Abstract
In Escherichia coli, the periplasmic protein disulfide isomerase, DsbC, is maintained reduced by transfer of electrons from cytoplasmic thioredoxin-1 (Trx1) via the cytoplasmic membrane protein, DsbD. The transmembrane domain of DsbD (DsbDβ), which comprises eight transmembrane segments (TMs), contains two redox-active cysteines (Cys-163 and Cys-285), each of which is water-exposed to both sides of the membrane. Cys-163 in TM1 and Cys-285 in TM4 can interact with cytoplasmic Trx1 and a periplasmic Trx-like domain of DsbD, respectively. When Cys-163 and Cys-285 are disulfide-bonded, the C-terminal halves of TM1 and TM4 are water-exposed, whereas the N-terminal halves of these TMs are not. To assess possible conformational changes of DsbDβ when its two cysteines are reduced, we have determined the accessibility of portions of TM1 and TM4. We substituted cysteines for amino acids in these TM segments and determined alkylation accessibility. We find that the alkylation accessibility of single Cys replacements in TM1 and TM4 is the same in oxidized and reduced DsbDβ, indicating a relatively static conformation of DsbDβ between the two redox states. We also find that the accessibility of amino acids of TM2 and TM3 when Cys-163 and Cys-285 are oxidized or reduced shows no change. Together, these results support a relatively static structure of DsbDβ in the switch between the oxidized and the reduced state but raise the possibility of conformational changes when interacting with Trx proteins. In addition, we also find water-exposed residues in the cytoplasmic proximal portion of TM3, allowing a more detailed characterization of the cavity in DsbDβ.
The cell envelope of most bacteria is an oxidizing environment. In many bacteria, the main oxidant system consists of DsbA and DsbB. DsbA introduces disulfide bonds into newly synthesized and secreted polypeptides containing cysteines and is regenerated as an oxidative enzyme by the membrane protein DsbB. Electrons are ultimately transferred from DsbB to the respiratory chain (1–3). However, there are also certain cell envelope proteins that require a reductive enzyme to act on them. This is the case for those proteins that contain multiple cysteines and that are often misoxidized by DsbA, thus generating non-native disulfide bonds. The protein DsbC, a protein disulfide isomerase, can promote rearrangement of such incorrect disulfide bonds, resulting in a correctly folded protein (4–7). It does this either by using the reduced cysteine in its active site to resolve non-native disulfide bonds and promoting the formation of the native pairs or simply by reducing the substrate protein, which may be correctly oxidized by DsbA and given a second chance (8). In the latter mechanism, DsbC becomes oxidized and must be reduced. This reduction is carried out by a cytoplasmic membrane protein, DsbD, which receives electrons for this purpose from thioredoxin-1 (Trx1)2 in the cytoplasm (5, 9).
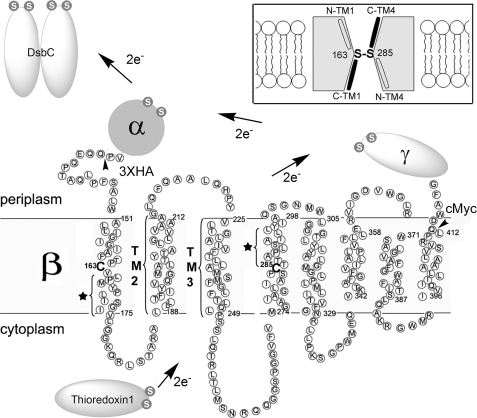
DsbD is composed of three domains, each containing two redox-active cysteines (Fig. 1). DsbDβ, the membrane-embedded domain containing eight transmembrane segments (TMs), receives electrons from Trx1 and then transfers them to the C-terminal periplasmic domain, DsbDγ, which contains a Trx-like fold (10–15). The N-terminal periplasmic domain, DsbDα, which contains an immunoglobulin-like fold, is then reduced by DsbDγ and transfers electrons to DsbC (13, 16, 17).
FIGURE 1.
Electron transfer pathway through transmembrane domain (β) of DsbD and its membrane topology predicted from the primary sequence. The topology of DsbDβ was predicted using HMMTOP. The essential two cysteines are shown in bold without a circle and numbered. The residues indicated with a star in TM1 and TM4 are water-exposed when the Cys-163 and Cys-285 are disulfide-bonded (19). Studies on the residues in TM2 and TM3 are shown in Fig. 4. The essential cysteines in the other domains (α and γ) and interacting proteins (Trx1 and DsbC) are shown in a white S (the sulfur of thiol) in gray circles. The tailless arrows indicate where a signal sequence of DsbD and three hemagglutinin (HA) epitopes are fused at the N terminus of DsbDβ, and a c-Myc epitope is fused at the C terminus of DsbDβ. The figure in the inset describes the model of DsbDβ, in which Cys-163 and Cys-285 form a disulfide bond in the middle of the protein and halves of C-terminal TM1 and TM4 are water-exposed (black; C-TM1 and C-TM4), whereas those of N-terminal ones are not (N-TM1 and N-TM4).
Electron transfer through the transmembrane domain DsbDβ is quite unusual when compared with that of other membrane electron transport proteins. Required extrinsic factors, for example, quinone, FAD, heme, or metal centers, which are often used as cofactors for electron transfer, have not been found (18). As a result, it is proposed that thiol-disulfide exchange reactions alone promote the transfer of electrons across the cytoplasmic membrane, utilizing the two cysteines, Cys-163 and Cys-285 of DsbDβ. Evidence for this mechanism comes from the detection of likely reaction intermediates including the Cys-163–Cys-285 disulfide and mixed disulfide complexes, Trx1-DsbDβ(Cys-163) and DsbDβ(Cys-285)-DsbDγ (13, 14, 19).
We have previously studied the accessibility to the aqueous environment of amino acids in TM1 and TM4 of DsbDβ, which contain Cys-163 and Cys-285, respectively. Our results, in conjunction with a comparison of the amino acid sequences of TM1–3 and TM4–6 of DsbDβ, suggest antiparallel and pseudosymmetrical properties of TM1 and TM4 (19, 20). Cys-163 in TM1 and Cys-285 in TM4 are water-exposed to both sides of the membrane when they are in the reduced state and suggested to be located in the middle of the membrane (helices). When Cys-163 and Cys-285 are disulfide-bonded, the proximal portion of the cytoplasmic side of TM1 at the C terminus of Cys-163 and that of the periplasmic side of TM4 at the C terminus of Cys-285 are highly water-exposed, whereas the other portion of each TM is not. Therefore, we proposed an hourglass-like model (Fig. 1, inset) and suggested that the water-exposed halves of TM1 and TM4 are cavity-located non-membrane-spanning helices and involved in the interactions of Trx1 and DsbDγ, respectively.
However, in our previous studies (19), we did not determine whether, when DsbDβ is in the reduced state, the arrangement of TM1 and TM4 or other TMs would be similar to that seen when Cys-163 and Cys-285 are disulfide-bonded. Doing this comparison is important because it has been proposed that alternative exposure of the cysteines to the aqueous environment depending on their redox states explains the electron transfer process across the membrane (Refs. 21–23; see “Discussion”). In addition, to further define the structural features of DsbDβ, we wished to determine how TM segments other than TM1 and TM4 are arranged in terms of their water accessibility. In this study, we examined the accessibility of many residues in TM1 and TM4, as well as studying the arrangements of TM2 and TM3, using site-directed cysteine alkylation in both redox states.
Our data show that the four TMs studied, TM1, TM2, TM3, and TM4, have similar accessibility properties whether DsbDβ is in the oxidized or reduced state. We also find additional water-exposed residues in the proximal portion of the cytoplasmic side of TM3.
EXPERIMENTAL PROCEDURES
Strains and Medium—MC1000 is a laboratory collection and a background Escherichia coli strain (araD139(araABC-leu)7679 galU galK Δ(lac)X74 rpsL thi) of this study. FED126 is MC1000 ΔdsbD (10), and FED513 is FED126 trxB::Kan (13). pAP06 plasmid is a pBAD18 derivative encoding DsbDβ containing c-Myc tag (19). E. coli cells were grown in NZ medium (10 g of N-Z-Amine A® (Kerry), 8 g of Nacl, and 5 g of yeast extract per liter) in the presence of 200 μg/ml ampicillin at 37 °C. Expression of the genes was induced for 1 h by adding 0.2% l-arabinose when the optical density of cells at 600 nm reached 0.1.
DNA Manipulation—The pAP06 plasmid and its derivatives were constructed before (19) or for the present study. The site-directed mutagenesis was performed using Pfu Turbo (Stratagene) according to the manufacturer's protocol. All the dsbD sequences in the plasmids in this report were verified by DNA sequencing by the Dana-Farber/Harvard Cancer Center DNA Resource Core, Harvard Medical School.
Membrane Protein Topology Prediction—Membrane topology of DsbDβ was predicted using HMMTOP (24), SOSUI (25), TMHMM Server version 2.0 (26), and TopPred (27), membrane protein topology prediction programs.
Antibodies—Anti-c-Myc (A-14) rabbit polyclonal antiserum was purchased from Santa Cruz Biotechnology, Inc.
AMS Alkylation and malPEG Counter-alkylation of Proteins—5 ml of cells were harvested at mid-log phase and resuspended in 1 ml of ice-cold buffer containing 50 mm Tris-HCl (pH 8.0), 1 mm CaCl2, 3 mm EDTA, 18% sucrose, 30 μg/ml lysozyme. Subsequently, the required concentrations of 4-acetamido-4′maleimidylstilbene-2,2′-disulfonic acid (AMS) (Invitrogen) were added, and the samples were sonicated for 10 s and incubated for 30 min at 4 °C. After incubation, samples were trichloroacetic acid-precipitated, washed with acetone, and resuspended in 50 μl of SDS/sample buffer containing 1.7 mm methoxypoly(ethylene glycol)-maleimide (malPEG) (NOF Corp.). 2-kDa malPEG was used in the analysis of TM1 and TM4; 5-kDa malPEG was used in that of TM2 and TM3.
In the previous analysis, we added AMS after sonication (19). However, we found that sonication before adding AMS rarely caused inefficient AMS accessibility when we tested A288C, the replacements in the periplasmic side of TM4. We believe that this is due to inverted membrane-vesicle formation in some fractions before adding AMS. Therefore, we changed the method by adding AMS before sonication. Nevertheless, the results from the previous study hold true because the positive control, A288C, showed complete AMS alkylation in the sonicated sample in the previous study, and the accessibility results for TM1 and TM4 in the fraction of oxidized proteins obtained in this study, are almost identical with the previous results (19).
In the analyses of TM2 and TM3 in Fig. 4, we added AMS after sonication. Nevertheless, the positive control, A288C, from four independent tests, showed complete AMS alkylation. In addition, the accessibility results for TM2 and TM3 in those fractions that were oxidized in supplemental Fig. 3 in which AMS was added before sonication are almost identical with the data from Fig. 4. Therefore, we believe that few membrane vesicles were made before adding AMS during the experiment performed for Fig. 4.
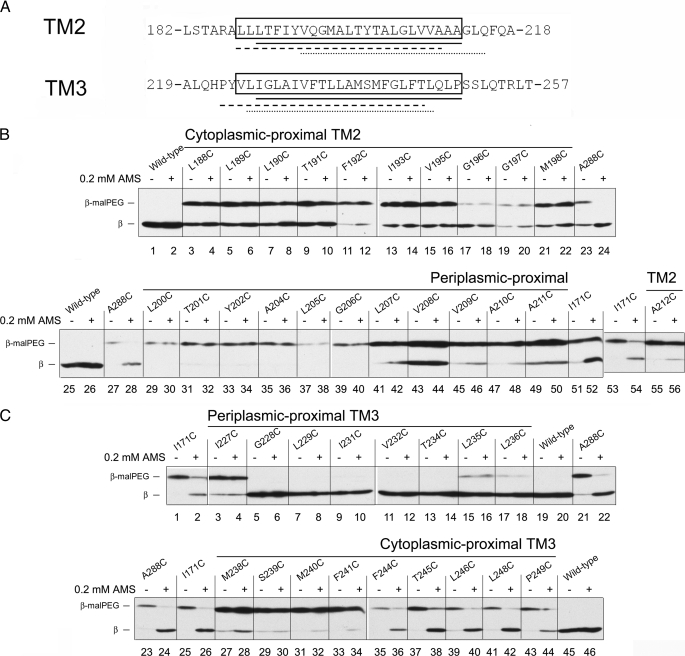
FIGURE 4.
AMS accessibility to the residues in TM2 and TM3 in the oxidized states. A, the amino acid sequence around predicted TM2 and TM3 using HMMTOP (□), SOSUI (——), TMHMM (–––), and TopPred (□). B, AMS alkylation and 5-kDa malPEG counter-alkylation of single Cys replacements within TM2. The samples were analyzed according to the procedure in Fig. 2A except that AMS was added after sonication (see “Experimental Procedures”) and the strain background was FED513 (the dsbD and trxB double mutant), where oxidized Trx1 is accumulated and Cys-163 and Cys-285 are disulfide-bonded. In the even-numbered lanes, 0.2 mm AMS was added, whereas no AMS was added in the odd-numbered lanes. Western blotting was performed using an antibody against c-Myc. C, AMS alkylation and malPEG counter-alkylation of single Cys residues replaced within TM3. Conditions are the same as in panel B.
RESULTS
The Arrangement of TM1 and TM4 in the Reduced Form—To determine whether we can detect any alterations in the conformation of DsbDβ when it is shifted from the oxidized to the reduced state, we have investigated the accessibility of the residues in TM1 and TM4 of reduced DsbDβ. This was done by introducing single Cys substitutions into the protein and subjecting the protein to alkylation under different conditions.
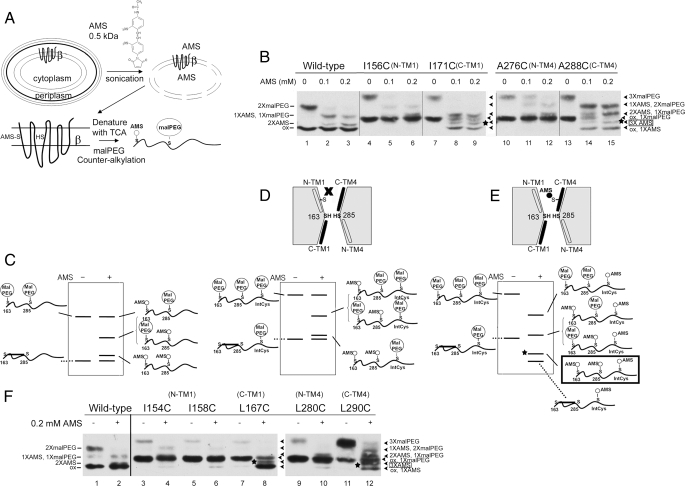
When the three domains of DsbD are split and expressed each from a different plasmid in the same cell, they reconstitute DsbC-reducing activity (13). Because each of the split domains is functional, we could use, for these experiments, only the DsbDβ domain. This domain could be detected by introducing a C-terminal c-Myc tag. The functionality of the protein with a single Cys replacing a native residue was tested in the previous study (19), and only functional ones were used in this study. As shown schematically in Fig. 2A, the accessibility of the cysteines was analyzed by first sonicating cells to generate solubilized membranes in a buffer containing AMS (a membrane-impermeable alkylating agent). Importantly, this step allows the protein to be accessible to both sides of the membrane and kinetically allows alkylation of free cysteines in the protein that are accessible to the aqueous environment. It is followed by denaturation of the extracts and counter-alkylation by malPEG. After this denaturation step, any water-inaccessible or partially accessible cysteines are counter-alkylated by malPEG, whereas the water-accessible cysteines in the native protein modified by AMS cannot be further alkylated. The addition of malPEG molecules to the protein decreases its mobility in SDS-PAGE due to its relatively high molecular weight. In our previous study, if the introduced cysteines were highly accessible, at 0.2 mm AMS, the AMS alkylation was complete (19). Thus, in this study, we used 0.2 mm AMS, and when we followed the change of alkylation of specific bands, we used also 0.1 mm AMS. It should be noted that Cys-163 and Cys-285, when they are reduced, can be partially alkylated at these concentrations (see below).
FIGURE 2.
AMS accessibility to the residues in TM1 and TM4 in both redox states. A, experimental scheme. Protein expression was induced by 0.2% arabinose, and cells were harvested at mid-log phase, treated with lysozyme and EDTA, and immediately subjected to sonication after adding AMS. After alkylation, cell lysates were precipitated with trichloroacetic acid (TCA) (denaturation) and subjected to malPEG alkylation. The cysteines in this figure for purposes of illustrating AMS and malPEG alkylation are arbitrarily chosen. B, AMS alkylation and 2-kDa malPEG counter-alkylation of wild type and single Cys replacements within TM1 and TM4. One sample was not treated with AMS (lanes 1, 4, 7, 10, and 13), and the others were treated with 0.1 mm AMS (lanes 2, 5, 8, 11, and 14) and 0.2 mm AMS (lanes 3, 6, 9, 12, and 15). Western blotting was performed using an antibody against c-Myc. — indicates the alkylated bands in wild type, whereas ◂ indicates those in single Cys replacements. The star indicates the triply AMS-alkylated band. N-TM1 and N-TM4 indicate N-terminal TMs of Cys-163 and Cys-285, respectively; C-TM1 and C-TM4, C-terminal ones, respectively. C, schematic band pattern of alkylation in wild-type protein in panel B. D, schematic band pattern of alkylation when the introduced cysteine is not accessible to AMS. E, schematic band pattern of alkylation when the introduced cysteine is accessible to AMS. The star indicates the triply AMS-alkylated band. See details for panels C, D, and E under “Results.” F, expanded analyses of panel B using other single Cys replacements in TM1 and TM4. No AMS and 0.2 mm AMS were added in the samples in odd- and even-numbered lanes, respectively.
In our previous study (19), we tested the accessibility of cysteine residues introduced into the oxidized (disulfide-bonded) DsbDβ by using a strain missing Trx reductase (TrxB). Without TrxB, Trx1 cannot be reduced, and the resultant oxidized Trx1 fully oxidizes Cys-163 and Cys-285 in the protein. In that situation, only the newly introduced cysteine residue in the protein is free, and its accessibility can be easily studied by itself. In contrast, in this study, we assessed the accessibility of the newly introduced cysteine in the reduced protein, using a background expressing wild-type TrxB. This approach complicates our analysis because, in principle, we have three reduced cysteines to analyze (the two redox-active cysteines and one introduced cysteine). In fact, this is not quite the case, as we have shown before (19) because a significant proportion of the reduced Cys-163–Cys-285 pair becomes oxidized during the preparation of samples. Nevertheless, the remaining portion of the reduced protein is sufficient for the analysis. Furthermore, the oxidized protein can even serve as an internal control for assessing alkylation of the protein in wild type and single Cys replacements. As a result, for the wild-type protein, we observe both a doubly malPEG-alkylated protein (from fully reduced DsbDβ) and the oxidized protein (Fig. 2, B, lane 1, and C) (19).
When the wild-type protein was incubated with AMS prior to sonication, the fraction of the two cysteines that had not become oxidized could be alkylated by AMS. Thus, the doubly malPEG-alkylated band disappeared, whereas a singly malPEG-alkylated band originated from a single AMS alkylation and a doubly AMS-alkylated band appeared (Fig. 2B, lanes 1–3). With increasing concentrations of AMS, the intensity of the singly malPEG-alkylated band decreased, and that of the doubly AMS-alkylated band increased. A schematic drawing to show various alkylated species is seen in Fig. 2C. The identification of the nature of the alkylation of each specific band with specific mobilities is based on control experiments with this protein previously published (19). These observations show that the two cysteines are accessible to a water environment, confirming previous results (19).
If we now analyze a protein into which we have introduced an additional cysteine residue into a position in a TM that is not water-accessible, as shown in Fig. 2D, that cysteine should not be alkylated by AMS. This cysteine could then be counter-alkylated by malPEG, and all the alkylated proteins would exhibit a mobility shift in the gel corresponding to the addition of a single molecule of malPEG (Fig. 2D). This pattern was observed with I156C, a residue in the periplasmic side of TM1, and A276C, a residue in the cytoplasmic side of TM4 (Figs. 1 and 2B, lanes 4–6 and 10–12). In our previous study with oxidized DsbDβ in which Cys-163 and Cys-285 are disulfide-bonded, these were residues that were inaccessible to AMS (19). Consistently, the oxidized band for these two mutants shows a mobility shift indicative of the addition of a single malPEG molecule, and this band remained unchanged when AMS had been added. The following presents our interpretation, based on our previous studies (19), of the products seen on gels when reduced species are analyzed. At 0.1 mm AMS, some doubly or triply malPEG-alkylated species were detected (Fig. 2B, lanes 5 and 11), indicating that either one or the other of the Cys-163 or Cys-285 residues can be partially alkylated by AMS and that a low amount of AMS-unalkylated protein remains. By increasing the AMS concentration, the intensity of the doubly AMS- and singly malPEG-alkylated bands increased, due presumably to increased alkylation of either Cys-163 or Cys-285 or both (Fig. 2B, lanes 6 and 12). An alternative explanation might be considered in which the results are due to other combinations of alkylation, for example, in which either Cys-163 or Cys-285 cannot be alkylated and the introduced cysteine can be alkylated. However, we think that this possibility is unlikely because the single Cys-replaced proteins used in this study are functionally active, and, thus, we presume that Cys-163 and Cys-285 would behave like the wild-type protein and follow similar alkylation kinetics (Fig. 2B, lanes 2 and 3).
If, in contrast to residues such as I156C, the additionally introduced single Cys is highly accessible, as shown in Fig. 2E, it would be alkylated by AMS and avoid counter-alkylation by malPEG. In this case, we would expect it to show a similar pattern to the wild type except for an additional single AMS modification under the condition where AMS is added (Fig. 2E). This pattern was observed for I171C, a residue on the cytoplasmic side of TM1, and A288C, a residue on the periplasmic side of TM4 (Figs. 1 and 2B, lanes 7–9 and 13–15). These were the same residues that were accessible residues when previously we analyzed oxidized DsbDβ in which Cys-163 and Cys-285 are disulfide-bonded (19). Consistent with these results is the finding that the oxidized band, which is singly malPEG-alkylated in the absence of AMS, became AMS-alkylated when AMS was added.
The reduced DsbDβ carrying either I171C or A288C is alkylated in four ways, as shown in Fig. 2E. The identification and verification of triply AMS-alkylated species are very important in the analysis of this particular kind of reduced protein. We draw conclusions about the nature of these different bands by the following reasoning. In all cases where no AMS is added (Fig. 2B, lanes 4, 7, 10, and 13), there are only two bands, an oxidized form of the protein with one malPEG added and a fully reduced form with three malPEGs added. (2-kDa malPEG was used to alkylate cysteines efficiently instead of 5-kDa malPEG used in the previous study (19). These results show that the free cysteines are fully alkylated by malPEG after denaturation and that none of the bands seen in these gels can contain free, reduced cysteines. This means that any bands that run faster than the oxidized singly malPEG-alkylated form do not contain any free cysteines, and from mobility, can be identified either as oxidized with one AMS or as a reduced form with three AMSs. The triply AMS-alkylated bands are a little closer to that of singly malPEG-alkylated oxidized bands than that of doubly AMS-alkylated bands in wild type (Fig. 2B, compare lanes 8, 9, 14, and 15 with lanes 2 and 3; lanes 1–9 are derived from a single gel). Similar arguments can be made for the identity of the remaining bands.
With I171C, by increasing AMS concentration, the intensity of the triply AMS-alkylated band (Fig. 2B, star) increased a little and that of doubly AMS- and singly malPEG-alkylated bands decreased, presumably due to the increase of AMS alkylation of both Cys-163 and Cys-285 (Fig. 2B, compare lane 8 with lane 9). With A288C, the intensity of the triply AMS-alkylated band (star) increased with increasing AMS concentration (Fig. 2B, compare lane 14 with lane 15). However, the intensity of doubly AMS- and singly malPEG-alkylated bands increased and that of singly AMS- and doubly malPEG-alkylated bands, which were invisible in I171C, decreased a little (Fig. 2B, compare lanes 14 and 15 with lanes 8 and 9). Because the A288C mutant has a partial defect in electron transfer activity (19), it is possible either that the two reactive cysteines are less accessible than those in wild type, unlike our presumption or, alternatively, that the introduced cysteine in the reduced protein is less accessible than that in I171C. Nevertheless, the important result is the detection of triple-AMS alkylation, showing clearly the accessibility of the introduced cysteine in both cases. Thus, in the reduced protein, similarly to the oxidized protein, we observed that the single Cys replacements placed to the N termini of the two redox-active cysteines are inaccessible to AMS, whereas those placed to the C termini of reactive cysteines are accessible in pseudo-symmetrical fashion.
We expanded our analysis by using many more single Cys mutants (Fig. 2F and supplemental Fig. 1). As in the above analysis, I154C and I158C from TM1 and L280C from TM4 (Fig. 1), the replacements positioned to the N termini of Cys-163 and Cys-285, showed almost no AMS alkylation of the introduced cysteine, whereas M167C from TM1 and L290C from TM4 (Fig. 1), the replacements in the C termini of Cys-163 and Cys-283, showed AMS alkylation both when DsbDβ was in the oxidized state and when DsbDβ was in the reduced state (Fig. 2F, compare even lanes with odd lanes). The above results suggest that the oxidized and reduced protein have similar conformations; the C-terminal portions of TM1 and TM4 are still water-exposed, whereas the N-terminal portions of them are not.
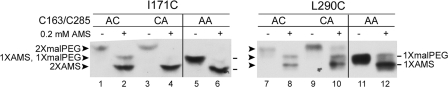
The Accessibility of I171C and L290C in a C163A or C285A Background—There is an alternative interpretation of our results according to which the AMS alkylation of the reactive cysteines causes an “artifactual” change in the conformation of the protein, which results in the introduced cysteine becoming accessible to AMS. To test for this possibility, we constructed mutants of DsbDβ where neither cysteine could be alkylated because they lack both Cys-163 and Cys-285 (Fig. 3, indicated by AA where A indicates replacement of Cys by Ala) or only one of these two cysteines (Fig. 3, indicated by AC and CA). The latter two constructs allow us to test for the effects of AMS alkylation of either cysteine on conformation. We combined the three sets of mutations with I171C and L290C, the two accessible replacements in the above analysis, and tested for accessibility of these introduced cysteines. Without prior treatment with AMS, with AC and CA, doubly malPEG-alkylated bands were observed, whereas in AA, only singly malPEG-alkylated bands were (Fig. 3, odd lanes). When AMS was added, with the I171C replacement, a majority of the cysteine(s) was doubly AMS-alkylated in AC and CA and singly AMS-akylated in AA (Fig. 3, lanes 2, 4, and 6). With the L290C replacement, a fraction of the cysteine(s) was partially AMS-alkylated in the three mutants, but significant portions were fully AMS-alkylated (Fig. 3, lanes 8, 10, and 12). The results show that I171C and L290C are accessible to AMS in all cysteine-less mutant backgrounds, suggesting that their accessibility is independent of AMS alkylation in the reactive cysteines.
FIGURE 3.
AMS accessibility to the I171C and L290C in C163A (AC), C285A (CA), and C163A/C285A (AA) backgrounds. The samples were analyzed according to the procedure in Fig. 2A. No AMS and 0.2 mm AMS were added in the samples in odd- and even-numbered lanes, respectively, and 2-kDa malPEG counter-alkylation was followed. Western blotting was performed using an antibody against c-Myc. — indicates the alkylated bands in AA mutant, whereas ▸ indicates those in AC or CA mutants.
The Arrangement of TM2 and TM3—We proposed that parts of TM1 and TM4 located to the C termini of Cys-163 and Cys-285 are involved in the formation of cytoplasmic and periplasmic cavities based on the water-exposed properties of the helices and the two cysteines (Fig. 1, inset). We proceeded to ask whether there might be water-exposed regions like those in TM1 and TM4 in other TMs of the protein. We have tested the AMS accessibility of single Cys replacements in TM2 and TM3, using the same AMS alkylation and malPEG counter-alkylation approaches. We performed separate experiments using wild-type (DsbDβ reduced) and trxB null (DsbDβ oxidized) strains to assess the accessibility of residues in two different redox states of the protein. In this section, we show the results using the trxB null strain, in which we need to consider only the introduced cysteine because Cys-163 and Cys-285 are disulfide-bonded. The results using strains with the wild-type trxB are shown in supplemental Fig. 3, showing similar accessibility of the introduced cysteines to that seen with trxB- strains.
We predicted the extent of the transmembrane helices corresponding to TM2 and TM3 using several prediction programs including the HMMTOP analysis, which predicts the location of Cys-163 and Cys-285 in the middle of the TM1 and TM4, respectively (Figs. 1 and 4A). Among them, SOSUI gives similar prediction for the location of the cysteines to that obtained with HMMTOP, and their predictions for TM2 and TM3 are similar (Fig. 4A). Relying on the coherence in predictions by the two programs, we used the HMMTOP analysis to choose the amino acid residues in TM2 and TM3 for accessibility testing. The prediction seems to fit well with potential models, at least for TM2, in terms of the accessibility studied below. We made single Cys replacements of the predicted TM residues, tested DsbC-reducing activity, and chose only the functional replacements to perform alkylation experiments (supplemental Fig. 2). Some of the cysteine replacements in TM3 that we use do have significantly diminished activity in reducing DsbC. However, the analysis of accessibility of the introduced cysteines is still possible because Cys-163 and Cys-285 are redox-active, although a greater portion of the protein seems to be oxidized (Fig. 4C and supplemental Fig. 3B). Here we used the approaches previously described (19) to measure the accessibility of the TM2 and TM3 residues from both sides of the membrane, first by doing alkylation experiments with spheroplasts of the bacteria where the periplasmic face of the cytoplasmic membrane is accessible. In the spheroplast sample, with prior treatment of AMS, only the positive control, A288C from TM4, which is water-accessible from the periplasmic side, showed AMS alkylation. Most of the tested residues in TM2 and TM3 were not alkylated in the spheroplast preparation, suggesting that these residues are not accessible from the periplasmic side (data not shown). In the cell lysate samples where both sides of the membrane become available (see “Experimental Procedures” for the timing of AMS addition), without or with prior treatment of AMS, wild-type protein did not show malPEG alkylation because there are no free cysteines (Fig. 4, B, lanes 1, 2, 25, and 26, and C, lanes 19, 20, 45, and 46). Without prior treatment of AMS, the positive controls, A288C, and I171C, a residue accessible from the cytoplasmic side (see above), showed single malPEG alkylation, whereas with prior treatment of AMS, they did not, suggesting that the introduced cysteines are accessible to AMS (Fig. 4, B, lanes 23, 24, 27, 28, and 51–54, and C, lanes 1, 2, and 21–26).
In the single Cys replacements in TM2, some degree of variations in malPEG alkylation was observed when no AMS was added, possibly due to incomplete malPEG alkylation in some replacements (Fig. 4B, odd lanes in lanes 3–21, 29–49, and 55). However, when AMS was added, although L190C, L207C, V209C, A210C, and A212C showed a small amount of AMS alkylation (Fig. 4B, lanes 8, 42, 46, 48, and 56), none of them showed significant AMS alkylation when compared with the positive controls (Fig. 4B, even lanes in lanes 4–22, 30–50, and 56). These results showed that tested TM2 residues are largely inaccessible to AMS.
In the single Cys replacements in TM3, the results are more interesting. Like the single Cys replacements in TM2, I227C, M238C, S239C, M240C, and F241C were not accessible to AMS (Fig. 4C, lanes 3, 4, and 27–34). G228C, L229C, I231C, V232C, T234C, L235C, and L236C showed no or very inefficient mal-PEG counter-alkylation regardless of prior treatment of AMS (Fig. 4C, lanes 5–18). Therefore, we could not assess their accessibility to AMS. These regions might be highly protected by native protein-protein interactions, which are somehow resistant to strong denaturing conditions, adding trichloroacetic acid and SDS. Alternatively, strong denaturing conditions might promote an artificial conformation of the protein that protects the introduced cysteine from malPEG alkylation. Finally, we found highly AMS-accessible single Cys replacements, F244C, T245C, L246C, L248C, and P249C, from the cytoplasmic proximal portion of TM3 (Fig. 4C, lanes 35–44). Thus, the cytoplasmic proximal portion of TM3, along with TM1, may be another component of the proposed water-exposed cavity in the cytoplasmic proximal portion of the protein.
DISCUSSION
DsbD is a member of a family of integral cytoplasmic membrane proteins that exhibit inverted symmetry in their membrane domains. These proteins carry out a variety of processes for transporting small molecules and even proteins across the membrane. However, DsbD is unusual in that one of the steps of transferring electrons from the cytoplasm to the periplasm involves the breakage by Trx of a covalent bond between two cysteines lying in different TMs. This reductive step frees up the redox-active cysteines to transfer electrons to a periplasmic domain of DsbD (DsbDγ), which is also a Trx-like protein. The inverted symmetry of the protein may reflect the requirement for both faces of the protein (cytoplasmic and periplasmic) to interact with Trx proteins.
We have previously shown that the two cysteines of DsbDβ when they are in the reduced state are accessible to the aqueous environment on either side of the membrane. Further, domains of the two TMs (TM1 and TM4) that lie to the C-terminal side of the two cysteines are also exposed to the aqueous environment when the two cysteines are disulfide-bonded. The domains of the TMs to the N-terminal side are not so exposed. Here we show that the same amino acid residues that are accessible or not to water in the oxidized protein are similarly accessible or not when the two cysteines are reduced. Further, we find no residues in TM2 that are exposed in either redox state of the protein, whereas a cytoplasmic proximal portion of TM3 is exposed under both conditions.
These results provide two findings important for understanding how DsbD works. First, remarkably, there is no indication from our results of any dramatic conformational change in the structure of the DsbDβ domain between its oxidized and reduced forms. Second, our analysis increases the detail with which we can picture the structure of DsbDβ in the membrane. In particular, we can place TM2 embedded in the lipid bilayer or within the protein, accessing only hydrophobic environments. In contrast, a portion of TM3, being open to the cytoplasmic environment, is likely a part of the channel that allows cytoplasmic Trx1 to interact with the disulfide bond in the membrane. Because of the inverted symmetry between TM1–3 and TM4–6, we hypothesize that the C-terminal domain of TM6 contributes to the presumed channel on the other side of the membrane from that of TM3.
In its normal mode of action, DsbD becomes oxidized, and thus inactive, as a result of its reduction of DsbC and other periplasmic substrates. The first step in the regeneration of active DsbD is the reduction of the intramembranous disulfide bond of DsbDβ domain by cytoplasmic Trx1. It had been proposed by us and others that the two cysteines of DsbDβ may be alternatively exposed to the cytoplasm and the periplasm by conformational changes such that the disulfide bond is more available to reduced Trx1 and, when reduced, the two cysteines are more available to oxidized periplasmic DsbDγ (21–23). However, our findings that the shift from the oxidized to the reduced state does not result in a change in the accessibility to either side of the membrane of the cysteines and of different portions of DsbDβ TMs suggest that those earlier proposals may not be correct. Further, our previous results that reduced DsbDβ can be oxidized by reverse electron transfer with oxidized cytoplasmic Trx1 (in the trxB mutant) indicate that the cytoplasmic cavity must also be open to accommodate oxidized Trx1 when Cys-163 and Cys-285 are reduced (28).
In fact, Rohzkova and Glockshuber (18) presented spectroscopic data assessing conformational changes of DsbD between the reduced and oxidized forms using purified but liposome-reconstituted protein that are consistent with the results presented here. They found that subtle changes do occur in the tertiary structure around some tryptophan residues but that they are not accompanied by any significant changes in the secondary structure. Furthermore, the experiments of Hiniker et al. (23) showing different proteolytic sensitivities between the reduced and oxidized forms of DsbD were done under conditions that may not reflect the native conformations of DsbDβ because the detergent used may not have provided the same lipid-mediated protein interactions in DsbDβ that may be important for its structure.
On the other hand, our evidence indicating little change in the conformation of the protein (the accessibility of the residues lining the cavities) is based on accessibility to alkylation by AMS, a small molecule of 0.5 kDa. Thus, the cysteine pair in the oxidized and reduced state could have differences in their accessibility to larger molecules such as Trx1. That such differences could occur and not be detected by using AMS is indicated by our earlier results (14). In those experiments, we found that Cys-163 and Cys-285 were accessible to the 5-kDa malPEG molecule but only from the cytoplasmic side, not the periplasmic side of the membrane. These results suggest that more subtle differences might be detected by using an array of alkylating agents of different sizes.
There is still likely a need for significant changes in conformation in order for the protein to interact productively with Trx1 or with DsbDγ. We think that such changes may be necessary to allow the active sites of these two Trx family members, which are 12–14-kDa proteins, to access the redox-active cysteines of DsbDβ through what may be small channels. For example, as shown in Fig. 5, in the case of Trx1, the interaction of Trx1 with amino acid residues on the cytoplasmic face of DsbDβ may open up the channel, allowing a direct reaction between the cysteine pairs of the two proteins. On the periplasmic side, it may be that after breaking of the disulfide bond between Cys-163 and Cys-285, the interaction with oxidized DsbDγ opens up further the channel, allowing Cys-285 and DsbDγ to come sufficiently close for the transfer of electrons to take place. Thus, it appears possible that, in contrast to the absence of difference we see in the purely oxidized or reduced states of DsbDβ, complexes trapped between one or the other of these Trx substrates of the protein may show more dramatic differences. We are currently exploring this possibility.
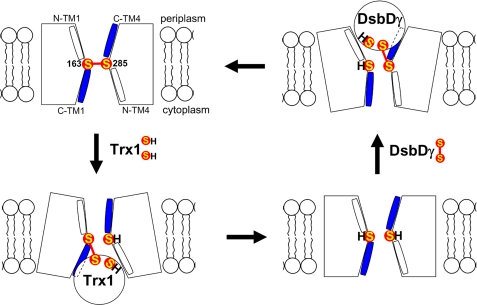
FIGURE 5.
Model of structural changes of DsbDβ during redox reactions. The model shows that DsbDβ adopts relative small openings toward both sides of the membrane when Cys-163 and Cys-285 are in the oxidized and reduced states, whereas when interacting with either partner Trx proteins, it adopts a relative large opening toward the interacting side of the membrane. S represents the sulfur of thiol in a cysteine residue. Halves of C-terminal TM1 and TM4 are water-exposed (blue; C-TM1 and C-TM4), whereas those of N-terminal ones are not (white; N-TM1 and N-TM4).
Our picture of the channels of DsbDβ still remains one of hourglass structures based on inverted symmetry. There are a number of other membrane transporters that exhibit inverted symmetry between TM domains and more or less hour-glass-like shapes. Among them are LeuT and NhaA, which are secondary carriers, for which it is proposed that ligand binding mediates alternating opening and closing toward both sides of the membrane (29, 30). In contrast, aquaporin-1 has only an open selective filter in the middle of the protein (31). In SecY, a protein-translocating channel, displacement of a plug segment in the pore is proposed to be involved in gating the pore while maintaining the hourglass-like shape (32). In the ClC chloride channel, a gating mechanism is proposed, but it is mediated by the motion of a single amino acid rather than the large conformational change of portions of the protein (33). It appears that DsbDβ and members of that family of proteins present yet a different variant on how this structure can be used for transport across the membrane. In the membrane transporter proteins, when the helices form the pore in the channel, an amphipathic helical property is observed. However, the water-exposed helices in the TMs of DsbDβ do not seem to show an amphipathic property but, rather, display a continuous water-exposed property. These findings suggest that these domains may form more or less unstructured loops rather than “α-helices.”
Finally, we point out that we have assumed the lengths of the TMs of DsbD and that the amino acid residues of which they are composed are those predicted by the HMMTOP program. However, it is possible that those predictions are incorrect for TM3 where we observe a portion of the TM that exhibits water accessibility. It could, instead, be the case that the residues of TM3 we find are actually part of a cytoplasmic domain of the protein. However, the rest of our analyses for TM1, TM2, and TM4 indicate that these TMs are correctly predicted by the program. The inverted symmetry of TM1 and TM4 both in sequence and in water accessibility and the positioning of the two cysteines in the middle of these two TM segments are consistent with those predictions. For TM2, the entire length of the predicted TM is AMS-inaccessible, consistent with a sequence of amino acids fully embedded in the membrane. Finally, it is hard to imagine that the water-exposed C-terminal domain of TM1 is sufficient to serve as an opening in the membrane for entry of Trx1. From inverted symmetry, it would appear that neither TM5 nor TM6 would have water-accessible portions that are cytoplasm-proximal. For these reasons, it appears likely that it is, in fact, the C-terminal portion of TM3 that is water-accessible and is contributing to the cavity that Trx1 must enter to reduce the disulfide bond in DsbDβ.
Supplementary Material
Acknowledgments
We thank members of Beckwith laboratory for helpful discussions and suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant GMO55090 (to J. B.).
The on-line version of this article (available at http://www.jbc.org) contains three supplemental figures.
Footnotes
The abbreviations used are: Trx, thioredoxin; TM, transmembrane segment; AMS, 4-acetamido-4′maleimidylstilbene-2,2′-disulfonic acid; malPEG, methoxypoly(ethylene glycol)-maleimide.
References
- 1.Bardwell, J. C., McGovern, K., and Beckwith, J. (1991) Cell 67 581-589 [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi, T., Kishigami, S., Sone, M., Inokuchi, H., Mogi, T., and Ito, K. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 11857-11862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bader, M., Muse, W., Ballou, D. P., Gassner, C., and Bardwell, J. C. (1999) Cell 98 217-227 [DOI] [PubMed] [Google Scholar]
- 4.Zapun, A., Missiakas, D., Raina, S., and Creighton, T. E. (1995) Biochemistry 34 5075-5089 [DOI] [PubMed] [Google Scholar]
- 5.Rietsch, A., Belin, D., Martin, N., and Beckwith, J. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 13048-13053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joly, J. C., and Swartz, J. R. (1997) Biochemistry 36 10067-10072 [DOI] [PubMed] [Google Scholar]
- 7.Berkmen, M., Boyd, D., and Beckwith, J. (2005) J. Biol. Chem. 280 11387-11394 [DOI] [PubMed] [Google Scholar]
- 8.Walker, K. W., and Gilbert, H. F. (1997) J. Biol. Chem. 272 8845-8848 [DOI] [PubMed] [Google Scholar]
- 9.Rietsch, A., Bessette, P., Georgiou, G., and Beckwith, J. (1997) J. Bacteriol. 179 6602-6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart, E. J., Katzen, F., and Beckwith, J. (1999) EMBO J. 18 5963-5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung, J., Chen, T., and Missiakas, D. (2000) Mol. Microbiol. 35 1099-1109 [DOI] [PubMed] [Google Scholar]
- 12.Gordon, E. H., Page, M. D., Willis, A. C., and Ferguson, S. J. (2000) Mol. Microbiol. 35 1360-1374 [DOI] [PubMed] [Google Scholar]
- 13.Katzen, F., and Beckwith, J. (2000) Cell 103 769-779 [DOI] [PubMed] [Google Scholar]
- 14.Katzen, F., and Beckwith, J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 10471-10476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, J. H., Kim, S. J., Jeong, D. G., Son, J. H., and Ryu, S. E. (2003) FEBS Lett. 543 164-169 [DOI] [PubMed] [Google Scholar]
- 16.Goulding, C. W., Sawaya, M. R., Parseghian, A., Lim, V., Eisenberg, D., and Missiakas, D. (2002) Biochemistry 41 6920-6927 [DOI] [PubMed] [Google Scholar]
- 17.Haebel, P. W., Goldstone, D., Katzen, F., Beckwith, J., and Metcalf, P. (2002) EMBO J. 21 4774-4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rozhkova, A., and Glockshuber, R. (2008) J. Mol. Biol. 380 783-788 [DOI] [PubMed] [Google Scholar]
- 19.Cho, S.-H., Porat, A., Ye, J., and Beckwith, J. (2007) EMBO J. 26 3509-3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimball, R. A., Martin, L., and Saier, M. H., Jr. (2003) J. Mol. Microbiol. Biotechnol. 5 133-149 [DOI] [PubMed] [Google Scholar]
- 21.Rozhkova, A., Stirnimann, C. U., Frei, P., Grauschopf, U., Brunisholz, R., Grütter, M. G., Capitani, G., and Glockshuber, R. (2004) EMBO J. 23 1709-1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porat, A., Cho, S.-H., and Beckwith, J. (2004) Res. Microbiol. 155 617-622 [DOI] [PubMed] [Google Scholar]
- 23.Hiniker, A., Vertommen, D., Bardwell, J. C., and Collet, J. F. (2006) J. Bacteriol. 188 7317-7320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tusnady, G. E., and Simon, I. (2001) Bioinformatics (Oxf.) 17 849-850 [DOI] [PubMed] [Google Scholar]
- 25.Hirokawa, T., Boon-Chieng, S., and Mitaku, S. (1998) Bioinformatics (Oxf.) 14 378-379 [DOI] [PubMed] [Google Scholar]
- 26.Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001) J. Mol. Biol. 305 567-580 [DOI] [PubMed] [Google Scholar]
- 27.Claros, M. G., and von Heijne, G. (1994) Comput. Appl. Biosci. 10 685-686 [DOI] [PubMed] [Google Scholar]
- 28.Cho, S.-H., and Beckwith, J. (2006) J. Bacteriol. 188 5066-5076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamashita, A., Singh, S. K., Kawate, T., Jin, Y., and Gouaux, E. (2005) Nature 437 215-223 [DOI] [PubMed] [Google Scholar]
- 30.Arkin, I. T., Xu, H., Jensen, M. Ø., Arbely, E., Bennett, E. R., Bowers, K. J., Chow, E., Dror, R. O., Eastwood, M. P., Flitman-Tene, R., Gregersen, B. A., Klepeis, J. L., Kolossváry, I., Shan, Y., and Shaw, D. E. (2007) Science 317 799-803 [DOI] [PubMed] [Google Scholar]
- 31.Murata, K., Mitsuoka, K., Hirai, T., Walz, T., Agre, P., Heymann, J. B., Engel, A., and Fujiyoshi, Y. (2000) Nature 407 599-605 [DOI] [PubMed] [Google Scholar]
- 32.Van den Berg, B., Clemons, W. M., Jr., Collinson, I., Modis, Y., Hartmann, E., Harrison, S. C., and Rapoport, T. A. (2004) Nature 427 36-44 [DOI] [PubMed] [Google Scholar]
- 33.Dutzler, R., Campbell, E. B., and MacKinnon, R. (2003) Science 300 108-112 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.