Abstract
The mitochondrial dimeric phospholipid cardiolipin is characterized by a high degree of unsaturation of its acyl chains, which is important for its functional interaction with mitochondrial enzymes. The unusual fatty acid composition of cardiolipin molecular species emerges from a de novo synthesized “premature” species by extensive acyl chain remodeling that involves as yet only partially identified acyltransferases and phospholipases. Recently, the yeast protein Taz1p was shown to function as a transacylase, which catalyzes the reacylation of monolysocardiolipin to mature cardiolipin. A defect in the orthologous human TAZ gene is associated with Barth syndrome, a severe genetic disorder, which may lead to cardiac failure and death in childhood. We now identified the protein encoded by reading frame YGR110W as a mitochondrial phospholipase, which deacylates de novo synthesized cardiolipin. Ygr110wp has a strong substrate preference for palmitic acid residues and functions upstream of Taz1p, to generate monolysocardiolipin for Taz1p-dependent reacylation with unsaturated fatty acids. We therefore rename the Ygr110wp as Cld1p (cardiolipin-specific deacylase 1).
Cardiolipin (CL)2 is a dimeric phospholipid specifically enriched in mitochondrial membranes (1). It plays an important role in mitochondrial structure and function and stabilizes respiratory chain super complexes and individual electron transport complexes (2-4). The CL biosynthetic pathway is well characterized in the yeast Saccharomyces cerevisiae, and the enzymes catalyzing the three sequential reactions involved are all associated with mitochondrial membranes (4, 5). CL synthesis shares the same precursor, CDP-diacylglycerol, with the main cellular phospholipids, yet it differs significantly from other phospholipids in its acyl-chain composition that is characterized by a high degree of unsaturated fatty acids. Although cardiolipin was believed to be essential to support mitochondrial function, mutants defective in cardiolipin synthase (Crd1p) are viable and display only moderate defects of mitochondrial function (6, 7). More severe is a defect in the first committed step of CL synthesis, catalyzed by phosphatidylglycerolphosphate synthase Pgs1p/Pel1p (8). pgs1 mutants are temperature-sensitive, unable to grow on non-fermentable carbon sources for growth, and petite lethal, i.e. dependent on intact mitochondrial DNA for survival (8). More subtle mitochondrial phenotypes emerge from alterations of cardiolipin acyl-chain remodeling. “Premature” cardiolipin synthesized by Crd1p undergoes significant remodeling of its acyl-chain composition (9). Mature CL in yeast contains mainly palmitoleic acid (C16:1) and oleic acid (C18:1), which distinguishes CL from most other phospholipids by its high degree of unsaturation (4, 5, 10). In humans, a severe genetic disorder, the Barth syndrome, is associated with defective CL acyl-chain remodeling (11, 12). This disease is caused by mutations in the TAZ gene encoding Tafazzin, for which a functional ortholog also exists in yeast (13). Tafazzin and its yeast counterpart Taz1p are shown to function as transacylases with substrate specificity for either lyso-PC or monolyso-CL (MLCL) (14, 15). The yeast mutant taz1 shows dramatic alterations in the cardiolipin fatty acid pattern, an accumulation of MLCL, and a decreased overall CL content (15). A model for CL acyl-chain remodeling postulated by Xu et al. (14, 16) suggests an active deacylation-reacylation cycle between CL and MLCL. The transacylase Taz1p catalyzes the reacylation of MLCL to CL, but it does not possess any phospholipase activity (14). Thus, the activity generating MLCL, presumably a mitochondrial CL-specific phospholipase A (PLA), remained to be identified.
In this work we show that the yeast open reading frame YGR110W encodes a mitochondrial protein, which functions as PLA in vitro and is involved in generating MLCL in vivo. Ygr110wp deacylates de novo synthesized CL with a clear substrate preference for palmitic acid residues. We hence term the gene CLD1, encoding a cardiolipin-specific deacylase.
EXPERIMENTAL PROCEDURES
Media and Growth Conditions—Yeast strains were grown in YPD medium at 30 °C on a rotary shaker with vigorous aeration. Standard YPD medium contained 1% yeast extract (Difco), 2% glucose (Merck), and 2% Bacto-peptone (Difco). For plate drop tests cells were grown in YPD for 16 h, and cell numbers were estimated using a Casy cell counter (Schärfe Systems). Serial dilutions of cell cultures were prepared in microtiter plates (5 × 105 to 5 × 102 cells per well). Cells were spotted on YPD, YNBLac (0.67% Yeast Nitrogen Base (Difco), vitamins, trace elements (17), and 3% lactic acid, Merck, adjusted to pH 5.5) or YNB synthetic medium containing 3% glycerol (Roth) and 1% ethanol (Merck). Yeast transformants carrying plasmids were grown on uracil-free YNB synthetic medium containing 2% glucose (Merck), 0.67% Yeast Nitrogen Base (Difco), vitamins, and trace elements (17).
Strains and Plasmids—Strains used in this study are listed in Table 1. The double mutant ygr110wΔtaz1Δ was constructed by standard genetic crosses and tetrad dissection and verified by colony PCR, using gene deletion-specific primers (18).
TABLE 1.
Strains and genotypes used in this study
| Strains | Genotype | Source |
|---|---|---|
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Euroscarf |
| taz1Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YPR140W::kanMX4 | Euroscarf |
| ygr110wΔ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YGR110W::kanMX4 | Euroscarf |
| ygr110wΔtaz1Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YGR110W::kanMX4 YPR140W::kanMX4 | This study |
For generation of the plasmid pYGR110W-GFP, the YGR110W gene was amplified from BY4742 chromosomal DNA using 5′-CGGAATTCATGTTCAAGTCAACTTTAAACTCC-3′ as forward and 5′-CAAGACGTCGACTATTTTTTGCATTTCTTTCGTAAGA-3′ as reverse primers. The EcoRI and SalI restriction sites are underlined. The restricted PCR fragment was inserted into the EcoRI/SalI sites of the pUG35 vector (provided by W. H. Hegemann, Institute of Microbiology, Düsseldorf, Germany). For the generation of plasmid pGST-YGR110W, YGR110W reading frame excised with EcoRI and SalI from plasmid pYGR110W-GFP was ligated into pYEX4T-1 (Clontech). All vectors were transformed into yeast strains using the lithium acetate protocol (19). Positive transformants were selected on uracil-free YNB synthetic medium.
Isolation of Mitochondria—Cells were grown overnight in YPD before inoculation of the main culture to an optical density of A600 nm = 0.1. Wild-type BY4742 and ygr110wΔ, taz1Δ, and ygr110wΔtaz1Δ mutant cells were incubated at 30 °C on a shaking platform for 16 h before harvesting. For overexpressing GST fusion proteins, wild-type BY4742, ygr110wΔ, and ygr110wΔtaz1Δ mutant cells harboring either pYEX4T-1 or pGST-YGR110W were inoculated into synthetic media without uracil to an optical density of A600 nm = 0.1. Cells were grown for 16 h at 30 °C before induction of expression with 0.5 mm CuSO4 for 4 h. Mitochondria were prepared from spheroplasts as described (20). Isolated mitochondria were resuspended in 10 mm Tris/HCl, pH 7.4, and stored at -80 °C until further analysis.
Proteins were precipitated with trichloroacetic acid and quantified according to Lowry et al. (21). Proteins from cell homogenate (6 μg) and mitochondria (5 μg) were separated on 10% SDS-polyacrylamide gels prior to Western blot analysis (22, 23). The GST fusion protein was detected using rabbit anti-GST antibody (Calbiochem), and porin was detected using a rabbit anti-porin antibody (generously provided by G. Daum, Technical University Graz). Luminescence was detected using Curix Ultra UV-G x-ray films (Agfa). The density of the immunoreactive bands was determined with Image Quant software (Amersham Biosciences). For calculating the relative enrichment of porin in mitochondria, the signal obtained for homogenate fractions was set to one.
Lipid Analysis—Total lipids were extracted from whole cell extracts and from mitochondria (equivalent to 2.5-5 mg of protein) isolated from BY4742, ygr110wΔ as well as from strains harboring either pYEX4T-1 or pGST-YGR110W with chloroform/methanol 2:1 (v/v) according to Folch et al. (24). The organic phase was dried under a stream of nitrogen, and dissolved in 400 μl of chloroform/methanol (2:1, v/v). Phospholipids were separated by two-dimensional TLC using silica gel 60 plates (Merck) as described before (25). Phospholipids were visualized by staining with iodine vapor and spots were scraped off for fatty acid determination. Fatty acids were converted to fatty acid methyl esters using a boron-trifluoride complex solution (14% solution in methanol, Sigma) as described (26). Fatty acid methyl esters were dissolved in petrol ether and analyzed by GC/MS on a Trace-GC Ultra-DSQ-MS system (Thermo-Electron). The GC conditions were as follows: splitless injection, injector temperature 250 °C, HP-5 MS column (30 m × 0.25 mm ID, 0.25-μm film, Agilent, Waldbronn, Germany); carrier gas helium 5.0, flow 1 ml/min, temperature gradient programmed from 60 to 300 °C at 20 °C/min after an initial time of 4 min. The MS conditions were as follows: positive electron impact ionization, ionization energy of 70 eV, ionization source temperature of 280 °C, emission current of 100 μA, full-scan mode, scan range m/z 50-800, and scan time of 0.29 s. Data acquisition and analysis were done with Xcalibur 2.0™ software. For quantitative analysis, the corresponding peaks of the fatty acid methyl esters in the extracted ion chromatogram of m/z 74 (McLafferty fragment of fatty acid methyl ester) were integrated, and the relative fatty acid distribution was determined.
One-dimensional separation of mitochondrial lipids was used for the analysis of MLCL levels. Lipid standards were purchased from Avanti Polar Lipids. Lipid extracts were applied onto silica gel plates treated with 1.8% boric acid using an automated sampler (Camag Automatic TLC Sampler 4), and lipids were separated using chloroform/ethanol/water/triethylamine (30/35/7/35, v/v) as the solvent as described by Vaden et al. (27). Lipids were visualized by carbonization at 120 °C for 10 min after dipping plates into 3.2% H2SO4 and 0.5% MnCl2 and subsequent staining with iodine vapor. Stained silica plates were scanned using an Epson Perfection 3200 Photo Scanner (Epson), and relative amounts of phospholipids were quantified using ImageJ software (National Institutes of Health).
Microscopy—BY4742 cells harboring the plasmid pYGR110W-GFP were cultivated overnight in YNB media without uracil. The cells were subsequently shifted to media lacking methionine for 3 h to induce expression of the GFP fusion protein. Microscopy was performed on a Leica SP2 confocal microscope using a 100× oil immersion objective (numerical aperture 1.4). For labeling of yeast mitochondria 1 μl of MitoTracker® Red CM-H2XRos (Invitrogen) was added to 1 ml of cell suspension in a 1.5-ml reaction tube (final concentration: 1 μg/ml). Labeling was performed for 10 min at room temperature without subsequent washing of cells. GFP was excited at 488 nm, and fluorescence emission was detected between 500 and 535 nm. MitoTracker® Red CM-H2XRos was excited at 543 nm, and fluorescence emission was detected between 550 and 650 nm. Fluorescence images were acquired simultaneously. Transmission images were recorded using differential interference contrast optics.
PLA Activity Assays—Mitochondria isolated from ygr110wΔ mutant cells harboring either the plasmid pYEX4T-1 or pGST-YGR110W and wild-type cells were used as enzyme source to analyze PLA2 activity by the cPLA2 Assay Kit from Cayman Biochemicals. Hydrolysis of the substrate arachidonoyl thiophosphatidylcholine was determined according to the manufacturer's protocol.
Cardiolipin from Escherichia coli (Avanti Polar Lipids) was used to assay mitochondrial fractions for CL-specific phospholipase activity. 400 μg of cardiolipin was suspended in 150 μl of reaction buffer (160 mm HEPES, pH 7.4, 300 mm NaCl, 20 mm CaCl2, 8 mm CHAPS, 60% glycerol) and mixed with mitochondria (350 μg of protein). Assays were performed at 30 °C for up to 4 h under vigorous shaking. Lipids were extracted and analyzed as described above.
RESULTS
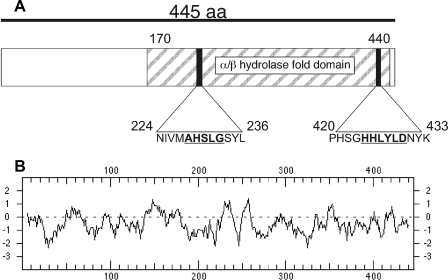
Sequence Features of Ygr110wp—Reading frame YGR110W encodes a 445-amino acid protein with a calculated molecular mass of 52 kDa. The protein shows distant homology to the mammalian protein CGI-58, which is an activator of the major mammalian triglyceride lipase ATGL (28) and was recently shown to act as a lysophosphatidic acyltransferase (29). The amino acid sequence of Ygr110wp contains an α/β hydrolase fold and features a conserved serine residue embedded in a 228AXSXG232 motif. This motif is a variation of GXSXG, which is commonly found in esterases, acyltransferases, and lipases. Sequence alignments of Ygr110wp unveiled similarities to bacterial lysophospholipase domains (COG 2267) and a hydrolase/acyltransferase domain (COG 0596). The structural motif 424HXXXXD429 is indicative of glycerolipid acyltransferase activity (30) and is also present in the homologous protein, Ict1p, which was recently described as an acyl-CoA-dependent lysophosphatidic acid acyltransferase (31) (Fig. 1A). Taken together, these data suggested a possible function for Ygr110wp as a lipase or acyltransferase. The hydrophobicity plot indicates a lack of transmembrane domains (Fig. 1B), and no specific subcellular targeting signals were predicted by in silico analyses.
FIGURE 1.
Sequence features of Ygr110wp. A, positions of conserved lipase and acyltransferase motifs are indicated by black boxes in the schematic illustration of Ygr110wp. The AXSXG lipase as well as the HXXXXD acyltransferase motifs are underlined. The striped box represents the α/β hydrolase fold domain. B, Kyte-Doolittle plot of the Ygr110wp primary amino acid sequence. No transmembrane regions were predicted by in silico analysis.
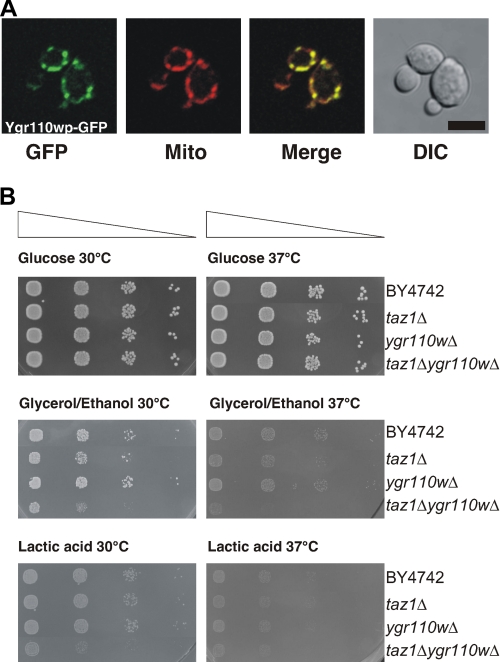
Ygr110wp Localizes Exclusively to Mitochondria but Is Not Essential for Mitochondrial Function—To obtain further insight into the function of Ygr110wp we investigated the subcellular localization by fluorescence microscopy of a C-terminal GFP fusion protein expressed from plasmid pYGR110W-GFP in wild-type cells. Ygr110wp-GFP was detected exclusively in mitochondria, as determined by co-staining with the mitochondria-specific vital dye, MitoTracker® Red (Fig. 2A). This specific mitochondrial localization of Ygr110wp was also confirmed by cell fractionation (see below).
FIGURE 2.
Subcellular localization of Ygr110wp and phenotypic analysis of mutant strains. A, subcellular localization of Ygr110wp. The expression of the C-terminal GFP fusion protein of Ygr110wp in BY4742 was induced by shifting the cells to synthetic medium lacking methionine for 3 h. Cells were incubated with MitoTracker® Red CM-H2XRos for 10 min, and images of red (MitoTracker) and green (GFP) fluorescence were collected by confocal microscopy. Mitochondrial localization of GFP fusion proteins was confirmed by merging of red and green fluorescence images. B, growth on fermentable and non-fermentable carbon sources. Serial dilutions of cells were prepared in microtiter plates (5 × 105 to 5 × 102 cells per well) and spotted on YPD, YNBLac, and YNB containing glycerol/ethanol as carbon sources, using a prong plunger. Experiments were performed three times. The double mutant ygr110wΔtaz1Δ displays a severe growth defect on non-fermentable carbon sources.
The mitochondrial localization of the protein prompted us to investigate the impact of a mutation on mitochondrial function. As shown in Fig. 2B, deletion of YGR110W did not affect growth on glucose or on non-fermentable carbon sources at various temperatures, unlike the taz1Δ mutant, which showed reduced growth at 37 °C on glycerol/ethanol-containing media (15). We conclude that YGR110W does not play an essential role in the cell, nor is it indispensable for mitochondrial function.
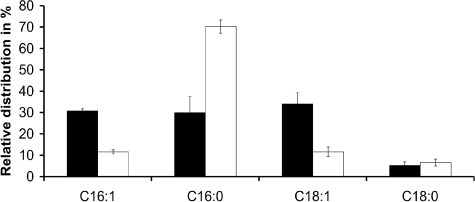
ygr110wΔ Mutants Do Not Show Gross Alterations of Cellular Phospholipids but Specific Defects in Mitochondrial Cardiolipin—Based on the putative function as a lipase or acyltransferase, as predicted by in silico analyses, we determined total cellular fatty acid and lipid composition of wild-type and ygr110wΔ mutant cells. Cells were grown to late exponential phase on glucose media, and lipids were extracted and analyzed as described under “Experimental Procedures.” No significant differences were observed in cellular lipid composition and fatty acid distribution between wild-type and mutant cells (data not shown). Because Ygr110wp is a mitochondrial protein, we then specifically focused our attention on mitochondrial lipids and determined the fatty acid composition of mitochondrial phospholipids individually. Mitochondria were isolated from the ygr110wΔ mutant and from wild-type cells, and phospholipids were extracted and separated by two-dimensional TLC. Lipid spots were scraped off for determining the acyl-chain composition by GC/MS. As shown in Fig. 3, the deletion of YGR110W had a dramatic and specific effect on the composition of fatty acids in cardiolipin. We found that the content of C16:0-acyl chains in CL was increased 2.4-fold in the mutant, compared with wild type, whereas the levels of C16:1- and C18:1-acyl chains were reduced by factors of 2.6 and 2.8, respectively (Fig. 3). Analysis of the fatty acid patterns of other mitochondrial phospholipids, phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, and phosphatidylinositol, did not unveil significant changes in the ygr110wΔ mutant (Table 2). Thus, deletion of gene YGR110W specifically affected acyl-chain composition of CL, suggesting that Ygr110wp is part of the cardiolipin remodeling process in yeast.
FIGURE 3.
Cardiolipin acyl chain composition of wild-type and ygr110wΔ mutant strains. Mitochondria from wild-type BY4742 and ygr110wΔ mutant cells were isolated after 16 h of growth in YPD. Total lipids were extracted from mitochondria and separated by two-dimensional TLC as described under “Experimental Procedures.” Fatty acid (FA) profiles of CL were analyzed by GC-MS from ygr110wΔ (white bars) and wild-type (black bars). Data are expressed as relative percentage of total acyl chains in CL and represent means ± S.D. of at least three independent experiments. The ygr110wΔ mutant shows a 2.4-fold increased level of C16:0 acyl chains in CL compared with wild type.
TABLE 2.
Fatty acid composition of phospholipids from mitochondria isolated from ygr110wΔ mutant and wild-type cells
Phospholipids were extracted, and fatty acid methyl esters were isolated and quantified as described under “Experimental Procedures.” Results represent the means ± S.D. of at least three analyzes performed with two separate mitochondrial preparations. PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine.
| C16:1 | C16:0 | C18:1 | C18:0 | |
|---|---|---|---|---|
| BY4742 | ||||
| PC | 33.67 ± 5.51 | 40.33 ± 5.13 | 15.67 ± 2.31 | 10.33 ± 2.52 |
| PE | 26.00 ± 4.32 | 48.25 ± 7.68 | 22.25 ± 4.50 | 3.25 ± 0.96 |
| PI | 8.00 ± 2.00 | 56.33 ± 8.96 | 12.00 ± 1.00 | 23.67 ± 7.64 |
| PS | 13.00 ± 1.00 | 59.00 ± 7.21 | 18.00 ± 2.00 | 10.33 ± 5.03 |
| ygr110wΔ | ||||
| PC | 34.67 ± 6.11 | 38.67 ± 7.37 | 16.67 ± 2.31 | 9.67 ± 1.15 |
| PE | 24.00 ± 5.10 | 51.25 ± 3.86 | 20.50 ± 1.29 | 4.00 ± 1.83 |
| PI | 8.00 ± 1.00 | 59.67 ± 2.31 | 12.00 ± 1.73 | 20.33 ± 2.08 |
| PS | 13.00 ± 1.00 | 58.00 ± 6.08 | 19.67 ± 0.58 | 9.33 ± 5.51 |
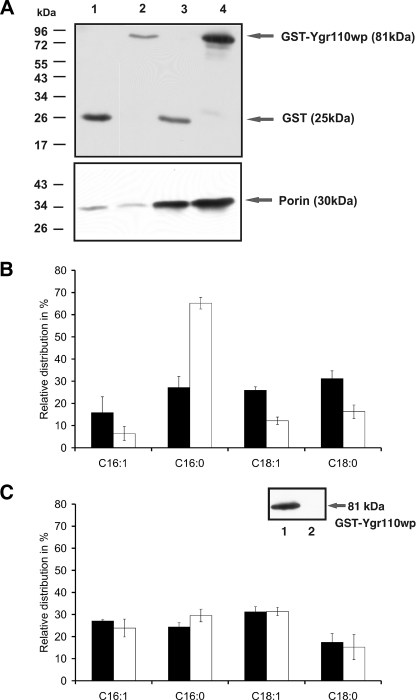
Expression of GST-Ygr110wp in ygr110wΔ Mutants Restores Wild-type C16:0 Level in CL—To exclude the possibility that the observed changes in CL composition are the combined results of the nuclear mutation and induced secondary defects of mitochondrial DNA, we expressed a plasmid-borne GST-Ygr110wp fusion construct in the ygr110w mutant and analyzed CL fatty acid profiles. First, we confirmed by cell fractionation and immunoblot analysis the mitochondrial localization of the fusion protein (Fig. 4A). Mitochondria were isolated from the mutant strain overexpressing the fusion protein or GST only, as described under “Experimental Procedures.” The relative enrichment of porin in mitochondrial fractions was greater than 10-fold, confirming the high quality of the mitochondrial fractions (32). Interestingly, the N-terminal GST tag did not impair mitochondrial localization of the protein, consistent with the lack of any predictable N-terminal mitochondrial targeting sequences (see above).
FIGURE 4.
Expression of GST-Ygr110wp in the ygr110wΔ mutant and in wild-type cells. A, Western blot analysis. Mitochondria were isolated as described under “Experimental Procedures,” and Western blots were probed with antibodies against GST and porin. Lane 1, homogenate (6 μg) obtained from the mutant strain ygr110wΔ-expressing GST only; lane 2, homogenate (6 μg) obtained from ygr110wΔ overexpressing GST-Ygr110wp; lane 3, mitochondria (5 μg) prepared from ygr110wΔ expressing GST only; lane 4, mitochondria (5 μg) prepared from ygr110wΔ overexpressing GST-Ygr110wp. Numbers and bars on the left indicate the molecular mass markers. Experiments were performed as triplicates with at least two independent preparations of mitochondria. A strong band at 81 kDa was detected with anti-GST antibodies in mitochondria expressing the fusion protein. B, fatty acid profile of cardiolipin from mutant cells expressing GST or GST-Ygr110wp. Fatty acid methyl esters were isolated and quantified as described under “Experimental Procedures.” White bars, FA profile of CL isolated from ygr110wΔ expressing GST only; black bars, FA profile of CL from ygr110wΔ overexpressing GST-Ygr110wp. Data are expressed as percentage of total acyl chains in CL and represent means ± S.D. of at least three independent experiments. The fusion protein GST-Ygr110wp suppressed the accumulation of C16:0 acyl chains in CL of ygr110wΔ to wild-type level. C, fatty acid profile of CL from mitochondria isolated from wild-type cells expressing GST-Ygr110wp (lane 1) or GST (lane 2). White bars, FA profile of CL isolated from wild-type expressing GST only; black bars, FA profile of CL from wild-type overexpressing GST-Ygr110wp. Data are expressed as percentage of total acyl chains in CL and represent means ± S.D. of at least three independent experiments. Overexpression of GST-Ygr110wp did not change the fatty acid profile of cardiolipin in wild type.
We next extracted and analyzed mitochondrial lipids from mutant cells expressing GST only, or transformed with the plasmid expressing the GST-Ygr110wp fusion. Overexpression of GST-Ygr110wp restored wild-type levels of C16:0-acyl residues in cardiolipin of the ygr110wΔ strain, whereas they remained at the high level in the mutant strain expressing GST only (Fig. 4B). Interestingly, overexpression of GST-Ygr110wp also increased the relative amount of C18:0 in CL, at the expense of C16:1, in the mutant (Fig. 4B), however, no changes in CL fatty acids were observed in the wild-type upon GST-Ygr110wp overexpression (Fig. 4C). These data support the notion of a CL remodeling complex that is dependent on stoichiometric balance of its components.
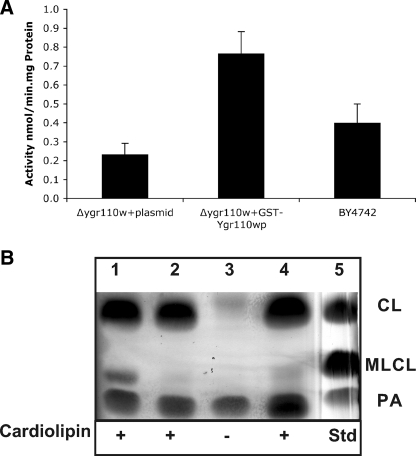
Ygr110wp Expressed in ygr110wΔ Shows PLA Activity in Vitro—The specific phospholipases A catalyzing the deacylation of newly synthesized cardiolipin are currently unknown. The altered fatty acid pattern of cardiolipin in ygr110wΔ mutants and structural motifs predicted from the sequence prompted us to test whether the protein may function as a phospholipase A in vitro. Despite the rather hydrophilic character of the protein we failed to isolate purified GST fusion protein from yeast or E. coli extracts due to its strong aggregation behavior (data not shown). Instead, we tested mitochondrial extracts prepared from the ygr110wΔ mutant strain overexpressing GST-Ygr110wp or expressing GST only and wild-type, for PLA2 activity, as described under “Experimental Procedures.” Mitochondria harboring the GST-Ygr110wp fusion protein showed a PLA2-specific activity of 0.767 nmol/min.mg of protein using phosphatidylcholine as the substrate (Fig. 5A). This PLA2 activity in mitochondria of YGR110W-expressing cells was 3.3-fold higher compared with extracts from control mitochondria and doubled compared with wild-type mitochondria (Fig. 5A).
FIGURE 5.
Phospholipase A activity of GST-Ygr110wp. Mitochondrial fractions were used for PLA activity assays as described under “Experimental Procedures.” A, the specific activity of GST-Ygr110wp toward sn2-arachidonoyl phosphatidylcholine was 0.767 nmol/min.mg of protein, which corresponds to a 3.3-fold induction over the control and is 2-fold higher compared with wild-type mitochondria. Data represent means ± S.D. of at least three independent experiments. B, CL-specific phospholipase A activity of GST-Ygr110wp. Lane 1, mitochondria from ygr110wΔ mutant cells harboring pGST-YGR110W. Lane 2, mitochondria from ygr110wΔ mutant cells harboring pYEX4T-1. Lane 3, mitochondria from ygr110wΔ mutant cells harboring pGST-YGR110W without exogenous CL. Lane 4, mitochondria from wild-type cells. Lane 5, lipid standards; CL, cardiolipin; MLCL, monolysocardiolipin; PA, phosphatidic acid. Experiments were performed three times with two independent mitochondrial preparations.
To confirm that Ygr110wp also acts as a CL-specific phospholipase A in vitro we analyzed its activity against bacterial cardiolipin, which contains 27% C16:0 (Avanti Polar Lipids). Mitochondrial extracts of mutant cells overexpressing GST-Ygr110wp led to a substantial conversion of CL to MLCL, which was not observed in the control extracts from ygr110wΔ cells expressing GST only (Fig. 5B). In the absence of bacterial cardiolipin MLCL was not detectable in mitochondrial extracts of mutant cells overexpressing GST-Ygr110wp. A weak band of MLCL was observed with mitochondria from wild-type cells under the assay conditions (Fig. 5B). These results further demonstrate that Ygr110wp indeed functions as a CL-specific phospholipase A.
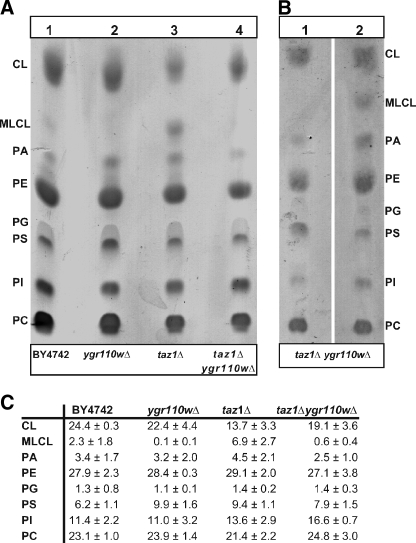
Ygr110wp Genetically and Biochemically Interacts with Tafazzin—Recently, it was shown that a deletion of the TAZ1 gene in yeast resulted in a decrease of unsaturated fatty acids and an accumulation of MLCL (15). Taz1p was therefore proposed to be involved in the reacylation of MLCL specifically with unsaturated fatty acids. Because ygr110wΔ mutants displayed a similar reduction in unsaturated fatty acids of CL (Fig. 3), we tested them for a potential accumulation of MLCL, as described by Vaden et al. (27). In contrast to the taz1Δ mutant, MLCL was not detectable in mitochondria of ygr110wΔ (Fig. 6A, lane 2), and even the low level of MLCL observed in wild-type mitochondria was absent in the mutant. Strikingly, the accumulation of MLCL in taz1Δ mutant was suppressed when both genes, YGR110W and TAZ1, were deleted (Fig. 6A, lane 4). Deletion of both genes resulted in a mitochondrial phenotype and growth of the double mutant was severely retarded on non-fermentable carbon sources at 30 °C (Fig. 2B). Because the level of CL was higher in the ygr110wΔtaz1Δ double mutant than in the taz1Δ single mutant, this phenotype is not due to an overall reduced CL content; the taz1Δ single mutant showed only a weak growth phenotype at 30 °C (Figs. 2B and 6C). These findings confirm that both proteins function in the same pathway.
FIGURE 6.
Determination of MLCL in BY4742 and ygr110wΔ, taz1Δ, and taz1Δygr110wΔ mutant cells. Lipid extracts of the mitochondrial fractions were subjected to one-dimensional TLC as described under “Experimental Procedures.” One representative experiment from triplicate determinations from at least two independent mitochondrial preparations is shown. A, accumulation of MLCL was observed in the mutant strain taz1Δ (lane 3). No MLCL was detectable in the mutant strains ygr110wΔ (lane 2) and taz1Δygr110wΔ (lane 4). Wild-type BY4742 (lane 1). B, overexpression of GST-Ygr110wp in the double mutant taz1Δygr110wΔ led to an accumulation of MLCL (lane 2). Double mutant taz1Δygr110wΔ cells expressing GST only (lane 1). C, lipid composition. Spots on TLC were quantified using ImageJ software (NIH). CL, cardiolipin; MLCL, monolysocardiolipin; PA, phosphatidic acid; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PS, phosphatidylserine; PI, phosphatidylinositol; PC, phosphatidylcholine. Data represent means ± S.D. of at least three independent experiments.
Lack of any MLCL in the ygr110wΔ mutant strongly suggests that it is not defective in a pathway that operates in parallel to Taz1p, as an acyltransferase. Moreover, expression of GST-Ygr110wp in the ygr110wΔtaz1Δ double mutant led to an accumulation of MLCL (Fig. 6B, lane 2) and thus provides further strong evidence that Ygr110wp indeed generates the substrate for the Taz1p reaction, in vivo.
DISCUSSION
In this work we show that Ygr110wp is part of the remodeling process of CL, which involves deacylation-reacylation of premature CL (9). The dimeric phospholipid CL is predominantly enriched in the inner membrane of mitochondria, and its fatty acid pattern displays a high degree of unsaturation compared with other phospholipids (4). Defects in CL remodeling were shown to be causative for the Barth syndrome, a severe disease that may lead to cardiac failure in childhood (13). The Barth syndrome results from mutations in the TAZ gene, which leads to a dramatic decrease in total CL, an accumulation of MLCL, and absence of unsaturated CL species (15).
Remodeling of acyl chains in phospholipids involves sequential actions of phospholipases and acyl transferases (33). The mitochondrial yeast protein Taz1p was shown to function as a transacylase with substrate specificity for either MLCL or lysophosphatidylcholine (14). Due to the fact that TAZ1 deletion mutants accumulate MLCL, Taz1p was proposed to catalyze the reacylation of MLCL (15). The first step in the remodeling cycle, however, requires a phospholipase activity that generates MLCL.
We found that the previously uncharacterized yeast protein Ygr110wp functions as mitochondrial phospholipase. Overexpression of a GST-Ygr110w fusion protein stimulated mitochondrial phospholipase activity >3-fold in a commercial in vitro assay that relied on sn2-arachidonoyl phosphatidylcholine as the substrate. Furthermore, Ygr110wp also showed significant CL-specific phospholipase activity against CL enriched in C16:0 in vitro. Additional evidence for this CL-specific PLA activity is derived from the following data: (i) The localization of Ygr110wp to mitochondria indicates that this enzyme is involved in a mitochondrial process. (ii) The mutant ygr110wΔ shows a specific deficiency in CL fatty acid composition that is not observed in other cellular or mitochondrial phospholipids. Thus mitochondrial phospholipids other than CL can be excluded as substrates for Ygr110wp. (iii) A deletion of YGR110W in a taz1Δ mutant background prevents the formation of MLCL that accumulates in the taz1Δ mutant lacking the CL specific transacylase. (vi) Overexpression of GST-Ygr110wp in a ygr110wΔtaz1Δ double mutant induces accumulation of MLCL, which requires PLA activity.
Furthermore, we show that Ygr110wp preferentially catalyzes the removal of C16:0 from premature CL in vivo. A deletion of YGR110W resulted in a 2.4-fold increase of palmitic acid in CL, on the expense of the unsaturated fatty acid species. In addition, overexpression of GST-Ygr110wp in ygr110wΔ mutant strains reduced the high amount of C16:0 in cardiolipin of mutant cells to wild-type level.
Interestingly, we detected an elevated relative amount of C18:0 of in CL, at the expense of C16:1, in ygr110wΔ mutant cells overexpressing GST-Ygr110wp. In contrast, no changes in CL fatty acids were observed in the wild-type upon GST-Ygr110wp overexpression. From these findings we assume that CL remodeling is dependent on a stoichiometric balance of all components involved, perhaps in a specific lipid remodeling complex.
A deletion of YGR110W resulted in an aberrant fatty acid pattern in CL, but total CL was not decreased and no accumulation of MLCL was detected, which is in contrast to taz1Δ mutants (15). In addition, we did not observe significant growth defects on media containing ethanol or glycerol as carbon sources for the ygr110wΔ single mutant, indicating that an increased level of saturated fatty acids in CL of the mutant is not sufficient to impair mitochondrial function. However, we observed a severe growth defect on non-fermentable carbon sources for the ygr110wΔtaz1Δ double mutant, which proves that Ygr110wp and Taz1p are involved in the same pathway.
We propose that Ygr110wp functions as the phospholipase upstream of Taz1p, generating MLCL for Taz1p-dependent reacylation with unsaturated fatty acids. The preferred removal of C16:0 from cardiolipin by Ygr110wp is contradictory to a recent model for the exchange of acyl groups in de novo synthesized CL (16). The authors suggested that specific fatty acids may accumulate in CL by concerted action of acyl-specific inflow and nonspecific outflow. On the basis of our in vivo results the outflow of acyl residues from CL has also to be considered a specific process. We propose a model in which enzymes with phospholipase A or B activity specifically detach saturated C16:0 or C18:0 acyl chains from de novo synthesized CL to generate MLCL, which will be subsequently reacylated specifically by transacylases, such as Taz1p. Thus the exchange of the CL acyl chains may require the coordinated action of multiple phospholipases and transacylases, each of them showing specificities for degree of unsaturation but also for acyl-chain length. Cld1 (cardiolipin-specific deacylase 1) encoded by gene YGR110W is the first phospholipase activity identified so far that preferentially removes C16:0 acyl chains from cardiolipin.
Acknowledgments
We thank W. H. Hegemann, Institute of Microbiology, Düsseldorf, Germany, for providing the plasmid pUG35 and G. Daum, Institute of Biochemistry, Technical University Graz, for providing anti-porin antibody.
This work was supported by the Austrian Science Fund (Project Number P19041 to R. L.) and Special Research Area lipotox (Grant F3005 to S. D. K.).
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: CL, cardiolipin; MLCL, monolysocardiolipin; PLA, phospholipase A; GFP, green fluorescent protein; ATGL, adipose triglyceride lipase; GST, glutathione S-transferase; PC, phosphatidylcholine; FA, fatty acid.
References
- 1.Wriessnegger, T., Guebitz, G., Leitner, E., Ingolic, E., Cregg, J., de la Cruz, B. J., and Daum, G. (2007) Biochim. Biophys. Acta 1771, 455-461 [DOI] [PubMed] [Google Scholar]
- 2.Zhang, M., Mileykovskaya, E., and Dowhan, W. (2005) J. Biol. Chem. 280, 29403-29408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfeiffer, K., Gohil, V., Stuart, R. A., Hunte, C., Brandt, U., Greenberg, M. L., and Schägger, H. (2003) J. Biol. Chem. 278, 52873-52880 [DOI] [PubMed] [Google Scholar]
- 4.Li, G., Chen, S., Thompson, M. N., and Greenberg, M. L. (2007) Biochim. Biophys. Acta 1771, 432-441 [DOI] [PubMed] [Google Scholar]
- 5.Schlame, M., Rua, D., and Greenberg, M. L. (2000) Prog. Lipid Res. 39, 257-288 [DOI] [PubMed] [Google Scholar]
- 6.Chang, S. C., Heacock, P. N., Mileykovskaya, E., Voelker, D. R., and Dowhan, W. (1998) J. Biol. Chem. 273, 14933-14941 [DOI] [PubMed] [Google Scholar]
- 7.Zhang, M., Su, X., Mileykovskaya, E., Amoscato, A. A., and Dowhan, W. (2003) J. Biol. Chem. 278, 35204-35210 [DOI] [PubMed] [Google Scholar]
- 8.Chang, S. C., Heacock, P. N., Clancey, C. J., and Dowhan, W. (1998) J. Biol. Chem. 273, 9829-9836 [DOI] [PubMed] [Google Scholar]
- 9.Schlame, M., and Rustow, B. (1990) Biochem. J. 272, 589-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlame, M., Ren, M., Xu, Y., Greenberg, M. L., and Haller, I. (2005) Chem. Phys. Lipids 138, 38-49 [DOI] [PubMed] [Google Scholar]
- 11.Vreken, P., Valianpour, F., Nijtmans, L. G., Grivell, L. A., Plecko, B., Wanders, R. J., and Barth, P. G. (2000) Biochem. Biophys. Res. Commun. 279, 378-382 [DOI] [PubMed] [Google Scholar]
- 12.Schlame, M., Towbin, J. A., Heerdt, P. M., Jehle, R., DiMauro, S., and Blanck, T. J. (2002) Ann. Neurol. 51, 634-637 [DOI] [PubMed] [Google Scholar]
- 13.Ma, L., Vaz, F. M., Gu, Z., Wanders, R. J., and Greenberg, M. L. (2004) J. Biol. Chem. 279, 44394-44399 [DOI] [PubMed] [Google Scholar]
- 14.Xu, Y., Malhotra, A., Ren, M., and Schlame, M. (2006) J. Biol. Chem. 281, 39217-39224 [DOI] [PubMed] [Google Scholar]
- 15.Gu, Z., Valianpour, F., Chen, S., Vaz, F. M., Hakkaart, G. A., Wanders, R. J., and Greenberg, M. L. (2004) Mol. Microbiol. 51, 149-158 [DOI] [PubMed] [Google Scholar]
- 16.Xu, Y., Kelley, R. I., Blanck, T. J., and Schlame, M. (2003) J. Biol. Chem. 278, 51380-51385 [DOI] [PubMed] [Google Scholar]
- 17.Sherman, F. (2002) Methods Enzymol. 350, 3-41 [DOI] [PubMed] [Google Scholar]
- 18.Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P., and Boeke, J. D. (1998) Yeast 14, 115-132 [DOI] [PubMed] [Google Scholar]
- 19.Gietz, R. D., and Schiestl, R. H. (1991) Yeast 7, 253-263 [DOI] [PubMed] [Google Scholar]
- 20.Zinser, E., Sperka-Gottlieb, C. D., Fasch, E. V., Kohlwein, S. D., Paltauf, F., and Daum, G. (1991) J. Bacteriol. 173, 2026-2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951) J. Biol. Chem. 193, 265-275 [PubMed] [Google Scholar]
- 22.Laemmli, U. (1970) Nature 227, 680-685 [DOI] [PubMed] [Google Scholar]
- 23.Haid, A., and Suissa, M. (1983) Methods Enzymol. 96, 192-205 [DOI] [PubMed] [Google Scholar]
- 24.Folch, J., Lees, M., and Sloane Stanley, G. H. (1957) J. Biol. Chem. 226, 497-509 [PubMed] [Google Scholar]
- 25.Schneiter, R., and Daum, G. (2006) Methods Mol. Biol. 313, 75-84 [DOI] [PubMed] [Google Scholar]
- 26.Morrison, W. R., and Smith, L. M. (1964) J. Lipid Res. 5, 600-608 [PubMed] [Google Scholar]
- 27.Vaden, D. L., Gohil, V. M., Gu, Z., and Greenberg, M. L. (2005) Anal. Biochem. 338, 162-164 [DOI] [PubMed] [Google Scholar]
- 28.Lass, A., Zimmermann, R., Haemmerle, G., Riederer, M., Schoiswohl, G., Schweiger, M., Kienesberger, P., Strauss, J. G., Gorkiewicz, G., and Zechner, R. (2006) Cell Metab. 3, 309-319 [DOI] [PubMed] [Google Scholar]
- 29.Ghosh, A. K., Ramakrishnan, G., Chandramohan, C., and Rajasekharan, R. (2008) J. Biol. Chem. 283, 24525-24533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heath, R. J., and Rock, C. O. (1998) J. Bacteriol. 180, 1425-1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh, A. K., Ramakrishnan, G., and Rajasekharan, R. (2008) J. Biol. Chem. 283, 9768-9775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zinser, E., and Daum, G. (1995) Yeast 11, 493-536 [DOI] [PubMed] [Google Scholar]
- 33.Tanaka, K., Fukuda, R., Ono, Y., Eguchi, H., Nagasawa, S., Nakatani, Y., Watanabe, H., Nakanishi, H., Taguchi, R., and Ohta, A. (2008) Biochim. Biophys. Acta 1781, 391-399 [DOI] [PubMed] [Google Scholar]