Abstract
Proper expression of the replication licensing factor Cdt1 is primarily regulated post-translationally by ubiquitylation and proteasome degradation. In a screen to identify novel non-histone targets of histone deacetylases (HDACs), we found Cdt1 as a binding partner for HDAC11. Cdt1 associates specifically and directly with HDAC11. We show that Cdt1 undergoes acetylation and is reversibly deacetylated by HDAC11. In vitro, Cdt1 can be acetylated at its N terminus by the lysine acetyltransferases KAT2B and KAT3B. Acetylation protects Cdt1 from ubiquitylation and subsequent proteasomal degradation. These results extend the list of non-histone acetylated proteins to include a critical DNA replication factor and provide an additional level of complexity to the regulation of Cdt1.
To maintain genomic integrity, DNA replication must be tightly controlled to ensure that each portion of the genome replicates once and only once per cell cycle (reviewed in Ref. 1). Replication licensing begins by the formation of the prereplication complex at multiple potential origins of replication. This is established sequentially, with the origin recognition complex (ORC)2 proteins binding first, followed by the recruitment of Cdc6 and Cdt1, which in turn recruit the MCM2–7 proteins. MCM proteins act as the replicative helicase. The licensed replication origins are activated by cyclin-dependent kinases at the start of S phase. Licensing occurs throughout the cell cycle once S phase is complete.
Cdt1 levels fluctuate throughout the cell cycle. It is destabilized at G1/S transition, and then levels begin to climb again upon S phase completion. To prevent licensing at inappropriate times, two separate processes regulate the inactivation or destruction of Cdt1. First, geminin negatively regulates Cdt1 function by prevention of the association of Cdt1 with MCM2–7 via steric hindrance (2). Interestingly, geminin also positively regulates Cdt1 by preventing its ubiquitylation, perhaps by prevention of its interaction with an E3 ligase. This allows Cdt1 to accumulate in G2 and M phases, to ensure adequate pools of Cdt1 to license the next cycle of replication (3). The ratio of geminin to Cdt1 likely determines whether geminin positively or negatively regulates Cdt1 (4). Second, Cdt1 is targeted for proteolysis by two distinct ubiquitin E3 ligases: the SCF-Skp2 complex and the DDB1-Cul4 complex (5). Phosphorylation by cyclin A/Cdk2 promotes interaction of Cdt1 with Skp2, leading to Cdt1 degradation during S phase (6–8). In addition, DDB1-Cul4 utilizes proliferating cell nuclear antigen as a binding platform to contact Cdt1, targeting the destruction of Cdt1 in S phase or following DNA damage (9, 10). Ubiquitylation by either of these E3 ligases promotes degradation of Cdt1 by the proteasome.
Ubiquitylation occurs primarily (but not exclusively) on the ε-amino group of lysine residues. Another prominent post-translational modification that occurs on that residue is acetylation. Acetylation and, correspondingly, deacetylation can modulate the function and activity of a variety of proteins (see Ref. 11 for review). Here, we report that Cdt1 physically interacts with HDAC11, a class IV histone deacetylase (12, 13), as well as with several lysine acetyltransferases (KATs). We show that Cdt1 is an acetylated protein and further show that acetylation protects Cdt1 from ubiquitylation and subsequent proteasomal degradation. This study uncovers yet another layer of complexity to the regulation of the critical licensing factor Cdt1.
EXPERIMENTAL PROCEDURES
Plasmids—HDAC11 (IMAGE clone 3906049) was obtained from Open Biosystems. Cdt1 cDNA, a kind gift of A. Dutta, has been described previously (14). cDNAs and corresponding deletions were shuttled into appropriate expression vectors by restriction digestion or PCR and verified by sequencing. Plasmids were purified by Qiagen maxipreps.
Cell Culture and Transfection—HeLa and 293T cells were grown in Dulbecco's modified Eagle's medium containing 10% heat-inactivated calf serum and antibiotics. Trichostatin A (TSA), MG132, and curcumin were obtained from Sigma. Transfection was performed using Lipofectamine 2000 (Invitrogen). To prepare cell lines stably overexpressing HDAC11, FLAG-tagged HDAC11 was transfected into HeLa cells. Colonies were selected in 500 μg/ml G418 and verified to express HDAC11 by Western blotting using anti-FLAG antibodies.
Protein Lysates, Immunoprecipitation, and Western Blotting—Whole cell lysates were prepared by resuspension of the cell pellet in phosphate-buffered saline lysis buffer containing 10% glycerol, 0.1% Nonidet P-40, and protease inhibitor mixture (Roche Applied Science). Extracts were quantified by Bradford assay (Bio-Rad). Immunoprecipitations of overexpressed proteins were carried out using anti-FLAG-agarose (Sigma), anti-Myc-agarose (Santa Cruz Biotechnology), or anti-HA-agarose (Sigma). Endogenous protein immunoprecipitations were performed with the indicated antibodies and protein A-agarose (Invitrogen). Following immunoprecipitation, beads were washed five times with lysis buffer, resuspended in sample buffer, resolved by SDS-PAGE, and then transferred to Immobilon C (GE Bioscience). Blots were blocked and then incubated with the appropriate primary antibodies overnight at 4 °C. Following incubation with horseradish peroxidase-labeled secondary antibodies, the blots were visualized by enhanced chemiluminescence (SuperSignal West Pico or Femto, Pierce). Inputs, where indicated, were 5–10% of the amount used for immunoprecipitation. Expression levels were quantified using ImageQuant. The following antibodies were used: FLAG, actin, and tubulin (Sigma); Myc (Santa Cruz Biotechnology); HA (Covance); Cdt1 (Abcam and Santa Cruz Biotechnology); HDAC11 (Santa Cruz Biotechnology); acetyllysine (Cell Signaling and Upstate); and ubiquitin (Upstate).
Glutathione S-Transferase (GST) Pulldown—GST, GST-HDAC11, and GST-geminin were expressed in BL21 cells. Bacterial lysates were prepared and bound to glutathione beads at 4 °C. Recombinant His6-tagged full-length Cdt1 (purified from BL21 cells by nickel affinity chromatography and eluted with imidazole) was incubated with various GST constructs bound to beads in binding buffer (phosphate-buffered saline containing 350 mm NaCl and 0.2% Nonidet P-40). Binding reactions were carried out at room temperature for 90 min. Samples were washed five times with binding buffer, resuspended in sample buffer, and processed for Western blotting.
In Vitro KAT Assay—KAT assays were performed in 1× KAT buffer (50 mm Tris-Cl, pH 8, 10% glycerol, 0.1 mm EDTA, 1 mm dithiothreitol, 10 mm sodium butyrate and 1× protease inhibitor mixture), containing [14C]acetyl-CoA. The His6-tagged Cdt1 substrates (expressed via pET vectors) were purified from bacteria by nickel chromatography and eluted with imidazole. Reactions were allowed to proceed for 1 h at 30 °C and then resolved by SDS-PAGE. Gels were stained with Coomassie Brilliant Blue to visualize the substrates and ensure equivalent loading. After destaining, gels were fixed in 10% acetic acid and 40% methanol and soaked first in EN3HANCE (PerkinElmer Life Sciences) and then in cold water. Gels were dried and exposed to x-ray film with intensifying screens. Recombinant KAT2B (PCAF) and KAT3B (p300) were obtained from Upstate. 0.5 μg of rKAT2B (amino acids 352–832) or 2.5 μg of rKAT3B (amino acids 1066–1707) were used in each reaction. KATs were also produced by overexpression of the appropriate tagged constructs in 293T cells and then immunoprecipitation. Immunoprecipitates were washed five times with lysis buffer and two times with 1× KAT buffer. KAT reactions were performed directly on the beads.
In Vivo Ubiquitylation Assay—Cells were transfected with His-ubiquitin and harvested after 20–24 h. 20 μm MG132 was added for the final 4 h of incubation where indicated. A whole cell extract was prepared from 20% of the cell pellet for a direct Western blot. 80% of the cell pellet was lysed under denaturing conditions using Buffer A (6 m guanidinium HCl, 100 mm sodium phosphate, pH 8, 10 mm 2-mercaptoethanol, and 10 mm imidazole) and then incubated with nickel-nitrilotriacetic acid agarose beads (Qiagen) while rotating at room temperature overnight. Beads were washed sequentially with Buffer A, Buffer B (8 m urea, 100 mm sodium phosphate, pH 8, and 10 mm 2-mercaptoethanol), Buffer C (8 m urea, 100 mm sodium phosphate, pH 6.3, and 10 mm 2-mercaptoethanol) containing 0.2% Triton X-100, and Buffer C. Samples were eluted with continuous shaking in elution buffer (200 mm imidazole, 150 mm Tris-Cl, pH 6.7, 30% glycerol, 0.72 m 2-mercaptoethanol, and 5% SDS). Following the addition of 6× loading dye, the samples were resolved by SDS-PAGE and processed for Western blotting.
RESULTS
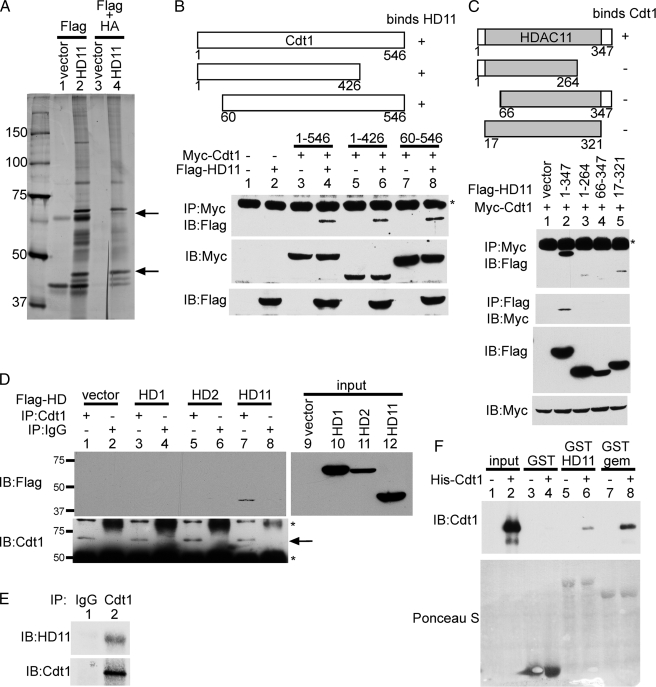
Cdt1 Copurifies with HDAC11—HDAC11 is the most recently described HDAC and one of the most poorly characterized. To identify potential non-histone targets of HDAC11, we made use of the fact that HDACs often physically interact with their substrates. To this end, we overexpressed HA-HDAC11-FLAG in HeLa cells. Whole cell extracts were purified sequentially on a FLAG column, and then FLAG eluates were purified on an HA column. Specific HDAC11-interacting proteins were excised from the stained polyacrylamide gel and analyzed by mass spectrometry. From this analysis, we obtained seven unique peptides, encompassing 100 amino acids, from the DNA replication licensing factor Cdt1 (Fig. 1A). We also identified several other HDAC11-interacting proteins that are currently under investigation.
FIGURE 1.
Cdt1 interacts specifically and directly with HDAC11. A, shown is a silver stain of the HDAC11-interacting proteins. An HA-HDAC11-FLAG expression plasmid or an empty vector were transfected into HeLa cells. Whole cell extracts were purified first on anti-FLAG beads, and then eluates were purified on anti-HA beads. The upper arrow indicates the band containing Cdt1, as identified by mass spectrometry. The lower arrow indicates HA-HDAC11-FLAG. B, shown is a verification of the Cdt1-HDAC11 interaction. 293T cells were transfected with the indicated plasmids, and whole cell extracts were immunoprecipitated (IP) with anti-Myc antibodies and then blotted (IB) with anti-FLAG antibodies. Direct Western blots were performed on whole cell extracts with the indicated antibodies to assess expression levels. The asterisk indicates an Ig heavy chain. C, the deletion of any part of HDAC11 impairs its ability to bind to Cdt1. The indicated FLAG-tagged HDAC11 deletion plasmids were cotransfected with a Myc-Cdt1 expression plasmid into 293T cells. Immunoprecipitations and Western blotting were performed as indicated. Direct Western blots of the whole cell extracts demonstrate expression levels. The shaded area represents the HDAC domain of HDAC11. D, Cdt1 interacts specifically with HDAC11. FLAG-tagged expression plasmids for the indicated HDACs were transfected into HeLa cells. Cell extracts were immunoprecipitated with anti-Cdt1 or normal goat IgG as indicated and then blotted with anti-FLAG antibodies. The blot was then stripped and reprobed with anti-Cdt1 antibodies. The extracts were also blotted directly with anti-FLAG to assess expression levels of the FLAG-tagged HDACs (input, right panel). The arrow indicates immunoprecipitated Cdt1. The lower asterisk indicates an Ig heavy chain, whereas the upper asterisk indicates a nonspecific band. E, the endogenous HDAC11 and Cdt1 interact. HeLa whole cell extracts were immunoprecipitated with either anti-Cdt1 or normal rabbit IgG and then blotted with anti-HDAC11 antibodies or anti-Cdt1 antibodies. F, Cdt1 directly interacts with HDAC11. Purified His-tagged recombinant Cdt1 was incubated with the indicated GST-tagged proteins attached to glutathione beads. After binding and washing, reaction mixtures were resolved on SDS-PAGE for analysis. The upper panel shows the Western blot using anti-Cdt1. The lower panel shows Ponceau S staining to visualize the GST fusion proteins. Input indicates an amount of recombinant Cdt1 equivalent to that used in the pulldowns that was directly loaded on the gel without incubation with GST fusion proteins. gem, geminin.
Cdt1 Interacts Specifically and Directly with HDAC11—To confirm the interaction between Cdt1 and HDAC11, we overexpressed Myc-tagged Cdt1 and FLAG-tagged HDAC11 in 293T cells. FLAG-HDAC11 co-immunoprecipitated with Myc-Cdt1 (Fig. 1B, lane 4). We also prepared N- and C-terminal deletions of Cdt1. Removal of either the first 59 amino acids (lane 8) or the last 120 amino acids (lane 6) had no effect on the ability of Cdt1 to bind to HDAC11. This indicates that the central portion of Cdt1 binds to HDAC11.
HDAC11 is the smallest member of the human HDAC family, containing only 347 amino acids. As such, the conserved HDAC domain (amino acids 17–321) comprises the majority of the protein. Deletion of any part of HDAC11 greatly impairs the ability of HDAC11 to bind to Cdt1 (Fig. 1C, top two panels, compare lane 2 with lanes 3–5). This suggests that the N and C termini, which are unique to HDAC11, play an important role in its interaction with Cdt1.
To explore the specificity of the HDAC11-Cdt1 interaction, we overexpressed a panel of FLAG-tagged HDACs in HeLa cells. We examined HDAC1 and HDAC2 because Cdt1 is a nuclear protein, and both HDAC1 and HDAC2 are primarily nuclear. Also, HDAC11 has been shown to localize to the nucleus (12). Following immunoprecipitation with anti-Cdt1, we could coprecipitate FLAG-HDAC11 (Fig. 1D, lane 7), but not FLAG-HDAC1 or FLAG-HDAC2. FLAG-HDAC3 also did not bind Cdt1 (data not shown). FLAG-HDAC1 and FLAG-HDAC11 were expressed at similar levels, yet only HDAC11 could bind to Cdt1. This suggests that Cdt1 interacts specifically with HDAC11, but not with other HDACs.
To show that endogenous HDAC11 and Cdt1 could also interact, we used anti-Cdt1 or normal rabbit IgG to immunoprecipitate HeLa whole cell extracts. Western blotting with an antibody directed against HDAC11 shows that HDAC11 is present in the Cdt1 immunoprecipitate (Fig. 1E, lane 2), but not in the IgG immunoprecipitate (lane 1). This confirms that physiologically relevant amounts of Cdt1 and HDAC11 can interact in HeLa cells.
Finally, to prove that HDAC11 and Cdt1 directly interact, in the absence of any other proteins, we performed GST pulldown assays. Bacterially expressed GST-HDAC11 could pull down bacterially expressed purified His-Cdt1 (Fig. 1F, lane 6). GST alone did not interact with His-Cdt1 (lane 4). GST-geminin, a known binding partner for Cdt1, was used as a positive control (lane 8).
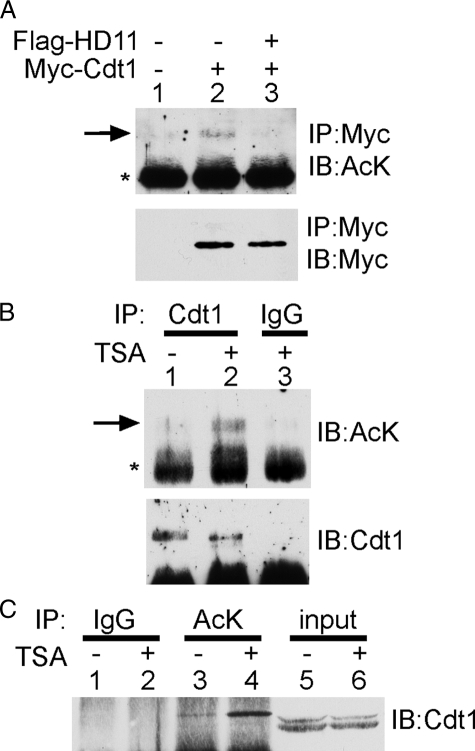
Cdt1 Is an Acetylated Protein—Because we found that Cdt1 could interact with HDAC11, we next wanted to determine whether Cdt1 was acetylated. To do this, we overexpressed Myc-Cdt1 in HeLa cells. Immunoprecipitation with anti-Myc beads followed by blotting with an anti-acetyllysine antibody showed that Myc-Cdt1 was acetylated (Fig. 2A, lane 2). The intensity of the acetylated band was severely diminished to background levels by cotransfection with FLAG-HDAC11 (lane 3), suggesting that HDAC11 could deacetylate Cdt1. To determine whether endogenous Cdt1 was also acetylated, we used antibodies to Cdt1 for immunoprecipitation from HeLa cell extracts. Western blotting with anti-acetyllysine shows a weak band corresponding to acetylated Cdt1 (Fig. 2B, lane 1). Treatment of the cells with the HDAC inhibitor TSA enhances the acetylation level of Cdt1 without affecting the expression of endogenous Cdt1 (lane 2). As a final confirmation, immunoprecipitation with an anti-acetyllysine antibody followed by blotting for Cdt1 revealed a Cdt1-specific band (Fig. 2C, lane 3). The intensity of this band was also enhanced by treatment with TSA (lane 4). Taken together, these results strongly indicate that Cdt1 is an acetylated protein.
FIGURE 2.
Cdt1 is acetylated. A, overexpressed Cdt1 is acetylated. Plasmids expressing the indicated proteins were transfected into 293T cells. Lysates were immunoprecipitated (IP) with anti-Myc antibodies and blotted (IB) with anti-acetyllysine antibodies (AcK). The blot was then stripped and blotted with anti-Myc antibodies. The arrow indicates acetylated Cdt1, whereas the asterisk indicates an Ig heavy chain. B and C, endogenous Cdt1 is acetylated. HeLa cell extracts were immunoprecipitated with anti-Cdt1 antibodies or normal goat IgG and then blotted with anti-acetyllysine antibodies. The blot was then stripped and blotted with anti-Cdt1 antibodies. C, HeLa cell extracts were immunoprecipitated with anti-acetyllysine antibodies or normal rabbit IgG and then blotted with anti-Cdt1 antibodies. Input indicates a direct Western blot on the whole cell lysate, using 10% of the amount used for immunoprecipitation. The doublet in the input lanes likely reflects the differentially phosphorylated forms of Cdt1. Where indicated, cells were treated with 400 ng/ml TSA for 24 h.
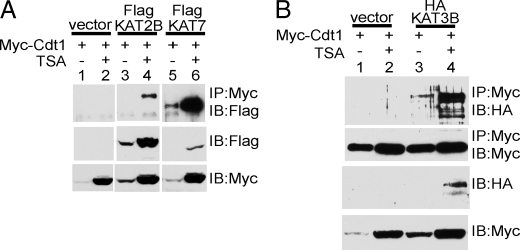
Lysine Acetyltransferases Associate with Cdt1—Having shown that Cdt1 is an acetylated protein and that it associates with HDAC11, we next determined whether it could associate with KATs. We examined several prototypic KATs from different families. This included KAT2B (PCAF), KAT3B (p300), and KAT7 (HBO1). KAT7 was of particular interest because it was identified as binding to the ORC (15), where Cdt1 also plays its critical role in the recruitment of MCM proteins. Myc-Cdt1 was cotransfected with each of the indicated tagged KATs, in the presence or absence of TSA. Cdt1 was immunoprecipitated with anti-Myc antibodies, and immunoprecipitated proteins were immunoblotted with anti-FLAG (Fig. 3A) or anti-HA (Fig. 3B). As shown in Fig. 3, each of the overexpressed KATs could be immunoprecipitated with Myc-Cdt1. In all cases, TSA treatment appeared to enhance the interaction; however, this was likely due to the fact that TSA treatment enhances the overall expression levels of both Cdt1 and KATs.
FIGURE 3.
Lysine acetyltransferases associate with Cdt1. A and B, expression plasmids encoding Myc-Cdt1 and the indicated tagged KATs were transfected into 293T cells. Where indicated, cells were treated with 400 ng/ml TSA for 16 h. Whole cell extracts were prepared and immunoprecipitated (IP) with anti-Myc antibodies. Western blots were performed using the indicated antibodies. IB, immunoblot.
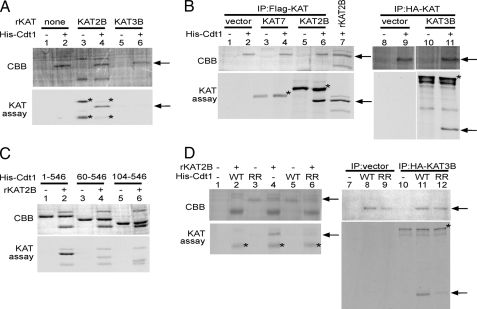
KAT2B and KAT3B Acetylate Cdt1—Having determined that all three tested KATs could interact with Cdt1, we next determined which KAT was responsible for acetylating Cdt1. To do this, we performed in vitro KAT assays, using bacterially purified His-tagged Cdt1 as the substrate. Recombinant KAT2B could acetylate Cdt1, but recombinant KAT3B could not (Fig. 4A, compare lanes 4 and 6). We next used immunoprecipitates of FLAG-KAT2B, FLAG-KAT7, or HA-KAT3B in the in vitro KAT assay to acetylate His-Cdt1. Surprisingly, FLAG-KAT2B (Fig. 4B, lane 6) and HA-KAT3B (lane 11), but not FLAG-KAT7 (lane 4), could acetylate Cdt1. Immunoprecipitates from cells transfected with vector alone also could not acetylate Cdt1 (lanes 2 and 9). This indicates that KAT2B and KAT3B acetylate Cdt1, but KAT7 does not, despite the localization of KAT7 to the ORC described previously (15, 16). The target of KAT7 at the ORC is likely histones. The discrepancy in the ability of the overexpressed KAT3B to acetylate Cdt1 (Fig. 4B, lane 11) versus the inability of the recombinant KAT3B (Fig. 4A, lane 6) to do so is likely due to the different portions of KAT3B expressed in each case. The noncatalytic domains of KAT3B may be required for protein-protein interaction or to affect secondary structure. Differing results dependent on the portion of KAT3B used have also been shown for the acetylation of MyoD (17). It is also formally a possibility that immunoprecipitation of KAT3B also brought down some KAT2B that is responsible for the acetylation we observe.
FIGURE 4.
KAT2B and KAT3B acetylate Cdt1 at the N terminus. Purified His-tagged recombinant Cdt1 and the indicated mutants were used as substrate in in vitro acetylation assays using recombinant KATs (A, C, and D), or tagged KATs expressed in 293T cells and immunoprecipitated (IP) with either anti-FLAG or anti-HA (B and D). Recombinant KAT2B is also included as a control in B (lane 7). The gels were stained with Coomassie Brilliant Blue (CBB) to verify loading. The arrows indicate the migration of Cdt1, and the asterisks indicate the autoacetylation of the KATs. WT, wild-type Cdt1; RR, K24R/K49R mutant.
Cdt1 Is Acetylated at the N Terminus—To delineate which of the 23 lysines in Cdt1 acetylation occurred, we performed in vitro KAT assays using Cdt1 deletions (Fig. 4C). Deletion of amino acids 1–59 causes a dramatic decrease in the amount of acetylation observed (compare lanes 2 and 4). Removal of an additional 44 amino acids did not have any further effect (lane 6). This suggests that the N terminus contains the major sites of acetylation. Two potential acetylated lysines reside within the first 59 amino acids: Lys24 and Lys49. Mutation of both lysines to arginines caused a significant decrease in acetylation observed in the in vitro KAT assay using either KAT2B (Fig. 4D, compare lanes 4 and 6) or KAT3B (compare lanes 11 and 12). It should be noted that both of these sites are neighbored by another basic residue three or four amino acids downstream of the acetylation site, which is typical of KAT3B substrates (18). KAT2B prefers a basic or aromatic residue three amino acids downstream of the acetylation site, which only Lys49 possesses (19).
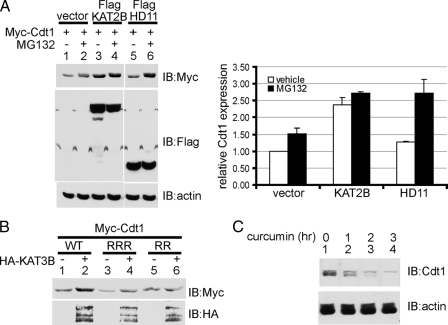
Acetylation Protects Cdt1 from Ubiquitylation and Subsequent Proteasomal Degradation—Cdt1 levels are regulated post-translationally by ubiquitylation and subsequent proteasome degradation (5). As such, treatment with the proteasome inhibitor MG132 would be predicted to increase Cdt1 protein levels. Indeed, addition of MG132 slightly increases the amount of Myc-Cdt1 protein in the absence of overexpressed acetyltransferase or deacetylase (Fig. 5A, compare lanes 1 and 2, and see quantification in right panel). Cotransfection of FLAG-KAT2B enhances the levels of Myc-Cdt1 (compare lanes 1 and 3, also compare lanes 1 and 3 in Fig. 3A). However, there is minimal further increase in Cdt1 levels upon the addition of MG132 (Fig. 5A, compare lanes 3 and 4). This indicates that Cdt1, acetylated by KAT2B, is already protected from proteasomal degradation. As such, addition of a proteasome inhibitor has little or no effect on Cdt1 protein levels. In contrast, treatment with MG132 enhances the level of Cdt1 considerably when coexpressed with HDAC11. This suggests that the deacetylated form of Cdt1 is prone to proteasomal degradation, and treatment with a proteasome inhibitor prevents degradation, leading to enhanced protein levels (lanes 5 and 6).
FIGURE 5.
Acetylation protects Cdt1 from proteasomal degradation. A, plasmids expressing Myc-tagged Cdt1 and the indicated FLAG-tagged proteins were transfected into 293T cells. Where indicated, 20 μm MG132 or an equivalent volume of Me2SO vehicle was added for the final 4 h of incubation. Whole cell extracts were prepared, and Western blotting (IB) was performed using the indicated antibodies to determine expression levels. Blots were stripped and reprobed with β-actin to ensure that MG132 did not affect overall protein levels. A representative blot is shown. Expression levels were quantified using ImageQuant and were shown normalized to the loading controlβ-actin (right panel). The data are the result of two independent experiments, shown with S.E. B, expression plasmids for Myc-Cdt1, or the indicated mutants, were transfected into 293T cells without or with a plasmid that expresses HA-KAT3B. All cells were treated with 400 ng/ml TSA. Whole cell extracts were prepared, and direct Western blots were performed to determine expression levels. WT, wild-type; RRR, K141R/K166R/K189R triple point mutant; RR, K24R/K49R double point mutant. C, inhibition of KAT activity diminishes endogenous Cdt1 levels. HeLa cells were treated with 100 μm curcumin for the indicated times. Whole cell lysates were examined for endogenous Cdt1 levels by Western blotting. β-Actin was used as the loading control.
Consistent with the idea that acetylation protects Cdt1 from degradation, overexpression of HA-KAT3B enhances wild-type Myc-Cdt1 levels (Fig. 5B, lanes 1 and 2). However, KAT3B does not affect the overall levels of the Myc-Cdt1(K24R/K49R) mutant (Myc-Cdt1RR, lanes 5 and 6), which showed greatly diminished acetylation in the in vitro KAT assay (Fig. 4D). To demonstrate specificity, we also mutated three irrelevant lysines (Lys141, Lys166, and Lys189) to arginines. Levels of this Myc-Cdt1RRR mutant were also enhanced upon cotransfection of HA-KAT3B (Fig. 5B, compare lanes 3 and 4). This supports the idea that acetylation at Lys24 and Lys49 can protect Cdt1 from proteasomal degradation.
To show that acetylation can protect endogenous Cdt1 from degradation, we made use of the KAT3 inhibitor curcumin (20). Treatment of HeLa cells with 100 μm curcumin diminished the amount of Cdt1 protein in a time-dependent manner, when compared with the untreated cells (Fig. 5C). Furthermore, treatment with TSA increases the amount of chromatin-bound Cdt1 (data not shown). Taken together, these data provide a compelling argument that acetylation of Cdt1 protects it from proteasomal degradation. Loss of acetylation, by pharmacologic inhibition of KAT or by overexpression of HDAC11, leads to loss of Cdt1 protein.
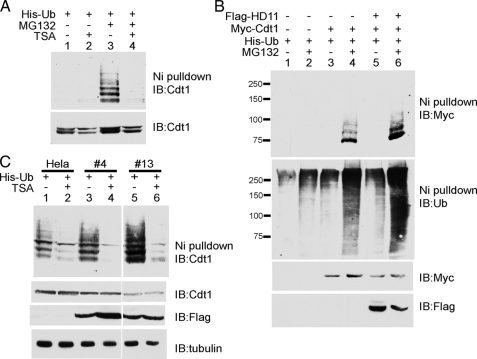
Acetylation Regulates Ubiquitylation of Cdt1—Having shown that acetylation modulates Cdt1 protein levels, we next examined its effect on ubiquitylation. To directly observe the effect of acetylation on ubiquitylation, we performed in vivo ubiquitylation assays by transfecting His-ubiquitin into HeLa cells and then performed nickel pulldowns under denaturing conditions to isolate the ubiquitylated proteins. This was followed by Western blotting, using antibodies against Cdt1 to identify the differently ubiquitylated forms of endogenous Cdt1. In the absence of MG132, ubiquitylated Cdt1 is not detected, likely because of the fact that it is rapidly degraded by the proteasome (Fig. 6A, lanes 1 and 2). However, in the presence of the proteasome inhibitor MG132, a ladder corresponding to the various polyubiquitylated forms of Cdt1 is clearly evident (lane 3). MG132 treatment correspondingly causes a small increase (1.5-fold) in the amount of endogenous Cdt1 as well (compare lanes 1 and 3, bottom panel). Treatment with TSA abrogates the level of endogenous Cdt1 ubiquitylation as demonstrated by decreased laddering (lane 4). When normalized for the amount of Cdt1 present, TSA caused a 91% reduction in laddering (compare lanes 3 and 4, upper panel).
FIGURE 6.
Acetylation regulates ubiquitylation of Cdt1. A, HDAC inhibition prevents endogenous Cdt1 ubiquitylation. HeLa cells were transfected with His-ubiquitin (Ub) and treated as indicated with 400 ng/ml TSA for 20 h, in the presence or absence of 20 μm MG132 for the final 4 h. Cells were harvested and lysed under denaturing conditions and then subjected to nickel (Ni) affinity chromatography. Samples were analyzed by Western blotting (IB) using anti-Cdt1 antibodies. A portion (20%) of the cells was set aside, and whole cell extracts were prepared to determine overall Cdt1 levels (lower panel). B, HDAC11 enhances Cdt1 ubiquitylation. 293T cells were transfected with plasmids expressing the indicated proteins. The cells were analyzed as described in A, using anti-Myc antibodies to detect the ubiquitylated Myc-Cdt1. The blot was stripped and then blotted with anti-ubiquitin. Whole cell extracts were blotted with anti-Myc and anti-FLAG to determine overall protein levels. C, stable overexpression of HDAC11 enhances endogenous Cdt1 ubiquitylation. Cells stably expressing FLAG-HDAC11 and parental HeLa cells were transfected with a plasmid expressing His-ubiquitin. Transfected cells were treated with 400 ng/ml TSA for 22 h where indicated, with 20 μm MG132 added for the final 4 h of incubation in all instances. Cells were then processed as described above. α-Tubulin was used as the control to ensure even loading of the whole cell extracts. The ability of TSA to enhance expression of FLAG-HDAC11 in clone 4, while having no effect on clone 13 is likely due to the site of DNA integration in the transfected cells.
To determine the specific effect of HDAC11 on Cdt1 ubiquitylation, we transfected 293T cells with Myc-Cdt1 in the presence or absence of FLAG-HDAC11. Transient overexpression of HDAC11 causes a 2-fold increase in the amount of ubiquitylated Myc-Cdt1 (Fig. 6B, compare lanes 4 and 6). In addition, the overall amount of polyubiquitylation as detected with an anti-ubiquitin antibody similarly increased. Correspondingly, overexpression of FLAG-HDAC11 also decreased by ∼50% the amount of Myc-Cdt1 detected in the whole cell extract (compare lanes 3 and 4 with lanes 5 and 6). These data support the idea that deacetylation of Cdt1 by HDAC11 leads to ubiquitylation and subsequent degradation.
To further explore the relationship between HDAC11 and Cdt1 ubiquitylation, we produced HeLa cell lines stably expressing FLAG-HDAC11. Consistent with the previous results, stable overexpression of HDAC11 enhanced endogenous Cdt1 ubiquitylation (Fig. 6C, compare lanes 3 and 5 with lane 1). This increase in ubiquitylation correlates with the amount of HDAC11 expressed. TSA treatment reduced the amount of laddering in all cases (lanes 2, 4, and 6). However, this reduction was more pronounced in the HDAC11-overexpressing cells, where laddering was reduced 27-fold for clone 4 and 19-fold for clone 13, as compared with the parental HeLa cells, where laddering was reduced ∼4-fold. Stable overexpression of HDAC11 also diminished the overall levels of Cdt1 in an HDAC11 dose-dependent manner, but TSA treatment did not appear to influence the HDAC11 effect on Cdt1 protein levels.
DISCUSSION
In this study, we have identified Cdt1 as an acetylated protein, adding another regulator of genomic integrity to the growing list of proteins modulated by acetylation and deacetylation. Similar to p53 (21, 22), we have found that acetylation protects Cdt1 from ubiquitylation and subsequent degradation. Both ubiquitylation and acetylation modify the ε-amino group of lysine residues. Although the sites of acetylation and ubiquitylation overlap in p53, this does not appear to be the case for Cdt1, as we have preliminary evidence that removal of the C-terminal 120 amino acids prevents ubiquitylation. Our findings apparently contradict a published report that Cdt1 can be N-terminally ubiquitylated in a lysine-independent manner (10). It should be noted, however, that Senga et al. (10) examined the degradation of the N-terminal 98 amino acids in which all lysines were mutated, but not the complete Cdt1 protein. In addition to Cdt1 and p53, acetylation appears to enhance the stability of multiple other proteins, including c-Myc, E2F1, and the androgen receptor, to name a few (23–25). This, then, raises the possibility that protein acetylation may be a common, although not universal, mechanism by which protein stability could be regulated.
The N terminus of Cdt1 clearly plays numerous regulatory roles. It regulates stability via two distinct regions (5) and also contains the nuclear localization signal (26). In addition, the N terminus plays a secondary, but critical, role in binding to the inhibitor geminin (2). We have shown that the N terminus of Cdt1 is dispensable for binding to HDAC11 and identified two acetylation sites (Lys24 and Lys49) in this region (Fig. 4). These lysines are conserved in humans, mice, and frogs, and both sites conform to the consensus for KAT3B substrates (18). Mutation of these sites has no effect on the ability of Cdt1 to bind to geminin (data not shown). Of note, K24 is in proximity to threonine 29. Phosphorylation of Thr29 is required for Cdt1 to bind to Skp2 (8). Conceivably, cross-talk may exist between acetylation and phosphorylation in Cdt1. It will be interesting to determine whether mutation of these lysine residues has any effect on binding to Skp2. Our preliminary evidence suggests that the K24R/K49R mutant is less stable over time. Without the protection that acetylation provides, the mutant is more likely to be degraded. This is consistent with our data showing that KAT3B cannot enhance the protein levels of the RR mutant.
Although we have shown that Lys24 and Lys49 are acetylated, it is also possible that Cdt1 has additional acetylation sites. In support of this idea is the fact that the 60–546 deletion mutant retained residual ability to be acetylated. Mass spectrometry to identify acetylation sites in Cdt1 has thus far been unsuccessful. It should be noted that due to sequence constraints, Lys24 was significantly under-represented in the peptides obtained. It is also highly likely, however, that acetylation occurs in a cell cycle-dependent manner. Our attempts at such analysis thus far have employed asynchronous cells overexpressing tagged Cdt1. Indeed, we have preliminary evidence that Cdt1 is acetylated 3 h past release from a nocodazole block, a time corresponding to the early G1 phase. Cdt1 stability is required at this time, which supports the idea that acetylation protects from degradation. Future experiments could examine Cdt1 acetylation in synchronized cells by mass spectrometry.
Of all human HDACs, HDAC11 is the least studied and is poorly characterized. We have recently found that HDAC11 regulates the expression of interleukin 10 and immune tolerance (27). Results from another recent study suggest that HDAC11 may be anti-proliferative. In the post-natal mouse brain, HDAC11 and the proliferation marker Ki67 are rarely observed co-localized in the same cell (28). Taken together with the fact that we obtained relatively few colonies when making FLAG-HDAC11 stable cell lines and that these cell lines grew slower than the parental cells, these data suggest that HDAC11 may diminish proliferation. The role of HDAC11 in the deacetylation and subsequent ubiquitylation of Cdt1 could begin to explain these observations.
In earlier studies, acetylation has been implicated in other aspects of DNA replication. For example, mutation of the HDAC Rpd3 promotes late replication origins to fire early in Drosophila follicle cells (29). Similarly, treatment of HeLa cells with TSA alters the choice of initiation sites as well as the timing of the onset of DNA synthesis (30). Although hyperacetylation of core histones is likely one of the major reasons for these observations, results from this current study open up the possibility that hyperacetylation of Cdt1, protecting it from degradation, might be an equally important contributory mechanism.
Acknowledgments
We thank Natalie Rezai-Zedah for technical assistance and A. Dutta (University of Virginia) for the Cdt1 cDNA. We acknowledge the support of the Molecular Biology and Molecular Imaging core facilities at Moffitt Research Institute. We also acknowledge the Microchemistry and Proteomics Analysis Facility at Harvard University for the mass spectrometry analysis.
This work was supported, in whole or in part, by National Institutes of Health Grant CA109699 (to E. S.). This work was also supported by American Heart Association Grant 0755298B and the Kaul Foundation (to E. S.).
Footnotes
The abbreviations used are: ORC, origin recognition complex; MCM, minichromosome maintenance; HDAC, histone deacetylase, KAT, lysine acetyltransferase; TSA, trichostatin A; GST, glutathione S-transferase; HA, hemagglutinin.
References
- 1.Machida, Y. J., Hamlin, J. L., and Dutta, A. (2005) Cell 123, 13-24 [DOI] [PubMed] [Google Scholar]
- 2.Saxena, S., Yuan, P., Dhar, S. K., Senga, T., Takeda, D., Robinson, H., Kornbluth, S., Swaminathan, K., and Dutta, A. (2004) Mol. Cell 15, 245-258 [DOI] [PubMed] [Google Scholar]
- 3.Ballabeni, A., Melixetian, M., Zamponi, R., Masiero, L., Marinoni, F., and Helin, K. (2004) EMBO J. 23, 3122-3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lutzmann, M., Maiorano, D., and Mechali, M. (2006) EMBO J. 25, 5764-5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishitani, H., Sugimoto, N., Roukos, V., Nakanishi, Y., Saijo, M., Obuse, C., Tsurimoto, T., Nakayama, K. I., Nakayama, K., Fujita, M., Lygerou, Z., and Nishimoto, T. (2006) EMBO J. 25, 1126-1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li, X., Zhao, Q., Liao, R., Sun, P., and Wu, X. (2003) J. Biol. Chem. 278, 30854-30858 [DOI] [PubMed] [Google Scholar]
- 7.Sugimoto, N., Tatsumi, Y., Tsurumi, T., Matsukage, A., Kiyono, T., Nishitani, H., and Fujita, M. (2004) J. Biol. Chem. 279, 19691-19697 [DOI] [PubMed] [Google Scholar]
- 8.Takeda, D. Y., Parvin, J. D., and Dutta, A. (2005) J. Biol. Chem. 280, 23416-23423 [DOI] [PubMed] [Google Scholar]
- 9.Arias, E. E., and Walter, J. C. (2006) Nat. Cell Biol. 8, 84-90 [DOI] [PubMed] [Google Scholar]
- 10.Senga, T., Sivaprasad, U., Zhu, W., Park, J. H., Arias, E. E., Walter, J. C., and Dutta, A. (2006) J. Biol. Chem. 281, 6246-6252 [DOI] [PubMed] [Google Scholar]
- 11.Glozak, M. A., Sengupta, N., Zhang, X., and Seto, E. (2005) Gene (Amst.) 363, 15-23 [DOI] [PubMed] [Google Scholar]
- 12.Gao, L., Cueto, M. A., Asselbergs, F., and Atadja, P. (2002) J. Biol. Chem. 277, 25748-25755 [DOI] [PubMed] [Google Scholar]
- 13.Gregoretti, I. V., Lee, Y. M., and Goodson, H. V. (2004) J. Mol. Biol. 338, 17-31 [DOI] [PubMed] [Google Scholar]
- 14.Wohlschlegel, J. A., Dwyer, B. T., Dhar, S. K., Cvetic, C., Walter, J. C., and Dutta, A. (2000) Science 290, 2309-2312 [DOI] [PubMed] [Google Scholar]
- 15.Iizuka, M., and Stillman, B. (1999) J. Biol. Chem. 274, 23027-23034 [DOI] [PubMed] [Google Scholar]
- 16.Miotto, B., and Struhl, K. (2008) Genes Dev. 22, 2633-2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polesskaya, A., and Harel-Bellan, A. (2001) J. Biol. Chem. 276, 44502-44503 [DOI] [PubMed] [Google Scholar]
- 18.Thompson, P. R., Kurooka, H., Nakatani, Y., and Cole, P. A. (2001) J. Biol. Chem. 276, 33721-33729 [DOI] [PubMed] [Google Scholar]
- 19.Zeng, L., Zhang, Q., Gerona-Navarro, G., Moshkina, N., and Zhou, M. M. (2008) Structure 16, 643-652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balasubramanyam, K., Varier, R. A., Altaf, M., Swaminathan, V., Siddappa, N. B., Ranga, U., and Kundu, T. K. (2004) J. Biol. Chem. 279, 51163-51171 [DOI] [PubMed] [Google Scholar]
- 21.Li, M., Luo, J., Brooks, C. L., and Gu, W. (2002) J. Biol. Chem. 277, 50607-50611 [DOI] [PubMed] [Google Scholar]
- 22.Tang, Y., Zhao, W., Chen, Y., Zhao, Y., and Gu, W. (2008) Cell 133, 612-626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel, J. H., Du, Y., Ard, P. G., Phillips, C., Carella, B., Chen, C. J., Rakowski, C., Chatterjee, C., Lieberman, P. M., Lane, W. S., Blobel, G. A., and McMahon, S. B. (2004) Mol. Cell. Biol. 24, 10826-10834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Balbas, M. A., Bauer, U. M., Nielsen, S. J., Brehm, A., and Kouzarides, T. (2000) EMBO J. 19, 662-671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaughan, L., Logan, I. R., Neal, D. E., and Robson, C. N. (2005) Nucleic Acids Res. 33, 13-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishitani, H., Lygerou, Z., and Nishimoto, T. (2004) J. Biol. Chem. 279, 30807-30816 [DOI] [PubMed] [Google Scholar]
- 27.Villagra, A., Cheng, F., Wang, H. W., Suarez, I., Glozak, M., Maurin, M., Nguyen, D., Wright, K. L., Atadja, P. W., Bhalla, K., Pinilla-Ibarz, J., Seto, E., and Sotomayor, E. M. (2009) Nat. Immunol. 10, 92-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, H., Hu, Q., Kaufman, A., D'Ercole, A. J., and Ye, P. (2008) J. Neurosci. Res. 86, 537-543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aggarwal, B. D., and Calvi, B. R. (2004) Nature 430, 372-376 [DOI] [PubMed] [Google Scholar]
- 30.Kemp, M. G., Ghosh, M., Liu, G., and Leffak, M. (2005) Nucleic Acids Res. 33, 325-336 [DOI] [PMC free article] [PubMed] [Google Scholar]