Abstract
Apoptosis is a highly organized, energy-dependent program by which multicellular organisms eliminate damaged, superfluous, and potentially harmful cells. Although caspases are the most prominent group of proteases involved in the apoptotic process, the role of lysosomes has only recently been unmasked. This study investigated the role of the lysosomal serine protease CLN2 in apoptosis. We report that cells isolated from patients affected with late infantile neuronal ceroid lipofuscinosis (LINCL) having a deficient activity of CLN2 are resistant to the toxic effect of death ligands such as tumor necrosis factor (TNF), CD95 ligand, or tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) but not to receptor-independent stress agents. CLN2-deficient cells exhibited a defect in TNF-induced Bid cleavage, release of cytochrome c, and caspase-9 and -3 activation. Moreover, extracts from CLN2-overexpressing cells or a CLN2 recombinant protein were able to catalyze the in vitro cleavage of Bid. Noteworthy, correction of the lysosomal enzyme defect of LINCL fibroblasts using a medium enriched in CLN2 protein enabled restoration of TNF-induced Bid and caspase-3 processing and toxicity. Conversely, transfection of CLN2-corrected cells with small interfering RNA targeting Bid abrogated TNF-induced cell death. Altogether, our study demonstrates that genetic deletion of the lysosomal serine protease CLN2 and the subsequent loss of its catalytic function confer resistance to TNF in non-neuronal somatic cells, indicating that CLN2 plays a yet unsuspected role in TNF-induced cell death.
Apoptosis is a highly organized, energy-dependent program by which multicellular organisms eliminate potentially harmful, superfluous, and damaged cells. Although caspases are the most prominent group of proteases involved in the apoptotic process, the role of lysosomes and more particularly of lysosomal cathepsins in cell death has only recently been unmasked (1-6). Release of cathepsins from the lysosomal lumen to the cytosol is a prerequisite for their participation in the regulation of apoptosis and has been described in response to a variety of death stimuli such as inducers of cell-surface TNF2 receptor family (7-10), chemotherapeutic drugs (11, 12), or nonreceptor-mediated apoptotic agents (13, 14). This lysosomal membrane permeabilization often relies on the activation of the intrinsic apoptosis pathway, which involves mitochondrial membrane permeabilization with the consequent release of the pro-apoptotic mitochondrial proteins into the cytosol. These factors lead to caspase activation and ultimately to cell death.
In addition to the cathepsin-dependent pathway, some reports have indicated that other lysosomal proteins can modulate or mediate some cell death programs. Indeed, overexpression of CLN1/palmitoyl protein thioesterase 1 (15) or CLN3/battenin (16) protected neuronal cells from stress-induced apoptosis. Moreover, we recently demonstrated that the apoptosis defect, reported in fibroblasts derived from patients affected with I-cell disease having a deficient activity of almost all lysosomal hydrolases (17, 18), could be partially corrected when the activity of CLN2/tripeptidyl peptidase 1 had been restored in these mutant cells (18). However, how this lysosomal protease is connected with the apoptotic machinery has never been studied.
The lysosomal serine protease CLN2 (EC 3.4.14.9) is the only hydrolase with tripeptidyl peptidase activity identified to date in the lysosomes of mammalian cells. This mannose 6-phosphorylated protease has an exopeptidase activity with an acidic pH optimum and cleaves off tripeptides sequentially from unsubstituted N termini of polypeptides or proteins. CLN2 also exhibits an endoprotease activity that may be important for the low pH-triggered intramolecular autoactivation of the inactive proenzyme to the mature form (19). Indeed, like caspases and cathepsins, CLN2 is synthesized as an inactive zymogen, and its activation involves proteolytic processing to yield a mature enzyme of 368 amino acid residues (∼48 kDa). Most mutations in the CLN2 gene lead to abolishment of the enzymatic activity and are the direct cause of a fatal childhood inherited neurodegenerative disease, the classical late-infantile form of neuronal ceroid lipofuscinosis (LINCL) (20-22). Although the structure of CLN2 has been recently studied (23, 24), its function and its biologically relevant substrates remain enigmatic. It was reported that in vitro, CLN2 is able to cleave peptide hormones such as angiotensin II (25), glucagon (26), substance P (25), sulfated cholecystokinin-8 (27), and neuromedin B (28), as well as synthetic amyloid-β peptides (25), and probably collagen (29). The proteolipid subunit c of ATP synthase has been also implicated as a potential biological target of the protease (30, 31). However, this protein/substrate not only accumulates in LINCL but also in a number of different forms of neuronal ceroid lipofuscinosis and other unrelated lysosomal storage diseases (32), suggesting that it is unspecific to CLN2.
To assess the yet undetermined role of CLN2 in apoptosis, we have used fibroblasts or lymphoblasts isolated from patients affected with LINCL having a catalytically inactive but stable CLN2 protein. The susceptibility of mutant cells to various apoptotic inducers was compared with that of cells derived from healthy patients, as were the cell death-signaling pathways. Here we show that apoptosis induced by TNF as well as related ligands was strongly inhibited in CLN2-deficient cells as compared with control cells suggesting a key role of this serine protease in cell death. We also demonstrate that the Bcl-2 family pro-apoptotic member Bid can be directly cleaved by CLN2.
EXPERIMENTAL PROCEDURES
Reagents—Human recombinant TNF and TRAIL were purchased from PeproTech-Tebu (Le Perray-en-Yvelines, France). Anti-CD95 (clone CH-11) was from Beckman-Coulter (Marseille, France), and CD95 ligand was recovered from the culture medium of transfected Neuro2a cells that overexpress a murine CD95 ligand (33). Purified recombinant human CLN2 proenzyme was kindly provided by Dr. P. Lobel (Piscataway, NJ). Recombinant human Bid and caspase-8 were obtained from R & D Systems Europe (Lille, France) and Abcys (Paris, France), respectively. Ac-Asp-Glu-Val-Asp-aminomethylcoumarin (Ac-DEVD-AMC) was from Coger (Paris, France). Ala-Ala-Phe-fluoromethyl ketone (AAF-cmk) was from Bachem (Voisins-Le-Bretonneux, France). DMEM, trypsin/EDTA, fetal calf serum, penicillin, and streptomycin were from Invitrogen. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride (AEBSF), N-acetyl-d-sphingosine (C2-ceramide), and staurosporine were supplied from Euromedex (Mundolsheim, France). Other reagents, including cycloheximide, were from Sigma.
Cell Lines—Transformed skin fibroblasts derived from control individuals or from patients affected with LINCL were obtained as described previously (34). Briefly, subconfluent primary cultures of skin fibroblasts were transfected with a plasmid (pAS, obtained from Dr. J. Feunteun, Villejuif, France) encoding the SV40 large T antigen, using electroporation. After 4-5 weeks, typical manifestation of unlimited growth potential without cell contact inhibition was obtained. Control and mutant fibroblasts were treated identically to achieve immortalization. These transformed cells were grown in a humidified 5% CO2 atmosphere at 37 °C in DMEM containing Glutamax (2 mm), penicillin (100 units/ml), streptomycin (100 μg/ml), and heat-inactivated FCS (10%). Human Epstein-Barr virus-transformed lymphoid cell lines were obtained from peripheral blood lymphocytes derived from control subjects or from patients affected with LINCL and were grown in RPMI 1640 medium containing 10% FCS. For this purpose, mononuclear cells were infected with Epstein-Barr virus (B95) supernatant.
CHO cells engineered to overexpress and secrete a wild-type (wt-) (35) or a mutant (S475L-) (24) form of CLN2 were kindly provided by Dr. P. Lobel (Piscataway, NJ) and Dr. A. Golabek (Staten Island, NY), respectively. The CHO cell line stably expressing CLN1 (36) was a gift from Dr. A. Jalanko (Helsinki, Finland). Control and modified CHO cells were cultured in DMEM.
Enzymatic Correction Assay—Control, CLN2wt-, or CLN2-S475L-CHO cells were incubated for 48 h in serum-free DMEM. Then the culture medium of these cells was harvested (centrifuged and filtrated) and added to LINCL fibroblasts for 24 h in the presence or absence of 50 μm AAF-cmk.
Cytotoxicity Assay—After treatment with cytotoxic agents, cell viability was evaluated by using the tetrazolium-based MTT assay (37), and the absorbance was measured at 560 nm.
Fluorogenic DEVD Cleavage Enzyme Assay—After incubation with TNF or anti-CD95 and cycloheximide, cells were sedimented and washed with phosphate-buffered saline (PBS). Cell pellets were homogenized in 10 mm HEPES (pH 7.4), 42 mm KCl, 5 mm MgCl2, 0.5% CHAPS, 1 mm dithiothreitol, 1 mm PMSF, and 2 μg/ml leupeptin. Reaction mixtures contained 100 μl of cell lysates and 100 μl of 40 μm Ac-DEVD-AMC. After 30 min of incubation at room temperature, the amount of the released fluorescent product aminomethylcoumarin was determined at 351 and 430 nm for the excitation and emission wavelengths, respectively.
Release of Cytochrome c—After incubation with TNF and cycloheximide, cells were sedimented and washed with PBS. Cell pellets were resuspended in 5 volumes of ice-cold homogenization buffer (20 mm HEPES/KOH (pH 7.4), 1 mm EDTA, 0.1% fatty acid-free bovine serum albumin, 250 mm sucrose, 1 mm dithiothreitol, 0.1 mm PMSF, 20 μg/ml leupeptin, 10 μg/ml aprotinin, and 10 μg/ml pepstatin A). After swelling for 10 min on ice, cells were homogenized by 15 strokes of a loose-fitting Dounce homogenizer. The suspension was then centrifuged at 750 × g for 5 min at 4 °C, and post-nuclear supernatants were centrifuged at 10,000 × g for 15 min at 4 °C. Equal amounts of protein were then analyzed by SDS-PAGE (15% gel) and Western blotting by using 1 μg/ml anti-cytochrome c mAb (BD Biosciences).
Western Blot Analyses—Equal amounts of proteins were separated in a 10-15% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane (Bio-Rad). Protein complexes were detected using an ECL detection system (Pierce). Polyclonal anti-FADD, polyclonal anti-Bid, monoclonal anti-caspase-8, monoclonal anti-caspase-3, polyclonal anti-caspase-9, polyclonal anti-cytochrome c, polyclonal anti-HSP70, and polyclonal anti-cIAP-1 antibodies were purchased from Cell Signaling Technology and used at 1/1000 dilution. CLN2 was detected by using a monoclonal antibody (mAbs 8C4) given by Dr. A. Golabek (Staten Island, NY); an anti-β-actin (Sigma) was used as a control for protein loading.
CLN2/Tripeptidyl Peptidase-1 Assay—CLN2 activity was assayed using H-Ala-Ala-Ala-Phe-aminomethylcoumarin (Bachem, Voisins-Le-Bretonneux), as reported (24). Cell pellets were homogenized in 600 μl of sodium acetate buffer, 50 mm (pH 4), with 0.1% Triton X-100, briefly sonicated, and centrifuged at 10,000 × g for 5 min at 4 °C. Reaction mixtures contained 150 μl of supernatant and 50 μl of substrate preparation (400 μm substrate in sodium acetate buffer, 80 mm (pH 4), with 20 mm EDTA). After 30 min of incubation at 37 °C, the reaction was stopped by adding 800 μl of sodium acetate buffer, 100 mm (pH 3.5). The amount of the released fluorescent product AMC was determined by fluorometry at 351 and 430 nm for the excitation and emission wavelengths, respectively. Protein concentration was measured with the Bio-Rad dye reagent using bovine serum albumin as a standard.
Fluorescence Microscopy—Transformed fibroblasts plated onto glass coverslips were fixed/permeabilized in methanol at -20 °C for 20 min and washed three times with PBS. Then the cells were incubated for 1 h with a mouse monoclonal anti-CLN2 antibody and washed before incubation for 1 h with species-specific secondary antibody conjugated with Alexa Fluor 488. The immunostaining protocol was repeated with a mouse monoclonal anti-LAMP1 (from the Developmental Studies Hybridoma Bank, University of Iowa) and then with a secondary antibody conjugated with Alexa Fluor 568. The coverslips were mounted with Prolong Gold (Invitrogen) and viewed with a Zeiss LSM 510 confocal microscope equipped with a 63 plan-apochromat objective. As a control, primary antibodies were omitted.
Transfection with siRNA and Treatment—To suppress the expression of Bid, duplexed siRNA targeting this human protein (sc-29800, Santa Cruz Biotechnology) were transfected into CLN2-corrected LINCL fibroblasts grown to 70-90% confluence using Hyperfect (Qiagen) reagent. For each transfection, 5 nm siRNA were used and incubated in DMEM without FCS for 24 h. After transfection, TNF (5 ng/ml) and cycloheximide (50 μg/ml) were added to the medium, and cells were further incubated for 24 h to determine cell viability.
In Vitro Bid Cleavage Assay—Cellular extracts from control, CLN2wt, or CLN1wt CHO cells were homogenized in 50 mm sodium formate buffer (pH 3.5) with 0.1% Triton X-100 and briefly sonicated at 4 °C. Reaction mixtures contained 25 μg of protein, preincubated or not with 1 mm PMSF, 1 mm AEBSF, or 250 μm AAF-cmk for 15 min at room temperature, and 1.5 μg of human recombinant Bid protein. After 30 min of incubation at 37 °C, reaction products were analyzed by SDS-PAGE (15% gel) and Western blotting by using an anti-Bid mAb. Similar experiments were performed by replacing cellular extracts by 1-2 μg of CLN2 recombinant proenzyme that was preincubated in 50 mm sodium formate buffer (pH 3.5) or in 50 mm HEPES buffer (pH 7.5) with 150 mm NaCl before its incubation with Bid.
TNFR1 Expression Analysis by Flow Cytometry—TNFR1 expression was analyzed on resting cells. Cells were detached using PBS containing 10 mm EDTA, sedimented at 4 °C and washed with PBS. Cells were incubated at 4 °C in the dark for 30 min with fluorescein isothiocyanate-conjugated mouse anti-human TNFR1 (R & D Systems, Lille, France). Isotype control monoclonal antibody (Immunotech) was used as a negative control. Cytometric analyses were performed on a FACScan (BD Biosciences) flow cytometer.
Statistical Analyses—Data are presented as means ± S.E. The Student's t test was used for statistical analysis (*, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control).
RESULTS
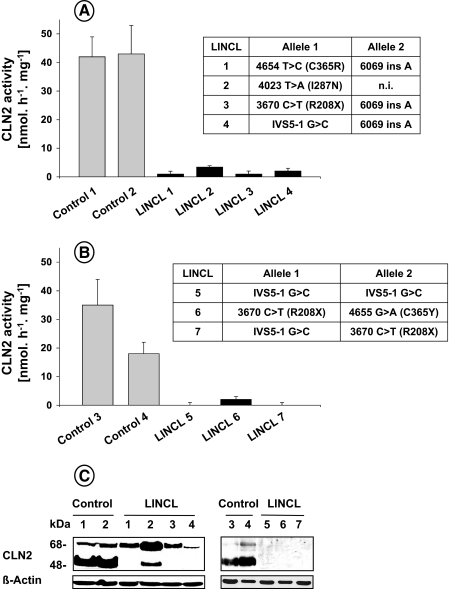
Death Receptor-induced Cell Killing Requires a Catalytically Active CLN2 Protein—To address the functional role of CLN2 in cell death, we first used a genetic approach by testing two cellular models, i.e. SV40-transformed fibroblasts and Epstein-Barr virus-transformed lymphoid cell lines derived from patients affected with LINCL. As compared with control fibroblasts (Fig. 1A) or lymphoblasts (Fig. 1B) derived from healthy individuals, the apparent CLN2 enzymatic activity in all LINCL cells was severely impaired independently of the nature of the mutations reported in the CLN2 gene (e.g. nonsense, missense, or splice-junction mutations). In contrast, the activities of other lysosomal enzymes (e.g. β-galactosidase and β-hexosaminidase) in LINCL cells were not different from those in controls (data not shown). Moreover, when lysates of mutant cells were analyzed on immunoblots using a mAb against the human CLN2 protein, the 48-kDa mature form was most often undetectable (Fig. 1C), demonstrating the absence of a catalytically active protein in these models.
FIGURE 1.
Deficient CLN2 activity in cell lines derived from patients affected with LINCL. CLN2 activity was determined in lysates of SV40-transformed fibroblasts (A) and Epstein-Barr virus-transformed lymphoid cell lines (B) derived from control subjects or different LINCL patients (means ± S.E. of 4-6 independent determinations). The insets correspond to the nature of the mutations reported for LINCL patients. Nucleotides are numbered according to AF039704; n.i., not identified. C, cellular lysates were prepared and analyzed by Western blotting using a monoclonal antibody against the CLN2 protein. Results are representative of three independent experiments.
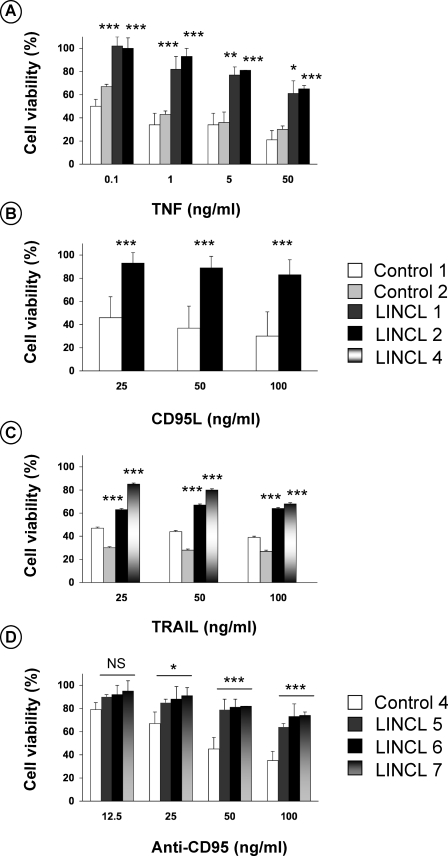
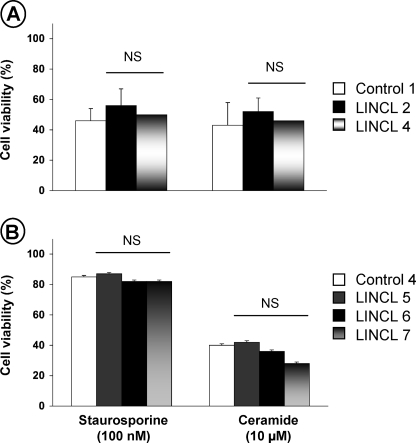
The sensitivity of CLN2-deficient cells to death ligands was evaluated. For this purpose, control and LINCL fibroblasts were exposed to TNF, CD95 ligand, or TRAIL. All these stimuli are known to trigger apoptosis in the presence of cycloheximide. Fig. 2 demonstrates that, after 16 h of exposure, these agents led to a dose-dependent reduction in the cell viability of control fibroblasts (Fig. 2, A-C, respectively). Under all concentrations tested, LINCL fibroblasts were more resistant to the lethal effects of death ligands than their normal counterparts. The resistance of CLN2-deficient cells was also observed on lymphoid cell lines treated with an agonistic anti-CD95 (CH-11) antibody (Fig. 2D). Altogether, these findings demonstrate the involvement of CLN2 in death receptor-mediated cytotoxicity. Finally, to assess the possible involvement of CLN2 in cell death induced by other cytotoxic drugs, control and mutant fibroblasts were exposed to the synthetic short chain C2-ceramide or to the protein kinase inhibitor staurosporine, which have been shown to induce apoptosis by activating the mitochondrial pathway. As illustrated in Fig. 3, A and B, control and LINCL cells were equally sensitive to the cytotoxic effects of these agents, indicating that CLN2 does not regulate receptor-independent cytotoxic signaling.
FIGURE 2.
CLN2-deficient cells are resistant to death receptor-mediated cytotoxicity. Control and LINCL-transformed fibroblasts were incubated for 16 h in DMEM containing 1% FCS and 50 μg/ml cycloheximide in the presence or absence of the indicated concentrations of TNF (A), CD95L (B), or TRAIL (C). Cell viability was assessed using the MTT assay and is expressed as percentage of the viability of cells treated with cycloheximide only. D, viability of control and LINCL Epstein-Barr virus-transformed lymphoid cell lines that were incubated for 24 h in RPMI medium containing 1% FCS in the presence of the indicated concentration of anti-CD95. Results are means ± S.E. of three independent experiments (all in triplicate).
FIGURE 3.
CLN2-deficient cells are not resistant to receptor-independent cytotoxic agents. Control and LINCL-transformed fibroblasts (A) or Epstein-Barr virus-transformed lymphoid cell lines (B) were incubated for 16 or 24 h, respectively, in medium containing 1% FCS with 100 nm staurosporine or 10 μm C2-ceramide. Cell viability was assessed using the MTT assay. Results are means ± S.E. of 3-6 independent determinations (all in triplicate). NS, not significant.
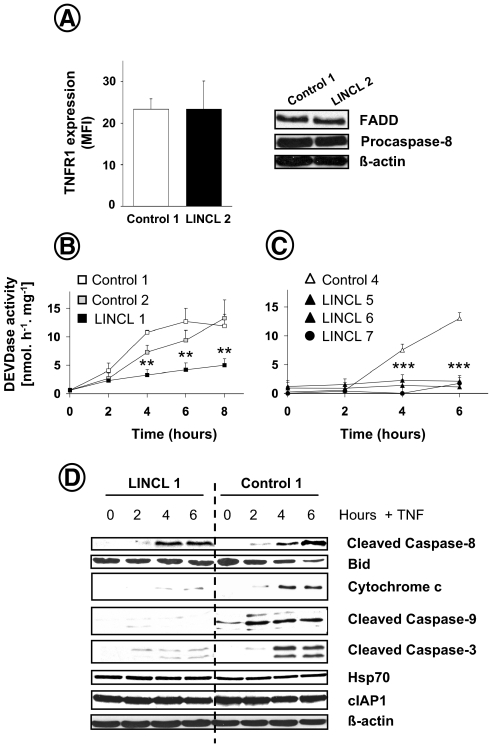
CLN2 Is Involved in Death Receptor Apoptotic Signaling—To substantiate the implication of the lysosomal serine protease CLN2 in ligand-induced cell death, the apoptotic cascade activated by TNF in LINCL-transformed fibroblasts was examined. Of note, cell surface TNFR1 levels, analyzed by flow cytometry, were similar in normal and LINCL cells (Fig. 4A, left). Moreover, the expression of the TNFR1-associated signaling proteins that form the apoptosis-inducing complex, such as the adaptor FADD and the initiator procaspase-8, was not perturbed in CLN2-deficient cells (Fig. 4A, right). By measuring caspase activity with the fluorogenic tetrapeptide substrate Ac-DEVD-AMC, which contains the cleavage site found in several caspase-3 and caspase-7 targets, we demonstrated that TNF-induced caspase processing was strongly impaired in CLN2-deficient cells (Fig. 4B). Consistent with these results, the time-dependent activation of effector caspases observed in anti-CD95-treated control lymphoblasts was also abrogated in three different LINCL lymphoid cells (Fig. 4C).
FIGURE 4.
TNF-induced apoptotic cascade is impaired in CLN2-deficient cells. A, expression of cell surface TNFR1, FADD, and procaspase-8 in control and LINCL fibroblasts, as assessed by flow cytometry and Western blot, respectively. Fluorescence intensity of individual cells is reported as means of arbitrary fluorescence units (mean fluorescence intensity (MFI)) ± S.E. of three independent experiments. Control and LINCL-transformed fibroblasts (B) for the indicated times or Epstein-Barr virus-transformed lymphoid cell lines (C) were incubated in medium containing 1% FCS with 5 ng/ml TNF and 50 μg/ml cycloheximide or 50 ng/ml anti-CD95, respectively. Cells were then harvested, and DEVDase activity was determined. Data are means ± S.E. of three independent experiments. D, control and LINCL fibroblasts were treated for the indicated periods with 50 μg/ml cycloheximide and 5 ng/ml TNF. Cell lysates were prepared and analyzed by Western blotting for Bid proform disappearance, cytochrome c release, caspase-8, -9, and -3 cleavage, Hsp70, and cIAP1 expression. An anti-β-actin was used as a control for protein loading. Results are representative of three independent experiments.
To further dissect the role of CLN2 in apoptosis, we performed Western blot analyses of apoptotic mediators. As illustrated in Fig. 4D, TNF-induced caspase-8 activation was not altered in CLN2-deficient fibroblasts. However, disappearance of the pro-apoptotic Bcl-2-like protein Bid and progressive release of cytochrome c into the cytosol were inhibited in mutant cells. In addition, the cleavage of caspase-9 and caspase-3 triggered by the cytokine was also considerably reduced in LINCL cells. Altogether, these observations demonstrate the involvement of CLN2 in the apoptotic cascade initiated by TNF with a site of action likely lying upstream of mitochondria.
Resistance of CLN2-deficient Cells to TNF Is Not Because of Up-regulation of cIAP1 and Hsp70 Anti-apoptotic Proteins—Because the inhibitor of apoptosis cIAP1 and the major heat shock protein Hsp70 are known to counteract the proapoptotic action of TNF by interacting with the tumor necrosis factor receptor-associated factor TRAF2 (38) or by inhibiting the lysosomal membrane destabilization (39), respectively, we examined whether the expression level of these proteins was affected by CLN2 inactivation. As shown in Fig. 4D, similar levels of cIAP1 and Hsp70 were detected in control and CLN2-deficient cells. These data indicate that the resistance of the mutant cells is not because of an increased expression of these anti-apoptotic proteins.
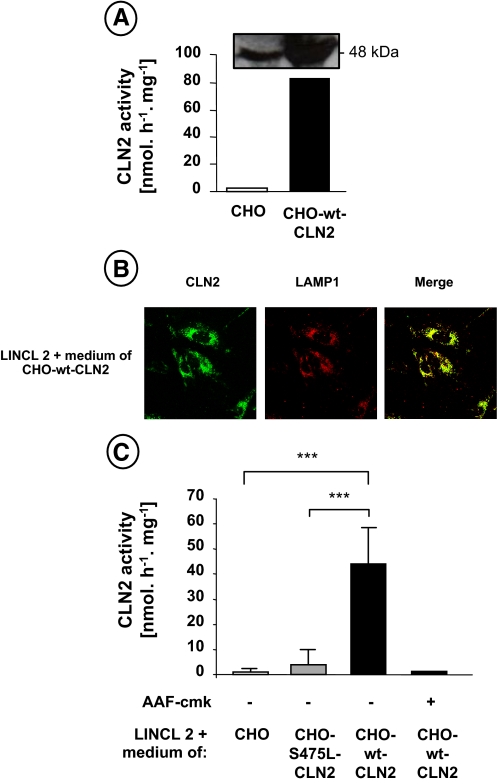
Restoration of TNF-induced Apoptotic Signaling in Catalytically Active CLN2-corrected LINCL Fibroblasts—To establish that the resistance of LINCL fibroblasts to TNF-triggered cell death was because of the loss of CLN2 catalytic function, we next investigated the effect of CLN2 enzymatic correction of these mutant cells on their sensitivity to TNF. To this end, CLN2-deficient fibroblasts were incubated with the extracellular medium of CHO cells overexpressing and secreting either a wild-type (35) (see Fig. 5A) or a mutated (S475L-) (24) form of CLN2. In this system, the extracellular glycoprotein CLN2 is efficiently endocytosed and targeted to the lysosome of mutant fibroblasts through mannose 6-phosphate receptor-mediated endocytosis (35). To confirm the lysosomal distribution of CLN2 in corrected cells, we analyzed by immunofluorescence the subcellular localization of CLN2 and LAMP-1, a lysosomal-associated membrane protein. Under basal conditions, CLN2 fluorescence was punctate with an intense co-localization with LAMP-1 staining (Fig. 5B). The same strong granular staining and co-localization were also obtained in LINCL fibroblasts that were stably transfected with a cDNA encoding human CLN2 (data not shown). Moreover, as shown in Fig. 5C, when CLN2-deficient cells were incubated for 48 h with the extracellular medium of CHO cells overexpressing the integral CLN2 protein, the intracellular activity of this enzyme was restored as compared with CLN2-deficient cells incubated with the medium of control CHO cells or CHO cells overexpressing an inactive mutant form of CLN2 (see Fig. 1A for comparison).
FIGURE 5.
Enzymatic correction of CLN2 activity in LINCL fibroblasts. A, expression and activity of CLN2 in the extracellular medium of control CHO cells (CHO) or CHO cells overexpressing a wild-type form of CLN2 (CHO-wt-CLN2). LINCL fibroblasts were incubated for 24 h with the extracellular medium of control (CHO) or CHO cells overexpressing a wild-type (CHO-wt-CLN2) or a mutated (CHO-S475L-CLN2) form of CLN2. Then intralysosomal localization of CLN2 was monitored by immunofluorescence microscopy in CLN2-corrected cells (B). Cells were fixed, permeabilized, incubated with anti-CLN2 and anti-LAMP1 antibodies, and stained with Alexa Fluor 488-labeled (green fluorescence) and Alexa Fluor 568-labeled (red fluorescence) secondary antibodies, respectively. Immunofluorescence images were recorded by laser scanning confocal microscopy. The merged fluorescence indicates the co-localization of the signals. Intracellular CLN2 activity was determined after preincubation of the cells in the presence or absence of 50 μm AAF-cmk (C).
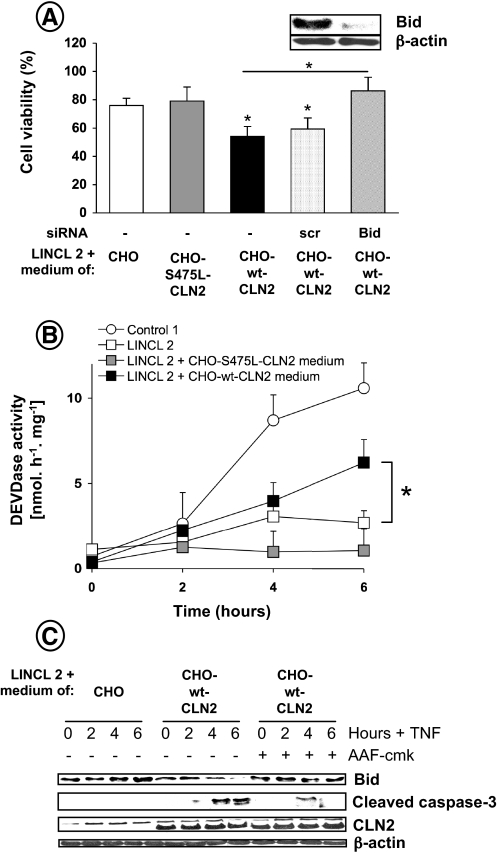
Of particular interest was the finding that CLN2-corrected mutant fibroblasts became more sensitive to TNF-induced cytotoxicity, as compared with CLN2-deficient cells incubated with the medium of control CHO cells or CHO cells overexpressing an inactive form of CLN2 (Fig. 6A). This sensitivity was further confirmed by examining caspase activity. As shown in Fig. 6B, the cleavage of Ac-DEVD-AMC was significantly increased in CLN2-corrected cells. These results were confirmed by Western blot analysis of caspase-3 processing (Fig. 6C). Moreover, correction of the lysosomal enzymatic defect of CLN2 fibroblasts restored Bid cleavage, whereas pharmacological inhibition of the enzyme with the tripeptidyl chloromethyl ketone AAF-cmk, a strong inhibitor of CLN2 activity (Fig. 5C) (26), abolished this processing (Fig. 6C). A similar phenomenon was observed for caspase-3 cleavage. Finally, transfection of CLN2-corrected cells with siRNA targeting Bid, which reduced Bid protein contents by ∼90% (Fig. 6A, inset), reduced TNF cytotoxicity (Fig. 6A). Altogether, these data indicate that CLN2 plays a critical role in TNF apoptotic signaling by regulating Bid activation.
FIGURE 6.
Enzymatic correction of CLN2 activity in LINCL fibroblasts restores TNF-induced apoptosis. A, TNF-induced cell death was monitored in cells transiently transfected or not with 5 nm Bid or scrambled (scr) siRNA and stimulated for 16 h with 50 μg/ml cycloheximide in the presence or absence of 5 ng/ml TNF. The effectiveness of Bid protein knockdown by siRNA was assessed by immunoblotting using a polyclonal anti-Bid antibody (inset). Cell viability was assessed by MTT assay. Results are means ± S.E. of 3-4 independent determinations (all in triplicate). Cells, preincubated or not with 50 μm AAF-cmk, were stimulated with 50 μg/ml cycloheximide in the presence or absence of 5 ng/ml TNF for the indicated times. Cells were then harvested, and DEVDase activity was determined (B). Data are means ± S.E. of 3-4 independent experiments. C, analysis of Bid cleavage, caspase-3 processing, and CLN2 expression by Western blot. An anti-β-actin was used as a control for protein loading.
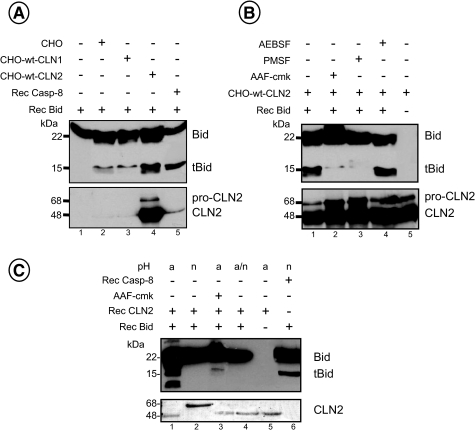
In Vitro Cleavage of Bid by CLN2—The proteolytic activation of Bid has been reported to represent a mechanism through which lysosomal proteases could trigger the mitochondrial membrane permeabilization (6). Because we observed that CLN2 preferentially acts upstream of mitochondrial dysfunction, we examined the effect of CLN2-enriched cellular extracts or a CLN2 recombinant protein on the cleavage of a recombinant full-length Bid protein, and we analyzed the products by Western blotting along with caspase 8-cleaved Bid for comparison. As shown in Fig. 7A, incubation of cellular extracts from CHO cells overexpressing the integral human CLN2 protein resulted in the cleavage of Bid (lane 4) leading to bands that co-migrated with those induced by recombinant caspase-8 (lane 5). Of note, the faint cleavage of Bid observed when the Bcl-2 homolog was incubated with extracts derived from control cells (Fig. 7A, lane 2) or CHO cells overexpressing the lysosomal palmitoyl protein thioesterase 1 (CLN1) (lane 3), as a negative control, was probably because of endogenous caspase-8 or other proteases than CLN1. Moreover, incubation of CLN2-enriched cellular extracts with AAF-cmk almost completely abolished Bid processing (Fig. 7B, lane 2). This cleavage was also blocked by the serine protease inhibitor PMSF (Fig. 7B, lane 3) but not by AEBSF (lane 4), which has been described to inhibit a serine protease implicated in the processing of the CLN2 proenzyme to the mature active form in vivo (40). However, CLN2-enriched cellular extracts already contained the 48-kDa mature enzyme as illustrated in Fig. 7B (lower panel).
FIGURE 7.
In vitro cleavage of Bid by CLN2. Cellular lysates (25 μg) from control CHO cells (CHO) or CHO cells overexpressing a wild-type form of CLN2 (CHO-wt-CLN2) or CLN1 (CHO-wt-CLN1) preincubated for 15 min in the absence (A) or presence (B) of PMSF (1 mm), AEBSF (1 mm), or AAF-cmk (250 μm) were incubated for 30 min at 37 °C with 1.5 μg of human recombinant Bid protein. Then Western blotting analyses were performed using either an anti-Bid or an anti-CLN2 mAb. A recombinant caspase-8 protein (1 μg) was used as a control for Bid cleavage. C, CLN2 recombinant proenzyme (1-2 μg) preincubated in acidic (a) or neutral (n) buffer was incubated for 30 min at 37 °C with 1.5 μg of human recombinant Bid protein and then analyzed by Western blot. a/n (lane 4) corresponds to CLN2 that was preincubated first in a small volume of an acidic buffer and then incubated with Bid under neutral conditions. Results are representative of at least three independent experiments.
To confirm that the effect of CLN2-overexpressing CHO cells on Bid proteolysis was mainly caused by CLN2, we tested whether a CLN2 recombinant protein purified from the medium of CLN2-overexpressing CHO cells (35) could cleave Bid. Upon acidification, this inactive soluble proenzyme (68 kDa) is autocatalytically processed to the mature form and acquires activity (Fig. 7C, lower panel, lanes 1 and 2). At acidic pH, several cleavage products of Bid were observed upon incubation with mature CLN2 (Fig. 7C, lane 1). This proteolysis can be inhibited by AAF-cmk (Fig. 7C, lane 3) and does not take place at neutral pH (lane 2). Finally, to determine whether CLN2-induced Bid cleavage needs to be carried out in acidic conditions, CLN2 recombinant protein was first preincubated at acidic pH and then mixed with Bid at neutral pH. Western blot analysis of Bid revealed that no cleavage occurred under these conditions (Fig. 7C, lane 4), suggesting that the effect of CLN2 on Bid processing requires acidic conditions.
DISCUSSION
In the last decade, accumulating evidence has been provided, suggesting that lysosomes play an important role as key organelles in the apoptotic process. Indeed, various stimuli were found to directly or indirectly target the lysosomal membrane, thereby inducing lysosomal permeabilization and the release of acidic proteases such cysteine or aspartic cathepsins into the cytosol. Once in the cytosol, these lysosomal enzymes can contribute to the execution of the apoptotic process. However, the precise mechanisms by which lysosomes or cathepsins are involved in cell death are still under investigation.
In this study, evidence for the participation of the lysosomal serine protease CLN2 in cell death comes from two observations. First, CLN2-deficient cells were more resistant than their normal counterparts to the cytotoxic effects of various death ligands. Second, correction of CLN2 enzyme activity in LINCL cells restored susceptibility to killing. The implication of CLN2 in apoptosis was further demonstrated by the fact that TNF-induced cleavage of Bid, release of mitochondrial cytochrome c into the cytosol, and the cleavage of initiator caspase-9 and effector caspases-3 were all strongly attenuated in CLN2-deficient cells. In contrast, cell death induced by the protein kinase inhibitor staurosporine or the proapoptotic sphingolipid ceramide, which are well established apoptosis inducers acting through mitochondrial dysfunction, was not blocked in LINCL cells. Moreover, the expression of cell-surface TNFR1, FADD, and the initiator procaspase-8, which formed the apoptosis-inducing complex, were not perturbed in CLN2-deficient cells suggesting that CLN2 preferentially acts upstream of and triggers the mitochondrial intrinsic apoptotic pathway. Importantly, using various cell lines harboring different defects in the CLN2 gene and a CHO cell line overexpressing a mutated, inactive CLN2, we show that the catalytic function of CLN2 (i.e. presumably its proteolytic activity) is a key determinant for modulating TNF-induced cell death.
An attractive candidate bridging the lysosome leakage to mitochondria might be the proapoptotic Bcl-2 homolog Bid, which is known to control the mitochondrial checkpoint of apoptosis in different cellular models (41-44). Interestingly, cleavage of Bid was inhibited in CLN2-deficient cells, whereas correction of the lysosomal enzymatic defect restored Bid processing. In addition, TNF-induced cell death was abolished in CLN2-corrected LINCL cells having reduced Bid levels, indicating that the pro-apoptotic Bcl-2-like protein is essential for CLN2-mediated apoptosis. This is consistent with observations that Bid plays an important role as the substrate/target of lysosomal cathepsins in cytochrome c-mediated cell death (45). The proteolytic activation of Bid can be catalyzed by several cellular proteases as follows: caspase-8, several isoforms of cathepsins, granzyme B, and calpains that cleave Bid at Asp59, Arg65, Asp75, and Gly70, respectively. Additionally, it has recently been reported that a chymotrypsin B, purified from rat liver lysosomes, might be involved in the regulation of apoptosis through cleavage of Bid mapped at Phe67 (46). The notion that Bid could be cleaved by some other lysosomal enzymes was also reinforced with the fact that in vitro incubation of lysosomal extracts with E-64 (45), a broad range inhibitor of cysteine cathepsins, H-Ala-Ala-Phe-chloromethyl ketone, an irreversible inhibitor of cathepsins B and L, or pepstatin A (47), a potent inhibitor of the aspartyl cathepsin D, did not impair formation of an active truncated Bid.
This study demonstrates that Bid can be directly cleaved by CLN2, generating an ∼15-kDa cleavage product of Bid. As the tripeptidyl peptidase CLN2 exhibits both endopeptidase and exopeptidase activity, it cannot be excluded that other forms of Bid are generated by sequential cleavages. Of note, the cysteine cathepsin B, reported to mediate apoptosis by acting on Bid, cleaves within polypeptide chains but also possesses a dipeptidyl-carboxypeptidase activity (48). However, which of these two hydrolytic activities is preferentially implicated during the apoptotic process remains to be clarified using additional approaches.
Moreover, we show that when tested in vitro the proteolytic activation of Bid by CLN2 requires an acidic pH. Several apoptosis-stimulating events, such as formation of the apoptosome, activation of caspases (49, 50), or stimulation of leukocyte elastase inhibitor/leukocyte-derived DNase II (51), are enhanced by decreasing cytosolic pH. Recently, it has been shown that treatment of U937 cells with TNF resulted in lysosomal membrane permeabilization that was associated with an increased release of cathepsin D and cytosol acidification (8). In addition, treatment of cancer cells with inhibitors of the vacuolar-type H+-ATPase (V-ATPase) have been shown to induce alkalinization of lysosomal pH, cytosol acidification, lysosomal membrane permeabilization, and caspase activation (52).
By analogy with other lysosomal proteases that escape the lysosomal compartment during cell death (1-3), CLN2 might be released into the cytosol and cleave Bid.3 Another possible mechanism to understand how CLN2 could interact with Bid during the apoptotic process is the translocation of Bid to lysosomes. Indeed, it has been reported that some proapoptotic members of the Bcl-2 family could form pores and permeabilize the lysosomal membrane (53, 54) as they do in mitochondria. Upon stimulation of fibroblasts with staurosporine or hepatocytes with TNF, Bax or Bid has been proposed to translocate to the lysosomal membrane leading to the release of cathepsin D (55) or B (56, 57), respectively. Moreover, studies to suppress Bax activation either by pharmacological approaches or small interfering RNA-mediated silencing have confirmed that lysosomal membrane permeabilization is Bax-dependent (10, 58). Conversely, Bcl-2, an anti-apoptotic protein, has been reported to exert a protective effect by inhibiting oxidant-mediated lysosomal rupture (53, 54). All these hypotheses need further examination.
The implications of the present observations to the pathophysiology of LINCL are still unclear. Like other forms of ceroid lipofuscinoses, targeted disruption of CLN2 in mice causes a neurological disorder closely resembling human LINCL. Neuronal loss and apoptosis were prominent in the brain of CLN2-deficient mice, which likely led to premature death of these animals (59). Our findings on the role of CLN2 in cell death may appear paradoxical when comparing the results obtained in vitro on CLN2-deficient cells treated by cytotoxic agents, with the brain pathology observed in LINCL. Similar opposing roles are observed for some other lysosomal proteases that can either prevent apoptosis as described under physiological conditions in knock-out mice (60-63) or promote apoptosis in cells treated with stress agents (64, 65). However, it is conceivable that the role played by these enzymes differs according to the tissue or cell type. For instance, differences in processing and trafficking have been reported for CLN1/PPT1 in neuronal versus non-neuronal cells (66). This phenomenon has also been observed for nonlysosomal apoptotic proteins, such as the serine protease HtrA2/Omi that functions as an anti-apoptotic protein in stress-induced neuronal cell death in contrast to its role in other somatic cells (67).
Finally, the possibility that the neuronal loss that characterizes LINCL is not only due to exacerbated apoptosis cannot be totally excluded. Indeed, it was shown that alterations of the macroautophagy pathway could mediate neuronal cell death in cathepsin D-deficient mice, which exhibit neuronal ceroid lipofuscinosis-like symptoms, in the absence of apoptosis (68).
To summarize, by demonstrating for the first time the implication of CLN2 in death receptor apoptotic signaling, our findings lend further credence to the notion that the lysosomal pathway is an important regulator of this cell death program.
Acknowledgments
We thank Dr. M. J. Warburton (St. George's Hospital Medical School, London, UK) for providing LINCL1 fibroblast cell line; Dr. P. Lobel (University of Medicine and Dentistry of New Jersey, Piscataway, NJ) for CLN2-overexpressing CHO cells and recombinant human CLN2 protein; Dr. A. Golabek (New York State Institute for Basic Research in Developmental Disabilities, Staten Island, NY) for CHO cells engineered to overexpress a mutant form of CLN2 and CLN2 monoclonal antibody; and Dr. A. Jalanko (National Public Health Institute and FIMM, Helsinki, Finland) for providing CLN1-overexpressing cells.
This work was supported by grants from INSERM, Université Paul Sabatier, the Association Vaincre les Maladies Lysosomales, the Fédération pour la Recherche sur le Cerveau, and Groupement d'Intérêt Scientifique Institut des Maladies Rares.
Footnotes
The abbreviations used are: TNF, tumor necrosis factor-α; TNFR, TNF receptor; AAF-AMC, H-Ala-Ala-Phe amino methylcoumarin; AAF-cmk, H-Ala-Ala-Phe-fluoromethyl ketone; Ac-DEVD-AMC, Ac-Asp-Glu-Val-Asp-aminomethylcoumarin; CLN, ceroid lipofuscinosis; FCS, fetal calf serum; LINCL, late-infantile neuronal ceroid lipofuscinosis; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; AEBSF, 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride; TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; CHO, Chinese hamster ovary; PBS, phosphate-buffered saline; PMSF, phenylmethanesulfonyl fluoride; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; DMEM, Dulbecco's modified Eagle's medium; mAb, monoclonal antibody; siRNA, small interfering RNA.
Upon incubation with TNF, CLN2 redistributed from lysosomes to the cytoplasm as indicated by a diffuse staining observed by immunocytochemical studies (V. Albinet, T. Levade, and N. Andrieu-Abadie, unpublished observation).
References
- 1.Leist, M., and Jaattela, M. (2001) Cell Death Differ. 8, 324-326 [DOI] [PubMed] [Google Scholar]
- 2.Guicciardi, M. E., Leist, M., and Gores, G. J. (2004) Oncogene 23, 2881-2890 [DOI] [PubMed] [Google Scholar]
- 3.Kroemer, G., and Jaattela, M. (2005) Nat. Rev. Cancer 5, 886-897 [DOI] [PubMed] [Google Scholar]
- 4.Chwieralski, C. E., Welte, T., and Buhling, F. (2006) Apoptosis 11, 143-149 [DOI] [PubMed] [Google Scholar]
- 5.Tardy, C., Codogno, P., Autefage, H., Levade, T., and Andrieu-Abadie, N. (2006) Biochim. Biophys. Acta 1765, 101-125 [DOI] [PubMed] [Google Scholar]
- 6.Stoka, V., Turk, V., and Turk, B. (2007) Biol. Chem. 388, 555-560 [DOI] [PubMed] [Google Scholar]
- 7.Foghsgaard, L., Wissing, D., Mauch, D., Lademann, U., Bastholm, L., Boes, M., Elling, F., Leist, M., and Jaattela, M. (2001) J. Cell Biol. 153, 999-1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nilsson, C., Johansson, U., Johansson, A. C., Kagedal, K., and Ollinger, K. (2006) Apoptosis 11, 1149-1159 [DOI] [PubMed] [Google Scholar]
- 9.Nagaraj, N. S., Vigneswaran, N., and Zacharias, W. (2007) Apoptosis 12, 125-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Werneburg, N. W., Guicciardi, M. E., Bronk, S. F., Kaufmann, S. H., and Gores, G. J. (2007) J. Biol. Chem. 282, 28960-28970 [DOI] [PubMed] [Google Scholar]
- 11.Emert-Sedlak, L., Shangary, S., Rabinovitz, A., Miranda, M. B., Delach, S. M., and Johnson, D. E. (2005) Mol. Cancer Ther. 4, 733-742 [DOI] [PubMed] [Google Scholar]
- 12.Paris, C., Bertoglio, J., and Breard, J. (2007) Apoptosis 12, 1257-1267 [DOI] [PubMed] [Google Scholar]
- 13.Brunk, U. T., and Svensson, I. (1999) Redox Rep. 4, 3-11 [DOI] [PubMed] [Google Scholar]
- 14.Bivik, C., Rosdahl, I., and Ollinger, K. (2007) Carcinogenesis 28, 537-544 [DOI] [PubMed] [Google Scholar]
- 15.Cho, S., and Dawson, G. (2000) J. Neurochem. 74, 1478-1488 [DOI] [PubMed] [Google Scholar]
- 16.Puranam, K. L., Guo, W. X., Qian, W. H., Nikbakht, K., and Boustany, R. M. (1999) Mol. Genet. Metab. 66, 294-308 [DOI] [PubMed] [Google Scholar]
- 17.Terman, A., Neuzil, J., Kagedal, K., Ollinger, K., and Brunk, U. T. (2002) Exp. Cell Res. 274, 9-15 [DOI] [PubMed] [Google Scholar]
- 18.Tardy, C., Autefage, H., Garcia, V., Levade, T., and Andrieu-Abadie, N. (2004) J. Biol. Chem. 279, 52914-52923 [DOI] [PubMed] [Google Scholar]
- 19.Ezaki, J., Takeda-Ezaki, M., Oda, K., and Kominami, E. (2000) Biochem. Biophys. Res. Commun. 268, 904-908 [DOI] [PubMed] [Google Scholar]
- 20.Sleat, D. E., Donnelly, R. J., Lackland, H., Liu, C. G., Sohar, I., Pullarkat, R. K., and Lobel, P. (1997) Science 277, 1802-1805 [DOI] [PubMed] [Google Scholar]
- 21.Rawlings, N. D., and Barrett, A. J. (1999) Biochim. Biophys. Acta 1429, 496-500 [DOI] [PubMed] [Google Scholar]
- 22.Lin, L., Sohar, I., Lackland, H., and Lobel, P. (2001) J. Biol. Chem. 276, 2249-2255 [DOI] [PubMed] [Google Scholar]
- 23.Liu, C. G., Sleat, D. E., Donnelly, R. J., and Lobel, P. (1998) Genomics 50, 206-212 [DOI] [PubMed] [Google Scholar]
- 24.Walus, M., Kida, E., Wisniewski, K. E., and Golabek, A. A. (2005) FEBS Lett. 579, 1383-1388 [DOI] [PubMed] [Google Scholar]
- 25.Junaid, M. A., Wu, G., and Pullarkat, R. K. (2000) J. Neurochem. 74, 287-294 [DOI] [PubMed] [Google Scholar]
- 26.Vines, D., and Warburton, M. J. (1998) Biochim. Biophys. Acta 1384, 233-242 [DOI] [PubMed] [Google Scholar]
- 27.Bernardini, F., and Warburton, M. J. (2001) Eur. J. Paediatr. Neurol 5, Suppl. A, 69-72 [DOI] [PubMed] [Google Scholar]
- 28.Kopan, S., Sivasubramaniam, U., and Warburton, M. J. (2004) Biochem. Biophys. Res. Commun. 319, 58-65 [DOI] [PubMed] [Google Scholar]
- 29.McDonald, J. K., Hoisington, A. R., and Eisenhauer, D. A. (1985) Biochem. Biophys. Res. Commun. 126, 63-71 [DOI] [PubMed] [Google Scholar]
- 30.Ezaki, J., Tanida, I., Kanehagi, N., and Kominami, E. (1999) J. Neurochem. 72, 2573-2582 [DOI] [PubMed] [Google Scholar]
- 31.Ezaki, J., Takeda-Ezaki, M., and Kominami, E. (2000) J. Biochem. (Tokyo) 128, 509-516 [DOI] [PubMed] [Google Scholar]
- 32.Elleder, M., Sokolova, J., and Hrebicek, M. (1997) Acta Neuropathol. 93, 379-390 [DOI] [PubMed] [Google Scholar]
- 33.Shimizu, M., Fontana, A., Takeda, Y., Yagita, H., Yoshimoto, T., and Matsuzawa, A. (1999) J. Immunol. 162, 7350-7357 [PubMed] [Google Scholar]
- 34.Chatelut, M., Harzer, K., Christomanou, H., Feunteun, J., Pieraggi, M. T., Paton, B. C., Kishimoto, Y., O'Brien, J. S., Basile, J. P., Thiers, J. C., Salvayre, R., and Levade, T. (1997) Clin. Chim. Acta 262, 61-76 [DOI] [PubMed] [Google Scholar]
- 35.Lin, L., and Lobel, P. (2001) Biochem. J. 357, 49-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehtovirta, M., Kyttala, A., Eskelinen, E. L., Hess, M., Heinonen, O., and Jalanko, A. (2001) Hum. Mol. Genet. 10, 69-75 [DOI] [PubMed] [Google Scholar]
- 37.Denizot, F., and Lang, R. (1986) J. Immunol. Methods 89, 271-277 [DOI] [PubMed] [Google Scholar]
- 38.Wang, C. Y., Mayo, M. W., Korneluk, R. G., Goeddel, D. V., and Baldwin, A. S., Jr. (1998) Science 281, 1680-1683 [DOI] [PubMed] [Google Scholar]
- 39.Nylandsted, J., Gyrd-Hansen, M., Danielewicz, A., Fehrenbacher, N., Lademann, U., Hoyer-Hansen, M., Weber, E., Multhoff, G., Rohde, M., and Jaattela, M. (2004) J. Exp. Med. 200, 425-435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Golabek, A. A., Kida, E., Walus, M., Wujek, P., Mehta, P., and Wisniewski, K. E. (2003) J. Biol. Chem. 278, 7135-7145 [DOI] [PubMed] [Google Scholar]
- 41.Cirman, T., Oresic, K., Mazovec, G. D., Turk, V., Reed, J. C., Myers, R. M., Salvesen, G. S., and Turk, B. (2004) J. Biol. Chem. 279, 3578-3587 [DOI] [PubMed] [Google Scholar]
- 42.Heinrich, M., Neumeyer, J., Jakob, M., Hallas, C., Tchikov, V., Winoto-Morbach, S., Wickel, M., Schneider-Brachert, W., Trauzold, A., Hethke, A., and Schutze, S. (2004) Cell Death Differ. 11, 550-563 [DOI] [PubMed] [Google Scholar]
- 43.Blomgran, R., Zheng, L., and Stendahl, O. (2007) J. Leukocyte Biol. 81, 1213-1223 [DOI] [PubMed] [Google Scholar]
- 44.Droga-Mazovec, G., Bojic, L., Petelin, A., Ivanova, S., Romih, R., Repnik, U., Salvesen, G. S., Stoka, V., Turk, V., and Turk, B. (2008) J. Biol. Chem. 283, 19140-19150 [DOI] [PubMed] [Google Scholar]
- 45.Stoka, V., Turk, B., Schendel, S. L., Kim, T. H., Cirman, T., Snipas, S. J., Ellerby, L. M., Bredesen, D., Freeze, H., Abrahamson, M., Bromme, D., Krajewski, S., Reed, J. C., Yin, X. M., Turk, V., and Salvesen, G. S. (2001) J. Biol. Chem. 276, 3149-3157 [DOI] [PubMed] [Google Scholar]
- 46.Miao, Q., Sun, Y., Wei, T., Zhao, X., Zhao, K., Yan, L., Zhang, X., Shu, H., and Yang, F. (2008) J. Biol. Chem. 283, 8218-8228 [DOI] [PubMed] [Google Scholar]
- 47.Reiners, J. J., Caruso, J. A., Mathieu, P., Chelladurai, B., Yin, X., and Kessel, D. (2002) Cell Death Differ. 9, 934-944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mort, J. S., and Buttle, D. J. (1997) Int. J. Biochem. Cell Biol. 29, 715-720 [DOI] [PubMed] [Google Scholar]
- 49.Matsuyama, S., Llopis, J., Deveraux, Q. L., Tsien, R. Y., and Reed, J. C. (2000) Nat. Cell Biol. 2, 318-325 [DOI] [PubMed] [Google Scholar]
- 50.Segal, M. S., and Beem, E. (2001) Am. J. Physiol. 281, C1196-C1204 [DOI] [PubMed] [Google Scholar]
- 51.Huc, L., Rissel, M., Solhaug, A., Tekpli, X., Gorria, M., Torriglia, A., Holme, J. A., Dimanche-Boitrel, M. T., and Lagadic-Gossmann, D. (2006) J. Cell. Physiol. 208, 527-537 [DOI] [PubMed] [Google Scholar]
- 52.De Milito, A., Iessi, E., Logozzi, M., Lozupone, F., Spada, M., Marino, M. L., Federici, C., Perdicchio, M., Matarrese, P., Lugini, L., Nilsson, A., and Fais, S. (2007) Cancer Res. 67, 5408-5417 [DOI] [PubMed] [Google Scholar]
- 53.Zhao, M., Eaton, J. W., and Brunk, U. T. (2000) FEBS Lett. 485, 104-108 [DOI] [PubMed] [Google Scholar]
- 54.Zhao, M., Eaton, J. W., and Brunk, U. T. (2001) FEBS Lett. 509, 405-412 [DOI] [PubMed] [Google Scholar]
- 55.Kagedal, K., Johansson, A. C., Johansson, U., Heimlich, G., Roberg, K., Wang, N. S., Jurgensmeier, J. M., and Ollinger, K. (2005) Int. J. Exp. Pathol. 86, 309-321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Werneburg, N., Guicciardi, M. E., Yin, X. M., and Gores, G. J. (2004) Am. J. Physiol. 287, G436-G443 [DOI] [PubMed] [Google Scholar]
- 57.Guicciardi, M. E., Bronk, S. F., Werneburg, N. W., Yin, X. M., and Gores, G. J. (2005) Gastroenterology 129, 269-284 [DOI] [PubMed] [Google Scholar]
- 58.Feldstein, A. E., Werneburg, N. W., Li, Z., Bronk, S. F., and Gores, G. J. (2006) Am. J. Physiol. 290, G1339-G1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sohar, I., Sleat, D. E., Jadot, M., and Lobel, P. (1999) J. Neurochem. 73, 700-711 [DOI] [PubMed] [Google Scholar]
- 60.Saftig, P., Hetman, M., Schmahl, W., Weber, K., Heine, L., Mossmann, H., Koster, A., Hess, B., Evers, M., von Figura, K., and Peters, C. (1995) EMBO J. 14, 3599-3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koike, M., Nakanishi, H., Saftig, P., Ezaki, J., Isahara, K., Ohsawa, Y., Schulz-Schaeffer, W., Watanabe, T., Waguri, S., Kametaka, S., Shibata, M., Yamamoto, K., Kominami, E., Peters, C., von Figura, K., and Uchiyama, Y. (2000) J. Neurosci. 20, 6898-6906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakanishi, H., Zhang, J., Koike, M., Nishioku, T., Okamoto, Y., Kominami, E., von Figura, K., Peters, C., Yamamoto, K., Saftig, P., and Uchiyama, Y. (2001) J. Neurosci. 21, 7526-7533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koike, M., Shibata, M., Ohsawa, Y., Nakanishi, H., Koga, T., Kametaka, S., Waguri, S., Momoi, T., Kominami, E., Peters, C., Figura, K., Saftig, P., and Uchiyama, Y. (2003) Mol. Cell. Neurosci. 22, 146-161 [DOI] [PubMed] [Google Scholar]
- 64.Wu, G. S., Saftig, P., Peters, C., and El-Deiry, W. S. (1998) Oncogene 16, 2177-2183 [DOI] [PubMed] [Google Scholar]
- 65.Conus, S., Perozzo, R., Reinheckel, T., Peters, C., Scapozza, L., Yousefi, S., and Simon, H. U. (2008) J. Exp. Med. 205, 685-698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lyly, A., von Schantz, C., Salonen, T., Kopra, O., Saarela, J., Jauhiainen, M., Kyttala, A., and Jalanko, A. (2007) BMC Cell Biol. 8, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu, M. J., Liu, M. L., Shen, Y. F., Kim, J. M., Lee, B. H., Lee, Y. S., and Hong, S. T. (2007) Biochem. Biophys. Res. Commun. 362, 295-300 [DOI] [PubMed] [Google Scholar]
- 68.Shacka, J. J., and Roth, K. A. (2007) Autophagy 3, 474-476 [DOI] [PubMed] [Google Scholar]