Abstract
Tumor necrosis factor-α (TNF-α), an inflammatory cytokine, has been shown to activate the small GTPase Rho, but the underlying signaling mechanisms remained undefined. This general problem is particularly important in the kidney, because TNF-α, a major mediator of kidney injury, is known to increase paracellular permeability in tubular epithelia. Here we aimed to determine the effect of TNF-α on the Rho pathway in tubular cells (LLC-PK1 and Madin-Darby canine kidney), define the upstream signaling, and investigate the role of the Rho pathway in the TNF-α-induced alterations of paracellular permeability. We show that TNF-α induced a rapid and sustained RhoA activation that led to stress fiber formation and Rho kinase-dependent myosin light chain (MLC) phosphorylation. To identify new regulators connecting the TNF receptor to Rho signaling, we applied an affinity precipitation assay with a Rho mutant (RhoG17A), which captures activated GDP-GTP exchange factors (GEFs). Mass spectrometry analysis of the RhoG17A-precipitated proteins identified GEF-H1 as a TNF-α-activated Rho GEF. Consistent with a central role of GEF-H1, its down-regulation by small interfering RNA prevented the activation of the Rho pathway. Moreover GEF-H1 and Rho activation are downstream of ERK signaling as the MEK1/2 inhibitor PD98059 mitigated TNF-α-induced activation of these proteins. Importantly TNF-α enhanced the ERK pathway-dependent phosphorylation of Thr-678 of GEF-H1 that was key for activation. Finally the TNF-α-induced paracellular permeability increase was absent in LLC-PK1 cells stably expressing a non-phosphorylatable, dominant negative MLC. In summary, we have identified the ERK/GEF-H1/Rho/Rho kinase/phospho-MLC pathway as the mechanism mediating TNF-α-induced elevation of tubular epithelial permeability, which in turn might contribute to kidney injury.
Tumor necrosis factor-α (TNF-α)2 is a pleiotropic proinflammatory cytokine that is synthesized as a membrane protein in response to inflammation, infection, and injury (1). Subsequently it is cleaved by the metalloprotease TNF-α convertase enzyme to release a 17-kDa soluble peptide (for a review, see Ref. 2). TNF-α has two receptors, the constitutively expressed, ubiquitous TNF receptor 1 and the inducible TNF receptor 2.
An increasing body of evidence supports a key role for TNF-α in both acute renal injury and chronic kidney diseases (for reviews, see Refs. 3 and 4). Although TNF-α is almost undetectable in normal kidneys, elevated intrarenal, serum, or urine concentrations have been reported in various pathological states including ischemia-reperfusion, endotoxinemia, and early diabetic nephropathy (5–8). Moreover kidney injury in various pathological states was prevented or mitigated by inhibition of TNF-α production, by addition of neutralizing antibodies, or in TNF receptor knock-out mice (for a review, see Ref. 3). The central role of TNF-α in mediating kidney injury is therefore well established. Importantly TNF-α can be produced in the kidney not only by infiltrating macrophages and lymphocytes but by resident cells including the tubular epithelium. For example, in reperfusion injury TNF-α expression precedes macrophage infiltration and localizes mostly to the tubules (3, 7). Tubular TNF-α production is also enhanced by endotoxin and hypoxia (9–12). Although effects of locally released TNF-α on the tubular epithelium could contribute to its deleterious actions, the underlying mechanisms have been incompletely explored.
Although a large number of studies have focused on the inflammatory and apoptotic signaling initiated by TNF-α in various cells, its cytoskeletal effects remain much less explored. In recent years Rho and its effector, Rho kinase (ROK), key regulators of both the actin cytoskeleton and myosin phosphorylation (13), have emerged as important mediators of TNF-α effects in endothelial cells (14–18). Similar effects in the tubular epithelium, however, have not been established. Even more importantly, the upstream signaling that connects the TNF receptor to activation of the Rho pathway remains completely unknown. Like other small GTPases, Rho cycles between an inactive (GDP-bound) and active (GTP-bound) form (13). The exchange of GDP to GTP during activation is stimulated by GDP-GTP exchange factors (GEFs). The diverse family of Rho GEFs contains >70 members in humans (19), making it challenging to identify the specific factors involved in mediating Rho activation through receptor-mediated stimuli. In the case of TNF-α, neither the particular Rho GEF involved nor the mechanism of its regulation has been identified in any of the cell systems studied.
A rise in epithelial paracellular permeability through the intercellular junctions is a prominent event during inflammation (“leaky epithelium”) (for reviews, see Refs. 20 and 21). In addition, the junctions maintain the polarized phenotype of epithelial cells that is necessary for directional transport processes and constitute an important signaling platform that transmits environmental cues to the cells. Therefore, the consequences of junction disruption during inflammation might go beyond the compromised barrier functions. Interestingly TNF-α has been reported to affect the permeability of the tubular epithelium. Mullin et al. (22) have reported that in a tubular cell line TNF-α induced a temporary elevation in transepithelial resistance followed by a drop in transepithelial resistance and increased paracellular permeability. The transepithelial resistance decrease was blocked by genistein, a general tyrosine kinase inhibitor; however, the exact mechanism underlying the observed permeability changes remained incompletely explored.
The actin cytoskeleton and especially phosphorylation of myosin light chain (MLC) was shown to be essential for the permeability increase caused by pathogens, cytokines, and growth factors in various epithelial and endothelial systems (for reviews, see Refs. 21, 23, and 24). Interestingly although myosin phosphorylation mediates the TNF-α-elicited permeability changes in intestinal cells (25, 26), phospho-MLC was reported not to be involved in the TNF-α-induced permeability rise in endothelial cells (17). The possible role of the Rho pathway and myosin phosphorylation in the TNF-α-induced permeability changes in the tubular epithelium therefore remains to be established.
The aim of this study was to explore the signaling pathways through which TNF-α causes cytoskeleton remodeling and elevates paracellular permeability in kidney tubular cells. Our findings show that TNF-α induces rapid activation of RhoA that leads to Rho/Rho kinase-dependent actin remodeling and myosin phosphorylation. Using an affinity precipitation assay followed by mass spectrometry, we identified GEF-H1 as a TNF-α-activated GEF. We showed that GEF-H1 mediates the TNF-α-induced stimulation of Rho and its effectors. In addition, activation of the GEF-H1/Rho pathway by TNF-α was downstream of ERK signaling and required GEF-H1 phosphorylation on Thr-678. Finally using a dominant negative MLC mutant, we showed that myosin phosphorylation is essential for the TNF-α-induced elevation in paracellular permeability.
EXPERIMENTAL PROCEDURES
Materials—Y-27632, PD98059, SB202474 (a negative control for PD98059), and U0126 were from EMD Biosciences (San Diego, CA). Human recombinant TNF-α and bovine serum albumin was from Sigma-Aldrich. The protease and phosphatase inhibitors used were: Complete Mini Protease Inhibitor tablet and PhosSTOP Phosphatase Inhibitor tablet (Roche Diagnostics), Protease Inhibitor Mixture (BD Pharmingen), and calyculin A (EMD Biosciences). Antibodies against the following proteins were used: RhoA, phospho-ERK1/2; phosphocofilin, cofilin, GEF-H1, and mono- and diphosphorylated MLC (Cell Signaling Technology, Danvers, MA); phosphothreonine (Invitrogen); HA (Covance, Emeryville, CA); Myc (9E10); ERK1/2, RhoA, NFκB p65, and green fluorescent protein (GFP) (Santa Cruz Biotechnology Inc., Santa Cruz, CA); β-actin and myosin light chain (Sigma-Aldrich); GFP (Nacalai USA, San Diego, CA); phospho (Thr)-MAPK/cyclin-dependent kinase substrate (Cell Signaling Technology); and glyceraldehyde-3-phosphate dehydrogenase (EMD Biosciences). For the detection of monophosphorylated MLC by Western blotting we used antibodies from Cell Signaling Technology (antimonophospho-MLC, serine 19) and Abcam (Cambridge, MA) (anti-phospho-MLC, Ser-20) with similar results. Rhodamine phalloidin was from Cytoskeleton Inc. (Denver, CO). Peroxidase, Cy3-, and FITC-labeled secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA). Anti-HA coupled to agarose was from Santa Cruz Biotechnology Inc.
Cell Treatment—LLC-PK1 (CL-101), a kidney proximal tubule epithelial cell line, and MDCK, a canine distal tubular epithelial cell line, obtained from the American Type Culture Collection (Manassas, VA) were maintained as in our previous studies (27). For all experiments, cells were grown to confluence. Treatment with 10 ng/ml TNF-α was performed in serum-free Dulbecco's modified Eagle's medium or Hanks' buffered salt solution. Where indicated, cells were preincubated with different inhibitors as described in the figure legends followed by the addition of TNF-α in the presence of the inhibitor.
Generation of a Polyclonal Stable AA-MLC Cell Line—The retroviral vector expressing a non-phosphorylatable, dominant negative version of MLC in which Thr-18 and Ser-19 were replaced with alanines (AA-MLC), was described previously (28). Viral particles were prepared, and LLC-PK1 cells were transduced as described previously (28). Control cells were transduced using the empty vector lacking the AA-MLC insert but harboring G418 resistance. Cells were selected using 1 mg/ml G418 starting at 48 h after transfection. AA-MLC expression was routinely checked by immunofluorescence using an anti-Myc antibody and was ≥80%.
Vectors and Transient Transfection—The vectors used were kind gifts from the following investigators: cDNAs encoding for C3 transferase (an enzyme that inactivates Rho via ADP-ribosylation (29)), the glutathione S-transferase-Rho binding domain (GST-RBD) portion of Rhotekin, and GST-RhoG17A (30) were from Dr. K. Burridge (University of North Carolina, Chapel Hill, NC); AA-MLC vector (28) was from Dr. H. Hosoya (Department of Biological Sciences, Hiroshima University); and HA-tagged GEF-H1 cDNA (31) was from Dr. G. Bokoch (Scripps Research Institute). The enhanced GFP vector, pEGFP, was from Clontech. The GFP-tagged wild type (WT) GEF-H1, the deletion mutant GEF-H1ΔC, and GEF-H1T678A were described previously (32). LLC-PK1 cells were transfected using the transfection reagent FuGENE 6 (Roche Applied Science) according to the manufacturer's instructions using 1 μg of DNA/well for 6-well plates and 5–10 μg of DNA for 10-cm dishes. For co-expression of C3 toxin and GFP, cells were co-transfected with cDNAs encoding for C3 transferase (1 μg) and GFP (0.2 μg). Transfection with GFP by itself had no effect on actin staining (not shown).
Obtaining a Partial Sequence of Porcine GEF-H1—To obtain a partial DNA sequence of porcine GEF-H1 for identification of short interfering RNA (siRNA) targets, LLC-PK1 cells were grown to confluence, and total cellular RNA was extracted using TRIzol (Invitrogen) according to the manufacturer's instructions. cDNA was synthesized from 5 μg of total RNA using SuperScript II reverse transcriptase (Invitrogen) and poly(A)+ oligo(dT)12–18 as a template primer. The cDNA was subsequently subjected to PCR using GEF-H1-specific primers that were designed for highly conserved regions identified by alignment of human, chimp, murine, and canine GEF-H1 cDNA sequences. The following primers were used: GEF-H1, 5′-GAT GAA GGA AGC CAA GGA TG-3′ and 5′-CCA GCA ATG TTG GTG GTA GA-3′. The PCR was performed using 2 μl of cDNA as described previously (28). The products were separated on a 0.8% agarose gel, and the amplified 389-bp product was purified from the gel using the PureLink™ Quick Gel Extraction kit (Invitrogen) and sent for sequencing.
Short Interfering RNA—The following sequences were targeted in porcine GEF-H1: siRNA 1, AAGCAGGGACTGCCGGAAGCT; siRNA 2, AACAAGAGCATCACAGCCAAG. The siRNAs were obtained from Applied Biosystems/Ambion Inc. (Austin, TX). Cells were transfected with 100 nm siRNA oligonucleotide using the Lipofectamine™ RNAiMAX Transfection Reagent (Invitrogen) according to the manufacturer's instructions. Control cells were transfected with 100 nm Silencer siRNA negative control 1 (non-related siRNA) (Ambion Inc.). Experiments were performed 48 h after transfection. The levels of GEF-H1 were routinely checked by Western blotting.
Western Blotting—Following treatment, cells were lysed on ice with cold lysis buffer (100 mm NaCl, 30 mm Hepes (pH 7.5), 20 mm NaF, 1 mm EGTA, 1% Triton X-100, 1 mm Na3VO4, and 1 mm phenylmethylsulfonyl fluoride supplemented with protease inhibitors). SDS-PAGE and Western blotting were performed as described previously (33). Blots were blocked in Tris-buffered saline containing either 5% bovine serum albumin or 5% milk and incubated with the corresponding primary antibody (overnight for antibodies from Cell Signaling Technology and 1 h at room temperature for other antibodies). Antibody binding was visualized with the corresponding peroxidase-conjugated secondary antibodies (1:5000 dilution) and the enhanced chemiluminescence method (kit from GE Healthcare). Equal loading was verified by reprobing blots using β-actin or glyceraldehyde-3-phosphate dehydrogenase antibodies. For the phosphospecific antibodies, loading was verified by reprobing with the corresponding total antibodies.
Preparation of GST-RBD and GST-RhoG17A Fusion Proteins—GST-RBD and GST-RhoG17A beads were prepared essentially as described previously (30, 34). Escherichia coli expressing a construct encoding for a GST fusion protein containing either the RBD (amino acids 7–89) of Rhotekin or GST-RhoG17A were induced with 0.1 mm isopropyl β-d-thiogalactoside for 16 h at room temperature. Next the bacteria were sedimented and resuspended in ice-cold lysis buffer containing 20 mm Hepes (pH 7.5), 150 mm NaCl, 5 mm MgCl2, 1% Triton X-100, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitors. The bacterial suspensions were sonicated on ice for 1 min and centrifuged at 13,500 rpm for 15 min (4 °C), and supernatants were added to pre-equilibrated glutathione-Sepharose beads (GE Healthcare). After rotating end over end for 45 min at 4 °C, the beads were washed twice in lysis buffer followed by two washes with Triton X-100-free lysis buffer. Bound protein was estimated by SDS-PAGE followed by Coomassie Blue staining, and the beads were kept at 4 °C for immediate use or stored frozen in the presence of glycerol.
Rho Activity Assay—The amount of active RhoA was determined using an affinity precipitation assay with the RBD of Rhotekin (GST-RBD) as in our earlier studies (27, 34). Briefly confluent LLC-PK1 cells grown in 6- or 10-cm dishes were treated as indicated in the respective figure legends. Cells were lysed with ice-cold lysis buffer containing 100 mm NaCl, 50 mm Tris base (pH 7.6), 20 mm NaF, 10 mm MgCl2, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 1 mm Na3VO4, and protease inhibitors. After centrifugation at 12,000 rpm (1 min at 4 °C), aliquots for determination of total RhoA were removed. The remaining supernatants were incubated at 4 °C for 45 min with 20–25 μg of GST-RBD beads followed by extensive washing. Total cell lysates and the pelleted beads were analyzed by Western blotting using RhoA antibody. Results were quantified by densitometry, and the active RhoA in each sample was normalized to the corresponding total RhoA. The data obtained in each experiment were expressed as -fold increase compared with the level of the control.
Affinity Precipitation of Activated GEFs and Mass Spectrometry—Active GEFs were affinity-precipitated from cell lysates using the RhoG17A mutant that cannot bind nucleotide and therefore has high affinity for GEFs (30). The precipitation was performed as described previously (30). For initial identification of activated GEFs, the precipitated proteins were separated by SDS-PAGE. The gel was stained with Coomassie Blue dye. Bands that appeared only in the precipitates obtained from TNF-α-activated cells (see Fig. 3) were excised from the gel and submitted for analysis to the Mass Spectrometry Facility of the Hospital for Sick Children, Toronto, Ontario, Canada. The peptide sequences were identified by screening against the canine genome.
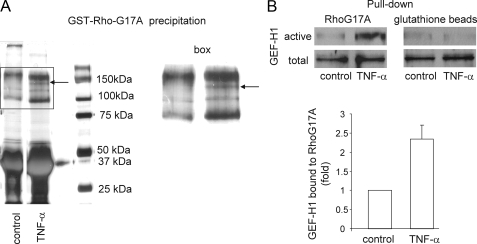
FIGURE 3.
Identification of GEF-H1 as a TNF-α-activated GEF. A, control or TNF-α-treated (10 ng/ml for 2 min) MDCK cells were lysed, and active GEFs were captured using GST-RhoG17A. The precipitated proteins were separated by SDS-PAGE. A typical gel stained with Coomassie Blue is shown. The two lanes were run on the same gel. Additional lanes running between them were removed from the image. The box, enlarged on the right, marks an area of the samples where TNF-α stimulation resulted in the appearance of well defined bands. The prominent band, indicated by the arrow, yielded peptide sequences corresponding to GEF-H1. B, LLC-PK1 cells were treated with 10 ng/ml TNF-α, lysed, and subjected to precipitation with either GST-RhoG17A (left) or control glutathione-Sepharose beads (right). GEF-H1 in the precipitates (active) and the cell lysates (total) was detected by Western blotting. The blots were analyzed by densitometry, and for each experiment the changes were expressed as -fold increase from the control. The graph shows mean ± S.E. (n = 11 independent experiments).
For detection of activated GEF-H1, affinity precipitation with GST-RhoG17A was followed by Western blotting using a GEF-H1-specific antibody or anti-GFP to detect the presence of the endogenous or GFP-GEF-H1 proteins, respectively. Precipitation with glutathione-Sepharose beads containing no fusion proteins resulted in no GEF-H1 precipitation (see Fig. 3B). GEF-H1 in total cell lysates was also detected for each sample (total GEF-H1). Precipitated (active) and total GEF-H1 was quantified by densitometry. The amount of active GEF-H1 in each sample was normalized to the corresponding total GEF-H1, and the data obtained in each experiment were expressed as -fold increase compared with the control taken as unity.
Immunofluorescence Microscopy—Confluent cells grown on coverslips were treated as indicated in the corresponding figure legends and fixed with 4% paraformaldehyde. Immunofluorescent staining was carried out as described previously (27). Briefly following permeabilization with Triton X-100, the coverslips were blocked with 3% bovine serum albumin in phosphate-buffered saline. Next cells were incubated with primary antibody (1:100). Bound antibody was detected using the corresponding fluorescent secondary antibody (1:1000). For visualizing actin following treatment, cells were fixed, permeabilized, and incubated with rhodamine-phalloidin for 1 h. All samples were viewed using an Olympus IX81 microscope (Melville, NY) coupled to an Evolution QEi Monochrome camera (Media Cybernetics, Bethesda, MD). Where indicated, out of focus fluorescent signal was removed by deconvolution using the Image Pro software.
Immunoprecipitation and Identification of the Threonine Phosphorylation Site by Mass Spectrometry—LLC-PK1 cells in 10-cm dishes were transfected with HA-GEF-H1 or GFP-GEF-H1 constructs as indicated in the figure legends. 24–48 h later the cells were treated and lysed with ice-cold RIPA buffer (50 mm Tris (pH 7.5), 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, and 0.1% SDS) supplemented with 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, 10 nm calyculin A, and Complete Mini Protease Inhibitor, and PhosSTOP Phosphatase Inhibitor tablets. Lysates were centrifuged at 12,000 rpm for 5 min. Samples from the supernatants (total cell lysate) were removed, and the remaining supernatants were precleared with protein G-Sepharose (Pierce) for 30 min. After removal of the beads by centrifugation, the lysates were incubated with 1 μg/ml anti-HA or anti-GFP antibody for 1 h at 4 °C (for HA) or overnight (GFP) followed by incubation with protein G-Sepharose (25 μl/sample) for 1 h. After washing with RIPA buffer containing 1 mm Na3VO4, the immunoprecipitated proteins were eluted from the beads by boiling for 5 min in Laemmli sample buffer. Samples were subjected to Western blot analysis, and membranes were probed with anti-phosphothreonine followed by stripping and reprobing with anti-HA or anti-GFP. Control experiments in which either the HA or GFP antibody was omitted during the immunoprecipitation or lysates from non-transfected cells were used verified the specificity of the immunoprecipitation (not shown). Blots were analyzed by densitometry, and the amount of phosphothreonine in each sample was normalized to the amount of the corresponding precipitated HA-GEF-H1 or GFP-GEF-H1 and expressed as -fold increase from the control.
To analyze phosphorylation site(s) in GEF-H1, LLC-PK1 cells transfected with HA-tagged GEF-H1 were pretreated for 30 min with 10 nm calyculin A and then treated with TNF-α for 2 min. Following lysis as above, samples from eight 10-cm dishes were pooled, and HA-GEF-H1 was precipitated using agarose-coupled anti-HA. The precipitated proteins were separated by SDS-PAGE. The gel was stained with Coomassie Blue dye, and the dominant band appearing around 130 kDa was excised from the gel and submitted for analysis to the Mass Spectrometry Facility of the Hospital for Sick Children. Aliquots of the samples were analyzed by Western blotting to verify low basal and increased TNF-α-induced threonine phosphorylation.
GEF Activity Assay—HA-GEF-H1 was precipitated from control or TNF-α-treated cells as described above using anti-HA-agarose. Protein was eluted from the beads using a buffer containing 500 mm NaCl and 0.025% SDS and quantitated, and aliquots were assessed by Western blotting to verify specific elution of GEF-H1. Remaining eluates were used to measure GEF activity with the RhoGEF Exchange Assay Biochem kit from Cytoskeleton Inc. Fluorescence was followed using a Fluoroskan Ascent FL (Thermo Scientific, Waltham, MA) plate reader.
Transepithelial Permeability Measurements—Control or AA-MLC-expressing LLC-PK1 cells were grown on Transwell filters (Costar; pore size, 0.4 μm) for 1 day after becoming confluent. The medium in both the top and bottom compartments was replaced with Hanks' buffered salt solution. Next the cells were exposed to 2 mg/ml FITC-labeled dextran (4 kDa; Sigma-Aldrich) added to the top compartment in the absence or presence of 10 ng/ml TNF-α. TNF-α was also added to the bottom compartment. 50-μl samples were taken from the bottom compartment at 30-min intervals, and the fluorescence was determined by a fluorescence microplate reader (Fluoroskan Ascent FL) using 480 and 518 nm as the excitation and emission wavelengths, respectively. To determine the size selectivity of the transepithelial transport, a characteristic feature of the paracellular transport pathway, similar experiments were carried out using a 70-kDa FITC-labeled dextran (Sigma-Aldrich). This large molecular mass dextran showed only negligible permeability that was unchanged by TNF-α treatment (not shown).
Protein Assay—Protein concentration was determined by the bicinchoninic acid assay (Pierce Biotechnology) with bovine serum albumin used as a standard.
Densitometry—Films with non-saturated exposures were scanned, and densitometry analysis was performed using the Image J software or a GS-800 calibrated densitometer and the “band analysis” option of the Quantity One software (Bio-Rad) (33).
Statistical Analysis—All blots shown and immunofluorescent pictures are representatives of at least three similar experiments. Data are presented as mean ± S.E. of the number of experiments indicated (n). Statistical significance was assessed by analysis of variance using the GraphPad InStat software.
RESULTS
TNF-α Activates RhoA in Kidney Tubular Epithelium—To assess whether TNF-α alters the cytoskeleton of tubular epithelium, LLC-PK1 proximal tubular cells were treated with 10 ng/ml TNF-α for 2 min, and F-actin was visualized using rhodamine-labeled phalloidin. As shown in Fig. 1A, top pictures, untreated epithelial cells in confluent monolayers had few stress fibers and a fine, even actin ring in the junctional area. In contrast, in TNF-α-treated cells F-actin was organized in numerous, well detectable stress fibers. The overall staining with phalloidin in these cells was also brighter, indicating elevated F-actin levels. Because such changes can be due to the activation of the Rho pathway, we next measured the levels of active RhoA. Control or TNF-α-treated cells were lysed, and active Rho was captured using GST-RBD that binds only GTP-bound Rho. As shown in Fig. 1B, the amount of the precipitated active RhoA rapidly increased when cells were stimulated with TNF-α. A substantial (on average 4-fold) activation of RhoA was detected as early as 1 min. In addition, RhoA activation was sustained as elevated levels of active RhoA were detected after 2 h of TNF-α treatment (Fig. 1B). To prove that the altered F-actin staining observed in TNF-α-treated cells was indeed due to Rho activation, we transfected cells with the Rho-inactivating C3 transferase toxin along with GFP to enable detection of cells expressing the untagged toxin. As expected, consistent with inactivation of the Rho pathway, the C3 toxin-expressing cells were larger and showed a more spread out morphology with diminished F-actin staining and no stress fibers (Fig. 1C, top row). Importantly the TNF-α-induced increase in phalloidin staining and stress fibers was prevented by C3 toxin (Fig. 1C, lower row, compare the C3 toxin-expressing cells and surrounding untransfected cells). However, long term inactivation of Rho by the expression of C3 toxin eliminated the basal stress fibers as well. To further test the role of the Rho pathway in the TNF-α-induced increase of stress fibers, we used the Rho kinase inhibitor Y-27632. Short incubation (20 min) of LLC-PK1 cells with 1 μm Y-27632 decreased but did not eliminate the basal stress fibers (Fig. 1A, lower row). However, in the presence of the drug, the TNF-α-induced increase in the stress fibers was largely prevented (Fig. 1A, lower row). These results point to the importance of the Rho/Rho kinase pathway in enhancing stress fibers following TNF-α stimulation. RhoA was also rapidly activated by TNF-α in the distal tubular epithelial cell line MDCK (Fig. 1D), showing that the Rho-activating effect of TNF-α is a general phenomenon and is not restricted to the proximal tubular cell line.
FIGURE 1.
TNF-α induces Rho-mediated cytoskeletal remodeling in tubular epithelium. A, LLC-PK1 cells, grown on coverslips, were pretreated with 1 μm Y-27632 for 20 min as indicated followed by addition of 10 ng/ml TNF-α for 2 min. The cells were fixed and permeabilized, and F-actin was visualized using rhodamine-labeled phalloidin. The images were enhanced by deconvolution to remove out of focus fluorescence. B and D, confluent LLC-PK1 (B) or MDCK (D) cells grown on 10-cm dishes were treated with 10 ng/ml TNF-α for the indicated times (B) or 2 min (D). The amount of active RhoA was determined using a GST-RBD precipitation assay. RhoA in the precipitates (active RhoA) and total cell lysates (total RhoA) was detected by Western blotting using RhoA antibody. The blots were quantified by densitometry, and the results were expressed as the -fold increase compared with the values obtained in control cells. The graph in B shows mean ± S.E. from n = 3 independent experiments. C, LLC-PK1 cells grown on coverslips were cotransfected with vectors for C3 transferase and GFP. After 48 h, the cells were treated with TNF-α for 2 min. The cells were fixed, and F-actin was visualized as in A. F-actin staining (left panels) and GFP fluorescence (right panels) in the same field are shown. con, control.
TNF-α Elevates Myosin Light Chain and Cofilin Phosphorylation through Rho Kinase—We have previously shown that the Rho/ROK pathway is the major regulator of MLC phosphorylation (cell contractility) in kidney tubular cells (27). Therefore, we next examined whether MLC phosphorylation was also altered by TNF-α in LLC-PK1 cells. Cells were treated for various times with TNF-α, and the levels of phospho-MLC were detected by Western blotting using an antibody against monophosphorylated MLC (pMLC). As shown in Fig. 2A, TNF-α induced a rapid and sustained increase in phospho-MLC. Moreover the kinetics of the appearance of pMLC correlated well with Rho activation (see Fig. 1B). Developing these blots with a diphospho-MLC-specific antibody revealed that the diphosphorylated form of MLC was also increased (not shown). We next examined the subcellular distribution of phosphorylated MLC by immunofluorescence. In resting cells the majority of pMLC was present in the cytosol with some fine staining localized at the cell periphery (Fig. 2B). Importantly TNF-α-stimulated cells showed stronger overall pMLC staining, which was organized in stress fibers and also appeared as enhanced peripheral labeling. The TNF-α-elicited elevation in MLC phosphorylation was mediated by ROK as the ROK inhibitor Y-27632 completely eliminated this response (Fig. 2C). Similar to Rho activation, the TNF-α-induced effects on MLC are not restricted to the proximal tubules as TNF-α also induced ROK-mediated elevation of pMLC in MDCK cells (not shown).
FIGURE 2.
TNF-α induces ROK-dependent MLC and cofilin phosphorylation in tubular epithelium. A, C, and D, confluent LLC-PK1 cells were treated with 10 ng/ml TNF-α for the indicated times. Cells were lysed, and phospho-MLC (A and C) or phosphocofilin (p-Cofilin) (D) was detected by Western blotting. Where indicated, cells were pretreated with 20 μm Y-27632 for 30 min. The inhibitor was present throughout the TNF-α treatment. The blots were redeveloped with β-actin, MLC (A and C), or cofilin (D) antibodies to demonstrate equal loading. Note that although the phospho-MLC antibody recognizes two isoform of MLC the total MLC antibody reacts with only one band. B, cells grown on coverslips were exposed to 10 ng/ml TNF-α for 2 min, fixed, and stained using the pMLC-specific antibody. The images were deconvolved as in Fig. 1A. con, control.
Phosphorylation of cofilin, an actin-binding and F-actin-severing protein, by LIM kinase inhibits its actin depolymerizing activity, thus promoting F-actin accumulation (35). LIM kinase can be downstream of Rho kinase (36). Modification of cofilin by TNF-α, however, has not yet been investigated. Therefore we next asked whether TNF-α also enhances cofilin phosphorylation. LLC-PK1 cells were treated with TNF-α, and the level of cofilin phosphorylation was determined using a phosphocofilin-specific antibody. As shown in Fig. 2D TNF-α elevated the phosphorylation of cofilin. Moreover Y-27632 completely abolished this reaction, showing that it is indeed downstream of ROK.
In summary, our data show that TNF-α induces a major increase and rearrangement of F-actin. This effect is likely due to Rho-ROK-mediated enhanced phosphorylation of MLC and cofilin.
Identification of GEF-H1 as a TNF-α-regulated Rho Exchange Factor—Having shown that TNF-α activates RhoA, our next aim was to gain insight into the mechanisms involved. To this end, we first wished to identify the GEF(s) that is activated by TNF-α. The large number of potential GEFs renders the identification of the particular GEF involved in a specific pathway tedious (19). To overcome this problem, we utilized an unbiased approach in which we captured activated GEF(s) using an affinity precipitation assay (30, 37, 38) and identified candidate GEFs using mass spectrometry. The GEF pull-down assay takes advantage of the Rho mutant RhoG17A. This mutant exerts high affinity toward activated GEFs as it binds neither GTP nor GDP and therefore mimics the nucleotide-free, intermediate state of Rho bound to the GEFs. Indeed RhoG17A has been shown to bind to GEF-H1 (39). A GST fusion protein of RhoG17A was used as a bait to capture activated GEFs from lysates of control and TNF-α-stimulated MDCK cells. We used MDCK cells for these experiments as the canine genome has been fully sequenced, thus allowing a data base search and easier identification of the proteins. Precipitated proteins were separated by SDS-PAGE and stained with Coomassie Blue dye. As shown in Fig. 3A, a small number of bands increased or became visible after TNF-α treatment. Bands that were present only in the TNF-α-treated samples were excised and submitted for mass spectrometry analysis. The band (indicated by the arrow in Fig. 3A) yielded peptides whose sequence corresponded to GEF-H1.
Having identified GEF-H1 as a candidate, we next used the RhoG17A affinity precipitation assay to verify and follow its activation by TNF-α in LLC-PK1 cells. GEFs from cell lysates were precipitated using GST-RhoG17A as described above, and the presence of GEF-H1 was detected by Western blotting. GST-RhoG17A showed increased association with GEF-H1 in the lysates obtained from TNF-α-stimulated cells, indicating activation of GEF-H1 (Fig. 3B, left). The right blot in Fig. 3B verifies that the glutathione beads alone failed to precipitate GEF-H1 from either untreated or TNF-α-treated cells. In 11 similar experiments, we found a consistent increase in GEF-H1 precipitated with GST-RhoG17A following TNF-α stimulation. The activation range was between 1.5- and 10-fold, averaging 2.3-fold (see Fig. 3B, graph). Taken together, our data show that TNF-α induces a rapid increase in the amount of GEF-H1 capable of associating with nucleotide-free Rho, probably reflecting activation of GEF-H1.
GEF-H1 Mediates TNF-α-induced Activation of the Rho Pathway—Having shown that TNF-α increases the association of GEF-H1 with nucleotide-free Rho, suggesting its activation, we next asked whether this exchange factor mediates TNF-α-induced Rho activation. To design siRNAs to down-regulate the protein, we first obtained a partial sequence of the porcine GEF-H1. We used primers against highly homologous regions to amplify a portion of the porcine GEF-H1 cDNA obtained from LLC-PK1 cells (see “Experimental Procedures”). As shown in Fig. 4A, both siRNAs used (designated as siRNA 1 and 2) induced a large (up to 80%) decrease in the levels of GEF-H1 with siRNA 2 resulting in a more potent and consistent down-regulation. We first investigated the effect of GEF-H1 down-regulation on the TNF-α-induced Rho activation. Cells were transfected with a nonrelated (NR) siRNA or one of the specific GEF-H1 siRNAs. As shown in Fig. 4, B and C, down-regulation of GEF-H1 resulted in the elimination of TNF-α-induced Rho activation: whereas in cells transfected with the NR siRNA TNF-α induced a 6-fold RhoA activation, TNF-α failed to increase RhoA activity in the absence of GEF-H1. Moreover there was no difference between siRNA 1 or 2, arguing against an off-target effect of the siRNA. To further substantiate the central role of GEF-H1 in mediating TNF-α-induced cytoskeleton rearrangement, we studied how changes in F-actin were affected by GEF-H1 down-regulation. As shown in Fig. 4D, the TNF-α-induced increase in F-actin staining and stress fiber abundance was readily recapitulated in cells transfected with the non-related siRNA. Down-regulation of GEF-H1 did not alter the basal F-actin staining and stress fiber structure. Importantly the TNF-α-elicited cytoskeleton remodeling and F-actin increase was strongly mitigated following down-regulation of GEF-H1 (Fig. 4D, compare second and fourth pictures) consistent with the failure of TNF-α to stimulate RhoA under such conditions. In addition, TNF-α also failed to enhance MLC phosphorylation in cells lacking GEF-H1 (Fig. 4E). GEF-H1 down-regulation, however, does not exert a global inhibitory effect on TNF-α signaling in tubular cells. Activation of the inflammatory transcription factor NFκB was largely unaffected by down-regulation of GEF-H1 as shown by similar TNF-α-induced nuclear translocation of p65 in the NR and GEF-H1 siRNA-treated cells (Fig. 4F). Taken together, these data show that activation of the Rho pathway by TNF-α and the downstream cytoskeletal effects are mediated by GEF-H1.
FIGURE 4.
GEF-H1 down-regulation prevents TNF-α-induced activation of the Rho pathway. A, LLC-PK1 cells were transfected with NR siRNA or two different siRNAs designed against porcine GEF-H1 (GEF-H1 number 1 and GEF-H1 number 2). 48 h after transfection cells were lysed, and GEF-H1 in the cell lysates was detected by Western blotting. The same blot was redeveloped with an antibody against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control. B–E, cells were transfected with NR or GEF-H1-specific siRNA 1 (B) or2(C–E). 48 h later the cells were treated with TNF-α for 2 min as indicated. Active RhoA (B and C), F-actin (D), and pMLC (E) were detected as in Fig. 1. In B, C, and E the blots with the total cell lysates were stripped and redeveloped with a GEF-H1 antibody. Glyceraldehyde-3-phosphate dehydrogenase served as loading control. The graphs in C and E show mean ± S.E. from at least three independent experiments. F, GEF-H1 down-regulation does not affect activation of NFκB. Cells grown on coverslips and transfected with NR or GEF-H1-specific siRNA 2 were left untreated or exposed to TNF-α for 30 min. The cells were fixed and stained with an antibody against NFκB p65. Note that TNF-α-induced p65 translocation to the nucleus following GEF-H1 down-regulation is unaltered, showing that Rho-independent TNF-α signaling is intact. con, control.
TNF-α Induces ERK-dependent Phosphorylation of GEF-H1—Our next aim was to gain insight into the mechanism whereby TNF-α induces GEF-H1 activation. Phosphorylation is an important regulator of GEF-H1 (32, 40–42). ERK1/2 has recently been shown to phosphorylate GEF-H1 on a threonine site and thereby enhance its activity toward Rho (32). Based on this, we next investigated whether the ERK pathway has a role in the TNF-α-induced GEF-H1 and Rho activation. Similar to other cells including fibroblasts (43), TNF-α induced strong ERK1 and -2 activation in LLC-PK1 cells (Fig. 5A). Moreover the rapid appearance of active ERK corresponded well with the kinetics of GEF-H1 and Rho pathway activation. To investigate the effect of TNF-α on GEF-H1 phosphorylation, we transfected LLC-PK1 cells with a construct encoding HA-tagged GEF-H1 and immunoprecipitated the molecule through the HA tag. Precipitated HA-GEF-H1 from control and TNF-α-treated cells was analyzed by Western blotting. As shown in Fig. 5B, probing the blots with an antibody against phosphothreonine revealed that GEF-H1 was rapidly phosphorylated in TNF-α-stimulated cells. Next we tested the effect of the MEK1/2 inhibitor PD98059, which efficiently eliminated ERK1/2 activation (Fig. 5A). When cells were treated with PD98059 alone, the basal level of GEF-H1 threonine phosphorylation was usually found to be slightly elevated. This elevation, however, was smaller than the level of the phosphorylation induced by TNF-α. Importantly PD98059 treatment completely prevented the TNF-α-induced rise in GEF-H1 threonine phosphorylation (Fig. 5B). This suggests that TNF-α is unable to enhance GEF-H1 phosphorylation in the absence of a functional ERK pathway.
FIGURE 5.
TNF-α induced phosphorylation of Thr-678 in GEF-H1. A, LLC-PK1 cells were treated with 10 μm PD98059 for 15 min as indicated followed by the addition of TNF-α for the indicated times. The cells were lysed, and phospho-ERK1/2 (pERK) was detected. B, cells were transfected with cDNA encoding for HA-tagged GEF-H1. 24 h later, the cells were left untreated or exposed to TNF-α for 2 min. Where indicated, cells were treated with 10 μm PD98059 for 15 min followed by additions of TNF-α in the presence of the drug. The cells were then lysed, and GEF-H1 was precipitated using an anti-HA antibody. The precipitates were subjected to Western blotting with anti-phosphothreonine (p-Thr). The same blot was stripped and reprobed with anti-HA to demonstrate equal precipitation of HA-GEF-H1. The graph summarizes the densitometric analysis of n = 3 independent experiments (mean ± S.E.). C, cells were transfected with cDNA encoding for GFP-tagged GEF-H1 WT, C terminus-deleted (ΔC), or T678A mutant. 48 h later, the cells were left untreated or exposed to TNF-α for 2 min. The cells were then lysed, and GEF-H1 was precipitated using an anti-GFP antibody. The precipitates were subjected to Western blotting with anti-phosphothreonine. The same blot was stripped and reprobed with anti-GFP to demonstrate equal precipitation of the proteins. The arrowhead points to the level of the WT and T678 GEF-H1, and the arrow shows the level of GEF-H1ΔC. The graph summarizes the densitometric analysis of n = 3 independent experiments analyzing threonine phosphorylation of the WT GEF-H1 (mean ± S.E.). D, cells were transfected with cDNA encoding for GFP-tagged GEF-H1 T678A mutant. The cells were treated with 10 μm PD98059 (PD) for 15 min followed by precipitation using anti-GFP and detection of phosphothreonine as in C. The graph summarizes the densitometric analysis of n = 3 independent experiments (mean ± S.E.). WB, Western blot; IP, immunoprecipitation; con, control.
To identify the threonine site(s) that are phosphorylated, we immunoprecipitated HA-tagged GEF-H1 from TNF-α-treated cells and had the samples analyzed by mass spectrometry. Ingel digestion of the sample resulted in recovery of 63% of the full-length GEF-H1 sequence. Importantly analysis of the sequences of the peptides revealed that Thr-678, the site shown to be a target of ERK1/2 (32), was phosphorylated in the TNF-α-treated sample. Based on these results, we next analyzed TNF-α-induced threonine phosphorylation of GFP-tagged mutant GEF-H1 proteins lacking Thr-678. Cells were transfected with either WT GEF-H1 or a deletion mutant lacking most potential phosphorylation sites (ΔC) or a point mutant lacking threonine 678 (T678A) (32). These proteins were then precipitated from control or TNF-α-treated cells and analyzed using Western blotting. Fig. 5C shows that TNF-α induced a marked increase in threonine phosphorylation in WT GEF-H1. The increased GEF-H1 phosphorylation was also detectable with a phospho (Thr)-MAPK/cyclin-dependent kinase substrate antibody, further suggesting a role for ERK (not shown). In contrast, TNF-α-induced threonine phosphorylation was undetectable in GEF-H1ΔC or T678A (Fig. 5C). These results clearly show that Thr-678 is a target of TNF-α-induced phosphorylation.
To gain further insight into the unexpected elevation in GEF-H1 threonine phosphorylation induced by PD98059, we examined whether a similar phenomenon could be detected in the case of the T678A mutant. GEF-H1T678A was precipitated from control or PD98059-treated cells. As shown in Fig. 5D, the drug caused an ≈2-fold increase in the threonine phosphorylation of GEF-H1T678A that is similar to the level observed in HA-GEF-H1 (Fig. 5B) or GFP-GEFH1 WT (Fig. 5D). These data suggest that inhibition of basal MEK1/2-ERK activity enhances the phosphorylation of GEF-H1 on a site other than the TNF-α-regulated Thr-678.
GEF-H1 Activation Requires the ERK Pathway—Having shown that TNF-α induces ERK pathway-dependent phosphorylation of GEF-H1, we next tested whether activation of the GEF-H1/Rho pathway is dependent on the ERK pathway. First we performed GST-RhoG17A precipitation experiments using PD98059. As shown earlier, TNF-α treatment increased the association of GEF-H1 to Rho (Fig. 6A). Importantly PD98059 completely prevented the TNF-α-induced elevation in active GEF-H1 levels (Fig. 6A, compare the second and fourth columns). These data imply that GEF-H1 activation is induced in an ERK pathway-dependent manner. We also tested the effect of eliminating the Thr-678 phosphorylation site on activation. Cells were transfected with WT or T678A GEF-H1, and association of the GFP-tagged proteins with RhoG17A was followed. As shown in Fig. 6B, the WT protein showed a TNF-α-induced increase in association with RhoG17A that was similar to that of the endogenous protein (see Fig. 6A). Association of the T678A mutant with RhoG17A in unstimulated cells was similar to that of the WT protein; however, the TNF-α-induced rise was substantially reduced. These data suggest a key role for TNF-α-induced, MEK1/2-ERK-dependent phosphorylation of Thr-678 in the enhanced GEF-H1 association to RhoAG17A.
FIGURE 6.
Role of the ERK pathway in activation of the GEF-H1/Rho pathway. In A, C, and D, LLC-PK1 cells were treated with 10 μm PD98059 for 15 min as indicated followed by the addition of TNF-α for 2 min. B, cells were transfected with GFP-tagged WT or T678A GEF-H1. In both A and B, active GEF-H1 was precipitated as described earlier, and samples were analyzed by Western blot using either GEF-H1 or GFP antibody. C, Active Rho was precipitated as in Fig. 1. D, cell lysates were probed for pMLC. The graphs summarize the densitometric analysis of n = 5(A), n = 4(B), n = 7(C), or n = 3(D) independent experiments (mean ± S.E.). con, control; WB, Western blot.
Next we investigated the ERK pathway dependence of the TNF-α-induced stimulation of the Rho pathway. As shown in Fig. 6C, addition of PD98059 alone caused a rise in basal RhoA activity. Importantly similar to GEF-H1 activity, TNF-α-induced RhoA activation was prevented by PD98059 (Fig. 6C). We also explored the effect of PD98059 on TNF-α-induced MLC and cofilin phosphorylation. Fig. 6D shows that TNF-α failed to increase pMLC levels in the presence of PD98059. Cofilin phosphorylation exhibited a similar pattern (not shown), further substantiating the absence of TNF-α-elicited activation of the Rho pathway when the ERK pathway was inhibited. To rule out any potential nonspecific effects of PD98059, we also used an unrelated MEK1/2 inhibitor, U126, that also prevented TNF-α-induced Rho activation and MLC phosphorylation. In contrast, SB202474 (a negative control for PD98059) failed to affect basal or TNF-α-induced MLC phosphorylation (not shown).
Taken together, these data show that the TNF-α-induced GEF-H1 phosphorylation and GEF-H1/Rho pathway activation require MEK1/2-ERK activity. Interestingly inhibition of MEK1/2 alone appeared to elevate activity of the GEF-H1/Rho pathway (see “Discussion”).
Myosin Phosphorylation Is Essential for TNF-α-induced Elevation of Paracellular Permeability—Having shown that in the tubular epithelium TNF-α induces MLC phosphorylation through the GEF-H1/Rho/ROK pathway, we next wished to investigate the functional significance of increased pMLC. To this end, we asked whether pMLC is necessary for the TNF-α-induced rise in the paracellular permeability. To assess TNF-α-induced changes in paracellular permeability we followed the transepithelial transport of small molecular mass (4 kDa) fluorescent dextran. Confluent LLC-PK1 layers grown on semipermeable filters were exposed to fluorescent dextran given from the apical side, and the transport of dextran through the epithelial layer was assessed by sampling the bottom compartment every 30 min. Each experiment was carried out in triplicates. Fig. 7A shows data from a typical experiment. The untreated cells in all three parallel measurements showed only a small increase in the level of fluorescent dextran in the bottom compartment throughout the 3-h detection period. This indicates that under the conditions used LLC-PK1 cells form a strong barrier against the 4-kDa dextran. In contrast, when cells were treated with TNF-α, fluorescence increased much more quickly, indicating elevated permeability. As described under “Experimental Procedures,” control experiments verified that high molecular mass (70-kDa) dextran had only negligible permeability through the epithelial layer, and TNF-α did not alter its transport. These findings are in good agreement with those reported by Mullin et al. (22, 44) and verify that our assay indeed measures transepithelial permeability through a size-selective, paracellular pathway as opposed to a transcellular pathway that would not show size selectivity. In addition, the exclusive permeability toward small molecular mass dextran also rules out any injury or loss of cells from the epithelial layer throughout the measurement. TNF-α also induced similar permeability elevation in MDCK cells (not shown). Based on these introductory experiments, we chose the 3-h time point to assess alterations in permeability. To compare the results obtained in individual experiments, the three parallel measurements were averaged and normalized to the control, and the -fold increase values obtained in the individual experiments were averaged. The graph in Fig. 7B depicts the combined results of nine similar experiments. TNF-α-treated cells exhibited a 2.4-fold (±0.73, n = 9) increase in their permeability.
FIGURE 7.
The TNF-α-induced rise in paracellular permeability is mediated by MLC phosphorylation. LLC-PK1 cells were grown to confluence on Costar Transwell filters. The medium in both the top and bottom compartments was replaced by Hanks' buffered salt solution, and the cells were exposed to 2 mg/ml FITC-labeled dextran (4 kDa) added apically in the absence or presence of 10 ng/ml TNF-α. A, FITC fluorescence was measured in samples taken from the bottom compartment at 30-min intervals. The graph represents data of three parallel measurements of a typical experiment for control (dark symbols) or TNF-α-treated cells (open symbols). B, the fluorescence values obtained at the 3-h time point in parallel measurements as in A were averaged and normalized to the control. The graph shows the mean ± S.E. from n = 9 experiments. C, where indicated, LLC-PK1 cells grown on Transwell filters were pretreated with 1 μm PD98059 for 15 min followed by the addition of TNF-α. The inhibitor was present throughout the whole measurement. Paracellular permeability was measured as in A. The graph shows data of fluorescence values obtained at the 3-h time point of n = 5 measurements. D, AA-MLC acts as a dominant negative MLC. Control (empty vector-expressing cells) or AA-MLC-expressing LLC-PK1 cells were grown on coverslips. The cells were treated with TNF-α for 30 min, fixed, and co-stained with anti-Myc and anti-pMLC. The pictures show Myc (top row) and pMLC (bottom row) staining of the same field. E, paracellular permeability across confluent monolayers of control or AA-MLC-expressing cells was measured as in A. A typical experiment is shown. Fluorescence values obtained at the 3-h time point of three parallel measurements were averaged and expressed as mean ± S.E. F, AA-MLC cells are responsive to TNF-α. AA-MLC cells were treated for the indicated times with TNF-α and lysed, and phospho-ERK1/2 (pERK) was detected. The same blot was redeveloped with anti-β-actin to demonstrate equal loading. AU, arbitrary units; con, control.
To test the role of the ERK/GEF-H1/Rho pathway in mediating TNF-α-induced permeability increase, we first used PD98059. However, at 10 μm concentration, this drug largely elevated the basal permeability. Therefore, we treated the cells with a lower concentration (1 μm) that caused partial inhibition of ERK activation (not shown). As shown in Fig. 7C, PD98059 largely prevented the TNF-α-induced permeability increase. We also attempted to measure the permeability changes following down-regulation of GEF-H1. However, GEF-H1-depleted LLC-PK1 or MDCK cells were unable to form tight monolayers on the filters, precluding this measurement.
To test the role of pMLC in the permeability changes, we used a mutant, non-phosphorylatable MLC in which threonine 18 and serine 19 were changed to alanines (AA-MLC). We have previously shown that this mutant does not disrupt the basal actin cytoskeleton but prevents MLC phosphorylation induced by TGF-β and depolarization (28). We established a polyclonal LLC-PK1 cell line expressing AA-MLC. First we verified that the mutant MLC indeed prevents the TNF-α-induced elevation in pMLC. We exposed AA-MLC-expressing and control cells to TNF-α and visualized pMLC in them. As shown in Fig. 7D, cells expressing AA-MLC failed to show elevated pMLC staining upon TNF-α treatment. This shows that AA-MLC prevents endogenous MLC from becoming phosphorylated and thus acts as a dominant negative. Next we measured how TNF-α affects paracellular permeability in the AA-MLC-expressing cells. Fig. 7E shows a typical experiment performed using control and AA-MLC-expressing cells. Interestingly the AA-MLC cells showed slightly elevated basal permeability. Importantly although TNF-α enhanced permeability in control cells, it induced only a marginal elevation in the AA-MLC cells. To rule out that the inhibition was due to an overall unresponsiveness to TNF-α, we checked the intactness of myosin-independent signaling in AA-MLC cells exposed to TNF-α. As shown in Fig. 7F, TNF-α induced activation of ERK1/2 in AA-MLC cells, and this reaction was sustained and readily detectable in the time frame used for the permeability measurements (3 h). Taken together, these data show that pMLC is a key mediator of the TNF-α-induced permeability increase.
DISCUSSION
In this study we have shown that TNF-α induces MLC and cofilin phosphorylation through the GEF-H1/RhoA/Rho kinase pathway, leading to cytoskeletal remodeling in tubular epithelium. We also provide evidence that the ERK pathway is upstream of TNF-α-induced GEF-H1 and RhoA activation and that GEF-H1 phosphorylation on Thr-678 is a key event in this process. Moreover pMLC is a key mediator of the TNF-α-induced paracellular permeability increase.
The effects of TNF-α on the Rho pathway and its role in mediating permeability increase have not been previously explored in tubular epithelial cells. In endothelial cells, TNF-α stimulation causes a fast RhoA activation (17), and it was shown to contribute to TNF-α-induced cytokine and adhesion molecule synthesis and apoptosis (14–18, 45). The upstream mechanism of Rho activation, however, remained unsolved in these studies. Identity of the GEFs involved in specific pathways and the mode of their regulation are central questions in understanding the signaling toward small GTPase activation. Here we show for the first time that TNF-α induces the active (RhoG17A-binding) conformation of GEF-H1 leading to activation of the Rho pathway. The identification of the exchange factor was made possible by an affinity precipitation assay, which allows the assessment of the active pool of GEFs (19, 37, 38). This assay is also a useful tool in investigating pathways that regulate GEF activation. Having shown that GEF-H1 is regulated by TNF-α, we also attempted to measure its activation using a fluorescent GEF assay. In this assay HA-tagged GEF-H1 immunoprecipitated from tubular cells showed a well detectable exchange activity toward Rho that was unaltered following TNF-α treatment (not shown). The lack of stimulation of GEF-H1 activity, however, might be due to the limitations of the assay used. The amount of precipitated GEF-H1 used in the assay is hard to standardize, and GEF-H1 might lose its activated state during the process of precipitation and elution from the beads. Moreover the activated GEF molecules likely represent only a small percentage of the total pool (which has basal activity). In this case a few-fold activation present in a small percentage of GEF-H1 molecules will not cause a detectable difference in the overall activity of the immunoprecipitate. In agreement with this possibility, in vitro (and thus stoichiometric) threonine phosphorylation of recombinant Tiam1 (a kinase-regulated Rac-GEF) caused only <2-fold increase in GEF activity (46). Finally if the phosphorylation alters the affinity of GEF-H1 to GDP-Rho (which is added to the assay at saturating levels) the immunoprecipitation assay will not be adequate to detect the change. These considerations clearly imply that the pull-down assay, which measures the changes in the amount of conformationally active GEF-H1 in situ, is a more sensitive and suitable method to detect GEF-H1 activation.
Using this approach, GEF-H1 clearly showed an increased association with Rho in TNF-α-treated cells that might represent a key step in activation. Interestingly this increased association was dependent on the ERK pathway and phosphorylation of Thr-678 site in GEF-H1. Our experiments using GEF-H1 down-regulation also substantiated the central role of GEF-H1 in TNF-α-induced Rho activation. Absence of GEF-H1 prevented the TNF-α-provoked activation of Rho and its effectors. Interestingly GEF-H1 was suggested not to be involved in the TNF-α-induced Rho activation in endothelial cells (17). This conclusion was based on its unaltered localization and protein levels in TNF-α-stimulated human umbilical vein endothelial cells. Although we also found that the overall localization of HA-tagged GEF-H1 expressed in LLC-PK1 cells was unaltered by TNF-α (not shown), our conclusion pointing to a key role of GEF-H1 in epithelial cells is based on the direct assessment of the active pool of GEF-H1 as well as down-regulation experiments.
GEF-H1 is a microtubule-bound exchange factor (Refs. 39, 40, and 47 and for a review, see Ref. 48) that mediates Rho activation and permeability changes induced by the microtubule-disrupting agent nocodazole (49, 50). It is therefore conceivable that TNF-α might activate GEF-H1 by altering the microtubules. The effects of TNF-α on the microtubules have not been extensively studied, and directly opposing effects were reported for different cell types. In pulmonary endothelial as well as HeLa cells TNF-α caused disassembly of the peripheral microtubular network and a decrease in acetylated tubulin, indicating microtubule destabilization (51, 52). In contrast, in a fibrosarcoma cell line TNF-α treatment led to the rapid phosphorylation and inactivation of the microtubule-destabilizing protein oncoprotein 18, resulting in elongated microtubules (53). Our own initial experiments in tubular cells failed to reveal major changes in the microtubules in TNF-α-stimulated cells (not shown). However, further studies will have to establish the potential role of microtubules in mediating TNF-α-induced GEF-H1 activation.
GEF-H1 was also shown to bind cingulin, which localizes it to the tight junction, thereby keeping it in an inactive form (54). Interestingly GEF-H1 translocates to the junctions and mediates pMLC accumulation upon Ca2+ removal-induced junction disruption (55). The existence of a similar mechanism in the TNF-α-elicited effect remains to be established.
In search for the mechanism of activation of GEF-H1 we found that TNF-α enhanced its threonine phosphorylation, and this was prevented by the MEK inhibitor PD98059. A number of kinases have been shown to phosphorylate GEF-H1 (32, 40–42). Most of these mediate its serine phosphorylation and reduce its GEF activity. In contrast, ERK1/2 can catalyze the phosphorylation of Thr-678 in GEF-H1 (32), and this phosphorylation activates the protein. Using mass spectrometry and a T678A mutant, we identified Thr-678 as a site of the TNF-α-induced phosphorylation. Importantly the TNF-α-induced activation of the T678A mutant was largely reduced, suggesting that phosphorylation of Thr-678 is a key step in activation. The remaining small activation detected using the T678A mutant, however, raises the possibility that additional TNF-α-regulated phosphorylation site(s) might also exist. Taken together, our findings strongly suggest that the ERK pathway is an upstream regulator of TNF-α-induced GEF-H1 and Rho activation. Interestingly sustained activation of ERK has been shown to cause an increase in active Rho in oncogenic Ras-transformed cells by inactivating p190RhoGAP (56). Our data suggest that the interaction between Rho and ERK signaling can also occur at the level of GEF-H1.
Our surprising finding that inhibition of the ERK pathway alone leads to a slight but consistent increase in GEF-H1 phosphorylation points to the possibility that the ERK pathway has a more complex role. The site phosphorylated following MEK1/2 inhibition, however, is different from the one regulated by TNF-α-induced pathways as PD98059 also enhanced the threonine phosphorylation of the T678A mutant, which shows no phosphorylation upon TNF-α treatment. These findings suggest that ERK could be differentially involved in controlling the basal and stimulus-induced phosphorylation of GEF-H1. Enhanced phosphorylation in the presence of PD98059 correlated with a slight elevation in GEF-H1 and Rho pathway activity, suggesting that the phosphorylation happens on activatory site(s). Identification of the exact phosphorylation sites and the responsible kinase(s) will require further studies. In this regard, it has recently been reported that inhibition of the ERK pathway greatly enhances growth factor-induced AKT activation (57).
Interestingly although PD98059 added alone enhanced the phosphorylation of GEF-H1, when both PD98059 and TNF-α were present the phosphorylation decreased to control levels (and in some experiments even below). These intriguing observations would be consistent with the activation of a GEF-H1 phosphatase induced by TNF-α, operating independently of the ERK pathway. The (as yet unidentified) GEF-H1 phosphatase(s) are bound to have an important role in regulating activation as dephosphorylation of the inhibitory sites was shown to be a key step in the localized activation of GEF-H1 during mitosis (42).
Finally we investigated the functional significance of the TNF-α-induced activation of the ERK/GEF-H1/Rho/MLC pathway. MLC phosphorylation has been previously associated with elevated paracellular permeability both in epithelial and endothelial cells (20, 21). Interestingly, however, the TNF-α-induced permeability increase was found not to be mediated by ROK or myosin light chain kinase in endothelial cells (17). Most previous studies, however, used indirect approaches to assess the role of pMLC by inhibiting myosin light chain kinase. In addition, most of the pharmacological agents used also have nonspecific effects. In our hands both the ROK inhibitor Y-27632 and GEF-H1 down-regulation significantly elevated the basal permeability of the epithelial layers (not shown). Although we observed no further increase upon the addition of TNF-α, the elevated basal permeability rendered these data hard to interpret. The elevation of the basal permeability might be due to the substantial disruption of the actin cytoskeleton caused by Y-27632. To overcome this problem and to avoid any possible nonspecific effects of the pharmacological agents, we used a more targeted genetics approach to assess directly the role of phospho-MLC. As the non-phosphorylatable AA-MLC does not disrupt the basal actin structure (28) but effectively prevents MLC phosphorylation induced by TNF-α, it is an excellent tool to test the involvement of pMLC. Our data showing a lack of permeability increase in the AA-MLC stable expressers suggest that pMLC itself is a key mediator of the effect.
TNF-α was shown to induce myosin-dependent junction disruption in intestinal epithelia, contributing to the pathogenesis of inflammatory bowel disease (58). The effects of TNF-α in the tubular and intestinal epithelia, however, differ in a number of ways, suggesting that conclusions from studies in the intestine cannot be automatically applied for the kidney. Barrier dysfunction appears in cultured intestinal epithelia after long term (24–48-h) TNF-α treatment (59–61), and it is due to enhanced myosin light chain kinase synthesis and the resulting MLC phosphorylation (61–63). In addition, decreased expression of junctional proteins, including occludin (64), and apoptosis (65) were also shown to contribute. Interestingly long term TNF-α treatment also induces occludin degradation in endothelial cells (17). As opposed to the observations made in intestinal epithelium and in agreement with earlier findings (22, 44), we found that TNF-α increases the permeability of tubular cells within 1–3 h. Although this effect also requires phospho-MLC, the major MLC regulator in the tubules is Rho kinase and not myosin light chain kinase (27, 34). In addition, we found that in tubular cells TNF-α does not induce apoptosis or occludin degradation (not shown). A similar lack of TNF-α-induced apoptosis in tubular cells has been reported earlier (66), and TNF-α was even found to be antiapoptotic in proximal tubule cells (67). Importantly although the upstream MLC regulatory steps differ in various organs, all findings suggest that contractility is a central common factor in tight junction regulation. On the other hand, the different signaling pathways regulating the cell junctions in the tubular and intestinal cells might offer unique therapeutic targets for these two organs. The molecular mechanisms connecting phospho-MLC and the junctional permeability changes, however, are still unsolved, and future studies are warranted to address this important issue.
This work was supported in part by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC Grant 480619) and the Banting Foundation (to K. S.) and by the Kidney Foundation of Canada (to A. K. and K. S.).
Footnotes
The abbreviations used are: TNF-α, tumor necrosis factor-α; GEF, GDP-GTP exchange factor; GFP, green fluorescent protein; MLC, myosin light chain; RBD, Rho binding domain; ROK, Rho kinase; pMLC, monophosphorylated MLC; NR, non-related; MDCK, Madin-Darby canine kidney; ERK, extracellular signal-regulated kinase; HA, hemagglutinin; MAPK, mitogen-activated protein kinase; FITC, fluorescein isothiocyanate; GST, glutathione S-transferase; WT, wild type; siRNA, short interfering RNA; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase.
References
- 1.Baud, V., and Karin, M. (2001) Trends Cell Biol. 11, 372-377 [DOI] [PubMed] [Google Scholar]
- 2.Wajant, H., Pfizenmaier, K., and Scheurich, P. (2003) Cell Death Differ. 10, 45-65 [DOI] [PubMed] [Google Scholar]
- 3.Vielhauer, V., and Mayadas, T. N. (2007) Semin. Nephrol. 27, 286-308 [DOI] [PubMed] [Google Scholar]
- 4.Pascher, A., and Klupp, J. (2005) BioDrugs 19, 211-231 [DOI] [PubMed] [Google Scholar]
- 5.Noiri, E., Kuwata, S., Nosaka, K., Tokunaga, K., Juji, T., Shibata, Y., and Kurokawa, K. (1994) Am. J. Pathol. 144, 1159-1166 [PMC free article] [PubMed] [Google Scholar]
- 6.Sato, H., Tanaka, T., Kita, T., Yamaguchi, H., and Tanaka, N. (2004) Int. J. Exp. Pathol. 85, 345-353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donnahoo, K. K., Meng, X., Ao, L., Ayala, A., Shames, B. D., Cain, M. P., Harken, A. H., and Meldrum, D. R. (2001) Immunology 102, 53-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Navarro, J. F., Milena, F. J., Mora, C., Leon, C., Claverie, F., Flores, C., and Garcia, J. (2005) Kidney Int. Suppl. 99, S98-S102 [DOI] [PubMed] [Google Scholar]
- 9.Jevnikar, A. M., Brennan, D. C., Singer, G. G., Heng, J. E., Maslinski, W., Wuthrich, R. P., Glimcher, L. H., and Kelley, V. E. (1991) Kidney Int. 40, 203-211 [DOI] [PubMed] [Google Scholar]
- 10.Yard, B. A., Daha, M. R., Kooymans-Couthino, M., Bruijn, J. A., Paape, M. E., Schrama, E., van Es, L. A., and van der Woude, F. J. (1992) Kidney Int. 42, 383-389 [DOI] [PubMed] [Google Scholar]
- 11.Biancone, L., Conaldi, P. G., Toniolo, A., and Camussi, G. (1997) Exp. Nephrol. 5, 330-336 [PubMed] [Google Scholar]
- 12.Zager, R. A., Johnson, A. C., Hanson, S. Y., and Lund, S. (2005) Am. J. Physiol. 289, F289-F297 [DOI] [PubMed] [Google Scholar]
- 13.Jaffe, A. B., and Hall, A. (2005) Annu. Rev. Cell Dev. Biol. 21, 247-269 [DOI] [PubMed] [Google Scholar]
- 14.Petrache, I., Crow, M. T., Neuss, M., and Garcia, J. G. (2003) Biochem. Biophys. Res. Commun. 306, 244-249 [DOI] [PubMed] [Google Scholar]
- 15.Nubel, T., Dippold, W., Kleinert, H., Kaina, B., and Fritz, G. (2004) FASEB J. 18, 140-142 [DOI] [PubMed] [Google Scholar]
- 16.VandenBerg, E., Reid, M. D., Edwards, J. D., and Davis, H. W. (2004) J. Cell. Biochem. 91, 926-937 [DOI] [PubMed] [Google Scholar]
- 17.McKenzie, J. A., and Ridley, A. J. (2007) J. Cell. Physiol. 213, 221-228 [DOI] [PubMed] [Google Scholar]
- 18.Mong, P. Y., Petrulio, C., Kaufman, H. L., and Wang, Q. (2008) J. Immunol. 180, 550-558 [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Mata, R., and Burridge, K. (2007) Trends Cell Biol. 17, 36-43 [DOI] [PubMed] [Google Scholar]
- 20.Nusrat, A., Turner, J. R., and Madara, J. L. (2000) Am. J. Physiol. 279, G851-G857 [DOI] [PubMed] [Google Scholar]
- 21.Kapus, A., and Szaszi, K. (2006) Biochem. Cell Biol. 84, 870-880 [DOI] [PubMed] [Google Scholar]
- 22.Mullin, J. M., Laughlin, K. V., Marano, C. W., Russo, L. M., and Soler, A. P. (1992) Am. J. Physiol. 263, F915-F924 [DOI] [PubMed] [Google Scholar]
- 23.Walsh, S. V., Hopkins, A. M., and Nusrat, A. (2000) Adv. Drug Delivery Rev. 41, 303-313 [DOI] [PubMed] [Google Scholar]
- 24.Turner, J. R. (2000) Adv. Drug Delivery Rev. 41, 265-281 [DOI] [PubMed] [Google Scholar]
- 25.Zolotarevsky, Y., Hecht, G., Koutsouris, A., Gonzalez, D. E., Quan, C., Tom, J., Mrsny, R. J., and Turner, J. R. (2002) Gastroenterology 123, 163-172 [DOI] [PubMed] [Google Scholar]
- 26.Petrache, I., Verin, A. D., Crow, M. T., Birukova, A., Liu, F., and Garcia, J. G. (2001) Am. J. Physiol. 280, L1168-L1178 [DOI] [PubMed] [Google Scholar]
- 27.Szaszi, K., Sirokmany, G., Di Ciano-Oliveira, C., Rotstein, O. D., and Kapus, A. (2005) Am. J. Physiol. 289, C673-C685 [DOI] [PubMed] [Google Scholar]
- 28.Di Ciano-Oliveira, C., Lodyga, M., Fan, L., Szaszi, K., Hosoya, H., Rotstein, O. D., and Kapus, A. (2005) Am. J. Physiol. 289, C68-C81 [DOI] [PubMed] [Google Scholar]
- 29.Arthur, W. T., and Burridge, K. (2001) Mol. Biol. Cell 12, 2711-2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Mata, R., Wennerberg, K., Arthur, W. T., Noren, N. K., Ellerbroek, S. M., and Burridge, K. (2006) Methods Enzymol. 406, 425-437 [DOI] [PubMed] [Google Scholar]
- 31.Iwasaki, T., Murata-Hori, M., Ishitobi, S., and Hosoya, H. (2001) Cell Struct. Funct. 26, 677-683 [DOI] [PubMed] [Google Scholar]
- 32.Fujishiro, S. H., Tanimura, S., Mure, S., Kashimoto, Y., Watanabe, K., and Kohno, M. (2008) Biochem. Biophys. Res. Commun. 368, 162-167 [DOI] [PubMed] [Google Scholar]
- 33.Fan, L., Sebe, A., Peterfi, Z., Masszi, A., Thirone, A. C., Rotstein, O. D., Nakano, H., McCulloch, C. A., Szaszi, K., Mucsi, I., and Kapus, A. (2007) Mol. Biol. Cell 18, 1083-1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Ciano-Oliveira, C., Sirokmany, G., Szaszi, K., Arthur, W. T., Masszi, A., Peterson, M., Rotstein, O. D., and Kapus, A. (2003) Am. J. Physiol. 285, C555-C566 [DOI] [PubMed] [Google Scholar]
- 35.Huang, T. Y., DerMardirossian, C., and Bokoch, G. M. (2006) Curr. Opin. Cell Biol. 18, 26-31 [DOI] [PubMed] [Google Scholar]
- 36.Scott, R. W., and Olson, M. F. (2007) J. Mol. Med. 85, 555-568 [DOI] [PubMed] [Google Scholar]
- 37.Garrett, T. A., Van Buul, J. D., and Burridge, K. (2007) Exp. Cell Res. 313, 3285-3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arthur, W. T., Ellerbroek, S. M., Der, C. J., Burridge, K., and Wennerberg, K. (2002) J. Biol. Chem. 277, 42964-42972 [DOI] [PubMed] [Google Scholar]
- 39.Ren, Y., Li, R., Zheng, Y., and Busch, H. (1998) J. Biol. Chem. 273, 34954-34960 [DOI] [PubMed] [Google Scholar]
- 40.Zenke, F. T., Krendel, M., DerMardirossian, C., King, C. C., Bohl, B. P., and Bokoch, G. M. (2004) J. Biol. Chem. 279, 18392-18400 [DOI] [PubMed] [Google Scholar]
- 41.Callow, M. G., Zozulya, S., Gishizky, M. L., Jallal, B., and Smeal, T. (2005) J. Cell Sci. 118, 1861-1872 [DOI] [PubMed] [Google Scholar]
- 42.Birkenfeld, J., Nalbant, P., Bohl, B. P., Pertz, O., Hahn, K. M., and Bokoch, G. M. (2007) Dev. Cell 12, 699-712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kohno, M., Nishizawa, N., Tsujimoto, M., and Nomoto, H. (1990) Biochem. J. 267, 91-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mullin, J. M., Marano, C. W., Laughlin, K. V., Nuciglio, M., Stevenson, B. R., and Soler, P. (1997) J. Cell. Physiol. 171, 226-233 [DOI] [PubMed] [Google Scholar]
- 45.Neumann, H., Schweigreiter, R., Yamashita, T., Rosenkranz, K., Wekerle, H., and Barde, Y. A. (2002) J. Neurosci. 22, 854-862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fleming, I. N., Elliott, C. M., Buchanan, F. G., Downes, C. P., and Exton, J. H. (1999) J. Biol. Chem. 274, 12753-12758 [DOI] [PubMed] [Google Scholar]
- 47.Krendel, M., Zenke, F. T., and Bokoch, G. M. (2002) Nat. Cell Biol. 4, 294-301 [DOI] [PubMed] [Google Scholar]
- 48.Birkenfeld, J., Nalbant, P., Yoon, S. H., and Bokoch, G. M. (2008) Trends Cell Biol. 18, 210-219 [DOI] [PubMed] [Google Scholar]
- 49.Chang, Y. C., Nalbant, P., Birkenfeld, J., Chang, Z. F., and Bokoch, G. M. (2008) Mol. Biol. Cell 19, 2147-2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Birukova, A. A., Adyshev, D., Gorshkov, B., Bokoch, G. M., Birukov, K. G., and Verin, A. D. (2006) Am. J. Physiol. 290, L540-L548 [DOI] [PubMed] [Google Scholar]
- 51.Petrache, I., Birukova, A., Ramirez, S. I., Garcia, J. G., and Verin, A. D. (2003) Am. J. Respir. Cell Mol. Biol. 28, 574-581 [DOI] [PubMed] [Google Scholar]
- 52.Domnina, L. V., Ivanova, O. Y., Cherniak, B. V., Skulachev, V. P., and Vasiliev, J. M. (2002) Biochemistry (Mosc.) 67, 737-746 [DOI] [PubMed] [Google Scholar]
- 53.Vancompernolle, K., Boonefaes, T., Mann, M., Fiers, W., and Grooten, J. (2000) J. Biol. Chem. 275, 33876-33882 [DOI] [PubMed] [Google Scholar]
- 54.Benais-Pont, G., Punn, A., Flores-Maldonado, C., Eckert, J., Raposo, G., Fleming, T. P., Cereijido, M., Balda, M. S., and Matter, K. (2003) J. Cell Biol. 160, 729-740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samarin, S. N., Ivanov, A. I., Flatau, G., Parkos, C. A., and Nusrat, A. (2007) Mol. Biol. Cell 18, 3429-3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen, J. C., Zhuang, S., Nguyen, T. H., Boss, G. R., and Pilz, R. B. (2003) J. Biol. Chem. 278, 2807-2818 [DOI] [PubMed] [Google Scholar]
- 57.Hayashi, H., Tsuchiya, Y., Nakayama, K., Satoh, T., and Nishida, E. (2008) Genes Cells 13, 941-947 [DOI] [PubMed] [Google Scholar]
- 58.Wang, F., Schwarz, B. T., Graham, W. V., Wang, Y., Su, L., Clayburgh, D. R., Abraham, C., and Turner, J. R. (2006) Gastroenterology 131, 1153-1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmitz, H., Fromm, M., Bentzel, C. J., Scholz, P., Detjen, K., Mankertz, J., Bode, H., Epple, H. J., Riecken, E. O., and Schulzke, J. D. (1999) J. Cell Sci. 112, 137-146 [DOI] [PubMed] [Google Scholar]
- 60.Marano, C. W., Lewis, S. A., Garulacan, L. A., Soler, A. P., and Mullin, J. M. (1998) J. Membr. Biol. 161, 263-274 [DOI] [PubMed] [Google Scholar]
- 61.Wang, F., Graham, W. V., Wang, Y., Witkowski, E. D., Schwarz, B. T., and Turner, J. R. (2005) Am. J. Pathol. 166, 409-419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Graham, W. V., Wang, F., Clayburgh, D. R., Cheng, J. X., Yoon, B., Wang, Y., Lin, A., and Turner, J. R. (2006) J. Biol. Chem. 281, 26205-26215 [DOI] [PubMed] [Google Scholar]
- 63.Ye, D., Ma, I., and Ma, T. Y. (2006) Am. J. Physiol. 290, G496-G504 [DOI] [PubMed] [Google Scholar]
- 64.Mankertz, J., Tavalali, S., Schmitz, H., Mankertz, A., Riecken, E. O., Fromm, M., and Schulzke, J. D. (2000) J. Cell Sci. 113, 2085-2090 [DOI] [PubMed] [Google Scholar]
- 65.Soler, A. P., Marano, C. W., Bryans, M., Miller, R. D., Garulacan, L. A., Mauldin, S. K., Stamato, T. D., and Mullin, J. M. (1999) Eur. J. Cell Biol. 78, 56-66 [DOI] [PubMed] [Google Scholar]
- 66.Patrick, D. M., Leone, A. K., Shellenberger, J. J., Dudowicz, K. A., and King, J. M. (2006) BMC Physiol. 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Papakonstanti, E. A., and Stournaras, C. (2004) Mol. Biol. Cell 15, 1273-1286 [DOI] [PMC free article] [PubMed] [Google Scholar]