrolysis did not result in the formation of a proton gradient. These deficiencies could be due to a combination of low abundance of enzyme on the membranes and low turnover rate, e.g. in ATP hydrolysis the membrane ATPase activity might be too low to compensate for the natural proton leak rate of the membranes. Alternatively, it is possible that these two mutants are uncoupled, although when NADH-driven proton pumping was measured using ACMA fluorescence, both mutants were able to generate a proton gradient of a magnitude comparable with that of the other mutants and the wild-type enzyme. In general, the deletion mutants seem to be less well coupled than the wild type, as evidenced by their less pronounced acceleration of membrane ATPase activities upon addition of an uncoupler and by their (with the exception of Δ388GMDE391) reduced ATP synthesis/hydrolysis ratio. Further indication of possible uncoupling was found for the Δ384IAIL387 and Δ381QDIIAIL387 mutants upon studying ATP-driven H+-pumping and growth yields in limiting glucose. Interestingly, the defect that abolished oxidative phosphorylation in vivo still allowed ATP synthesis in vitro.
The ATPase activities of the individual deletion mutants differ widely. Δ395EDKL398 and Δ399VVHR402 have the lowest activities. It should be noted that these two deletions are the only ones that do not eliminate residues that, according to the available crystal structures, make contact with γ. Either mutation abolishes a turn in the C-terminal helix of the DELSEED-loop downstream of the γ interaction site. It seems possible that the deletions might affect the orientation of the side chain of residue βR404, which is part of a functionally important hydrogen-bonding network at the βDP/αDP interface (43). It should be mentioned, however, that this hypothesis is not supported by the modeling results, which show the side chain of βR404 in the same position in the mutants and in the wild-type enzyme.
At the other extreme, as shown in Table 3, some of the mutants have a significantly higher ATPase activity than the wild-type enzyme (although if one extrapolates the Arrhenius plots, at the optimal growth temperature of Bacillus PS3, 65 °C, these differences will be less pronounced). The increased activity could be due to perturbation of the interaction with the ε subunit. ε exists in two different conformations, “up” and “down.” In the up conformation it acts as intrinsic inhibitor of ATPase activity. In the wild-type enzyme, the up conformation of ε interacts with the βDELSEED-loop (44–46). If the relevant interactions are reduced in the deletion mutants, this might shift the equilibrium to the non-inhibitory down conformation. Obviously, increased ATPase activities without concomitant increase in ATP synthesis activity, as observed here, results in reduced synthesis/hydrolysis ratios and therefore in decreased apparent coupling efficiencies. If the increased ATPase activities in the mutants should be due to changes in interaction with ε, as suggested here, this would support the notion of a role of ε in coupling, as proposed for the PS3 as well as the E. coli enzyme (47, 48).
The Arrhenius analysis reflects the reduction of energetically meaningful interactions between β and γ in the deletion mutants. In the tested cases of mutants with deletions of 3–4 amino acids (excluding Δ395EDKL398 and Δ399VVHR402), the activation energy, Ea, and consequently of ΔH‡, was reduced by 18–22 kJ/mol, in the mutant with 7 deleted residues by 36 kJ/mol. The results indicate a decreased number of bond rearrangements that contribute to the rate-limiting step of the overall reaction, strongly supporting the notion of a pivotal role for the DELSEED-loop in coupling catalysis and γ rotation. Although this interpretation seems straightforward, it is remarkable how well the decrease in Ea and ΔH‡ correlates with the length of the deletion, seemingly independently of the character of the amino acids that were removed.
Although only two of the deletion mutants could be isolated in the form of an α3β3γ subcomplex suitable for a Trp fluorescence-based nucleotide binding assay, these studies gave a remarkable result. In both mutants, Δ388GMDE391 and Δ392LSD394, the affinity of the high affinity site was drastically decreased. Either deletion removes a residue (βE391 and βD394, respectively) that in the βTP subunit, which carries the high affinity binding site, interacts with γ. Interestingly, a similarly pronounced reduction in affinity of this site was recently described for a βD390C point mutation (42). Nevertheless, at this point in time it seems premature to ascribe the decrease in affinity to loss of a specific interaction between the βTPDELSEED-loop and γ.A 390AALSAAA396 quintuple point mutant, where all the negative charges of the DELSEED motif are removed (including those contained in Δ388GMDE391, Δ392LSD394, and βD390), could still bind MgATP and MgADP with high affinity.4 It appears possible that the deletion mutants Δ388GMDE391 and Δ392LSD394 as well as the βD390C point mutation cause larger scale conformational rearrangements that affect nucleotide binding to the high affinity catalytic site. Modeling of the deletion mutants did not reveal a specific pattern of conformational changes common to both mutants.
The reduction in affinity of the high affinity site had been observed previously, but mostly for mutations of residues involved directly in substrate binding and/or turnover (49–51). These mutants had very low residual enzymatic activity, and it is not clear whether they hydrolyze ATP by the normal rotational mechanism. In contrast, both deletion mutants described here, as well as the βD390C point mutation, display normal enzymatic activity using rotation, as evidenced by the observation of oxidative phosphorylation in vivo, ATP synthesis in vitro, and ATP-driven proton pumping. These findings seem to contradict the assumption contained in several models of the catalytic mechanism (see the Introduction) that tightening of the binding site around the newly bound MgATP, from “low affinity” to “high affinity,” might be responsible for driving the 80° rotation substep. Using the data given in Table 5, the increase in MgATP binding energy upon transition from low affinity (Kd3) to high affinity (Kd1) drops from 20.3 kJ/mol in the wild-type PS3 enzyme to less than half that value in the deletion mutants (5.0 kJ/mol for Δ388GMDE391, 8.3 kJ/mol for Δ392LSD394). A comparison of 1/K1 observed under uni-site conditions (corresponding to Kd1 measured here) and Km for multi-site catalysis (corresponding to Kd3) in wild-type and mutant E. coli enzyme (42) gives a similar decrease for the βD390C mutant. However, before it can be concluded that the mechanistic models are incorrect in this aspect, significantly more information is needed about the changes of ligand binding affinities of the catalytic sites as a function of the rotational angle of γ.
This work was supported, in whole or in part, by National Institutes of Health Grant GM071462 (to J. W.).
Footnotes
The abbreviations used are: AMP-PNP, adenosine 5′-(β,γ-imino)triphosphate; ACMA, 9-amino-6-chloro-2-methoxyacridine; CCCP, carbonyl cyanide m-chlorophenylhydrazone; CDTA, trans-1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid.
Bacillus PS3 numbering is used throughout, except where noted.
N. Mnatsakanyan, H. Z. Mao, and J. Weber, unpublished results.
References
- 1.Noji, H., and Yoshida, M. (2001) J. Biol. Chem. 276, 1665-1668 [DOI] [PubMed] [Google Scholar]
- 2.Weber, J., and Senior, A. E. (2003) FEBS Lett. 545, 61-70 [DOI] [PubMed] [Google Scholar]
- 3.Wilkens, S. (2005) Adv. Protein Chem. 71, 345-382 [DOI] [PubMed] [Google Scholar]
- 4.Dimroth, P., von Ballmoos, C., and Meier, T. (2006) EMBO Rep. 7, 276-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber, J. (2006) Biochim. Biophys. Acta 1757, 1162-1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamoto, R. K., Baylis Scanlon, J. A., and Al-Shawi, M. K. (2008) Arch. Biochem. Biophys. 476, 43-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber, J., Wilke-Mounts, S., Lee, R. S. F., Grell, E., and Senior, A. E. (1993) J. Biol. Chem. 268, 20126-20133 [PubMed] [Google Scholar]
- 8.Weber, J., and Senior, A. E. (2004) Methods Enzymol. 380, 132-152 [DOI] [PubMed] [Google Scholar]
- 9.Abrahams, J. P., Leslie, A. G. W., Lutter, R., and Walker, J. E. (1994) Nature 370, 621-628 [DOI] [PubMed] [Google Scholar]
- 10.Bowler, M. W., Montgomery, M. G., Leslie, A. G. W., and Walker, J. E. (2007) J. Biol. Chem. 282, 14238-14242 [DOI] [PubMed] [Google Scholar]
- 11.Menz, R. I., Walker, J. E., and Leslie, A. G. W. (2001) Cell 106, 331-341 [DOI] [PubMed] [Google Scholar]
- 12.Ketchum, C. J., Al-Shawi, M. K., and Nakamoto, R. K. (1998) Biochem. J. 330, 707-712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oster, G., and Wang, H. (1999) Structure (Lond.) 7, R67-R72 [DOI] [PubMed] [Google Scholar]
- 14.Hara, K. Y., Noji, H., Bald, D., Yasuda, R., Kinosita, K., Jr., and Yoshida, M. (2000) J. Biol. Chem. 275, 14260-14263 [DOI] [PubMed] [Google Scholar]
- 15.Gao, Y. Q., Yang, W., and Karplus, M. (2005) Cell 123, 195-205 [DOI] [PubMed] [Google Scholar]
- 16.Mao, H. Z., and Weber, J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104, 18478-18483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yasuda, R., Noji, H., Yoshida, M., Kinosita, K., Jr., and Itoh, H. (2001) Nature 410, 898-904 [DOI] [PubMed] [Google Scholar]
- 18.Gao, Y. Q., Yang, W., Marcus, R. A., and Karplus, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100, 11339-11344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishizaka, T., Oiwa, K., Noji, H., Kimura, S., Muneyuki, E., Yoshida, M., and Kinosita, K., Jr. (2004) Nat. Struct. Mol. Biol. 11, 142-148 [DOI] [PubMed] [Google Scholar]
- 20.Pu, J., and Karplus, M. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 1192-1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki, T., Ueno, H., Mitome, N., Suzuki, J., and Yoshida, M. (2002) J. Biol. Chem. 277, 13281-13285 [DOI] [PubMed] [Google Scholar]
- 22.Klionsky, D. J., Brusilow, W. S. A., and Simoni, R. D. (1984) J. Bacteriol. 160, 1055-1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsui, T., and Yoshida, M. (1995) Biochim. Biophys. Acta 1231, 139-146 [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., and Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., p. A2.4, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 25.Senior, A. E., Latchney, L. R., Ferguson, A. M., and Wise, J. G. (1984) Arch. Biochem. Biophys. 228, 49-53 [DOI] [PubMed] [Google Scholar]
- 26.Senior, A. E., Langman, L., Cox, G. B., and Gibson, F. (1983) Biochem. J. 210, 395-403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taussky, H. H., and Shorr, E. (1953) J. Biol. Chem. 202, 675-685 [PubMed] [Google Scholar]
- 28.Al-Shawi, M. K., and Senior, A. E. (1988) J. Biol. Chem. 263, 19640-19648 [PubMed] [Google Scholar]
- 29.Ren, H., Bandyopadhyay, S., and Allison, W. S. (2006) Biochemistry 45, 6222-6230 [DOI] [PubMed] [Google Scholar]
- 30.Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951) J. Biol. Chem. 193, 265-275 [PubMed] [Google Scholar]
- 31.Bradford, M. M. (1976) Anal. Biochem. 72, 248-254 [DOI] [PubMed] [Google Scholar]
- 32.Gibbons, C., Montgomery, M. G., Leslie, A. G. W., and Walker, J. E. (2000) Nat. Struct. Biol. 7, 1055-1061 [DOI] [PubMed] [Google Scholar]
- 33.Ono, S., Sone, N., Yoshida, M., and Suzuki, T. (2004) J. Biol. Chem. 279, 33409-33412 [DOI] [PubMed] [Google Scholar]
- 34.Mitome, N., Ono, S., Suzuki, Y., Shimabukuro, K., Muneyuki, E., and Yoshida, M. (2002) Eur. J. Biochem. 269, 53-60 [DOI] [PubMed] [Google Scholar]
- 35.Furuike, S., Adachi, K., Sakaki, N., Shimo-Kon, R., Itoh, H., Muneyuki, E., Yoshida, M., and Kinosita, K., Jr. (2008) Biophys. J. 95, 761-770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber, J., Dunn, S. D., and Senior, A. E. (1999) J. Biol. Chem. 274, 19124-19128 [DOI] [PubMed] [Google Scholar]
- 37.Löbau, S., Weber, J., and Senior, A. E. (1998) Biochemistry 37, 10846-10853 [DOI] [PubMed] [Google Scholar]
- 38.Dou, C., Fortes, P. A., and Allison, W. S. (1998) Biochemistry 37, 16757-16764 [DOI] [PubMed] [Google Scholar]
- 39.Bandyopadhyay, S., Valder, C. R., Huynh, H. G., Ren, H., and Allison, W. S. (2002) Biochemistry 41, 14421-14429 [DOI] [PubMed] [Google Scholar]
- 40.Ono, S., Hara, K. Y., Hirao, J., Matsui, T., Noji, H., Yoshida, M., and Muneyuki, E. (2003) Biochim. Biophys. Acta 1607, 35-44 [DOI] [PubMed] [Google Scholar]
- 41.Lowry, D. S., and Frasch, W. D. (2005) Biochemistry 44, 7275-7281 [DOI] [PubMed] [Google Scholar]
- 42.Baylis Scanlon, J. A., Al-Shawi, M. K., and Nakamoto, R. K. (2008) J. Biol. Chem. 283, 26228-26240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao, H. Z., Abraham, C. G., Krishnakumar, A. M., and Weber, J. (2008) J. Biol. Chem. 283, 24781-24788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hara, K. Y., Kato-Yamada, Y., Kikuchi, Y., Hisabori, T., and Yoshida, M. (2001) J. Biol. Chem. 276, 23969-23973 [DOI] [PubMed] [Google Scholar]
- 45.Suzuki, T., Murakami, T., Iino, R., Suzuki, J., Ono, S., Shirakihara, Y., and Yoshida, M. (2003) J. Biol. Chem. 278, 46840-46846 [DOI] [PubMed] [Google Scholar]
- 46.Feniouk, B. A., Suzuki, T., and Yoshida, M. (2007) J. Biol. Chem. 282, 764-772 [DOI] [PubMed] [Google Scholar]
- 47.Rondelez, Y., Tresset, G., Nakashima, T., Kato-Yamada, Y., Fujita, H., Takeuchi, S., and Noji, H. (2005) Nature 433, 773-777 [DOI] [PubMed] [Google Scholar]
- 48.Cipriano, D. J., and Dunn, S. D. (2006) J. Biol. Chem. 281, 501-507 [DOI] [PubMed] [Google Scholar]
- 49.Senior, A. E., and Al-Shawi, M. K. (1992) J. Biol. Chem. 267, 21471-21478 [PubMed] [Google Scholar]
- 50.Löbau, S., Weber, J., Wilke-Mounts, S., and Senior, A. E. (1997) J. Biol. Chem. 272, 3648-3656 [DOI] [PubMed] [Google Scholar]
- 51.Nadanaciva, S., Weber, J., and Senior, A. E. (1999) Biochemistry 38, 7670-7677 [DOI] [PubMed] [Google Scholar]
- 52.Goelz, S. E., and Cronan, J. E., Jr. (1982) Biochemistry 21, 189-195 [DOI] [PubMed] [Google Scholar]