Abstract
The MDM2 oncoprotein plays multiple regulatory roles in the control of p53-dependent gene expression. A picture of MDM2 is emerging where structurally discrete but interdependent functional domains are linked through changes in conformation. The domain structure includes: (i) a hydrophobic pocket at the N terminus of MDM2 that is involved in both its transrepressor and E3-ubiqutin ligase functions, (ii) a central acid domain that recognizes a ubiquitination signal in the core DNA binding domain of p53, and (iii) a C-terminal C2H2C4 RING finger domain that is required for E2 enzyme-binding and ATP-dependent molecular chaperone activity. Here we show that the binding affinity of MDM2s hydrophobic pocket can be regulated through the RING finger domain and that increases in pocket affinity are reflected by a gain in MDM2 transrepressor activity. Thus, mutations within the RING domain that affect zinc coordination, but not one that inhibits ATP binding, produce MDM2 proteins that have a higher affinity for the BOX-I transactivation domain of p53 and a reduced I0.5 for p53 transrepression. An allosteric model for regulation of the hydrophobic pocket is supported by differences in protein conformation and pocket accessibility between wild-type and the RING domain mutant MDM2 proteins. Additionally the data demonstrate that the complex relationship between different domains of MDM2 can impact on the efficacy of anticancer drugs directed toward its hydrophobic pocket.
The tumor suppressor protein p53 is a transcription factor that plays a key role in the control of pathways that protect cells from malignant transformation (1, 2). As such, p53 is the most frequently inactivated protein in human cancers (3). In response to cellular stress, p53 protein levels are elevated by a decrease in its rate of degradation, and this increase in levels is accompanied by changes in the status of p53 post-translational modifications. Together, elevated levels of p53 and activating post-translational modifications induce its sequence-specific DNA binding and transcriptional activity leading to changes in gene expression that classically result in either cell cycle arrest or apoptosis (4).
Under non-stressed conditions p53 is tightly controlled by the MDM2 oncoprotein through an autoregulatory feedback loop (5-8). MDM2 plays multiple regulatory roles in the control of p53-dependent gene expression. Originally identified as a transrepressor of p53-mediated transcription (5, 8), MDM2 was subsequently shown to control the steady-state levels of p53 in unstressed cells by acting as a RING finger domain E34-ubiquitin ligase (9, 10). More recently MDM2 has also been implicated in the control of p53 translation (11) and in addition has been shown to possess an ATP-dependent molecular chaperone activity toward p53 (12, 13). MDM2 is a multidomain protein comprising: (i) A hydrophobic pocket at its N terminus that binds with a high affinity to the BOX-I transactivation domain of p53 (14). This interaction is required for both the E3-ligase and transrepressor functions of MDM2 (15). (ii) A central acid domain that recognizes and binds with a relatively low affinity to a ubiquitination signal in the core of p53 (16-18). (iii) A C-terminal C2H2C4 RING finger domain (19-21). This domain is essential for zinc coordination and is involved in E2-binding and the formation of RING domain dimers (22). In addition the RING domain houses a P-Walker motif that is required for MDM2 to act as an ATP-dependent molecular chaperone (12, 23).
Negative regulation of p53 transcription by MDM2 involves competition for binding to p53s N-terminal LXXLL transactivation domain (BOX-I domain), thus binding of MDM2 can occlude the binding of transcription components like the coactivator p300 preventing the formation of a pre-initiation complex (8, 24-27). Mutations in the p53 activation domain that prevent MDM2 binding can attenuate MDM2-catalyzed transrepression (15, 28), and peptide ligands mimicking the activation domain of p53 that bind to the N-terminal domain of MDM2 and disrupt the MDM2:p53 protein complex in vitro can stimulate p53-dependent gene expression in cells (27, 28). Together these data suggest that binding of the BOX-I activation domain of p53 to the hydrophobic pocket of MDM2 is required for MDM2s oncogenic functions. Recent studies have identified a second MDM2 interaction site within the core domain (BOX-V) of p53 (16-18, 29). The BOX-V domain binds to the acid domain of MDM2, and, although this interaction has a relatively low affinity, it is key in determining the rate of p53 ubiquitination as it comprises part of the p53 ubiquitination signal (16).
The current study has used point mutations within critical C2H2C4 resides of the MDM2 RING domain to uncover cross-talk between the zinc coordinating structure and the hydrophobic pocket of MDM2. A gain in the ability of MDM2 to transrepress p53-dependent transcription is seen when zinc-coordinating residues are mutated, whereas mutation of a residue within the RING required for ATP binding has no effect on MDM2 mediated transrepression. Studies on purified MDM2 and p53 provide a mechanism for changes in transrepressor activity by demonstrating that the C2H2C4 RING mutants have a higher affinity for hydrophobic pocket interacting ligands and proteins. Thus, the RING finger domain of MDM2 is linked to the hydrophobic pocket through conformational changes that affect MDM2 transrepressor activity. In addition, the efficacy of small molecules, targeted to the hydrophobic pocket of MDM2, can be determined by the status of the C2H2C4 structure.
EXPERIMENTAL PROCEDURES
Reagents and Plasmids—Antibodies used were: DO-1 (p53) and 2A10/3G5/4B2/SMP14 (MDM2) produced in-house, p21 was detected using AB1 (Calbiochem). Peptides, N-terminal biotin-labeled plus an SGSG space (Chiron Mimotopes), were dissolved at 10 mg/ml in DMSO as previously described (16). The BOX-I and 12.1 sequences are PPLSQETFSDLWKLLP and MPRFMDYWEGLN, respectively. Nutlin-3 was from Alexis Biochemicals. Ubiquitin (U-100), UbcH5a (E2-616), and E1 (E-301) were from Boston Biochem. pcDNA3-p53 and His-ubiquitin were a gift from David Lane, pCMV-mycMDM2wt, pCMV-mycMDM2ΔN, and pCMV-MDM2ΔAc were provided by Aart Jochemsen. pcDNA3-MDM2 with the human MDM2 cDNA was used as a template to generate pcDNA3-MDM2C464A, pcDNA3-MDM2C478S, and pcDNA3-MDM2K454A using a QuikChange mutagenesis kit (Stratagene). The primers were as follows: K454A, GCATTGTCCATGGCGCAACAGGACATC; K454Arev, GATGTCCTGTTGCGCCATGGACAATGC; C464A, GGACATCTTATGGCCTGCTTTACAGCGGCAAAGAAGCTAAAGAAAAGG; C464Arev, CCTTTTCTTTAGCTTCTTTGCCGCTGTAAAGCAGGCCATAAGATGTCC; C478S, GGAATAAGCCCTGCCCAGTAAGCAGACAACCAATTCAAATGATTGTG; and C478Srev, CACAATCATTTGAATTGGTTGTCTGCTTACTGGGCAGGGCTTATTCC.
Cell Culture, Reporter Assays, and Immunoblots—H1299 cells were maintained in RPMI medium supplemented with 10% (v/v) fetal bovine serum and incubated at 37 °C with 5% CO2. A375 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum and incubated at 37 °C with 10% CO2. MDM2-/-p53-/- cells were maintained in Dulbecco's modified Eagle's medium with 10% (v/v) fetal bovine serum and 10% CO2. Upon reaching 80-90% confluency the cells were transfected using Lipofectamine 2000 (Invitrogen) with the total amount of DNA for all wells kept constant at 0.8 μg. A solution was formed with (30 ng) pGL4.10[luc2] bearing a 44-base stretch of the p21 promoter and with 70 ng of the phRL-CMV plus p53 and MDM2 plasmids as detailed in the figure legends. Twenty-four hours post-transfection, the cells were washed once in ice-cold phosphate-buffered saline and lysed with 1× Passive Lysis Buffer (supplied with the Dual-Luciferase Reporter Assay System from Promega). Alternatively, Nutlin was added after 24 h (as detailed in the figure legends), and the incubation continued for a further 6 h. Afterward the Dual-Luciferase Reporter Assay was performed in accordance with the manufacturer's instructions. For immunoblot analysis transfected cells were lysed in Nonidet P-40 buffer (25 mm HEPES, pH 7.5, 0.1% (v/v) Nonidet P-40, 150 mm KCl, 5 mm dithiothreitol, 50 mm NaF), and lysates were separated on 4-12% NuPAGE (to detect p53 modification) or 10% SDS-PAGE and transferred to nitrocellulose (16).
Ubiquitination Assay—The in vitro assay was carried out as previously described (16). Reactions contained 25 mm HEPES, pH 8.0, 10 mm MgCl2, 4 mm ATP, 0.5 mm dithiothreitol, 0.05% (v/v) Triton X-100, 0.25 mm benzamidine, 10 mm creatine phosphate, 3.5 units/ml creatine kinase, ubiquitin (2 μg), E1 (50-200 nm), E2s (0.1-1 μm), plus p53 purified from Escherichia coli (100 ng) and were initiated by the addition of purified human MDM2 (50 ng) in the presence or absence of either BOX-I or Nutlin as detailed in the figure legends. p53 ubiquitination in p53-/-MDM2-/- mouse embryonic fibroblasts was determined using a previously described method (16).
Protein Binding Assays—Recombinant human full-length untagged MDM2 proteins and p53 protein were purified from E. coli as previously described (12, 30). For peptide and protein binding assays the microtiter wells were adsorbed with streptavidin overnight, washed 6× with phosphate-buffered saline-Tween with biotinylated peptides added for 1 h alternatively the wells were coated with purified p53 (150 ng) as previously described (16). Following extensive washing with phosphate-buffered saline-Tween increasing amounts of MDM2 were added either in the absence or presence of Nutlin (as detailed in the figure legends). Following washing (6× washes with phosphate-buffered saline-Tween) MDM2 was detected using the monoclonal antibody 2A10 and a secondary rabbit anti-mouse horseradish peroxidase antibody the wells were developed using ECL. The results were quantified using Fluoroskan Ascent FL equipment (Labsystems) and analyzed with Ascent Software.
Tryptic Digestion—wt or mutant MDM2 protein (1 μg) plus trypsin (10 ng) was incubated at 30 °C in 50 mm ammonium bicarbonate buffer for up to 30 min. Aliquots were removed, and the reaction was stopped by the addition of SDS sample buffer and heating at 80 °C for 4 min. The samples were analyzed by 4-12% gradient gels (Invitrogen) and immunoblotted.
Intrinsic Fluorescence Assay—Fluorescence emission spectra were recorded on a SPEX FLUOROMAX-3 (Jobin Yvon Horiba) spectrofluorometer. The bandwidths for excitation and emission were set at 5 nm, and an excitation wavelength of 295 nm was used. Fluorescence spectra were recorded from 320 to 450 nm in 0.5 nm steps, with an integration time of 1 s. All experiments were carried out at 4 °C in buffer containing: 25 mm HEPES (pH 7.5), 50 mm KCl, 5% (v/v) glycerol, 10 μm ZnSO4, and 2 mm dithiothreitol. MDM2wt, MDM2C464A, and MDM2C478S used in this assay were initially preincubated on ice for 5 min at final concentrations raging from 20 nm to 1 μm (Vtotal = 0.5 ml). Each spectrum produced is the average of three emission scans minus the average of the three blanks (buffer) scans.
RESULTS
C2H2C4 MDM2 Mutant Proteins Have a Gain of Transrepressor Activity—We have recently shown that the E3-ligase activity of MDM2 requires interactions at both the hydrophobic pocket and the acid domain (16). These interactions are linked by an allosteric mechanism, with binding at the hydrophobic pocket favoring recognition of a p53 ubiquitination signal from its core DNA binding domain (BOX-V). This prompted us to ask whether similar mechanisms linked other MDM2 domains and how cooperation between domains impacted on the proteins multifunctional nature. The RING finger domain of MDM2 is required for both its E3-ligase and ATP-dependent chaperone activities through binding to an E2-ubiquitin conjugating enzyme and to ATP, respectively (12, 22). However, whether conformational changes in the RING domain are transmitted to other MDM2 functional domains affecting their activity remains unclear.
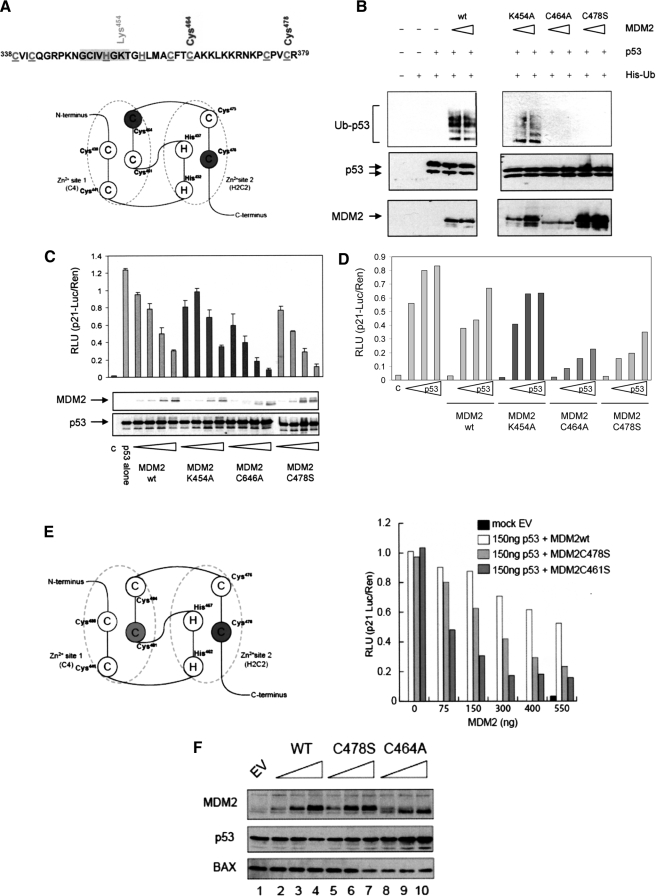
To study the RING domain in the context of full-length MDM2 we first generated a number of RING finger domain mutant constructs (Fig. 1A). Two of these, Cys464A (MDM2C464A) and Cys478S (MDM2C478S), introduced mutations into the C4 and H2C2 coordination sites of the C2H2C4 structure (21), respectively, and a third, Lys454A (MDM2K454A), is a RING domain residue, which is not required for zinc coordination but is essential for MDM2 to bind ATP (12, 23). The Cys464A mutant was chosen because it is the most widely studied E3-ligase-dead MDM2 RING mutant, in addition this cysteine lies within an α-helix, and structure predication using PyMOL suggests that mutation to Ala has little effect on the global structure of the RING or on the extent of solvent exposure. The Cys478S mutant is similarly predicted to have a minimal effect on the local environment, because it lies within an irregular loop structure. To characterize these mutants we first determined the effect of introducing the RING mutations on E3-ligase activity using a cell-based assay. When p53-/-MDM2-/- mouse fibroblasts were transfected with p53 and MDM2 together with His-tagged ubiquitin it was demonstrated that the Lys454A mutant retained the ability to ubiquitinate p53, as determined by the isolation of His-ubiquitin-conjugated p53 protein (Fig. 1B), whereas, as expected, the C2H2C4 mutants were no longer able to mediate p53 modification by ubiquitin. Although the introduction of the Cys mutations is sufficient to inactivate MDM2 as an E3-ligase, they do not affect the overall integrity of the C terminus as we have shown that the Cys464A and Cys478S MDM2 mutants retain the ability to bind ATP, while ATP-binding activity is lost in the Lys454A mutant (12). Thus, mutation of Cys residues in the RING appears to specifically inactivate RING function during ligation of ubiquitin to target substrates while leaving other RING domain functions intact.
FIGURE 1.
Mutation of the C2H2C4 RING gives a gain of MDM2 transrepression. A, schematic showing the RING finger domain of MDM2. The zinc conjugating residues are highlighted and underlined, the ATP binding site is shaded gray, and the residues picked out (above) are those that were mutated to generate the panel of mutants studied. B, immunoblot of p53 from p53-/-MDM2-/- mouse embryonic fibroblasts transfected with p53 (150 ng) and MDM2 (wt and mutant constructs as indicated;150 and 250 ng) plus His-Ub (200 ng). His-conjugated proteins were isolated using nickel-agarose and analyzedona 4-12% gradient gel. p53 was detected using DO-1. The data are representative of two independent experiments. In C: upper panel, H1299 cells were transfected with p53 alone (150 ng) or with a titration (75, 150, 300, 400, and 550 ng) of MDM2wt, MDM2K454A, MDM2C464A, or MDM2C478S plus the reporter plasmids. Total DNA was normalized using the vector control, and c is a vector only control. Post transfection (24 h) the Dual Luciferase Assay was performed. The results are normalized by expressing p21-Luciferase/Renilla activity in relative light units (RLU) and are the mean ± S.D. (lower panel). Immunoblot showing the levels of p53 with MDM2wt and mutant proteins, p53 was detected using DO-1 and MDM2 using 2A10. D, as above except that the cells were transfected with a p53 titration (75, 150, and 300 ng) alone or in the presence of a fixed concentration of wt and mutant MDM2 as indicated (400 ng). The vector-only control is shown as c, and total DNA was normalized as above the data are representative of two individual experiments. E, schematic depicting zinc coordination scheme of MDM2 (left panel). The key residues that were mutated are highlighted. The histogram represents data obtained from Dual Luciferase Assay carried out as described above, with MDM2wt MDM2C478S and MDM2C461S. F, MCF7 cells were transfected with plasmids encoding MDM2WT, MDM2C464A, and MDM2C478S (0-4 μg). Total DNA amount was normalized using vector control. Twenty-four hours post transfection immunoblot analysis was carried out, p53, MDM2, and BAX were detected using DO-1, 2A10, and anti-BAX, respectively. The results are representative of at least three independent experiments.
The various MDM2 RING finger domain mutants were tested in a p53-dependent transcription assay to determine if inactivation of either its E3-ligase or ATP-binding activities affected the ability of MDM2 to repress p53-dependent transcription. The ability of p53 to activate transcription was determined using the p53-responsive region of the p21-promoter to drive luciferase expression. When human p53 was transfected into H1299 cells (human p53 null non-small cell lung carcinoma cells) along with the p21-reporter a characteristic activation of the promoter was detected (Fig. 1C), coexpression of wild-type MDM2 in a titrative manner led to a progressive decrease in reporter activity indicative of transrepression, with an I0.5 in the region of 300 ng of MDM2 plasmid (150 ng of p53 plasmid (Fig. 1C)). All the RING mutants were expressed to a similar level and were able to inhibit p53-dependent transcription, however, both the Cys mutants displayed a gain of transrepressor function with an I0.5 of <75 ng for MDM2C464A and <150 ng for MDM2C478S. In contrast, mutation of Lys454 did not enhance the repressor activity of MDM2, and like the wild-type protein, this mutant had an I0.5 of around 300 ng. Similarly, although a titration of p53 could overcome transrepression imposed by a fixed amount of wild-type MDM2 and the Lys454A mutant, there was a significant reduction in the ability of p53 to overcome the effects of the Cys464A and Cys478S mutants (Fig. 1D). To determine whether comparison of an Ala-substituted Cys to a Ser-substituted Cys underlined the apparent difference in potency of the MDM2C464A and MDM2C478S mutants we used a second mutant from within the C4 coordination (Fig. 1E; C461S) where we mutated Cys461 to Ser. Analysis of this mutant demonstrated that it was also more potent than the MDM2C478S mutant. These results suggest that the difference in activity of the RING mutants is unlikely to be due primarily to the choice of the substitution residue.
To determine if endogenous p53-responsive promoters respond to the MDM2 RING domain mutant proteins in a similar way to that observed using p53-responsive reporter constructs we used BAX protein expression as a measure of its promoter activity, because BAX is a well characterized target for p53 (31). For this assay we used MCF7 cells because they have a wild-type p53 pathway. Fig. 1F shows that BAX protein levels are unaffected by the expression of wt MDM2 at the concentrations used in the experiment (Fig. 1F, lanes 2-4), however under the same conditions the MDM2C464A mutant suppressed BAX protein expression even at the lowest amount used (lanes 8-10), and consistent with the reporter assays, the MDM2C478S mutant had intermediate activity, because it suppressed BAX protein expression when present at the highest amount (lane 7).
Together the data presented above suggest that mutation of residues within the RING finger domain that directly affect MDM2 E3-ligase activity facilitate transrepression of p53 transcriptional activity. In contrast, loss of MDM2 molecular chaperone activity in the ATP-binding mutant (Lys454A) had no effect on transrepression of p53. Further, the data demonstrate that MDM2 E3-ligase activity can be completely uncoupled from its ability to repress p53 transcription.
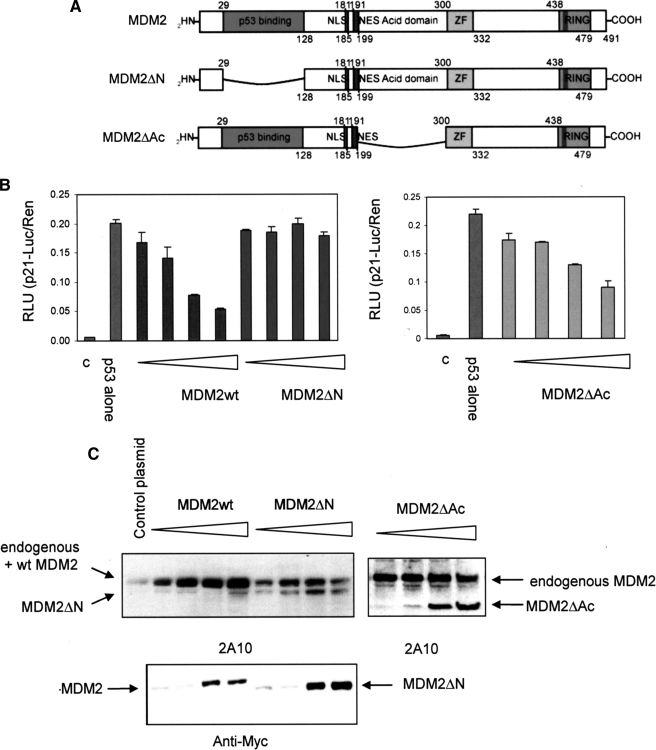
The MDM2 Hydrophobic Pocket but Not the Acid Domain Is Required for Efficient Transrepression of p53—Previous studies have suggested that the hydrophobic pocket of MDM2 is critical for its transrepressor activity (15, 28), however these studies were carried out before the complexity of the interaction between MDM2 and p53 was fully appreciated. Because the acid domain of MDM2 is now known to bind to the BOX-V domain of p53 (16, 18) we sought to use MDM2 deletion mutants to determine which interactions were salient for MDM2-mediated transrepression. To investigate the influence of MDM2 domain structure on p53 transrepression we used mutant forms of murine MDM2 where one or other of the two known p53 interacting domains had been deleted (Fig. 2A, MDM2ΔN and MDM2ΔAc). MDM2ΔN prevents the interaction between the transactivation domain of p53 (BOX-I) and the hydrophobic pocket in the N terminus of MDM2 (14), whereas the MDM2ΔAc precludes binding of the core domain of p53 (BOX-V) and the acid domain of MDM2 that is essential for MDM2-mediated ubiquitination of p53 (16).
FIGURE 2.
The hydrophobic pocket of MDM2 is essential for transrepression of p53. A, a schematic showing murine MDM2 deletion constructs. MDM2 ΔN is missing the MDM2 hydrophobic pocket, which binds the BOX-I domain of p53, and MDM2 ΔAc does not have the acid domain, which binds the BOX-V domain of p53. B, H1299 cells were transfected with p53 alone (150 ng) or with a titration (75, 150, 300, and 550 ng) of MDM2wt, MDM2ΔN, or MDM2ΔAc plus the reporter plasmids. Total DNA was normalized using the vector control. Post transfection (24 h) the Dual Luciferase Assay was performed. The results are normalized by expressing p21-Luciferase/Renilla activity in relative light units (RLU), are the mean ± S.D., and c is the vector-only control. C, immunoblot showing the levels of mycMDM2wt and mycMDM2ΔN detected using 2A10 (top panel) and anti-myc antibody (bottom panel), and MDM2ΔAc detected using 2A10 (top right panel).
When the deletion mutants were analyzed in the p53-reporter assay (Fig. 2B) loss of the hydrophobic pocket essentially inactivated MDM2 as a transrepressor of p53-mediated transcription although expressed at a similar level to the wt protein (Fig. 2C), whereas deletion of the acid domain did not have a major impact on this activity of MDM2. The data suggest that the interaction between the acid domain of MDM2 and the BOX-V region of the p53 core domain is not essential for MDM2 mediated transrepression, whereas binding of the hydrophobic pocket to the BOX-I transactivation domain of p53 is absolutely required because the ΔN mutant is essentially dead in this assay. Together with data in the previous section our results suggest that RING domain mutants that loose the ability to ubiquitinate p53 but gain p53 repressor activity do so, most likely, through modulation of the MDM2-hydrophobic pocket: p53-BOX-I interaction.
RING Mutations Affect the Efficacy of Nutlin as an Inhibitor of MDM2-mediated Repression of p53—The data presented above establish that the hydrophobic pocket of MDM2 is essential for transrepression of p53 (Fig. 2B) and that the ability of MDM2 to transrepress p53-dependent transcription can be uncoupled from MDM2 E3-ligase activity (Fig. 1C). Furthermore, the C2H2C4 RING mutants have a gain of transrepressor activity when compared with the wt protein (Fig. 1C). To further investigate the link between hydrophobic pocket binding and RING domain function we used the cell-permeable MDM2 hydrophobic pocket binding molecule Nutlin as a tool (32, 33).
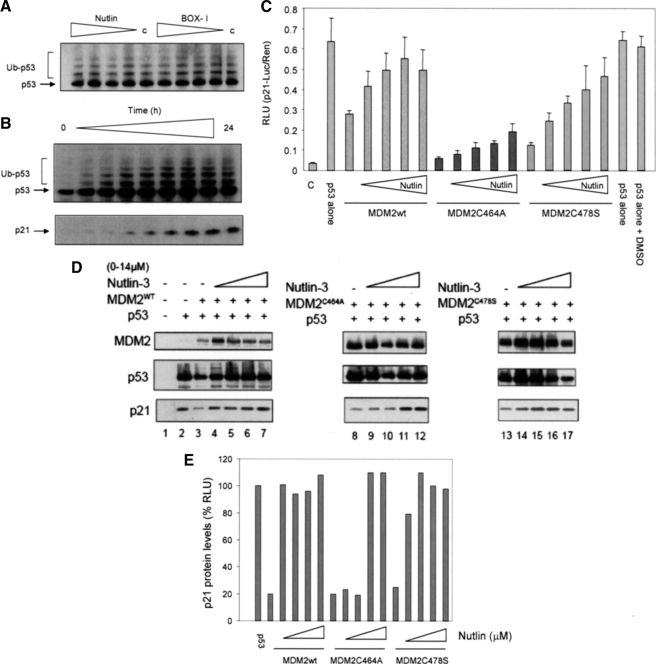
We have previously suggested that Nutlin most likely functions as an anticancer agent by affecting the ability of MDM2 to act as a transrepressor of p53 transcription rather than as an inhibitor of MDM2-dependent p53 ubiquitination (16). This hypothesis was based on observations, confirmed here, that Nutlin is not able to inhibit the ubiquitination of p53 in vitro using purified proteins (Fig. 3A) and that the addition of Nutlin to A375 cells does not decrease the number of modified p53 forms detected (Fig. 3B). In fact in the current experiments we show that an increase in p53 modification was proportionate to increases in total p53 protein levels (see Fig. 5B, upper panel) and to the activation of p53, as assessed by an increase in p21 protein levels (Fig. 3B, lower panel). Together these results suggest that modification of existing or newly synthesized p53 protein continues in the presence of Nutlin.
FIGURE 3.
The efficacy of Nutlin in cells is dependent on the C2H2H4 RING. A, ubiquitination reactions were assembled with p53, E1, E2, and MDM2 in the presence or absence (c) of either Nutlin or BOX-I peptide (0, 10, 20, 40, and 60 μm). Ubiquitination was analyzed by immunoblot using a 4-12% gradient gel, p53 was detected using DO-1. Unmodified (p53) and ubiquitinated p53 (Ub-p53) are shown. The data are representative of at least four separate experiments. B, Nutlin (10 μm) was added to A375 cells, and they were harvested over a 24-h time course (0, 1, 2, 3, 4, 6, 8, 12, and 24 h). The lysates were analyzed by immunoblot using either a 4-12% gradient gel (upper panel) or a 15% SDS-PAGE gel (lower panel) p53 was detected using DO-1 and p21 using AB-1. Modified forms of p53 are given as Ub-p53. The data are representative of three such experiments. C, H1299 cells were transfected with p53 alone (150 ng) or together with MDM2 (400 ng) after 24 h Nutlin (0, 1.5, 3, 5, and 14 μm) was added, and the incubation continued for a further 6 h. Total DNA was normalized using the vector control. The cells were harvested and the Dual Luciferase Assay was performed. The results are normalized by expressing p21-Luciferase/Renilla activity in relative light units (RLU) and are the mean ± S.D., and c is the vector-only control. A DMSO control is included for the Nutlin carrier. D, H1299 cells were transfected with p53 alone (150 ng) or together with (300 ng) of MDM2wt, MDM2C464A, or MDM2C478S. Total DNA was normalized using the vector control (c). Post transfection (24 h) cells were treated with Nutlin (0, 1.5, 3, 5, and 14 μm) for a further 6 h. Immunoblot analysis of MDM2, p53, and p21 was carried out using 2A10, DO-1, and Ab-1, respectively. E, densitometry analysis of the p21 immunoblot; p21 levels are given as a percentage of protein detected in the presence of p53 alone.
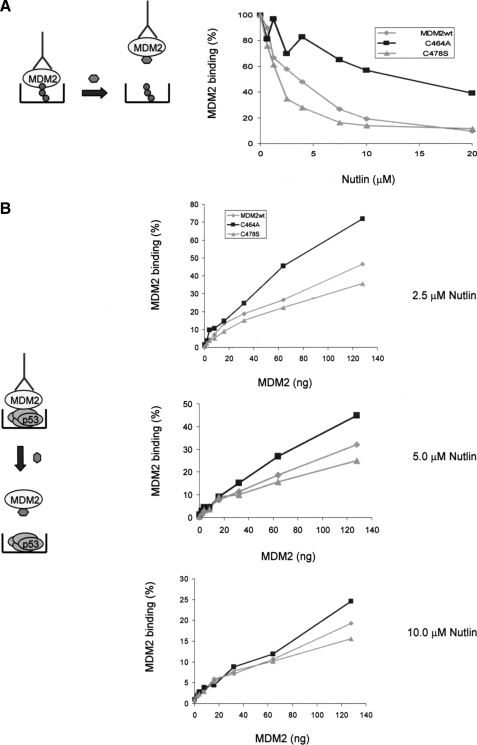
FIGURE 5.
Nutlin has a differential effect on the C2H2H4 MDM2 mutants. A, a schematic (left panel) shows the assay format with BOX-I peptide Nutlin and the detecting antibody in blue. Right panel, BOX-1 peptide (5 μm) was captured on streptavidin-coated wells and incubated with the indicated form of MDM2 (50 ng), which had been preincubated (5 min) with a titration of Nutlin (μm). MDM2 binding was detected using 2A10 and is given as relative binding expressed as a percentage where 100% binding is that measured in the absence of Nutlin. B, p53 protein (100 ng/well) was coated onto microtiter wells and incubated with a titration of wt and mutant MDM2 as indicated. The MDM2 had been preincubated with Nutlin (2.5, 5, and 10 μm as indicated). MDM2 binding was detected using 2A10 and is normalized to maximal binding to p53 in the presence of saturating amounts of each mutant in %. The experiment is representative of at least three independent experiments where each condition was carried out in duplicate. In the schematic the detecting antibody 2A10 and Nutlin are depicted.
If the above hypothesis is correct we would predict that Nutlin binding to MDM2 would be sufficient to relieve transrepression imposed by both wt and E3-inactive MDM2 protein. Using the p53-reporter assay the ability of Nutlin to reverse transrepression imposed by a fixed amount of wt MDM2 was shown to be efficient (Fig. 3C). Furthermore, Nutlin had a striking effect on the transrepressor activity of the Cys478S protein with p53 activity recovering to near the level seen in the absence of MDM2. This result lends strong support to the idea that Nutlin-dependent inhibition of MDM2 as a transrepressor occurs independently of MDM2 E3-ligase function and that this could be sufficient to explain the activating effect of Nutlin on p53.
Interestingly, although Nutlin was active against the Cys478S MDM2 mutant protein it had only weak activity against the Cys464A protein and even at 14 μm Nutlin did not inhibit this mutant. To confirm this result using a different assay system we looked at the ability of Nutlin to overcome wt and mutant MDM2-imposed transrepression of an endogenous p53 promoter using p21 protein levels as a downstream readout for transfected p53 (Fig. 3, D and E). An amount of transfected MDM2 wt and mutant constructs was chosen that gave maximal repression of endogenous p21 protein expression in H1299 cells (p21 levels decreased by ∼80%), Nutlin was then added to the cells in a titrative manner, and the effect on p21 levels was determined. Quantitation of the data generated in Fig. 3D shows that Nutlin can relieve transrepression imposed by both the wt and Cys478S mutant forms of MDM2 at the lowest concentration used (1.5 μm). However at 1.5 μm, Nutlin had no measurable effect on the levels of p21 protein in the presence of MDM2C464A, in fact it required 7 μm Nutlin to relief transrepression by the Cys464A protein on p21 protein expression.
The differential effect of Nutlin, dependent on the status of the C2H2C4 RING structure, suggests that the introduction of mutations that directly impact on RING structure can differentially impact on the affinity or availability of MDM2's hydrophobic pocket for p53 BOX-I binding.
MDM2 Cys RING Finger Mutant Proteins Bind with a Higher Affinity to the Transactivation Domain of p53—The cell-based experiments described above show that (i) C2H2C4 RING mutants have a gain of transrepressor activity and (ii) the hydrophobic pocket, but not the acid domain, is essential for MDM2 repressor activity. These results suggested to us that the affinity of the hydrophobic pocket for the BOX-I domain of p53 might be sufficient to dictate the potency of MDM2 as a transrepressor and may also determine the efficacy of hydrophobic pocket binding drugs such as Nutlin.
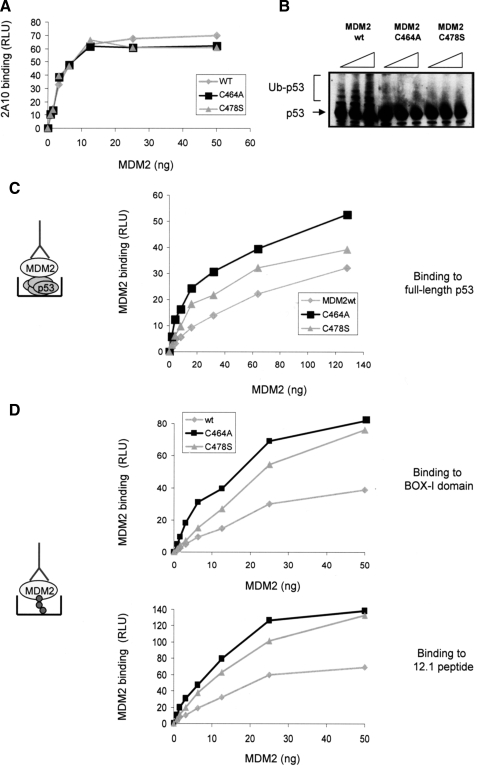
If the above hypothesis is correct we would expect the C2H2C4 RING domain MDM2 mutant proteins to bind BOX-I with a higher affinity than the wild-type protein. To address this we first purified full-length untagged human MDM2, as well as the Cys464A and Cys478S mutant MDM2 proteins from an E. coli expression system. The protein concentrations were normalized and the quantitation was confirmed using an enzyme-linked immunosorbent assay (Fig. 4A). E3-ligase activity was assayed using a purified assay system (16) and as expected (based on the results presented in Fig. 1B), whereas the wt protein was an active E3-ligase, neither the Cys464A nor the Cys478S mutant were able to mediate ubiquitination of p53 (Fig. 4B). Next the ability of the proteins to bind to full-length purified p53 was determined (Fig. 4C). In this assay a constant amount of full-length untagged p53, purified form E. coli, was captured onto microtiter wells and incubated with a titration of wt or mutant MDM2 protein. Following extensive washing bound MDM2 was detected using a monoclonal antibody (2A10). This showed that both of the MDM2 C2H2C4 mutant proteins bound with a higher affinity to full-length p53 than did wt MDM2, however, consistent with its lower I0.5 in the transrepressor assay (Fig. 1C), the Cys464A mutant bound better to p53 than the Cys478S mutant protein.
FIGURE 4.
C2H2C4 mutant MDM2 binds with a higher affinity to p53. A, to check the accuracy of protein normalization for the purified MDM2wt, MDM2C464A, and MDM2C478S proteins an enzyme-linked immunosorbent assay was performed in which a titration of MDM2 protein was coated onto the microtiter plate wells, following extensive washing, and MDM2wt and mutant protein levels were detected using 2A10. B, ubiquitination reactions were assembled with p53, E1, E2, and MDM2 wt or mutant proteins as indicated. Ubiquitination was analyzed by immunoblot using a 4-12% gradient gel, p53 was detected using DO-1. Unmodified (p53) and ubiquitinated p53 (Ub-p53) is shown. C, p53 (100 ng/well) was coated onto microtiter wells and incubated with wt or mutant MDM2 as indicated. MDM2 binding was detected using 2A10 and is expressed as relative light units (RLU) against the MDM2 amount. The experiment is representative of three independent experiments where each condition was carried out in duplicate. In the schematic the detecting antibody 2A10 is depicted. D, BOX-I (upper panel) or 12.1 (lower panel) peptides (5 μm) were captured onto streptavidin-coated wells, and MDM2 binding was determined as described in C. In the schematic BOX-I/12.1 are depicted and the detecting antibody is 2A10.
Although the interaction between the acid domain of MDM2 and the BOX-V domain of p53 is not essential for MDM2 mediated transrepression (Fig. 2B) it is likely to play a role in binding of MDM2 to full-length p53 protein (16). Binding to the isolated BOX-I domain was therefore used to determine whether the increase in affinity of the Cys464A and Cys478S mutants for full-length p53 reflected a change in affinity for the p53 BOX-I domain. Binding of wt and the C2H2C4 mutant MDM2 proteins to a peptide based on the BOX-I domain of p53, or to an optimized hydrophobic pocket binding peptide, 12.1 (34), was determined. Biotinylated BOX-I domain or 12.1 peptide were immobilized on streptavidin-coated wells and incubated with a titration of wt MDM2 or the two C2H2C4 mutant proteins. Consistent with the data presented above, showing that the mutants bind better to full-length p53, both the Cys464A and Cys478S proteins bound with a higher affinity to BOX-I and 12.1 than wt MDM2 protein. Mirroring its decreased I0.5 for transrepression and increased affinity for full-length p53 the Cys464A mutant consistently bound better to BOX-I or the BOX-I mimetic (12.1) than the Cys478 mutant protein. The data presented above suggest that the increased transrepressor activity of the MDM2 C2H2C4 RING mutants is dictated by an increase in the affinity of their hydrophobic pockets for the BOX-I transactivation domain of p53.
The MDM2 RING Mutant Proteins Differ in Their Sensitivity to the Hydrophobic Pocket Binding Drug Nutlin—To determine whether the differential effect of Nutlin on the C2H2C4 MDM2 mutants observed in a cellular environment represented a quantitative difference in the affinity of the mutants for Nutlin binding, we determined the ability of Nutlin to compete with BOX-I for binding to MDM2. In this assay equal amounts of biotinylated BOX-I peptide were captured onto streptavidincoated microtiter wells and incubated with a fixed amount of wt or mutant MDM2 in the presence of increasing Nutlin concentrations (Fig. 5A). Consistent with the results observed in cells (Fig. 3C) this assay demonstrated an apparent difference in the ability of the RING mutants to bind Nutlin, suggesting that mutations within the C2H2C4 structure have a direct influence on Nutlin binding that is not mediated by other cellular proteins. Thus, Nutlin is a weak inhibitor of BOX-I binding to the Cys464A mutant, relative to wild-type MDM2 protein, with an I0.5 4× higher than wt MDM2. This suggests that the Cys464A protein has a lower affinity for Nutlin than the wild-type protein and binds with a higher affinity to BOX-I resulting in preferential BOX-I binding. Conversely, the Cys478S mutant appears to bind Nutlin with a higher affinity than the wt and Cys464A proteins. Thus, in this case Nutlin competes more easily with BOX-I for binding to the hydrophobic pocket. To determine if the differences observed in Nutlin binding are reflected in the ability of Nutlin to disrupt the interaction between full-length p53 and MDM2 the following experiment was performed. Full-length p53 was coated onto the microtiter well and incubated with a titration of MDM2 in the presence of various fixed concentrations of Nutlin (Fig. 5B; 2.5, 5, and 10 μm). Again there was a clear difference in Nutlin-dependent disruption of full-length p53 binding to the MDM2 proteins, with the p53-Cys478S mutant complex showing increased Nutlin sensitivity compared with the p53-Cys464A and the p53-MDM2wt interactions. Once again this suggests that the Cys478S mutant has a higher affinity for Nutlin than does the Cys464A protein.
In conclusion, although both the Cys464A and Cys478S proteins bind with a higher affinity to hydrophobic pocket interacting ligands than the wt protein, resulting in a gain of transrepressor activity (Fig. 2C), they do so differentially. Thus, whereas the Cys478S protein binds preferentially to Nutlin, the Cys464A protein binds with a higher affinity to BOX-I (Fig. 5) resulting in differential Nutlin sensitivity in cells (Fig. 2C). Moreover, mutations within the zinc coordinating structure of the RING are not synonymous.
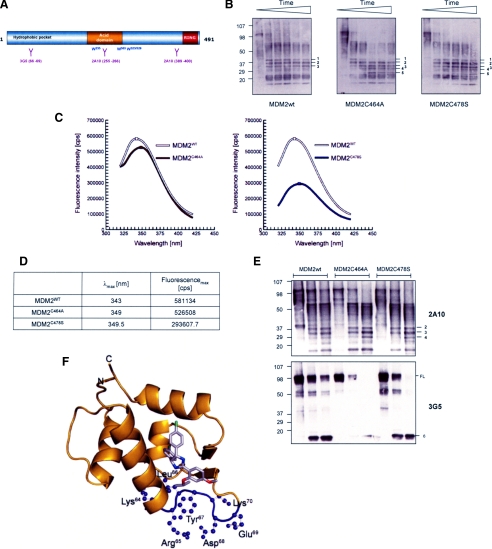
RING-generated Conformational Changes in MDM2 Are Transmitted to the Hydrophobic Pocket through the Central Domain—To support our hypothesis that hydrophobic pocket affinity can be modulated by the RING domain, evidence of a RING-dependent conformational change in full-length MDM2 was sought. Firstly, limited proteolysis was used to probe for changes in the accessibility of trypsin cleavage sites in the MDM2 Cys mutants verses the wt protein. Purified wt or mutant MDM2 was incubated with trypsin at a ratio of 1:100 (protein) for up to 30 min, the samples were then analyzed by gradient SDS-PAGE/immunoblot and cleavage fragments identified using a mixture of MDM2 monoclonal antibodies (Fig. 6B). The results show that the wt protein rapidly formed (within 5 min) a stable core comprising bands 1, 2, and 3, which was maintained throughout the course of the experiment. Although the MDM2C464A mutant initially formed a similar banding pattern to the wt protein, with bands 1, 2, and 3 being detected, it was then further processed so that by 15 min a new stable pattern had emerged comprising bands 2, 3, 4, and 5 with band 1 no longer being detected. Consistent with earlier results the MDM2C478S mutant appeared to be intermediate between wt and the Cys464A protein as at 30 min all five bands were readily detected. These results suggest that the Cys464A and Cys478S mutants have a different conformation to the wt protein leading to the exposure of trypsin-sensitive cleavage sites that are not accessible in wt MDM2.
FIGURE 6.
Conformational differences between wt and RING domain mutants of MDM2. A, a schematic representation of MDM2 showing the epitopes for 3G5 and 2A10; the position of the 4 tryptophan residues in MDM2 is marked. B, immunoblot showing wt or mutant forms of MDM2 following limited proteolysis with trypsin. The blot was developed using a mixture of the MDM2 monoclonal antibodies 2A10, 4B2, 3G5, and SMP14. The numbers are used to label the banding pattern; time points used were 0, 5, 10, 15, 20, and 30 min. The data are representative of three separate experiments. C, resolution of fluorescence emission spectra of MDM2wt (white) and the MDM2C464A (brown) and MDM2C478S (blue) RING mutants. The spectra were corrected for associated buffer background signals. D, maxima with corresponding wavelength of MDM2 fluorescence emission spectra. The values were calculated with mathematical n-polynomial based algorithm, where R2 ≥ 0.999. E, a sin B except the time course was 0, 5, and 20 min. The blots were developed using 2A10 (upper panel) and 3G5 (lower panel). F, localization of the 3G5 antibody epitope within the MDM2 hydrophobic pocket (pocket shown in ribbon representation, colored gold). Nutlin-3 bound to the pocket in stick representation (chlorine in green, carbon in gray, nitrogen in blue, and oxygen in red). The epitope of 3G5 antibody consists of four critical residues; Leu66, Tyr67, Asp68, and Glu69, surrounded by trypsin cleavable Lys and Arg residues (highlighted in purple). This figure was prepared using PyMOL (W. L. DeLano (2002) PyMOL, DeLano Scientific, San Carlos, CA), and structural data were from file 1TTV-RCSB PDB.
Further support for a difference in conformation between the RING mutants and wt MDM2 was provided by investigating the intrinsic fluorescence properties of the protein (Fig. 6, C and D). Although all aromatic amino acids can contribute to protein fluorescence, tryptophan is the dominant intrinsic fluorophore. MDM2 contains a total of four tryptophan residues (Fig. 6A) located in the central region of the protein, in and around the acid domain. Because the emission observed reflects the average environment surrounding each individual tryptophan and the tryptophans within MDM2 are not in identical local environments, each of these residues will contribute unequally to the emission spectrum. The λmax of the emission spectrum of a tryptophan will be shifted to longer wavelengths (i.e. red-shift) if it is involved in hydrogen bonding and/or is exposed to buffer relative to the lower wavelength λmax (blue shift) of a tryptophan buried within the hydrophobic core of the protein. Fig. 6C compares the emission fluorescence spectra of MDM2 wt and the two Cys mutants. For both of the mutant proteins we observe a red shift with λmax at 349-349.5 nm in contrast to that of the wt protein of λmax at 343 nm (Fig. 6D). Moreover the differential quenching of the emission spectrum (Fig. 6C) for the two mutants indicates that their conformation changes are not equivalent. Thus, the data show differences in conformation between wt and mutant forms of MDM2 and between the two mutants. The intrinsic fluorescence measurements are therefore consistent with the partial proteolytic digestion data (Fig. 6B) and with the differential binding affinity of the Cys464A and Cys478S mutants for BOX-I and Nutlin (Figs. 4 and 5).
The experiments presented above using limited proteolysis (Fig. 6B) and intrinsic fluorescence (Fig. 6C) suggest that the introduction of point mutations in key RING domain Cys residues generates a measurable conformational change in full-length MDM2 protein. To establish whether these conformational changes have a direct impact on the structure of the hydrophobic pocket we employed a monoclonal antibody (3G5), which binds to an epitope in the center of the hydrophobic pocket (Fig. 6A) (35). Using limited proteolysis we determined whether changes in the RING domain had an effect on the ability of 3G5 to bind to the hydrophobic pocket of MDM2. As a control for this experiment we used the monoclonal antibody 2A10, which has been mapped to two epitopes within the central and C-terminal domains of MDM2 (Fig. 6A). The 2A10 data (Fig. 6E, upper panel) suggests that the core fragments identified in Fig. 6B (in particular bands 2-4) are comprised largely of the central domain of MDM2 together with C-terminal truncations. In contrast 3G5 (Fig. 6E, lower panel) suggests that the hydrophobic pocket is rapidly cleaved from the core of MDM2 and that its removal generates a fast migrating 3G5 reactive product (band 6). The MDM2C478S mutant appears to be more sensitive to this type of cleavage, because 3G5-positive full-length protein is lost more rapidly than it is for wt MDM2. Of particular interest in this assay is the MDM2C464A mutant, as incubation of this protein leads to a dramatic loss of the 3G5 epitope suggesting that the hydrophobic pocket in this protein is more “relaxed” and therefore more accessible to trypsin digestion during which the 3G5 epitope is cleaved leading to a loss of antibody binding.
Together the data presented in this section support the conclusion that the RING domain of MDM2 can influence the activity of the N-terminal hydrophobic pocket by producing long range conformational changes that are transmitted through the central acid core of MDM2 and which lead to varying degrees of relaxation in the hydrophobic pocket.
DISCUSSION
Studies addressing how MDM2 structure supports its functional diversity have begun to provide insight into the proteins conformational flexibility. Thus, a picture is immerging of interdependent functional domains linked through changes in conformation, emphasizing the need to study the domain structure of MDM2 within the context of the whole protein. In the current study we have used point mutations within the RING finger domain to uncover cross-talk between the C terminus and interactions taking place at the N-terminal hydrophobic pocket of MDM2. Our results show that RING-mediated allosteric modulation of the hydrophobic pocket has implications for both MDM2 transrepressor activity and for the efficacy of MDM2-targeted therapy (Fig. 7).
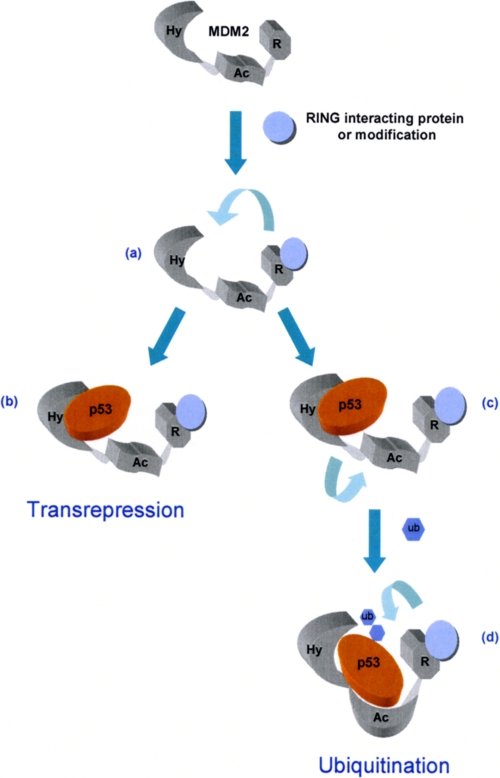
FIGURE 7.
Model for the allosteric regulation of MDM2. Our data suggest the binding of interacting proteins and ligands or post-translational modification within the RING domain of MDM2 (a), which affect RING architecture, will cause conformational changes leading to changes in the affinity of the hydrophobic pocket for BOX-I domain of p53 and other ligands. This can result in formation of a transcriptionally inactive complex (b), or, based on our previous studies (16), could result in a transition complex (c) where hydrophobic pocket binding leads the acid domain to bind to the core BOX-V site in p53-signaling p53 ubiquitination (d).
We previously described a link between the hydrophobic pocket of MDM2 and its acid domain (16). In this case occupation of the hydrophobic pocket promoted conformational changes in MDM2 that favored acid domain binding to a ubiquitination signal in the core DNA binding domain of p53. This allosteric mechanism promotes MDM2 function as an E3-ubiquitin ligase by stabilizing a low affinity interaction between the BOX-V domain of p53 and the acid domain of MDM2. The current data add to this model for the regulation of one MDM2 domain through modulation of a second by demonstrating that the RING finger domain, and more specifically residues required to form the C2H2C4 RING structure, can allosterically modulate hydrophobic pocket interactions. In this case the result is an increase in the affinity of the hydrophobic pocket, which produces a gain in transrepressor function in cells. Interestingly, mutations of Cys464 and Cys478 residues are not equivalent in terms of their effect on the hydrophobic pocket. Thus, although both mutants display increased binding to the BOX-I domain of p53 (Fig. 4), the Cys478S mutant binds Nutlin in preference to BOX-I, whereas the Cys464A protein preferentially binds to BOX-I (Fig. 5).
Structural and computational analysis of the MDM2 hydrophobic pocket interaction with p53 has revealed that this domain has a high degree of plasticity and has suggested that the shape of the binding cleft can change significantly (36, 37). A mechanism has been proposed where binding to the BOX-I domain of p53 requires a progressive opening up of the binding cleft to reveal a hydrophobic interface. p53 binding then proceeds through an intermediate complex where the cleft gradually adopts a more open conformation eventually accommodating Thr18-Asp29 of BOX-I. Based on this type of study it has been hypothesized that the cleft has enough plasticity to allow a range of low energy states rather than a single “open” or “closed” conformation. As a result the pocket can accommodate a range of peptides and small molecules that might not have any obvious structural similarity (32, 36-39). It is likely therefore that the differential binding preferences of our two Cys mutants are a result of different degrees of cleft accessibility. This hypothesis is supported by data showing that, although the Cys464A and Cys478S mutant proteins both adopt a different stable conformation from the wt protein (Fig. 6), their conformation is not equivalent. The intrinsic fluorescence of tryptophan residues within the central domain of the Cys478S mutant is quenched relative to those in the Cys464A mutant, suggesting that they are more solvent-exposed or are involved in additional interactions with nearby residues. Further evidence for a difference in the conformation of the hydrophobic cleft dependent on the status of the RING domain comes from the use of the monoclonal antibody 3G5 to probe for differences in conformation following limited tryptic digestion (Fig. 6E). This antibody recognizes an epitope in the hydrophobic pocket of MDM2, where residues 66LYDE69 are essential but not sufficient for binding, i.e. peptides containing the LYDE motif but not other surrounding residues are not sufficient for antibody recognition (35). The LYDE motif maps to a loop structure in the hydrophobic pocket that lies between two of the α-helical structures that form the BOX-I binding cleft (Fig. 6F) and is flanked by lysine and arginine residues that provide tryptic cut sites when exposed (61IMTKRLYDEKQQHIV75). Our data show that the LYDE motif is more accessible in the Cys464A mutant than in the wt or the Cys478S protein (Fig. 6F), suggesting that the cleft is more relaxed and therefore more readily able to accommodate the BOX-I peptide leading to an increase in BOX-I affinity.
Our data, showing that mutations in the C-terminal C2H2C4 RING of MDM2 can allosterically modulate not only the affinity of the hydrophobic pocket but also its specificity (Fig. 5), suggest that in cells the binding of interacting proteins and/or ligands to the RING is likely to impact on both MDM2 transrepressor activity and on the efficacy of pocket binding drugs in tumor cells (Fig. 6). This hypothesis is supported by data showing that binding of ligands such as zinc and ATP to the RING finger domain of MDM2 generate conformational changes (17, 23). In fact, we have previously shown that zinc binding can affect the interaction of MDM2 with the BOX-I domain of p53; in this case zinc binding leads to a decrease in BOX-I binding (17, 23). Together with our current data this suggests that modulation of the RING domain has the potential to both negatively and positively regulate binding of BOX-I to the hydrophobic pocket. As the Cys mutants used in the current study are E3-ligase dead, it is not possible to determine if C2H2C4 RING interacting factors could also promote MDM2 E3-ligase activity, because increased binding to hydrophobic pocket binding ligands could potentially favor acid domain binding to the BOX-V domain of p53 signaling ubiquitination (16). Thus, it will be interesting to determine whether RING domain-interacting factors, or modifications, such as acetylation (40), within the RING domain act in a concerted manner to promote both transrepression and ubiquitination of p53 or whether changes in RING conformation might act as a switch favoring one MDM2 activity over another.
Recent genetic studies have suggested that the primary physiological role of MDM2 is the regulation of p53 protein levels mediated by its E3-ligase activity (41, 42). These experiments were largely carried out in mouse embryonic fibroblasts and developing mouse embryos where MDM2 activity has been lost or reduced and is therefore rate-limiting. However, this situation is unlikely to reflect the environment encountered in most tumor cells. In fact current estimates suggest that MDM2 levels are elevated in ∼10% of all human tumors (43) due for example to enhanced translation or transcription in addition to amplification of the MDM2 gene (44). In contrast to the studies in non-transformed mouse cell models mentioned above work in tumor cells endogenously overexpressing MDM2 protein suggests that its ability to act as a transrepressor contributes significantly to the impaired p53-response seen in these cells. Thus, in cells that are homozygous for a polymorphism (SNP309) that enhances SP1-dependent MDM2 transcription the MDM2 protein is found in association with p53 resulting in transcriptionally inactive complexes (25, 45). Depletion of MDM2 by siRNA in these cells results in activation of p53-dependent transcription in the absence of increase p53 protein levels supporting the idea that, at least in some tumor cells, MDM2 acts predominantly as a transrepressor of p53 function (25). Furthermore, it has been demonstrated that tumor cells that do not have increased MDM2 levels also operate an MDM2-dependent mechanism for controlling p53 transcription in the absence of changes in p53 protein levels (46). Thus, the data presented here, showing that C2H2C4 RING mutants that loose E3-ubiquitin ligase activity preferentially bind to p53 inhibiting its transactivation activity, may be pertinent to the physiological environment of a tumor cell where MDM2 is controlling p53 activity primarily through transrepression.
We have shown that Nutlin does not inhibit MDM2 E3-ligase activity neither in vitro nor in cell models (Fig. 3; 16). However, despite its lack of E3-ligase inhibitory activity Nutlin can function in cells and animal models of cancer to activate p53 and to elevate p53 protein levels (32, 33). Evidence presented here showing that Nutlin can reverse transrepression imposed by both the wild-type and the Cys478S mutated form of MDM2 suggests that it functions primarily by relieving MDM2-imposed transrepression allowing binding to coactivators such as p300. This conclusion is supported by the observation that Nutlin promotes rapid activation of p53-dependent transcription before changes in p53 protein level are recorded (46). The fact that Nutlin is proving to be an effective drug in some models of human cancer provides further evidence for the role of MDM2 transrepressor activity in tumor development.
Acknowledgments
We thank Malcolm Walkinshaw for stimulating discussion and advice and Martin Wear (Center for Translation and Chemical Biology) for his expertise and helpful discussions on the biophysics of MDM2.
This work was supported by Cancer Research UK (to S. P. and B. W.).
Footnotes
The abbreviations used are: E3, ubiquitin-protein isopeptide ligase; CMV, cytomegalovirus; wt, wild type; E1, ubiquitin-activating enzyme; E2, ubiquitin carrier protein.
References
- 1.Vogelstein, B., Lane, D., and Levine, A. J. (2000) Nature 408, 307-310 [DOI] [PubMed] [Google Scholar]
- 2.Levine, A. J. (1997) Cell 88, 323-331 [DOI] [PubMed] [Google Scholar]
- 3.Hollstein, M., Rice, K., Greenblatt, M. S., Soussi, T., Fuchs, R., Sorlie, T., Hovig, E., Smith-Sorensen, B., Montesano, R., and Harris, C. C. (1994) Nucleic Acids Res. 22, 3551-3555 [PMC free article] [PubMed] [Google Scholar]
- 4.Vousden, K. H. (2006) J. Cell Sci. 119, 5015-5020 [DOI] [PubMed] [Google Scholar]
- 5.Oliner, J. D., Pietenpol, J. A., Thiagalingam, S., Gyuris, J., Kinzler, K. W., and Vogelstein, B. (1993) Nature 362, 857-860 [DOI] [PubMed] [Google Scholar]
- 6.Wu, X., Bayle, J. H., Olson, D., and Levine, A. J. (1993) Genes Dev. 7, 1126-1132 [DOI] [PubMed] [Google Scholar]
- 7.Brooks, C. L., and Gu, W. (2004) Cell Cycle 3, 895-899 [PubMed] [Google Scholar]
- 8.Momand, J., Zambetti, G. P., Olson, D. C., George, D., and Levine, A. J. (1992) Cell 69, 1237-1245 [DOI] [PubMed] [Google Scholar]
- 9.Honda, R., Tanaka, H., and Yasuda, H. (1997) FEBS Lett. 420, 25-27 [DOI] [PubMed] [Google Scholar]
- 10.Kubbutat, M. H., Jones, S. N., and Vousden, K. H. (1997) Nature 387, 299-303 [DOI] [PubMed] [Google Scholar]
- 11.Yin, Y., Stephen, C. W., Luciani, M. G., and Fahraeus, R. (2002) Nat. Cell Biol. 4, 462-467 [DOI] [PubMed] [Google Scholar]
- 12.Wawrzynow, B., Zylicz, A., Wallace, M., Hupp, T., and Zylicz, M. (2007) J. Biol. Chem. 282, 32603-32612 [DOI] [PubMed] [Google Scholar]
- 13.Burch, L., Shimizu, H., Smith, A., Patterson, C., and Hupp, T. R. (2004) J. Mol. Biol. 337, 129-145 [DOI] [PubMed] [Google Scholar]
- 14.Kussie, P. H., Gorina, S., Marechal, V., Elenbaas, B., Moreau, J., Levine, A. J., and Pavletich, N. P. (1996) Science 274, 948-953 [DOI] [PubMed] [Google Scholar]
- 15.Lin, J., Chen, J., Elenbaas, B., and Levine, A. J. (1994) Genes Dev. 8, 1235-1246 [DOI] [PubMed] [Google Scholar]
- 16.Wallace, M., Worrall, E., Pettersson, S., Hupp, T. R., and Ball, K. L. (2006) Mol. Cell 23, 251-263 [DOI] [PubMed] [Google Scholar]
- 17.Shimizu, H., Burch, L. R., Smith, A. J., Dornan, D., Wallace, M., Ball, K. L., and Hupp, T. R. (2002) J. Biol. Chem. 277, 28446-28458 [DOI] [PubMed] [Google Scholar]
- 18.Yu, G. W., Rudiger, S., Veprintsev, D., Freund, S., Fernandez-Fernandez, M. R., and Fersht, A. R. (2006) Proc. Natl. Acad. Sci. U. S. A. 103, 1227-1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang, S., Jensen, J. P., Ludwig, R. L., Vousden, K. H., and Weissman, A. M. (2000) J. Biol. Chem. 275, 8945-8951 [DOI] [PubMed] [Google Scholar]
- 20.Boddy, M. N., Freemont, P. S., and Borden, K. L. (1994) Trends Biochem. Sci. 19, 198-199 [DOI] [PubMed] [Google Scholar]
- 21.Kostic, M., Matt, T., Martinez-Yamout, M. A., Dyson, H. J., and Wright, P. E. (2006) J. Mol. Biol. 363, 433-450 [DOI] [PubMed] [Google Scholar]
- 22.Linke, K., Mace, P. D., Smith, C. A., Vaux, D. L., Silke, J., and Day, C. L. (2008) Cell Death Differ. 15, 841-848 [DOI] [PubMed] [Google Scholar]
- 23.Poyurovsky, M. V., Jacq, X., Ma, C., Karni-Schmidt, O., Parker, P. J., Chalfie, M., Manley, J. L., and Prives, C. (2003) Mol. Cell 12, 875-887 [DOI] [PubMed] [Google Scholar]
- 24.Knights, C. D., Liu, Y., Appella, E., and Kulesz-Martin, M. (2003) J. Biol. Chem. 278, 52890-52900 [DOI] [PubMed] [Google Scholar]
- 25.Arva, N. C., Gopen, T. R., Talbott, K. E., Campbell, L. E., Chicas, A., White, D. E., Bond, G. L., Levine, A. J., and Bargonetti, J. (2005) J. Biol. Chem. 280, 26776-26787 [DOI] [PubMed] [Google Scholar]
- 26.Thut, C. J., Goodrich, J. A., and Tijan, R. (1997) Genes Dev. 11, 1974-1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dornan, D., and Hupp, T. R. (2001) EMBO Rep. 2, 139-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, W. L., Midgley, C., Stephen, C., Saville, M., and Lane, D. P. (2001) J. Mol. Biol. 313, 711-731 [DOI] [PubMed] [Google Scholar]
- 29.Yu, D., Jing, T., Liu, B., Yao, J., Tan, M., McDonnell, T. J., and Hung, M. C. (1998) Mol Cell 2, 581-591 [DOI] [PubMed] [Google Scholar]
- 30.Hupp, T. R., Meek, D. W., Midgley, C. A., and Lane, D. P. (1992) Cell 71, 875-886 [DOI] [PubMed] [Google Scholar]
- 31.Miyashita, T., and Reed, J. C. (1995) Cell 80, 293-299 [DOI] [PubMed] [Google Scholar]
- 32.Vassilev, L. T., Vu, B. T., Graves, B., Carvajal, D., Podlaski, F., Filipovic, Z., Kong, N., Kammlott, U., Lukacs, C., Klein, C., Fotouhi, N., and Liu, E. A. (2004) Science 303, 844-848 [DOI] [PubMed] [Google Scholar]
- 33.Vassilev, L. T. (2004) Cell Cycle 3, 419-421 [PubMed] [Google Scholar]
- 34.Bottger, V., Bottger, A., Howard, S. F., Picksley, S. M., Chene, P., Garcia-Echeverria, C., Hochkeppel, H. K., and Lane, D. P. (1996) Oncogene 13, 2141-2147 [PubMed] [Google Scholar]
- 35.Bottger, A., Bottger, V., Garcia-Echeverria, C., Chene, P., Hochkeppel, H. K., Sampson, W., Ang, K., Howard, S. F., Picksley, S. M., and Lane, D. P. (1997) J. Mol. Biol. 269, 744-756 [DOI] [PubMed] [Google Scholar]
- 36.Espinoza-Fonseca, L. M., and Garcia-Machorro, J. (2008) Biochem. Biophys. Res. Commun. 370, 547-551 [DOI] [PubMed] [Google Scholar]
- 37.Espinoza-Fonseca, L. M., and Trujillo-Ferrara, J. G. (2006) Biopolymers 83, 365-373 [DOI] [PubMed] [Google Scholar]
- 38.Bowman, A. L., Nikolovska-Coleska, Z., Zhong, H., Wang, S., and Carlson, H. A. (2007) J. Am. Chem. Soc. 129, 12809-12814 [DOI] [PubMed] [Google Scholar]
- 39.Stoll, R., Renner, C., Hansen, S., Palme, S., Klein, C., Belling, A., Zeslawski, W., Kamionka, M., Rehm, T., Muhlhahn, P., Schumacher, R., Hesse, F., Kaluza, B., Voelter, W., Engh, R. A., and Holak, T. A. (2001) Biochemistry 40, 336-344 [DOI] [PubMed] [Google Scholar]
- 40.Wang, X., Taplick, J., Geva, N., and Oren, M. (2004) FEBS Lett. 561, 195-201 [DOI] [PubMed] [Google Scholar]
- 41.Itahana, K., Mao, H., Jin, A., Itahana, Y., Clegg, H. V., Lindstrom, M. S., Bhat, K. P., Godfrey, V. L., Evan, G. I., and Zhang, Y. (2007) Cancer Cell 12, 355-366 [DOI] [PubMed] [Google Scholar]
- 42.Toledo, F., Krummel, K. A., Lee, C. J., Liu, C. W., Rodewald, L. W., Tang, M., and Wahl, G. M. (2006) Cancer Cell 9, 273-285 [DOI] [PubMed] [Google Scholar]
- 43.Toledo, F., and Wahl, G. M. (2006) Nat. Rev. Cancer 6, 909-923 [DOI] [PubMed] [Google Scholar]
- 44.Ganguli, G., and Wasylyk, B. (2003) Mol. Cancer Res. 1, 1027-1035 [PubMed] [Google Scholar]
- 45.Bond, G. L., Hu, W., Bond, E. E., Robins, H., Lutzker, S. G., Arva, N. C., Bargonetti, J., Bartel, F., Taubert, H., Wuerl, P., Onel, K., Yip, L., Hwang, S. J., Strong, L. C., Lozano, G., and Levine, A. J. (2004) Cell 119, 591-602 [DOI] [PubMed] [Google Scholar]
- 46.White, D. E., Talbott, K. E., Arva, N. C., and Bargonetti, J. (2006) Cancer Res. 66, 3463-3470 [DOI] [PubMed] [Google Scholar]