Abstract
Quality control mechanisms during protein synthesis are essential to fidelity and cell survival. Leucyl-tRNA synthetase (LeuRS) misactivates non-leucine amino acids including isoleucine, methionine, and norvaline. To prevent translational errors, mischarged tRNA products are translocated 30Å from the canonical aminoacylation core to a hydrolytic editing-active site within a completely separate domain. Because it is transient, the tRNA translocation mechanism has been difficult to isolate. We have identified a “translocation peptide” within Escherichia coli LeuRS. Mutations in the translocation peptide cause tRNA to selectively bypass the editing-active site, resulting in mischarging that is lethal to the cell. This bypass mechanism also rescues aminoacylation of an editing site mutation that hydrolyzes correctly charged Leu-tRNALeu. Thus, these LeuRS mutants charge tRNALeu but fail to translocate these products to the hydrolytic site, where they are cleared to guard against genetic code ambiguities.
Quality control during translation depends on the family of aminoacyl-tRNA synthetases (aaRSs),2 which is responsible for the first step of protein synthesis. Each aaRS selectively aminoacylates just one of the 20 standard amino acids to its cognate tRNA (1). About half of this family of enzymes ensures fidelity by employing a “double sieve model” that relies on two active sites (2, 3). One sieve is synthetic and produces charged tRNA. The other is a hydrolytic editing-active site that clears mistakes. Defects in the editing mechanism cause cell death (4, 5) and also neurological disease in mammals (6).
The aminoacylation site in the ancient canonical core of the aaRS activates its cognate amino acid but can also misactivate structurally similar amino acids (1). The editing-active site blocks the correctly charged amino acid (7, 8) and hydrolyzes mischarged amino acids from the tRNA. Amino acid editing destroys mistakes before they can be incorporated by the ribosome, which would result in the production of statistical proteins (1).
Amino acid proofreading requires that the charged tRNA transiently migrates between two enzyme domains that are responsible for aminoacylation and editing. For leucyl-tRNA synthetase (LeuRS) and the homologous isoleucyl-(IleRS) and valyl-tRNA synthetases (ValRS), the editing domain resides in a structural insertion called CP1 (9) that splits the Rossmann ATP binding fold. The insert folds independent of the canonical core (10–12). The isolated CP1 domains from LeuRS, ValRS, and IleRS can independently and specifically hydrolyze mischarged amino acid from its cognate tRNA (13–15).
The aminoacylation and editing-active sites of LeuRS are separated by about 30 Å. Thus, the charged 3′ end of the tRNA must be faithfully translocated a significant distance for proofreading and then hydrolysis if it is mischarged (16). It has also been suggested that the tRNA 3′ end binds initially near the editing-active site and requires translocation to the aminoacylation site (17).
We hypothesized that flexible molecular hinges might facilitate conformational changes between the aminoacylation and the editing complexes (18). Two putative hinge sites were predicted by computational analysis of Thermus thermophilus LeuRS. One hinge at Ser-227 was located in the N-terminal β-strand that links the aminoacylation and CP1 editing domains (18). Mutations at the predicted hinge site in the β-strand linker of Escherichia coli LeuRS abolished aminoacylation activity and significantly decreased amino acid editing activity (18).
A second hinge site at Glu-393 was identified in a flexible peptide within the CP1 domain of T. thermophilus LeuRS (18). Here, we describe results at a homologous Asp-391 site in E. coli LeuRS that demonstrate that this hinge comprises a portion of a translocation peptide. Unlike the predicted β-strand hinge mutation, the aminoacylation and editing activities of the CP1 domain-based hinge mutants in LeuRS were intact. Surprisingly however, mutations within the translocation peptide yield mischarged tRNA despite a robust deacylation activity. We hypothesize that impairing the LeuRS translocation peptide causes the charged tRNA 3′ end to bypass the editing sieve prior to product release. Defects in the translocation peptide and its mechanism result in amino acid toxicities that are lethal to the cell.
EXPERIMENTAL PROCEDURES
Materials—T7 RNA polymerase (19) was used to transcribe tRNALeu in vitro via run-off transcription (20). The tRNA product was purified by urea-containing polyacrylamide gel electrophoresis (20) and quantitated based on absorbance at 260 nm using an extinction coefficient of 840,700 liters/mol·cm (21). Purified tRNALeu was denatured at 80 °C for 1 min followed by an addition of 1 mm MgCl2 and quick cooling on ice for refolding.
Protein Mutagenesis and Preparation—Mutations were introduced into the wild type E. coli leuS gene by PCR according to a previously described protocol (13) using plasmid p15EC3-1 (22) to generate the following plasmids: pRHFN1 (F382A/N383A), pRHFN2 (F387A/N388A), pRHHE (H392A/E393A), pRHFN3 (F396A/N397A), pRHD391A (D391A), pRHH392A (H392A), pRHE393A (E393A), pRHDH (D391A/H392A), pRHE386A (E386A), pRHF387A (F387A), and pRHN388A (N388A). These plasmids encoding a mutant LeuRS were used to introduce a second T252A mutation.
Wild type and mutant proteins were expressed in E. coli BL21 (DE3) codon PLUS (Stratagene). Harvested cells were lysed by sonication, and each LeuRS, which contained an N-terminal six-histidine tag, was purified by affinity purification using HIS-Select HF nickel affinity resin (Sigma) as described before (18). The purified proteins were concentrated via a Centricon-50 (Amicon) and quantitated using the Bio-Rad protein assay according to the commercial protocol.
Complementation of E. coli KL231—Competent E. coli KL231 cells (23) were co-transformed with pGP1-2 (KanR), which encodes T7 RNA polymerase (24), and with plasmids encoding wild-type or mutant E. coli LeuRSs (AmpR). Transformants were selected and grown as described previously (4). The cells were shifted to 42 °C for 30 min followed by a 2-h incubation at 40 °C for induction of T7 RNA polymerase expression. The cells were then harvested and resuspended in 5 ml of MS-Amp/Kan/Str medium. A 500-μl aliquot of the cell suspension was mixed with 3 ml of 0.7% soft agar and spread evenly on the MS-Amp/Kan/Str medium plates. An aliquot of 100 μl of 100 mm norvaline was incorporated into a bored out well in the center of the plate. The plates were incubated at 42 °C for 48 h.
Aminoacylation Assays—Leucylation reactions contained 60 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 1 mm dithiothreitol, 4 μm folded tRNALeu, 21 μm l-[3H]leucine (150 μCi/ml), 50 nm enzyme and were initiated by 4 mm ATP. The reaction was carried out at 30 °C. Isoleucylation reactions substituted 21 μm l-[3H]isoleucine. Aliquots of 10 μl were quenched on trichloroacetic acid-soaked filter pads, washed, and quantitated via scintillation counting as described before (18).
Pre-steady state kinetic analysis of the aminoacylation activity of wild type and mutant LeuRS enzymes was carried out using a quenched-flow RQF-3 KinTek instrument (KinTek Corp., Austin, TX) as described previously (25). Multiple turnover experiments included saturating concentrations of 25 μm tRNALeu with 5 μm wild type or mutant LeuRS at 37 °C.
Single turnover experiments were conducted under excess enzyme to substrate concentrations with 40 μm wild type or mutant LeuRS enzyme and 4 μm tRNALeu. The LeuRS-adenylate complex was formed at 30 °C for 30 min using 40 μm LeuRS, 2.5 mm ATP, and 50 μm [14C]-leucine (2.7 μCi/ml) in 60 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 1 mm dithiothreitol, and 0.2 units of yeast inorganic pyrophosphatase (26). The LeuRS-adenylate complex was purified and eluted using a pre-equilibrated Quick Spin Sephadex G-25 column (Roche Applied Science) (26). The entire eluent was loaded into one syringe on the quenched-flow instrument and then combined with 4 μm tRNALeu contained within a second syringe. Reactions were quenched with 3 m sodium acetate (pH 5.0) (25).
The data were analyzed using GraphPad Prism 4 (y = y0 + A × (1 - e-k ×t1) + k2 × E0 × t) to identify a burst and calculate rate constants or the first order exponential equation (y = A(1 - e-kobsk) + C). The kinetic data were fit to obtain R-values > 0.99.
Charged tRNA Preparation and Enzymatic Deacylation—The [3H]Leu-tRNALeu was generated in a 1-h incubation of the leucylation reaction described above. The [3H]Ile-tRNALeu was prepared using an editing-defective T252Y LeuRS mutant in an aminoacylation reaction that was incubated at 30 °C for 3 h. Reactions were stopped using 0.18% acetic acid and extracted using two equal volumes of phenol/chloroform/isoamyl alcohol, pH 4.3 (125:24:1) (27). A one-half volume of 4.6 m NH4OAc, pH 5.0, was added followed by an ethanol precipitation. The dried aminoacylated tRNA pellets were resuspended in 10 mm KH2PO4, pH 5.0. Hydrolytic editing assays were carried out at room temperature in 60 mm Tris, pH 7.5, 10 mm MgCl2, and 0.8 μm [3H]Leu-tRNALeu or 0.8 μm [3H]Ile-tRNALeu and was initiated with 100 nm enzyme (7). At specific time points, 5-μl aliquots were spotted onto filter pads, washed, and quantitated.
RESULTS
Mutations at a Predicted Hinge Result in a Mischarging Phenotype in Vivo—Previously, we compared x-ray crystal structures of T. thermophilus LeuRS using Morph Server (28) within the data base of molecular motions (29) to identify flexible regions within LeuRS (18). A molecular hinge, which could confer flexibility, was predicted at two sequentially and spatially distinct sites: Ser-227 and Glu-393. The predicted hinge at Ser-227 is located within the N-terminal β-strand linker and significantly impaired aminoacylation and editing activities when mutated in E. coli LeuRS as reported previously (18).
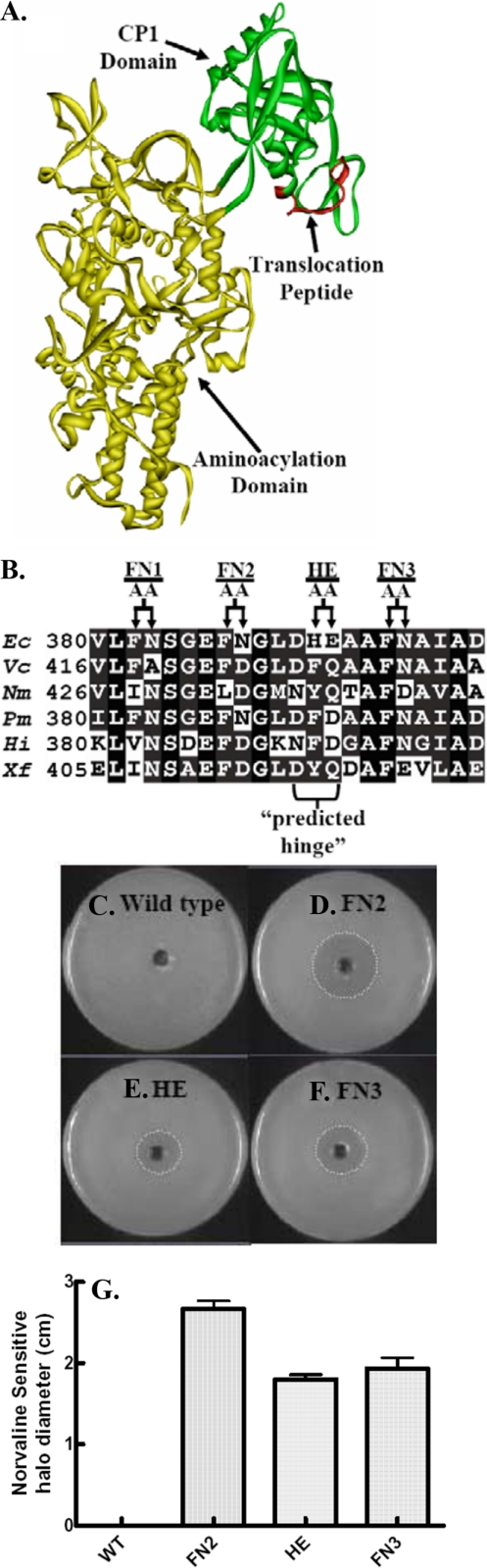
The second predicted hinge site is characterized herein. The T. thermophilus LeuRS x-ray crystal structures show that the Glu-393 predicted hinge is located in a flexible peptide in the CP1 domain with B-factors of 127.18 and 108.06 for the editing and exit complexes, respectively (30). This peptide is distal to the hydrolytic site and conserved among LeuRS from Gram-negative bacteria (Fig. 1). The corresponding site in E. coli LeuRS has a homologous aspartate (at position 391), which was mutationally targeted for analysis.
FIGURE 1.
Primary and tertiary structure of the E. coli LeuRS translocation peptide. A, E. coli LeuRS homology model based on the x-ray crystal structure of T. thermophilus (36). The CP1 domain (green) is inserted into the catalytic aminoacylation core (yellow) via twoβ-strand linkers. A putative translocation peptide is highlighted in red. B, sequence alignment for the region comprising the translocation peptide. Arrows point to residues that were targeted for mutagenesis. Shaded and black boxes represent conserved and homologous residues. The bracket refers to a putative hinge site on the protein. AA, alanine/alanine. Organism abbreviations are as follows: E. coli (Ec), Vibrio cholera (Vc), Pasteurella multocida (Pm), Haemophilus influenzae (Hi), Neisseria meningitides (Nm), and Xylella fastidiosa (Xf). C–F, E. coli KL231 complementation and norvaline sensitivity for wild type LeuRS (C) and the LeuRS mutants FN2 (D), HE (E), and FN3 (F). G, a histogram quantifying the halo diameters for norvaline sensitivities. The standard deviation (error bars) was based on three replicates.
The predicted Asp-391 hinge in E. coli LeuRS resides in a repeating pattern of aromatic amino acids followed by amino acids that can form hydrogen bonds as either a proton donor or an acceptor (Fig. 1B). Based on the x-ray co-crystal structure of T. thermophilus LeuRS bound to tRNALeu, it has been proposed that this analogous region could interact with the 3′ acceptor stem of the tRNA (30). However, in the static editing complex structure, the tRNA 3′ end and the predicted hinge are separated by about 12 Å.
We hypothesized that this CP1-based flexible peptide might undergo dynamic conformational changes to assist transient tRNA movement between the aminoacylation and editing domains. We introduced a series of double alanine substitutions at each of the repeating pairs in the LeuRS flexible peptide to probe for effects on aminoacylation and/or editing activity. The HE mutant LeuRS is the H392A/E393A double substitution at the predicted C-terminal side of the hinge site. The FN1, FN2, and FN3 LeuRS mutants represent F382A/N383A, F387A/N388A, and F396A/N397A double alanine substitutions, respectively.
As a first step, we screened each of these mutants for mischarging phenotypes in vivo (4) using E. coli KL231, which has a temperature-sensitive mutation in LeuRS (23). E. coli KL231 can be complemented by LeuRS editing-defective mutants. At 42 °C, the endogenous LeuRS is inactivated and the cells are sensitive to amino acid toxicities (4).
Competent E. coli KL231 cells were transformed with plasmids carrying the FN1, FN2, FN3, and HE LeuRS mutants, as well as the wild type gene. The FN2, HE, and FN3 LeuRS mutants rescued the temperature-sensitive E. coli KL231 at 42 °C similar to wild type. The FN1 LeuRS mutant did not complement the cells (data not shown).
The FN2, HE, and FN3 mutations were further tested for mischarging phenotypes using halo assays. Norvaline, a biosynthetic intermediate that is missing the branched methyl group of the leucine side chain, can be mischarged by editing-defective LeuRSs. This leucine analog was introduced into a central bored out well on the plates and resulted in a clear halo or zone of inhibition around the central well for the FN2, HE, and FN3 mutants (Fig. 1, D–G) in contrast to cells that were complemented by the wild type LeuRS, which did not produce a halo (Fig. 1C). We hypothesized that the LeuRS FN2, HE, and FN3 mutations impaired enzymatic hydrolysis of mischarged tRNA products to cause cell toxicity and death.
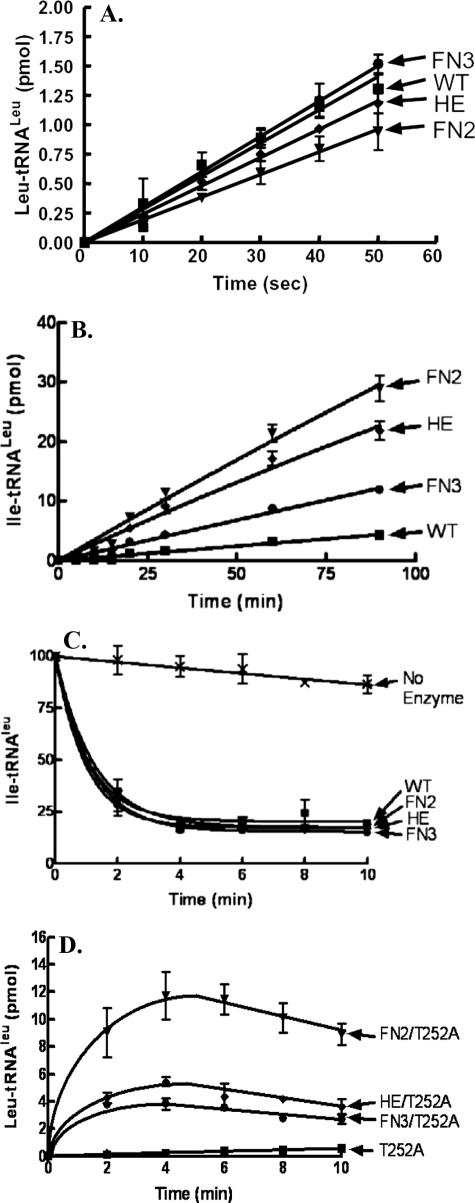
Isolation of the Transient tRNA Translocation Step for Editing—We purified the E. coli LeuRS FN2, HE, and FN3 mutants to investigate alterations in their enzymatic activities. The aminoacylation activity for each mutant was similar to the wild type enzyme (Fig. 2A). The respective initial velocities were 0.028 ± 0.0014, 0.0193 ± 0.0001, 0.0241 ± 0.0007, and 0.03 ± 0.001 pmol/s for the wild type enzyme and FN2, HE, and FN3 mutant LeuRSs.
FIGURE 2.
Enzymatic activities of wild type (WT) LeuRSs and translocation peptide mutants. A, leucylation reactions were carried out with 10 nm LeuRS, 4 μm tRNALeu transcript, and 22 μm [3H]leucine (167 μCi/ml). B, isoleucine mischarging reactions substituted 1 μm LeuRS enzyme and 22 μm [3H]isoleucine (167 μCi/ml). C, deacylation reactions included 1.5 μm [3H]Ile-tRNALeu and 5 nm enzyme. D, leucylation rescue activity of T252A utilized 50 nm LeuRS, 4 μm tRNALeu transcript, and 22 μm [3H]leucine (167 μCi/ml). Symbols represent the following: wild type or single T252A mutant (▪); FN2 or FN2/T252A, (▾); HE or HE/T252A, (♦); FN3 or FN3/T252A, (•); and no enzyme controls (×). Error bars are present for all points and represent reactions that were repeated at least in triplicate.
As would be expected based on the cellular toxicity assays, each of the mutants also mischarged tRNALeu (Fig. 2B). The initial velocities for mischarging activities were 0.332 ± 0.0097, 0.256 ± 0.009, and 0.135 ± 0.003 pmol/min, respectively, for the FN2, HE, and FN3 mutant LeuRSs when compared with 0.048 ± 0.002 pmol/s for the wild type enzyme. Surprisingly however, Ile-tRNALeu deacylation activity remained intact (Fig. 2C). Thus, we hypothesized that these mischarging mutations in the flexible CP1-based peptide that is marked by a predicted hinge site (18) impacted translocation of the mischarged tRNA from the aminoacylation to the editing site.
To test this hypothesis, we combined each of the putative translocation mutants with T252A substitutions in the hydrolytic editing-active site. The T252A mutation uncouples specificity and hydrolyzes correctly charged Leu-tRNALeu (7). Each of the FN2, HE, and FN3 substitutions rescued the T252A mutation to yield Leu-tRNALeu (Fig. 2D). This supports that mutations in the flexible peptide disrupt the transient tRNA translocation step. Differences in the rescue levels could be due to idiosyncratic effects of each mutant that include rebinding of charged tRNALeu for deacylation and/or equilibrium or kinetic shifts in one of the multiple physical and chemical steps that comprise the aminoacylation reaction.
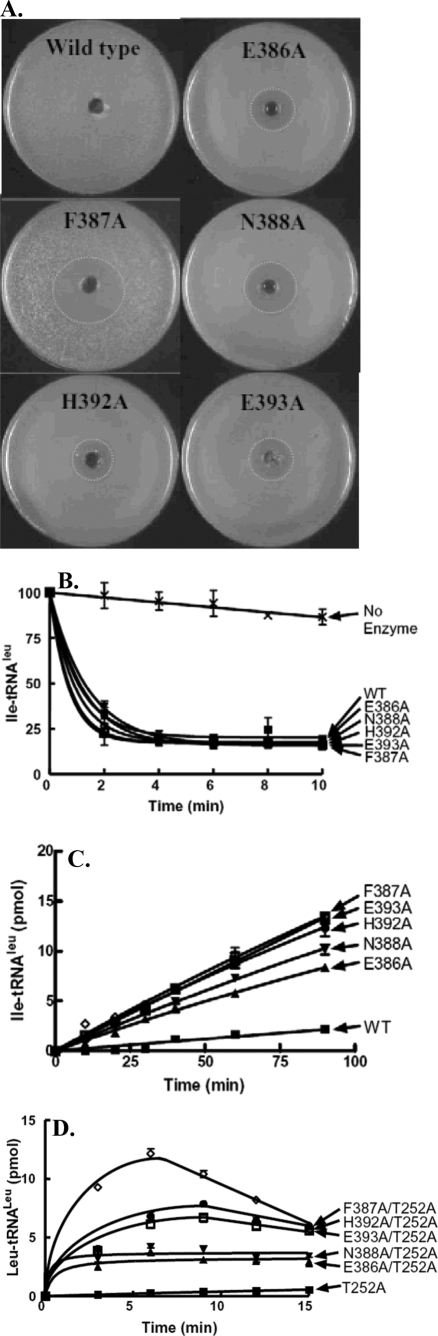
To localize specific amino acid sites that might be involved in transient tRNA translocation, we further probed the LeuRS translocation peptide by alanine-scanning mutagenesis of individual amino acid sites within the translocation peptide to generate E386A, F387A, N388A, D391A, H392A, and E393A. With one exception (D391A, data not shown), complementation assays with E. coli KL231 showed clear halos around the wells containing 100 mm norvaline (Fig. 3A). This suggested that each of these single mutations in the translocation peptide yielded mischarged tRNALeu.
FIGURE 3.
Characterization of single mutations in the E. coli LeuRS translocation peptide. A, E. coli KL231 complementation and norvaline sensitivity assay for the wild type LeuRS and the E386A, F387A, N388A, H392A, and E393A translocation peptide mutants. B, deacylation reactions were carried out with 1.5 μm [3H]Ile-tRNALeu and 5 nm enzyme. C, isoleucylation reactions contained 1 μm LeuRS enzyme and 22 μm [3H]isoleucine (167 μCi/ml). D, leucylation rescue activity of T252A utilized 50 nm LeuRS, 4 μm tRNALeu transcript, and 22 μm [3H]leucine (167 μCi/ml). Symbols represent the following: wild type (WT) or single mutant T252A, (▪); E386A or E386A/T252A, (▴); F387A or F387A/T252A, (⋄); N388A or N388A/T252A, (▾); H392A or H392A/T252A, (•); and E393A or E393A/T252A, (□). Error bars are present for all points and represent reactions that were repeated at least in triplicate.
We carried out in vitro enzymatic analysis on the LeuRS mutants that produced a halo. Each mutant exhibited leucylation activity similar to wild type (data not shown). Mischarged tRNALeu deacylation activities for these translocation mutants was robust and comparable with the wild type enzyme (Fig. 3B). This second set of mutant LeuRSs also mischarged isoleucine to tRNALeu (Fig. 3C), which is consistent with the intracellular halo assays that we hypothesize were sensitive to mischarging of norvaline to tRNALeu. The initial velocities of mischarging by the E386A, F387A, N388A, H392A, and E393A LeuRS mutants were respectively 0.0958 + 0.002, 0.1538 + 0.0033, 0.1173 + 0.002, 0.1421 + 0.0031, and 0.1487 + 0.0028 pmol/min. These single LeuRS mutants were combined with the T252A substitution as described above. In each case, leucylation activity was rescued (Fig. 3D) despite the fact that all of these T252A-based double mutants retained Leu-tRNALeu deacylation activity (data not shown). As indicated above, the decrease in charged product near the end of the time course in Figs. 2D and 3D is likely due to the release and rebinding of the charged Leu-tRNALeu in the editing complex. Thus, a competent editing complex is formed by directly binding the mischarged or charged tRNALeu, but translocation of charged tRNALeu from the synthetic site to the editing site is impaired.
We propose that mutants within the flexible peptide and at the predicted hinge site constitute a novel class of fidelity-deficient LeuRSs that occur because of defects in translocation. It is possible that mutations in the translocation peptide cause the charged tRNA to undergo premature product release. Alternatively or in addition, the 3′ end of the mischarged tRNA simply bypasses the hydrolytic editing-active site to form a so-called exit complex (30) that is positioned for product release to EF-Tu.
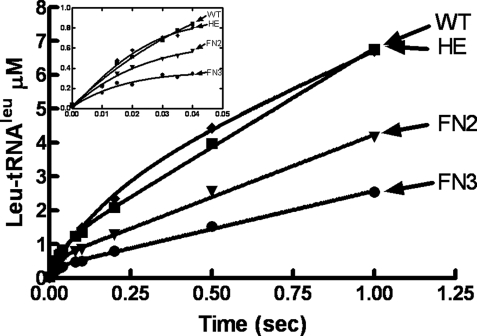
Translocation Peptide Defects Maintain LeuRS Exit Complex—It has been proposed that product release is the rate-limiting step for class I aaRSs (31). We employed rapid quench kinetic approaches to probe the mechanistic effects of each of these translocation-defective mutants on product release. As would be expected, under pre-steady state conditions, the aminoacylation activity of wild type LeuRS exhibited a burst during multiturnover kinetic analysis (Fig. 4). The k1 and k2 for the biphasic kinetic profile (Table 1) were similar to those measured for other class I aaRSs (31). Single turnover kinetic experiments showed that ktrans for LeuRS is consistent with k1 that was measured in the multiturnover experiments (Table 1). This supports that product release is the rate-limiting step (k2) for LeuRS as found for other class I aaRSs.
FIGURE 4.
Pre-steady state kinetic analysis of wild type (WT) and translocation-defective LeuRS mutants. Multiple turnover reactions contained 5 μm LeuRS and 25 μm tRNALeu in 60 mm Tris-HCl (pH 7.5), 10 mm MgCl2, and 1 mm dithiothreitol. The inset amplifies the initial time points, which have a burst at 0.04 s for the wild type and HE LeuRS mutant and 0.03 s for the FN2 and FN3 mutant LeuRSs. All data points represent the average of at least three separate experiments and were processed using Graph Pad Prism 4. The R-values were >0.99.
TABLE 1.
Pre-steady state kinetic parameters
| LeuRS | k1 | k2 | ktrans |
|---|---|---|---|
| s–1 | s–1 | s–1 | |
| Wild type | 21.6 ± 0.02 | 2.0 ± 0.3 | 22.0 ± 0.03 |
| FN2 | 37.7 ± 0.2 | 1.39 ± 0.01 | 42.5 ± 0.04 |
| HE | 5.7 ± 0.6 | 1.44 ± 0.04 | 6.2 ± 0.3 |
| FN3 | 34.2 ± 0.01 | 2.27 ± 0.01 | 31.1 ± 0.2 |
We hypothesize that the charged tRNA bypasses the editing-active site for these translocation mutations. The k1 and ktrans also correlated for each of the LeuRS translocation mutants and changed by only a small amount when compared with the wild type LeuRS (Table 1). This suggests that the mutants maintain the chemical reaction and competently transfer amino acid to tRNA. The most dramatic effect was about a 4-fold reduction for the HE LeuRS mutant. In this case, we hypothesized that translocation from the tRNA entrance complex to the aminoacylation complex could also be compromised by this translocation-defective mutant. Thus, although the HE mutation might not significantly change the rate-limiting product release step, this mutation could be impaired in forming the aminoacylation complex.
In contrast, k1 and ktrans increased slightly for the FN2 and FN3 LeuRS mutants (Table 1), indicating that bypassing the editing-active site in translocation-deficient mutants could also speed the process to forming the exit complex for product release. In addition to the rate-limiting product release step, the small changes in these kinetic values for the translocation-defective mutants suggest that the integrity of the chemical reactions remains intact.
Pre-steady state multiturnover analysis of each of the mutant LeuRSs showed that a burst was maintained (32), but its amplitude was reduced (Fig. 4, inset). In general, a decrease in burst amplitude has been proposed to be caused by three scenarios (32): 1) enzyme inactivation, 2) increased product release rate, and/or 3) reversal of the chemical reaction. Because the mutant enzymes exhibited robust behavior under long term steady-state analysis, it is unlikely that the protein has simply died. It is possible that a fraction of the charged tRNA product is released prematurely and that this may even be due to disruptions in tRNA translocation. However, it is striking that each mutant exhibited a k2 that is similar to the wild type LeuRS (Table 1) and supports that a rate-limiting product release step is intact. Thus, we hypothesize that at least some of the mutant population bypasses the editing site to form the exit complex prior to undergoing product release.
If the product release mechanism is maintained, then decreases in the amplitude of the burst could indicate that the internal equilibrium of the multistep aaRS reaction has shifted. However, it seems unlikely that the formation of charged tRNA from the adenylate intermediate is chemically reversed. This is particularly true because pyrophosphatase was included in the reaction to cleave PPi. Alternatively, it is possible that the effective adenylate intermediate concentration is lowered via a pretransfer editing activity that partially clears the adenylate intermediate. This might also account for the varying efficiencies by which the translocation mutants rescue the T252A mutation, which hydrolyzes correctly charged Leu-tRNALeu (7).
Previously, we showed that the canonical core of E. coli LeuRS has an inherent pretransfer editing activity that is dependent on the tRNA (33). It is masked by a dominant post-transfer editing activity. Disruption of the tRNA translocation mutation could destabilize the adenylate intermediate. In the case of this scenario, it might suggest that physical steps such as tRNA translocation from the entry complex (17) to form an aminoacylation complex could also be affected.
DISCUSSION
The aminoacylation and editing sites for LeuRS and the homologous IleRS and ValRS are separated in two distinct protein domains (10, 12) as originally predicted by the double sieve model (2, 3). Because of different molecular recognition strategies in the synthetic and hydrolytic active sites, amino acid fidelity for the synthetase is increased significantly. This fidelity increase is necessary to attain threshold levels that are required for cell survival.
Amino acid movement between the aminoacylation and editing-active sites is dependent on the tRNA. The tRNA translocation mechanism is transient and has been obscure. We hypothesized that the enzyme and tRNA would undergo a series of conformational changes throughout its multiple step reaction cycle. These conformational changes would rely on integrated chemical and physical steps. Over a dozen structures for LeuRS (reviewed in Ref. 34) show some of these changes, but the molecular determinants that facilitate transient tRNA translocation ∼30 Å between the aminoacylation and editing sites remain undefined.
Computational analysis compared the available LeuRS structures and predicted that a molecular hinge was located within a mobile peptide on the surface of the CP1 domain (18). Mutational analysis of this peptide in E. coli LeuRS demonstrated that the transient translocation step was selectively disrupted and produced mischarged tRNALeu. The aminoacylation and hydrolytic editing activity of the enzyme remained intact.
The translocation deficiency of the LeuRS mutant allowed the tRNA to effectively bypass a competent editing-active site. These mutations also rescued a T252A mutant phenotype for LeuRS that cleaved correctly charged Leu-tRNALeu. This suggests that overlapping mechanisms transiently translocate both charged and mischarged tRNAs from the aminoacylation to the editing-active site. During chemical proofreading, leucine is blocked by the critical Thr-252 specificity determinant (7), whereas non-leucine amino acids bind to the editing-active site and are hydrolyzed from mischarged tRNALeu.
The transient nature of the tRNA-protein translocation intermediate complex or complexes has likely impeded structural analysis. Disruption of one or more of these direct or indirect protein interactions with the tRNA would undermine its translocation mechanism to the editing site and allow it to proceed directly to the exit complex of the enzyme. It has been proposed that the charged tRNA in the exit complex could then be directly transferred from the class I synthetase to EF-Tu for transport to the ribosome (31).
The evolutionary addition of the CP1 domain to the canonical aminoacylation core provided a hydrolytic site capable of an editing activity to increase enzyme fidelity (13, 31). However, fidelity is also dependent on the accessibility and transfer of the mischarged tRNA between two enzyme domains to the editingactive site prior to product release for protein synthesis at the ribosome. For example, IleRS, which relies on tRNA-dependent pretransfer editing, utilizes Lys-183 and Trp-421 to facilitate translocation of the adenylate intermediate (35). For posttransfer editing, the LeuRS translocation peptide provides a mechanism to ensure faithful screening of all charged tRNAs in the editing site to prevent errors that would lead to genetic code ambiguity.
Acknowledgments
We thank Dr. V. Karkhanis for advice on complementation assays as well as Drs. M. Boniecki and M. Vu for assistance with rapid quench kinetic experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant GM 063789 (to S. A. M.).
Footnotes
The abbreviations used are: aaRS, aminoacyl-tRNA synthetase; LeuRS, leucyl-tRNA synthetase; IleRS, isoleucyl-tRNA synthetase; ValRS, valyl-tRNA synthetases.
References
- 1.Hendrickson, T., and Schimmel, P. (2003) in Translation Mechanisms (Lapointe, J., and Brakier-Gingras, L., eds), pp. 34-64, Landes Bioscience and Kluwer Academic/Plenum Publishers
- 2.Fersht, A. R., and Dingwall, C. (1979) Biochemistry 18, 2627-2631 [DOI] [PubMed] [Google Scholar]
- 3.Fersht, A. R. (1998) Science 280, 541. [DOI] [PubMed] [Google Scholar]
- 4.Karkhanis, V. A., Boniecki, M. T., Poruri, K., and Martinis, S. A. (2006) J. Biol. Chem. 281, 33217-33225 [DOI] [PubMed] [Google Scholar]
- 5.Nangle, L. A., Zhang, W., Xie, W., Yang, X. L., and Schimmel, P. (2007) Proc. Natl. Acad. Sci. U. S. A. 104, 11239-11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee, J. W., Beebe, K., Nangle, L. A., Jang, J., Longo-Guess, C. M., Cook, S. A., Davisson, M. T., Sundberg, J. P., Schimmel, P., and Ackerman, S. L. (2006) Nature 443, 50-55 [DOI] [PubMed] [Google Scholar]
- 7.Mursinna, R. S., Lincecum, T. L., Jr., and Martinis, S. A. (2001) Biochemistry 40, 5376-5381 [DOI] [PubMed] [Google Scholar]
- 8.Swairjo, M. A., and Schimmel, P. R. (2005) Proc. Natl. Acad. Sci. U. S. A. 102, 988-993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Starzyk, R. M., Burbaum, J. J., and Schimmel, P. (1989) Biochemistry 28, 8479-8484 [DOI] [PubMed] [Google Scholar]
- 10.Silvian, L. F., Wang, J., and Steitz, T. A. (1999) Science 285, 1074-1077 [PubMed] [Google Scholar]
- 11.Cusack, S., Yaremchuk, A., and Tukalo, M. (2000) EMBO J. 19, 2351-2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukai, S., Nureki, O., Sekine, S., Shimada, A., Tao, J., Vassylyev, D. G., and Yokoyama, S. (2000) Cell 103, 793-803 [DOI] [PubMed] [Google Scholar]
- 13.Betha, A. K., Williams, A. M., and Martinis, S. A. (2007) Biochemistry 46, 6258-6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, L., Hale, S. P., and Schimmel, P. (1996) Nature 384, 33-34 [DOI] [PubMed] [Google Scholar]
- 15.Zhao, M. W., Zhu, B., Hao, R., Xu, M. G., Eriani, G., and Wang, E. D. (2005) EMBO J. 24, 1430-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lincecum, T. L., Jr., Tukalo, M., Yaremchuk, A., Mursinna, R. S., Williams, A. M., Sproat, B. S., Van Den Eynde, W., Link, A., Van Calenbergh, S., Grøtli, M., Martinis, S. A., and Cusack, S. (2003) Mol. Cell 11, 951-963 [DOI] [PubMed] [Google Scholar]
- 17.Rock, F. L., Mao, W., Yaremchuk, A., Tukalo, M., Crepin, T., Zhou, H., Zhang, Y. K., Hernandez, V., Akama, T., Baker, S. J., Plattner, J. J., Shapiro, L., Martinis, S. A., Benkovic, S. J., Cusack, S., and Alley, M. R. (2007) Science 316, 1759-1761 [DOI] [PubMed] [Google Scholar]
- 18.Mascarenhas, A. P., and Martinis, S. A. (2008) Biochemistry 47, 4808-4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellinger, T., and Ehricht, R. (1998) BioTechniques 24, 718-720 [DOI] [PubMed] [Google Scholar]
- 20.Sampson, J. R., and Uhlenbeck, O. C. (1988) Proc. Natl. Acad. Sci. U. S. A. 85, 1033-1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puglisi, J. D., and Tinoco, I., Jr. (1989) Methods Enzymol. 180, 304-325 [DOI] [PubMed] [Google Scholar]
- 22.Martinis, S. A., and Fox, G. E. (1997) Nucleic Acids Symp. Ser. 36, 125-128 [PubMed] [Google Scholar]
- 23.Low, B., Gates, F., Goldstein, T., and Söll, D. (1971) J. Bacteriol. 108, 742-750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabor, S., and Richardson, C. C. (1985) Proc. Natl. Acad. Sci. U. S. A. 82, 1074-1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Francklyn, C. S., First, E. A., Perona, J. J., and Hou, Y. M. (2008) Methods (Amst.) 44, 100-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guth, E., Connolly, S. H., Bovee, M., and Francklyn, C. S. (2005) Biochemistry 44, 3785-3794 [DOI] [PubMed] [Google Scholar]
- 27.Schreier, A. A., and Schimmel, P. R. (1972) Biochemistry 11, 1582-1589 [DOI] [PubMed] [Google Scholar]
- 28.Krebs, W. G., and Gerstein, M. (2000) Nucleic Acids Res. 28, 1665-1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerstein, M., and Krebs, W. (1998) Nucleic Acids Res. 26, 4280-4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tukalo, M., Yaremchuk, A., Fukunaga, R., Yokoyama, S., and Cusack, S. (2005) Nat. Struct. Mol. Biol. 12, 923-930 [DOI] [PubMed] [Google Scholar]
- 31.Zhang, C. M., Perona, J. J., Ryu, K., Francklyn, C., and Hou, Y. M. (2006) J. Mol. Biol. 361, 300-311 [DOI] [PubMed] [Google Scholar]
- 32.Johnson, K. A. (2005) in Encyclopedia of Life Sciences (Cox, M. M., and Phillips, G. N., eds) Vol. 1, pp. 1-7, John Wiley and Sons, Inc., Hoboken, NJ [Google Scholar]
- 33.Boniecki, M. T., Vu, M. T., Betha, A. K., and Martinis, S. A. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 19223-19228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mascarenhas, A. P., Rosen, A., S., Martinis, S. A., and Musier-Forsyth, K. (2007) Protein Engineering (Musier-Forsyth, K., Ed.), Springer-Verlag New York Inc., New York
- 35.Bishop, A. C., Beebe, K., and Schimmel, P. R. (2003) Proc. Natl. Acad. Sci. U. S. A. 100, 490-494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, K. W., and Briggs, J. M. (2004) Proteins 54, 693-704 [DOI] [PubMed] [Google Scholar]