Abstract
Arginine methylation is a widespread post-translational modification of proteins catalyzed by a family of protein arginine methyltransferases (PRMTs). The ancient protozoan parasite, Trypanosoma brucei, possesses five putative PRMTs, a relatively large number for a single-celled eukaryote. Trypanosomatids lack gene regulation at the level of transcription, instead relying on post-transcriptional control mechanisms that act at the levels of RNA turnover, translation, and editing, all processes that likely involve multiple RNA-binding proteins, which are common targets of arginine methylation. Here, we report the characterization of a trypanosome PRMT, TbPRMT7, which is homologous to human PRMT7. Interestingly, trypanosomatids are the only single-celled eukaryotes known to harbor a PRMT7 homologue. TbPRMT7 differs dramatically from all known metazoan PRMT7 homologues in lacking the second AdoMet binding-like domain that is required for activity of the human enzyme. Nevertheless, bacterially expressed TbPRMT7 exhibits robust methyltransferase activity toward multiple targets in vitro. High resolution ion exchange chromatography analysis of methylated substrates reveals that TbPRMT7 is a type III PRMT, catalyzing the formation of only monomethylarginine, thereby representing the only exclusively type III PRMT identified to date. TbPRMT7 is expressed in both mammalian and insect stage T. brucei and is apparently dispensable for growth in both life cycle stages. The enzyme is cytoplasmically localized and is a component of several higher order complexes in vivo. Together, our studies indicate that TbPRMT7 is a Type III PRMT, and its robust activity and presence in numerous complexes suggest it plays multiple roles during the complex T. brucei life cycle.
Arginine methylation is a common post-translational modification of proteins that plays a role in numerous cellular functions including chromatin remodeling, RNA processing, DNA repair, and cell signaling (1-7). The transfer of methyl groups from the methyl donor, S-adenosyl-methionine (AdoMet),2 to the arginine residues of proteins is carried out by a class of enzymes known as protein arginine methyltransferases (PRMTs). The PRMTs are further classified into four classes, depending on the extent and specific placement of the methyl group(s) on the arginine amino acid. The most common type of PRMT is the Type I class of enzymes, which catalyze the formation of monomethylated arginine (MMA) on the terminal ω-nitrogen, followed by addition of a second methyl group on the same ω-nitrogen, yielding asymmetric dimethylarginine (ADMA). The Type II PRMTs also catalyze synthesis of MMA on the terminal ω-nitrogen, but subsequently add a second methyl group to the adjacent terminal ω-nitrogen, resulting in symmetric dimethylarginine (SDMA). The majority of PRMTs fall into the Type I or Type II designation. Type III PRMTs catalyze production of solely MMA, and the only previously reported Type III PRMT, yeast Hsl7, was recently shown to have Type II activity (8, 9). Finally, the Type IV PRMT, of which yeast Rmt2 seems to be the only example (10, 11), catalyzes MMA on the δ-nitrogen of arginine. PRMTs are absent from prokaryotes, but genes encoding varying numbers and classes of PRMT homologues are present in all eukaryotes examined, with the exception of the very early branching, Giardia lamblia (1, 12, 13). Common PRMT substrates include chromatin-associated proteins, signaling proteins, and a large number of RNA-binding proteins (RBPs) (1). Many of these substrates are methylated within glycine/arginine-rich (GAR) regions, although methylation of arginine residues in non-GAR regions is becoming more apparent as more substrates are identified.
Trypanosoma brucei is a parasitic protozoan that is the etiological agent of African sleeping sickness. It is consistently identified as being an early branching eukaryote (14). Trypanosomes have a variety of unique features, one of the most striking of which is the absence of gene regulation at the level of transcription (15, 16). Instead, control of gene expression in these organisms is mediated through several post-transcriptional processes including RNA stability, translation, and RNA editing. Thus, gene regulation presumably depends heavily on a multitude of RBPs, a few of which have been identified (17-23). In keeping with this model, the T. brucei genome encodes a large number of RBPs, including many that contain GAR motifs, and which may thus be targets of regulation by arginine methylation (24).3
Previous studies in T. brucei indicated that the parasite contains five putative PRMTs in its genome, one of the highest numbers for a single-celled eukaryote (13, 25). By comparison, there are only three PRMTs in the genomes of Saccharomyces cerevisiae and Schizosaccharomyces pombe (13). We previously reported that two of the trypanosome PRMTs are homologues of the human Type I PRMT, PRMT1, and the Type II PRMT, PRMT5 (25, 26). The other three T. brucei PRMTs remained uncharacterized, and in this study, we present the in vitro and in vivo characterization of one of these trypanosome PRMTs. We coin this enzyme TbPRMT7, as it exhibits the highest sequence identity to the human PRMT7 (Fig. 1). Human PRMT7 contains two AdoMet binding domains, both of which are required for its activity (27). The type of activity catalyzed by human PRMT7 is currently unclear. It has been reported to catalyze the production of either solely MMA on peptide substrates or SDMA on peptide and protein substrates (27, 28). In either case, the activity of human PRMT7 is reportedly weak. Here, we show that TbPRMT7 has a severely truncated structure compared with its human homologue, lacking the second AdoMet binding domain. In vitro assays demonstrate that, in contrast to human PRMT7, TbPRMT7 possesses robust PRMT activity toward multiple substrates. Despite its high level of activity, HPLC analysis revealed that TbPRMT7 catalyzes the formation of only MMA, classifying it as a Type III PRMT. TbPRMT7 may be the only exclusively Type III PRMT identified to date, as the designation of human PRMT7 as a Type II or Type III enzyme is not resolved (27, 28). In vivo, TbPRMT7 is expressed in both mammalian and insect life cycle stages of the parasite, and studies in which TbPRMT7 levels were depleted by RNAi suggest it is not essential for growth of either stage. TbPRMT7 is cytoplasmically localized, and it is a component of several higher ordered complexes. Interestingly, PRMT7 homologues are not identifiable in the genomes of any single-celled eukaryotes outside of trypanosomes and their relatives (13). Our studies indicate that TbPRMT7 is a Type III PRMT, and its robust activity and presence in numerous macromolecular complexes suggest that it may play multiple roles during the complex life cycle of T. brucei.
FIGURE 1.
Sequence analysis of TbPRMT7. A, ClustalW derived cladogram of TbPRMT7 compared with the seven active human PRMTs as well as the uncharacterized PRMT9(4q31) protein. The human PRMTs are grouped as Type I (ADMA catalyzing), Type II (SDMA catalyzing), and the less definitively characterized Type II or III (solely MMA catalyzing) PRMT7. TbPRMT7 is indicated by the arrow. B, domain structure of human PRMT7 compared with that of TbPRMT7. Four closely spaced vertical lines indicate AdoMet binding (or AdoMet binding-like, see text) domains; single vertical line indicates “THW” loops. The degree of homology between TbPRMT7 and human PRMT7 in the most conserved region (spanning 248 amino acids; indicated by the horizontal line) is shown above the diagram. The percent identity and similarity in this region is indicated. C, ClustalW alignment of TbPRMT7 with the N-terminal methyltransferase motifs of PRMT7 homologues from humans (HsPRMT7N (GenBank™ accession number NP_061896)) and Drosophila (DmDart7N (GenBank™ accession number NP_611753)). White letters on black background, identical; white letters on gray background, conserved. Asterisks indicate the conserved glutamate residues of the double E loop.
EXPERIMENTAL PROCEDURES
Cloning and Expression of TbPRMT7—The gene encoding the TbPRMT7 open reading frame (Tb927.7.5490) was PCR-amplified from oligo(dT)-primed cDNA derived from procyclic form (PF) T. brucei (strain 927 Eatro 1.1) RNA using the primers: PRMT7-5, 5′-GCGAATTCATGAAGCGCACACCTGTTAG-3′ and PRMT7-3, 5′-GGAAGCTTTTCCTTCTGACTGGCATC-3′. The resultant product was cloned into pCR2.1 (TA-TOPO kit, Invitrogen). TbPRMT7 was excised from pCR2.1-TbPRMT7 and ligated into the BamHI and XhoI sites of pGEX4T-1 (Novagen). The resultant pGEX-TbPRMT7 was then transformed into Rosetta strain Escherichia coli cells (Novagen) for expression. GST-tagged TbPRMT7 was purified using single step glutathione-agarose (Invitrogen) and a standard GST purification protocol. Removal of the GST tag of GST-TbPRMT7 was carried out by thrombin (Sigma) digestion overnight on ice. Complete digestion was confirmed by Coomassie Blue staining. TbPRMT7 with a C-terminal 6× histidine tag was produced by cloning the BamHI and XhoI digest of pGEX-TbPRMT7 into the BamHI and XhoI sites of pET21A (Novagen), resulting in pET21a-TbPRMT7. TbPRMT7-His was purified using a standard His purification protocol using TALON resin (Clontech).
Antibodies—Monoclonal antibodies against protein C were purchased from Sigma. Antibodies against T. brucei Hsp70 and the CTD of RNA polymerase II were generously provided by James Bangs (University of Wisconsin) and Vivian Bellafatto (University of Medicine and Dentistry of New Jersey), respectively.
Cell Cultures and RNA Interference—PF T. brucei strain 29-13 (provided by Dr. George A. M. Cross, Rockefeller University), which contains integrated genes for the T7 RNA polymerase and the tetracycline repressor, were grown in SDM-79 media supplemented with 15% fetal bovine serum as indicated previously (29). Bloodstream form (BF) single marker T. brucei cells (also provided by Dr. George A. M. Cross) were cultured in HMI-9 media supplemented with 10% fetal bovine serum and 10% serum plus (29). For creation of cells expressing dsRNA interference against TbPRMT7, the full-length TbPRMT7 open reading frame was excised from pGEX4T-1 using BamHI and XhoI and ligated into the BamHI-XhoI sites of the tetracycline-inducible RNAi vectors p2T7-177 (30) or pHD1621(31), yielding p2T7-177-TbPRMT7 and pHD1621-TbPRMT7, respectively. NotI-linearized p2T7-177-TbPRMT7 was transfected in to PF cells, and cells harboring this construct were selected with 2.5 μg/ml phleomycin. pHD1621-TbPRMT7 was transfected in BF cells, and cells harboring this construct were selected with 20 μg/ml blasticidin. Clones were obtained by serial dilution and grown over the indicated time periods in the absence or presence of 2.5 μg/ml tetracycline. Knockdown of TbPRMT7 was verified by either Northern blotting or qRT-PCR. For integration of the tandem purification PTP tag (32) into the endogenous TbPRMT7 locus, primers PRMT7-5′-ApaI (5′-GTGGGCCCGCTATTCAGAGTCGACTTTAGC-3′) and PRMT7-3′-NotI (5′-CAGCGGCCGCGTTGTTTTGCCCTCGC-3′, encompassing nucleotides 830-1170 of the TbPRMT7 open reading frame and introducing ApaI and NotI restriction sites, were used to amplify the 3′-end of TbPRMT7 and introduce it into the ApaI-NotI restriction sites of pC-PTP (32), which we modified to contain the puromycin resistance gene. pC-PRMT7-PTP was linearized using the unique HpaI restriction site within TbPRMT7 and transfected into 29-13 PF T. brucei cells. Transgenic cells were selected with 1 μg/ml puromycin and cloned by limiting dilution. TbPRMT7-PTP expression was verified by Western blotting against the protein C tag within PTP, which detected only the expected band at 63 kDa.
In Vitro Methylation Assays—S-adenosyl-[methyl-3H]methionine ([3H]AdoMet) (65-82 Ci/mmol) was purchased from Amersham Biosciences and PerkinElmer Life Sciences. Methylation assays were performed essentially as in Refs. 25, 26, with the following modifications. Purified GST-TbPRMT7 (amounts as indicated in figure legends) was incubated in 50 μl of PBS buffer with specified substrate (typically 3 μg, unless indicated) and 2 μCi of [3H]Ado Met for 1-14 h at room temperature (22 °C). The substrates used in this study are as follows: the synthetic peptide H-CGRGRGRGRGRGRGRG-NH2 (33), myelin basic protein (MBP) (Sigma), bovine histones (IIAS, Sigma), His-RBP16 (18), GST-TbRGG1 (26), GST-TbRGG2(23), GST-MRP2 (34), and GST protein alone purified using a standard glutathione purification protocol as above.
High Resolution Ion Exchange Chromatographic Analysis of TbPRMT7 Substrates—Reactions were carried out overnight as indicated above using either 3 μg or 6 ng of GST-TbPRMT7 alone, GST-TbPRMT7 with 3 μg of His-RBP16, or GST-Tb-PRMT7 with 10 μg of bovine histones. Reactions with thrombin-digested TbPRMT7 to remove the GST tag were carried out with only 3 μg of enzyme. Reactions with TbPRMT7-ProtC purified from T. brucei were done with ∼100 ng of TbPRMT7. Methylated substrates were precipitated with 50% trichloro-acetic acid, washed with acetone, and resultant trichloroacetic acid pellet was acid-hydrolyzed to component amino acids under vacuum for 20 h at 110 °C. The samples were then dried and resuspended in 50 μl of water, followed by addition of 500 μl of sodium citrate buffer (pH 2.2), and 1.0 μmol each of unlabeled standards of ω-NG-monomethylarginine (acetate salt) (MMA), ω-NG, NG-dimethylarginine (hydrochloride) (ADMA), and ω-NG, N′G-dimethylarginine (di(p-hydroxyazobenzene-p′-sulfonate salt) (SDMA) (all purchased from Sigma) in a total of 1 ml. The reaction was then analyzed by cation exchange chromatography as detailed previously (35).
Fractionation of TbPRMT7-PTP Cells and Glycerol Gradients—PF TbPRMT7-PTP cells were fractionated into cytoplasmic and nuclear fractions using methods described previously (36). To establish the efficiency of fractionation, Western blots were performed with antibodies specific for cytoplasmic Hsp70 and nuclear RNA polymerase II CTD. For glycerol gradient analysis, cytoplasmic lysates from PF TbPRMT7-PTP cells (equivalent of 5 × 109 cells) were purified and separated on a 5-20% glycerol gradient in buffer (25 mm Tris, pH 8.0, 50 mm KCl, 10 mm MgOAc, 100 μm ATP, 5-20% glycerol, one-half tablet of complete protease inhibitor (Roche)) at 38,000 × g for 5 h at 4 °C. 500-μl fractions were subsequently collected from the top of the gradient, and 10 μl of each fraction were analyzed by Western blot using antibodies recognizing the protein C tag.
Northern Blotting and Quantitative RT-PCR—Total RNA was extracted from uninduced and induced TbPRMT7 RNAi cells using TRIzol reagent (Invitrogen), at day 5 post-induction for both BF and PF cells. Ten micrograms of RNA was treated with a DNA-free DNase kit (Ambion) to remove any residual DNA. RNA was reverse-transcribed using random hexamer primers and the Taq-Man reverse transcription kit (Applied Biosciences). Resultant cDNA was used in RT-PCR reactions using the TbPRMT7 specific primers TbPRMT7-fwd 5′-GCGTGATTCTTCACATGC-3′ and TbPRMT7-rev 5′-CACACTGTTCACCCTCATTTC-3′. 25 μl of quantitative RT-PCR (qRT-PC) reactions were set up as established in Carnes et al. (37) and the cDNA amplified using a MyiQ single color real-time PCR detection system (Bio-Rad). Results were analyzed using iQ5 software (Bio-Rad) and compared with levels of steady state β-tubulin RNA using the standard curve method. Similar results were also obtained with an 18 S rRNA standard. The levels of each mRNA are represented as the mean and standard error of six determinations.
RESULTS
TbPRMT7 Is a C-terminally Truncated PRMT7 Homologue—Analysis of the T. brucei genome reveals the presence of at least five distinct PRMTs (25). We previously reported the characterization of two of these enzymes, the type I PRMT1 homologue, TbPRMT1, and the type II PRMT5 homologue, TbPRMT5 (25, 26). In this report, we present the in vitro and in vivo analysis of a third T. brucei PRMT (GeneDB Tb927.7.5490). Phylogenetic comparison of Tb927.7.5490 with all human PRMTs demonstrates that it clusters with PRMT7 and the related PRMT9(4q31) (Fig. 1A). Pairwise BLAST analysis revealed slightly higher homology of Tb927.7.5490 to PRMT7 than to PRMT9(4q31), with 27.7% identity and 44.9% similarity over the 248-amino acid region encompassing the AdoMet binding domain and the “THW” loop (Fig. 1, B and C). Based on this homology, we have termed the T. brucei enzyme, TbPRMT7. Human PRMT7 (like PRMT9(4q31)) possesses two regions with high homology to AdoMet binding domains (Fig. 1B). While the C-terminal domain of human PRMT7 does not bind AdoMet, it is required for the overall activity of the full-length enzyme (27). The most striking difference between the 692-amino acid human PRMT7 and the 390-amino acid TbPRMT7 is the absence of a second AdoMet binding-like domain in TbPRMT7 (Fig. 1B). The TbPRMT7 amino acid sequence is more homologous to the “active” N terminus of PRMT7 than to the “inactive” PRMT7 C terminus (Fig. 1B). ClustalW alignment of TbPRMT7 with the N termini of human PRMT7 and the Drosophila homologue, Dart7 (38), illustrates significant homology across all characteristic methyltransferase motifs I, post I, II, and III and the double E loop (Fig. 1C). As in all known PRMT7 sequences, the threonine in the “THW” loop of TbPRMT7 is substituted by an aspartic acid. Interestingly, while all higher eukaryotic PRMT7 homologues possess the duplicated AdoMet binding-like domain in their C termini, the genomes of all kinetoplastid parasites (trypanosomes and Leishmania spp.) contain the shortened PRMT7 homologue (data not shown). Conservation of TbPRMT7 among kinetoplastids along with its similarity to the active AdoMet binding domain of metazoan PRMT7 homologues strongly suggests that TbPRMT7 is an active methyltransferase.
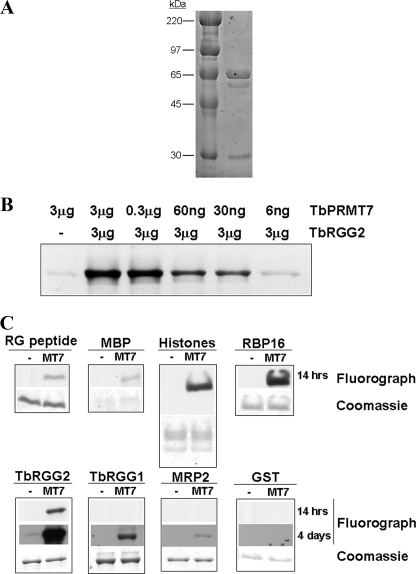
TbPRMT7 Is an Active PRMT with a Broad Substrate Range—Because TbPRMT7 lacks the C-terminal AdoMet binding-like domain that is required for activity of its human homologue, it was imperative to first establish that it is an active enzyme. To this end, we expressed full-length TbPRMT7 as a GST fusion in E. coli and purified the recombinant protein by affinity chromatography as described under “Experimental Procedures.” A major band of 71 kDa was observed, as expected for a fusion of GST with the 390-amino acid TbPRMT7 protein (Fig. 2A). We initially tested the enzymatic activity of TbPRMT7 by titrating increasing amounts of enzyme into in vitro methylation reactions containing [3H]AdoMet and the RG-rich T. brucei protein, TbRGG2 (23), as a substrate. Methylated substrate was then resolved by SDS-PAGE and visualized by fluorography. As shown in Fig. 2B, TbPRMT7 methylated TbRGG2 in reactions containing as little as 6 ng of enzyme, demonstrating that TbPRMT7 possesses robust methyltransferase activity. Additionally, recombinant TbPRMT7 with a C-terminal His6 tag displayed in vitro activity similar to that of the GST-TbPRMT7 construct, indicating that the tag did not appreciably affect the methyltransferase activity of the enzyme (data not shown). Therefore, despite the absence of a second AdoMet binding-like domain, TbPRMT7 is a highly active methyltransferase.
FIGURE 2.
Enzymatic activity of recombinant TbPRMT7. A, Coomassie Blue-stained SDS-PAGE gel of GST-TbPRMT7 purified from E. coli. B, in vitro methylation reaction with a titration of GST-TbPRMT7 and GST-TbRGG2 as substrate. Indicated amounts of enzyme and substrate were incubated for 14 h at 25 °C in the presence of 2 μCi of [3H]AdoMet in PBS. Products were separated by SDS-PAGE, and the gels treated with En3Hance, dried, and exposed to film for 14 h. The faint band in the reaction lacking substrate reflects SDS-PAGE stable binding of [3H]AdoMet by TbPRMT7, whose mobility is similar to, but slightly faster than that of the TbRGG2 substrate. C, methylation of multiple substrates by GST-TbPRMT7. Three micrograms of the indicated substrates were incubated overnight at 22 °C either in the absence or presence of GST-TbPRMT7 (6 ng), followed by visualization as in B. For several weakly methylated substrates (MRP2, TbRGG1, and GST) a 4-day exposure is also shown. The strongly methylated substrate, TbRGG2, is shown at the same 4-day exposure for comparison.
We next assessed the substrate specificity of TbPRMT7, using a panel of substrates including an RG-rich peptide, myelin basic protein (MBP) and bovine histones, which are known substrates of mammalian and trypanosome PRMTs, as well as four trypanosome RNA binding proteins (RBP16-His, GST-TbRGG2, GST-TbRGG1, and GST-MRP2) (20, 21, 23, 39) (Fig. 2C). TbPRMT7, at 6 ng per reaction, exhibited substantial methyltransferase activity toward all the assayed target proteins (Fig. 2C). GST was not methylated by TbPRMT7, indicating that methylation of the recombinant proteins did not take place on the GST tag (Fig. 2C). From these data, we conclude that not only is TbPRMT7 highly active, but it also exhibits a broad substrate range, methylating arginine residues in both arginine-glycine-rich context (RG peptide, RBP16, TbRGG2, TbRGG1) and glycine-poor context (MBP, histones, MRP2). TbPRMT7 does manifest substrate specificity; however, as it appears to methylate only one histone in the bovine histone pool (Fig. 2C, Histones). The broad substrate range and robust activity of TbPRMT7 are in contrast to the properties of human PRMT7, which has been reported to possess weak activity toward either solely peptide (27) or peptide and protein (28) substrates.
TbPRMT7 Is a Type III PRMT—Of the PRMTs that are active in vitro, the activity of PRMT7 is the most provisional. The enzyme has been reported to catalyze both Type II (SDMA) and Type III (MMA) PRMT activity, depending on the substrate and the reaction time (27, 28). Reaction conditions such as using proteins in lieu of peptides, using a high PRMT to substrate ratio, and extending the reaction time may favor production of Type II products over Type III products in vitro (8, 27, 28). To analyze the type of methylarginine synthesized by TbPRMT7, in vitro methylation assays were performed using 3 μg of GST-TbPRMT7 per reaction, ∼50 times more enzyme than used in Fig. 2C, to favor potential formation of SDMA. Two different protein substrates, bovine histones and RBP16, were used, and reactions were carried out for 14 h at 22 °C. Reaction products were then acid-hydrolyzed to free amino acids and subjected to high resolution cation exchange chromatography along with standards for ADMA, SDMA, and MMA. Little or no methylarginine derivatives were observed in reactions containing GST-TbPRMT7 alone (Fig. 3A, left panel), indicating that the enzyme does not catalyze automethylation. Surprisingly, HPLC analysis of reactions with either histone or RBP16 substrates revealed the presence of very large amounts of MMA but no discernable signal for either ADMA or SDMA (Fig. 3, B and C, left panels), despite high enzyme concentrations and long reaction times. In these experiments, the peak of radioactivity eluted approximately 1 min earlier than the MMA standard detected by ninhydrin analysis. Tritiated amines and amino acids have been shown to elute slightly earlier on high resolution cation exchange chromatography than their hydrogen counterparts due to mass and pKa differences (40-43). The elution of radioactivity here matches that shown previously for tritiated MMA (9, 44) and we conclude that the major methylated species is in fact MMA. Close analysis indicated that we would be able to readily detect the presence of SDMA or ADMA as products in these reactions if they were present at levels of only 0.03% of the MMA product. Reactions using lower amounts of TbPRMT7 (6 ng per reaction) gave similar results, with only MMA being catalyzed (data not shown).
FIGURE 3.
High resolution ion exchange chromatography analysis of methylarginine derivatives catalyzed by TbPRMT7. A, three micrograms of GST-TbPRMT7 in the absence of additional substrate, was incubated in the presence of [3H]AdoMet in PBS for 14 h at 22 °C. Protein was precipitated with 50% trichloroacetic acid and digested into amino acids by acid hydrolysis. Amino acids were analyzed by cation exchange chromatography in the presence of unlabeled ADMA, SDMA, and MMA standards.200 μl of each fraction(1/5 of the total fraction) was removed for radioactivity analysis and 100 μl was removed for ninhydrin analysis, and the fractions were counted three times for 3 min each. B, 10 μg of bovine histones were incubated with 3 μg of GST-TbPRMT7 in the presence of [3H]AdoMet as in A, and reactions were analyzed as in A. C, 3 μg of RBP16 were incubated with 3 μg of GST-TbPRMT7 in the presence of [3H]AdoMet as in A, and reactions were analyzed as in A. D-F, in vitro reactions were carried out as in A, B, and C using 3 μg of TbPRMT7 that was treated with thrombin to remove the GST tag.
The methyltransferase activity seen here for GST-TbPRMT7 is much more robust than that observed for the human GST-PRMT7. If we compare the methylation of the best substrate for GST-TbPRMT7 (histones, Fig. 3B) with the methylation of the best substrate for human GST-PRMT7 (R2 peptide, Fig. 6B of Ref. 27), there is a 30-fold enhancement in the number of methyl groups transferred/h/μg enzyme for the trypanosomal enzyme over the human enzyme. This estimate of the catalytic rate enhancement between these enzymes is probably a minimal one, because the GST-TbPRMT7 reaction was carried out at 22 °C in 0.48 μm AdoMet compared with 37 °C in 0.60 μm AdoMet for the human PRMT7 reaction.
FIGURE 6.
TbPRMT7 forms a dimer in vitro and is present in higher order complexes in vivo. A, anti-Prot C Western blot analysis of cytoplasmic extracts from cells expressing TbPRMT7-PTP (equivalent of 5 × 105 cells) and parental 29-13 cells under denaturing conditions (10% SDS-PAGE). B, Western blot analysis of TbPRMT7-PTP cells, parental 29-13 cells, and 1 μg of recombinant TbPRMT7-His under nondenaturing conditions (4-20% PAGE). C, cytoplasmic extracts from TbPRMT7-PTP expressing cells were fractionated on a 5-20% glycerol gradient, and the positions of TbPRMT7-PTP revealed by anti-Prot C Western blot analysis. The peak position of size markers separated on a parallel gradient are indicated above by arrows.
Several reports have shown that the N-terminal sequence of PRMTs, due to recombinant tags or different native isoforms can alter their inherent methyltransferase activity (35, 45). To address the possibility that the GST may affect the type of methylation catalyzed by recombinant TbPRMT7 we removed the GST tag by thrombin cleavage, resulting in a non-tagged enzyme (data not shown). Non-tagged TbPRMT7 exhibited in vitro methyltransferase activity toward several substrates, including bovine histones, RBP16, and TbRGG2, at levels similar to that of GST-TbPRMT7 (data not shown). To determine if non-tagged TbPRMT7 exhibited Type III methyltransferase activity, in vitro reactions using 3 μg of enzyme were analyzed by high resolution cation exchange as above (Fig. 3, D-F). As with the GST-tagged enzyme, non-tagged TbPRMT7 alone exhibits no appreciable methylation in the absence of substrate (Fig. 3D). More importantly, the non-tagged TbPRMT7 catalyzed the formation of only MMA on both bovine histones and RBP16, and no SDMA or ADMA was evident (Fig. 3, E and F). Thus, TbPRMT7 appears to be exclusively a Type III methyltransferase, as it did not catalyze the production of any SDMA on the target proteins analyzed. Given the high level of [3H]AdoMet incorporation in these reactions, the absence of SDMA cannot be attributed to weak enzyme activity, but is likely an intrinsic property of TbPRMT7.
TbPRMT7 Is Expressed in Two T. brucei Life Cycle Stages and Its Depletion Does Not Affect Growth—Once we established that TbPRMT7 is a bona fide arginine methyltransferase, we wanted to examine its properties in vivo. During its life cycle, T. brucei shuttles between its mammalian host and the tse tse fly insect vector, manifesting dramatic changes in gene expression, metabolism, and morphology. Both mammalian bloodstream (BF) and procyclic insect (PF) life cycle stages of the parasite can be grown in the laboratory, and here we examined the relative TbPRMT7 mRNA levels in the two life cycle stages by qRT-PCR. As shown in Fig. 4A, TbPRMT7 is expressed in both PF and BF stages of the parasite, with expression in BF slightly lower than in PF (∼75% of PF levels).
FIGURE 4.
TbPRMT7 is expressed in two life cycle stages and its depletion does not affect growth. A, qRT-PCR analysis of the relative amounts of TbPRMT7 mRNA in procyclic (PF) and bloodstream (BF) form T. brucei. TbPRMT7 levels were normalized to β-tubulin and RNA levels in PF were set to one. B, clonal procyclic (upper) and bloodstream (lower) form T. brucei cell lines expressing tetracycline-regulated TbPRMT7 RNAi were established. Production of dsRNA was induced using 2.5 μg/ml tetracycline on Day 0, and total cells were counted thereafter until day 10 (PF) or 12 (BF). Depletion of TbPRMT7 mRNA was confirmed using qRT-PCR (PF) or Northern blotting (BF).
We next wanted to determine the effects of TbPRMT7 depletion on parasite growth. To this end, TbPRMT7 RNAi constructs in either p2T7-177 or pHD1621 (30, 31) were generated and transfected into PF 29-13 or BF single marker cells, respectively, to allow tetracycline regulation of RNAi (46). Clonal cell lines were established, and depletion of TbPRMT7 mRNA in the presence of tetracycline was confirmed by either qRT-PCR (Fig. 4B, upper) or Northern blot (Fig. 4B, lower). We then monitored cell growth in the absence and presence of tetracycline for either 10 days (PF) or 12 days (BF). In both life cycle stages, TbPRMT7-depleted cells grew at the same rate as the uninduced control cells (Fig. 4B). Thus, wild-type levels of TbPRMT7 are not essential for growth in either life cycle stage in culture.
TbPRMT7 Is Cytoplasmically Localized—To gain further insight into the potential in vivo functions of TbPRMT7, we investigated the subcellular localization of the enzyme. Because an antibody to TbPRMT7 is unavailable, endogenous TbPRMT7 was tagged on one allele with a C-terminal PTP tag, which adds both protein A and protein C moieties separated by a TEV protease cleavage site (32). PF T. brucei was transfected with the pC-TbPRMT7-PTP plasmid and selected with puromycin, and stable clonal cell lines expressing the TbPRMT7-PTP construct were generated by limiting dilution. Western blot analysis of whole cell lysates with anti-ProtC, which detects the protein C portion of the PTP tag, revealed a major band of ∼63 kDa, as expected for the 19-kDa PTP tag linked to TbPRMT7 (Figs. 5 and 6A). We also observed a more slowly migrating minor band that may represent a modified form of the protein. We then fractionated cells expressing TbPRMT7-PTP into cytoplasmic and nuclear fractions, and confirmed efficient fractionation by Western blot analysis of Hsp70 (cytoplasmic) and RNA Pol II CTD (nuclear). Analysis of TbPRMT7-PTP in subcellular fractions revealed that it is exclusively localized to the cytoplasm under steady state conditions (Fig. 5). This is in contrast to the human PRMT7 and Drosophila DART7, which display both cytoplasmic and nuclear localizations (38, 47).
FIGURE 5.
TbPRMT7 is localized to the cytoplasm. Procyclic form T. brucei were transfected with pC-PRMT7-PTP to create a clonal cell line expressing C-terminal PTP-tagged TbPRMT7 expressed from one endogenous allele. Cells were fractionated into cytoplasmic and nuclear fractions and the equivalent of 5 × 105 cells was analyzed with anti-Prot C antibodies, which recognize the PTP tag. Hsp70 and the CTD of RNA polymerase II were used as cytoplasmic and nuclear markers, respectively. WCL, whole cell lysate.
TbPRMT7 Is Present in Higher Ordered Complexes—Many PRMTs are found in higher ordered complexes in vivo (25, 48-53). PRMT7 isoforms from hamsters are reportedly present in higher ordered complexes; however, human PRMT7 was found in several studies to be primarily monomeric (49, 54). Therefore, we wanted to examine whether TbPRMT7 was present in vivo as a monomer or as a component of larger complexes. We began by subjecting cytoplasmic extract from TbPRMT7-PTP-expressing cells to Western blot analysis under both denaturing and non-denaturing conditions. In the presence of SDS, TbPRMT7-PTP is present as a single species of ∼63 kDa (Fig. 6A). However, on a non-denaturing gel, TbPRMT7-PTP was present in higher order complexes greater than 170 kDa (Fig. 6B). In comparison, recombinant TbPRMT7-His was observed only at the expected size of a dimer (∼93 kDa) for TbPRMT7 (Fig. 6B). Because the expected size of a TbPRMT7-PTP dimer is ∼126 kDa, these data suggest that TbPRMT7 is likely associated with other proteins in vivo in addition to itself. To further analyze TbPRMT7 containing complexes, we fractionated cytoplasmic extract from TbPRMT7-PTP cells on a 5-20% glycerol gradient and detected TbPRMT7-PTP by anti-ProtC Western blot. TbPRMT7-PTP was present in a broad smear ranging up to ∼20S, again suggesting an interaction with additional cellular components (Fig. 6C).
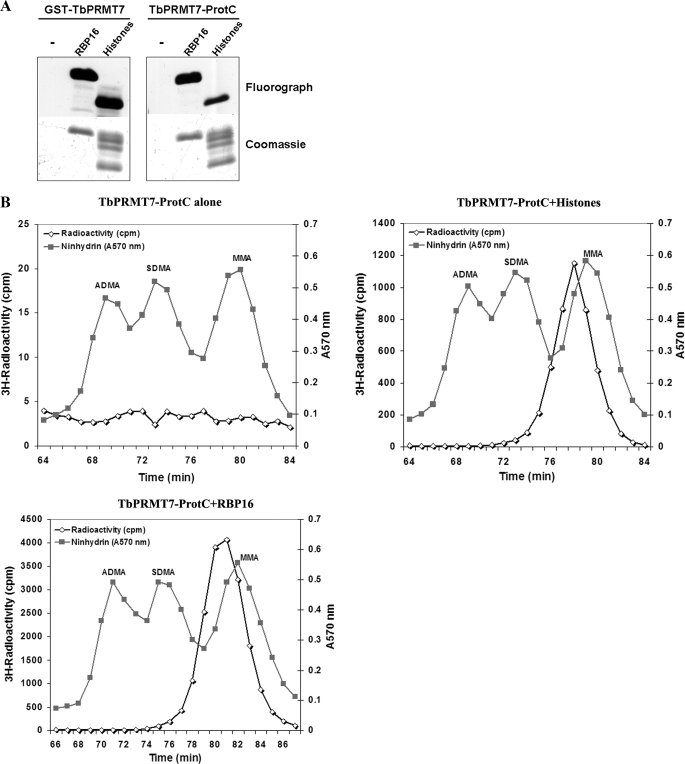
The association of TbPRMT7 with in vivo binding partners could change its Type III activity. To determine whether TbPRMT7 purified from T. brucei retains Type III activity, we purified TbPRMT7-PTP from PF cytoplasmic extracts using the tandem affinity purification method (32), resulting in TbPRMT7 with a C-terminal protein C tag (TbPRMT7-ProtC). The enzymatic activity of TbPRMT7-ProtC was determined in vitro either in the absence of substrate and either bovine histones or trypanosomal RBP16 as substrates. Recombinant GST-TbPRMT7 was used as a positive control (Fig. 7A). TbPRMT7-ProtC strongly methylated both histones and RBP16, demonstrating that the in vivo C-terminally tagged enzyme is active. The reactions shown in Fig. 7A were also subjected to high resolution cation exchange chromatography to determine whether the TbPRMT7-ProtC exhibited Type III methyltransferase activity as did the recombinant TbPRMT7. As shown in Fig. 7B, TbPRMT7-ProtC did not appreciably methylate itself (Fig. 7B). Moreover, the enzyme purified from T. brucei exhibited solely Type III PRMT activity toward both histones and RBP16 (Fig. 7B), with no appreciable SDMA or ADMA formed. These results confirm that the highly active TbPRMT7 catalyzes only Type III PRMT activity on all substrates analyzed, regardless if the enzyme is recombinant or purified from the parasite.
FIGURE 7.
Endogenously tagged TbPRMT7 exhibits type III PRMT activity. A, endogenously tagged TbPRMT7 has in vitro activity comparable to recombinant GST-TbPRMT7. Right panel, the indicated substrates were incubated with 100 ng of purified TbPRMT7-ProtC from T. brucei incubated with either PBS buffer alone (-), 3 μg of RBP16, or 10 μg of bovine histone in the presence of 2 μCi of [3H]AdoMet in PBS overnight at room temperature. Left panel, three micrograms of GST-PRMT7 was used as a positive control with each substrate for activity in vitro. For each reaction the Coomassie Blue stain of the substrate is shown below the resultant fluorograph of activity. B, purified TbPRMT7-ProtC from T. brucei still exhibits Type III PRMT activity. The reactions from A were subjected to high resolution cation exchange chromatography as in Fig. 3.
DISCUSSION
In this report, we describe a PRMT from the early branching eukaryote, T. brucei. Compared with the nine human PRMTs, this T. brucei enzyme is most homologous to the N-terminal domain of PRMT7, and thus is termed TbPRMT7. Notwith-standing this sequence conservation, TbPRMT7 displays a range of features that distinguish it from its metazoan homologues. The first remarkable difference is the lack of genetic duplication of the AdoMet binding-like domain in TbPRMT7. A duplicated C-terminal AdoMet binding-like domain is a hallmark of all metazoan PRMT7 enzymes, and this domain is required for the activity of human PRMT7 (27). Despite the absence of this domain, however, TbPRMT7 is highly active in vitro and has a broad substrate range. Second, the strong in vitro activity of TbPRMT7 seen here contrasts even with the relatively weak activity of the human PRMT7 enzyme (27, 28). Third, we show that TbPRMT7 exhibits solely Type III PRMT activity, catalyzing the production of only MMA even under conditions designed to favor formation of DMA. This again distinguishes TbPRMT7 from human PRMT7 (see below). Finally, in vivo, TbPRMT7 differs from its metazoan homologues in that it is exclusively cytoplasmic and apparently not essential for growth.
The type of methyltransfer reaction catalyzed by human PRMT7 is not entirely resolved. Previous studies reported either Type II or Type III enzymatic activity, although the discrepancy may have been due to different reaction times, the type of substrate (peptide versus protein) and the apparent low intrinsic in vitro activity of the recombinant PRMT7 protein (27, 28). Similarly, the yeast enzyme Hsl7 was originally classified as a Type III PRMT, but Type II activity became evident by extending the reaction time (8). Therefore, we wanted to carefully assess TbPRMT7 activity by establishing in vitro reactions with several enzyme concentrations, the use of protein substrates and long reactions times, which should favor SDMA production. Under these conditions, we observed very high levels of AdoMet incorporation, and high resolution cation exchange chromatography analysis revealed that this was entirely in the form of MMA. Therefore, TbPRMT7 represents the only exclusively Type III PRMT definitively identified to date. TbPRMT7 may in fact represent an evolutionary precursor enzyme to the genetically duplicated PRMT7 present in higher eukaryotes. The duplicated PRMT7 may then have acquired limited Type II activity due to this duplication event through modification of the enzymatic process and/or additional protein-protein interactions. This potential acquisition of Type II activity seems to have taken place at the expense of overall AdoMet incorporation levels.
T. brucei and many of its kinetoplastid relatives are parasites with complex life cycles in which they continually move between mammalian hosts and insect vectors. Therefore, these organisms must have mechanisms for rapidly adapting to very different environments. Intriguingly, kinetoplastids are the only known single-celled eukaryotes with PRMT7 homologues identifiable in their genomes (13). This enzyme is clearly lacking in yeasts and other investigated protozoa, implying that TbPRMT7 has functions specific to kinetoplastid biology. Furthermore, the T. brucei genome contains a total of five putative PRMTs, which is among the largest number of PRMTs identified in any single-celled eukaryote (13), suggesting a potentially heightened importance of protein arginine methylation in general in these organisms. One unique facet of kinetoplastid biology is that gene regulation is effected almost entirely at the post-transcriptional level, thereby presumably requiring large numbers of RNA binding proteins to facilitate or inhibit decay and/or translation of specific RNAs in response to changing environmental pressures. In this regard, it is of interest that RNA-binding proteins are a major class of methylarginine containing proteins in yeast and mammals (1, 55). The cytoplasmic localization of TbPRMT7 is consistent with a role in the regulation of RNA stability or translation, and studies aimed at identifying cytoplasmic TbPRMT7 substrates are underway. Moreover, cytoplasmic extracts contain mitochondria,4 and we have previously shown that arginine methylation regulates mitochondrial RNA stability in T. brucei, in part through the actions of the RNA-binding protein, RBP16 (56). Several proteins that serve as TbPRMT7 substrates in vitro, including RBP16, are known to function in mitochondrial RNA metabolism, and future experiments will address whether TbPRMT7 methylates these proteins in vivo and thereby modulates their functions.
The existence of an exclusively Type III PRMT in T. brucei presents different models for the function of this class of enzyme. The first is that TbPRMT7 may act coordinately with one or more of the four other T. brucei PRMTs. We have demonstrated that TbPRMT1 and TbPRMT5 are Type I and Type II enzymes, respectively (25, 26). The other two PRMTs are homologous to human PRMT3 and PRMT6, suggesting likely Type I activity, although this remains to be established. Current models for the mechanism of protein arginine methylation suggest that each Type I or Type II PRMT catalyzes MMA on a given substrate prior to catalyzing the subsequent ADMA or SDMA reaction on the same arginine residue. However, a highly active Type III TbPRMT7 could feasibly add the first methyl group to substrates, followed by a subsequent synthesis of DMA by TbPRMT1 or TbPRMT5. In support of this hypothesis, in vitro studies have recently shown that human PRMT6 displays a preference for monomethylated substrate, displaying both a lower Km and higher Vmax than observed for the corresponding unmethylated substrate (57). It will be of interest to determine whether this is also true for any of the trypanosome PRMTs, and whether dual RNAi knockdowns of TbPRMT7 with other TbPRMTs reveals synthetic lethality. A second model, which is not mutually exclusive, is that MMA serves as a distinct signal from the further modified ADMA or SDMA. In this case, TbPRMT1, TbPRMT5, and TbPRMT7 would serve separate roles in methylation-dependent signaling. This model would also predict that MMA-dependent signaling is especially predominant in trypanosomes compared with higher eukaryotes. Future biochemical and genetic analyses of trypanosome PRMTs and their substrates will provide insight into the functions of arginine methylation in an important human pathogen, as well as into the evolution of this vital and widespread process.
Acknowledgments
We thank Christine Clayton for the pHD1621 RNAi vector, Jay Bangs and Vivian Bellofatto for antibodies, and George Cross for cell lines.
This work was supported, in whole or in part, by National Institutes of Health Grant R01AI060260 (to L. K. R.) and Grant R37GM026020 (to S. G. C.). This work was also supported by American Heart Association Postdoctoral Fellowship 0725984T (to S. M.).
Footnotes
The abbreviations used are: AdoMet, S-adenosyl-methionine; PBS, phosphate-buffered saline; GST, glutathione S-transferase; PRMT, protein arginine methyltransferase; ADMA, asymmetric dimethylarginine; SDMA, symmetric dimethylarginine; MMA, monomethylated arginine.
L. K. Read, unpublished results.
J. C. Fisk and M. Pelletier, unpublished research.
References
- 1.Bedford, M. T. (2007) J. Cell Sci. 120, 4243-4246 [DOI] [PubMed] [Google Scholar]
- 2.Pahlich, S., Zakaryan, R. P., and Gehring, H. (2006) Biochim. Biophys. Acta 1764, 1890-1903 [DOI] [PubMed] [Google Scholar]
- 3.Wysocka, J., Allis, C. D., and Coonrod, S. (2006) Front. Biosci. 11, 344-355 [DOI] [PubMed] [Google Scholar]
- 4.Bedford, M. T., and Richard, S. (2005) Mol. Cell 18, 263-272 [DOI] [PubMed] [Google Scholar]
- 5.Boisvert, F.-M., Chenard, C. A., and Richard, S. (2005) Science's Stke [Electronic Resource]: Signal Transduction Knowledge Environment, re2. [DOI] [PubMed]
- 6.Lake, A. N., and Bedford, M. T. (2007) Mutat. Res. 618, 91-101 [DOI] [PubMed] [Google Scholar]
- 7.Bedford, M. T., and Clarke, S. G. (2009) Mol. Cell 33, 1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sayegh, J., and Clarke, S. G. (2008) Biochem. Biophys. Res. Commun. 372, 811-815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miranda, T. B., Sayegh, J., Frankel, A., Katz, J. E., Miranda, M., and Clarke, S. (2006) Biochem. J. 395, 563-570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McBride, A. E., Zurita-Lopez, C., Regis, A., Blum, E., Conboy, A., Elf, S., and Clarke, S. (2007) Eukaryot. Cell 6, 1119-1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niewmierzycka, A., and Clarke, S. (1999) J. Biol. Chem. 274, 814-824 [DOI] [PubMed] [Google Scholar]
- 12.Krause, C. D., Yang, Z.-H., Kim, Y.-S., Lee, J.-H., Cook, J. R., and Pestka, S. (2007) Pharmacol. Therap. 113, 50-87 [DOI] [PubMed] [Google Scholar]
- 13.Bachand, F. (2007) Eukaryot. Cell 6, 889-898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dacks, J. B., and Doolittle, W. F. (2001) Cell 107, 419-425 [DOI] [PubMed] [Google Scholar]
- 15.Clayton, C., and Shapira, M. (2007) Mol. Biochem. Parasitol. 156, 93-101 [DOI] [PubMed] [Google Scholar]
- 16.Clayton, C. E. (2002) EMBO J. 21, 1881-1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sbicego, S., Alfonzo, J. D., Estevez, A. M., Rubio, M. A. T., Kang, X., Turck, C. W., Peris, M., and Simpson, L. (2003) Eukaryot. Cell 2, 560-568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayman, M. L., and Read, L. K. (1999) J. Biol. Chem. 274, 12067-12074 [DOI] [PubMed] [Google Scholar]
- 19.Koller, J., Muller, U. F., Schmid, B., Missel, A., Kruft, V., Stuart, K., and Goringer, H. U. (1997) J. Biol. Chem. 272, 3749-3757 [DOI] [PubMed] [Google Scholar]
- 20.Vondruskova, E., van den Burg, J., Zikova, A., Ernst, N. L., Stuart, K., Benne, R., and Lukes, J. (2005) J. Biol. Chem. 280, 2429-2438 [DOI] [PubMed] [Google Scholar]
- 21.Vanhamme, L., Perez-Morga, D., Marchal, C., Speijer, D., Lambert, L., Geuskens, M., Alexandre, S., Ismaili, N., Goringer, U., Benne, R., and Pays, E. (1998) J. Biol. Chem. 273, 21825-21833 [DOI] [PubMed] [Google Scholar]
- 22.Madison-Antenucci, S., and Hajduk, S. L. (2001) Mol. Cell 7, 879-886 [DOI] [PubMed] [Google Scholar]
- 23.Fisk, J. C., Ammerman, M. A., Presnyak, V., and Read, L. K. (2008) J. Biol. Chem. 283, 23016-23025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Gaudenzi, J., Frasch, A. C., and Clayton, C. (2005) Eukaryot. Cell 4, 2106-2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasternack, D. A., Sayegh, J., Clarke, S., and Read, L. K. (2007) Eukaryot. Cell 6, 1665-1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelletier, M., Pasternack, D. A., and Read, L. K. (2005) Mol. Biochem. Parasitol. 144, 206-217 [DOI] [PubMed] [Google Scholar]
- 27.Miranda, T. B., Miranda, M., Frankel, A., and Clarke, S. (2004) J. Biol. Chem. 279, 22902-22907 [DOI] [PubMed] [Google Scholar]
- 28.Lee, J.-H., Cook, J. R., Yang, Z.-H., Mirochnitchenko, O., Gunderson, S. I., Felix, A. M., Herth, N., Hoffmann, R., and Pestka, S. (2005) J. Biol. Chem. 280, 3656-3664 [DOI] [PubMed] [Google Scholar]
- 29.Pelletier, M., and Read, L. K. (2003) Rna-A Publ. Rna Society 9, 457-468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wickstead, B., Ersfeld, K., and Gull, K. (2002) Mol. Biochem. Parasitol. 125, 211-216 [DOI] [PubMed] [Google Scholar]
- 31.Luu, V.-D., Brems, S., Hoheisel, J. D., Burchmore, R., Guilbride, D. L., and Clayton, C. (2006) Mol. Biochem. Parasitol. 150, 340-349 [DOI] [PubMed] [Google Scholar]
- 32.Schimanski, B., Nguyen, T. N., and Gunzl, A. (2005) Eukaryot. Cell 4, 1942-1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cimato, T. R., Tang, J., Xu, Y., Guarnaccia, C., Herschman, H. R., Pongor, S., and Aletta, J. M. (2002) J. Neurosci. Res. 67, 435-442 [DOI] [PubMed] [Google Scholar]
- 34.Ammerman, M. A., Fisk, J. C., and Read, L. K. (2008) RNA 14, 1069-1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sayegh, J., Webb, K., Cheng, D., Bedford, M. T., and Clarke, S. G. (2007) J. Biol. Chem. 282, 36444-36453 [DOI] [PubMed] [Google Scholar]
- 36.Zeiner, G. M., Sturm, N. R., and Campbell, D. A. (2003) Eukaryot. Cell 2, 222-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carnes, J., Trotter, J. R., Ernst, N. L., Steinberg, A., and Stuart, K. (2005) Proc. Natl. Acad. Sci. U. S. A. 102, 16614-16619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulanger, M.-C., Miranda, T. B., Clarke, S., Di Fruscio, M., Suter, B., Lasko, P., and Richard, S. (2004) Biochem. J. 379, 283-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goulah, C. C., and Read, L. K. (2007) J. Biol. Chem. 282, 7181-7190 [DOI] [PubMed] [Google Scholar]
- 40.Gottschling, H., and Freese, E. (1962) Nature 196, 829-831 [DOI] [PubMed] [Google Scholar]
- 41.Klein, P. D., and Szczepanik, P. A. (1967) Anal. Chem. 39, 1276-1281 [DOI] [PubMed] [Google Scholar]
- 42.Kleene, S. J., Toews, M. L., and Adler, J. (1977) J. Biol. Chem. 252, 3214-3218 [PubMed] [Google Scholar]
- 43.Xie, H., and Clarke, S. (1993) J. Biol. Chem. 268, 13364-13371 [PubMed] [Google Scholar]
- 44.Lin, W. J., Gary, J. D., Yang, M. C., Clarke, S., and Herschman, H. R. (1996) J. Biol. Chem. 271, 15034-15044 [DOI] [PubMed] [Google Scholar]
- 45.Goulet, I., Gauvin, G., Boisvenue, S., and Cote, J. (2007) J. Biol. Chem. 282, 33009-33021 [DOI] [PubMed] [Google Scholar]
- 46.Wirtz, E., Leal, S., Ochatt, C., and Cross, G. A. (1999) Mol. Biochem. Parasitol. 99, 89-101 [DOI] [PubMed] [Google Scholar]
- 47.Gonsalvez, G. B., Tian, L., Ospina, J. K., Boisvert, F.-M., Lamond, A. I., and Matera, A. G. (2007) J. Cell Biol. 178, 733-740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rho, J., Choi, S., Seong, Y. R., Cho, W. K., Kim, S. H., and Im, D. S. (2001) J. Biol. Chem. 276, 11393-11401 [DOI] [PubMed] [Google Scholar]
- 49.Gros, L., Renodon-Corniere, A., de Saint Vincent, B. R., Feder, M., Bujnicki, J. M., and Jacquemin-Sablon, A. (2006) Biochim. Biophys. Acta 1760, 1646-1656 [DOI] [PubMed] [Google Scholar]
- 50.Herrmann, F., Lee, J., Bedford, M. T., and Fackelmayer, F. O. (2005) J. Biol. Chem. 280, 38005-38010 [DOI] [PubMed] [Google Scholar]
- 51.Weiss, V. H., McBride, A. E., Soriano, M. A., Filman, D. J., Silver, P. A., and Hogle, J. M. (2000) Nat. Struct. Biol. 7, 1165-1171 [DOI] [PubMed] [Google Scholar]
- 52.Tang, J., Gary, J. D., Clarke, S., and Herschman, H. R. (1998) J. Biol. Chem. 273, 16935-16945 [DOI] [PubMed] [Google Scholar]
- 53.Teyssier, C., Chen, D., and Stallcup, M. R. (2002) J. Biol. Chem. 277, 46066-46072 [DOI] [PubMed] [Google Scholar]
- 54.Herrmann, F., Pably, P., Eckerich, C., Bedford, M. T., and Fackelmayer, F. O. (2009) J. Cell Sci. 122, 667-677 [DOI] [PubMed] [Google Scholar]
- 55.Godin, K. S., and Varani, G. (2007) Rna Biology 4, 69-75 [DOI] [PubMed] [Google Scholar]
- 56.Goulah, C. C., Pelletier, M., and Read, L. K. (2006) Rna-A Publ. Rna Soc. 12, 1545-1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lakowski, T. M., and Frankel, A. (2008) J. Biol. Chem. 283, 10015-10025 [DOI] [PubMed] [Google Scholar]