Abstract
Septins are filament-forming GTPases implicated in several cellular functions, including cytokinesis. We previously showed that SEPT2, SEPT9, and SEPT11 colocalize with several bacteria entering into mammalian non-phagocytic cells, and SEPT2 was identified as essential for this process. Here, we investigated the function of SEPT11, an interacting partner of SEPT9 whose function is still poorly understood. In uninfected HeLa cells, SEPT11 depletion by siRNA increased cell size but surprisingly did not affect actin filament formation or the colocalization of SEPT9 with actin filaments. SEPT11 depletion increased Listeria invasion, and incubating SEPT11-depleted cells with beads coated with the Listeria surface protein InlB also led to increased entry as compared with control cells. Strikingly, as shown by fluorescence resonance energy transfer, the InlB-mediated stimulation of Met signaling remained intact in SEPT11-depleted cells. Taken together, our results show that SEPT11 is not required for the bacterial entry process and rather restricts its efficacy. Because SEPT2 is essential for the InlB-mediated entry of Listeria, but SEPT11 is not, our findings distinguish the roles of different mammalian septins.
Septins were discovered in the budding yeast Saccharomyces cerevisiae (1) where they organize into a ring at the mother-bud neck during cell division (2). Septins are GTPases of 30-65 kDa found in most eukaryotes, except plants, sharing an essential role in cytokinesis (3, 4). Fourteen septins have been identified in humans and classified on the basis of sequence identity into four distinct groups (3, 5). Septins from different groups polymerize into hetero-oligomeric protein complexes and filaments and may associate with cellular membranes, actin filaments, and microtubules (6, 7). Septins are increasingly regarded as novel cytoskeletal elements (8), but their role in post-mitotic events remains poorly understood.
The crystal structure of the SEPT2-SEPT6-SEPT7 complex recently highlighted that septins, as opposed to actin and microtubules, form non-polar filaments (9). In the SEPT7-SEPT6-SEPT2-SEPT2-SEPT6-SEPT7 complex, SEPT2 has a central role in filament formation (9), whereas SEPT6 is thought to be replaceable with other SEPT6 group members, including SEPT11 (3). Widely expressed in mammalian tissues (10), SEPT11 may also be a substitute for SEPT6 in other mammalian septin complexes such as SEPT7-SEPT9-SEPT11 (10) or SEPT5-SEPT7-SEPT11 (11). Because other septins homologous to SEPT11 might compensate for its deficiency (12), the degree to which SEPT11 is required for septin filament structure and function is not yet known. Listeria monocytogenes is an invasive bacterium that enters into most mammalian cells in vitro through the interaction of the bacterial surface protein InlB with its host cellular receptor Met, the hepatocyte growth factor receptor (13). We originally identified SEPT9 associated with phagosomes containing latex beads coated with InlB (14). Given the association of septins with the cytoskeleton, and the importance of the cytoskeleton in bacterial invasion, we have started investigating septin function during infection of invasive bacteria in non-phagocytic cells. We have discovered that SEPT9, and its interacting partners SEPT2 and SEPT11, are recruited as 0.6-μm collars next to actin at the site of entry of invasive bacteria (15). Although functional studies using siRNA3 have revealed an essential role for SEPT2 in regulating bacterial entry, the role of SEPT11 has not yet been investigated. We thus addressed SEPT11 function in the context of Listeria infection.
EXPERIMENTAL PROCEDURES
Mammalian Cells, Bacterial Strains, and Culture Conditions—HeLa human cervix carcinoma cells (ATCC CCL-2) were cultured in DMEM plus GlutaMAX (Amersham Biosciences) supplemented with 1% sodium pyruvate (Amersham Biosciences) and 10% fetal calf serum. Cells were grown at 37 °C in a 10% CO2 atmosphere. L. monocytogenes type strain EGD (BUG 600) was grown overnight at 37 °C in brain heart infusion media (Difco Laboratories), diluted 15× in fresh brain heart infusion, and cultured until A600 nm = 0.8.
Gentamicin Survival Assays—Gentamicin survival assays were performed as previously described (16). In brief, cells were incubated in DMEM with Listeria at a multiplicity of infection of 50 for 1 h at 37 °C and 10% CO2, washed with DMEM without antibiotic, and subsequently incubated with fresh gentamicin-containing complete media (10 μg/ml) for an additional 1 h. Cells were washed and then lysed with distilled H2O. The number of viable bacteria released from the cells was assessed by plating on brain heart infusion agar plates. Each experiment was done in triplicate, and triplicates were performed at least three times independently.
Survival assays involving drug treatments were performed as described for other Listeria in vitro systems (17, 18), where cells were treated with various concentrations of drugs 30 min prior to infection. Upon infection, host cells were incubated in drugs (at molar concentrations varying from 200 to 0.01 μm) throughout the gentamicin survival assays as detailed above. Final values are expressed as the number of intracellular bacteria per drug treatment relative to DMSO-treated cells. Forchlorfeneuron, cytochalasin D, latrunculin B, nocodazole, and paclitaxel were all suspended in DMSO and handled as suggested by the manufacturer (Sigma).
Antibodies and Reagents—Primary antibodies used in this study included rabbit polyclonals anti-SEPT9 (R69) (14), anti-SEPT11 (gift from Makoto Kinoshita), and anti-SEPT2 (gift from William Trimble). Mouse monoclonal anti-InlB has been described elsewhere (19). Mouse monoclonal anti-α-tubulin was purchased from Molecular Probes. Mouse monoclonal anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase), used as a control throughout Western blot experimentation, was purchased from AbCam (6C5).
Secondary antibodies used were Cy5-conjugated (Jackson ImmunoResearch Laboratories), Alexa 488-conjugated, and Alexa 546-conjugated goat anti-rabbit and goat anti-mouse antibodies (Molecular Probes). F-actin was labeled with Alexa 488-fluorescein isothiocyanate and Alexa 546-phalloidin (Molecular Probes). For immunoblotting, total cellular extracts were blotted with the above-mentioned antibodies, followed by peroxidase-conjugated goat anti-mouse and anti-rabbit antibodies (Biosys Laboratories). Proteins were run on 10% acrylamide gels.
Preparation of InlB-coated Beads—Coating of latex beads with purified InlB and incubation of host cells with InlB-coated latex beads were performed as previously described (14, 20), where 1-μm diameter latex beads were coated following the manufacturer's instructions (Molecular Probes). Prior to addition to siRNA-treated HeLa cells, InlB-coated beads were briefly sonicated, diluted in DMEM, and added to HeLa cells at a multiplicity of ∼100 beads per cell.
For kinetic analysis by immunofluorescence and scanning electron microscopy, host cells were stored at 4 °C for 5 min before incubation with beads, then beads diluted in DMEM were added and cells were centrifuged at 1000 × g for 1 min at 4 °C. Medium was removed and replaced with fresh DMEM. Cells were then incubated at 37 °C and 10% CO2 for 5 min, after which they were washed twice with DMEM and processed for immunofluorescence or scanning electron microscopy analysis.
Immunofluorescence and Confocal Microscopy—Immunofluorescence analysis was performed as previously described (16). Briefly, 105-106 HeLa cells were plated onto glass coverslips in 6-well plates (Techno Plastic Products) and used for experiments 48 h later. Cells on coverslips were fixed 15 min in 4% paraformaldehyde, then washed with 1× phosphate-buffered saline, and processed for immunofluorescence. After 10 min of incubation in 50 mm ammonium chloride, cells were permeabilized 4 min with 0.1% Triton X-100, and then incubated in 1× phosphate-buffered saline. Hybridization of primary and secondary antibodies was performed in 1× phosphate-buffered saline. Extracellular InlB beads were sometimes labeled prior to permeabilization. Vectashield hard set mounting medium with or without DAPI was applied (Vector Laboratories).
Images were acquired on a fluorescence inverted microscope Axiovert 200M (Carl Zeiss MicroImaging, Inc.) equipped with a cooled digital charge-coupled device camera (Cool SNAPHQ, Photometrics) driven by Metamorph Imaging System software (Universal Imaging Corp). Confocal images were acquired using a Leica TCS SP5 (Leica Microsystems). Immunofluorescence and confocal images were visualized and processed using the ImageJ public domain image processing software (rsb.info.nih.gov/ij/).
To localize and track changes in septin filament distribution and to segment cells after siRNA treatment, a journal was created using MetaMorph (version 7.1, Universal Imaging). Confocal Z-stack images of siRNA-treated cells were catalogued where images from sets of at least two biological replicates per siRNA treatment were analyzed. Endogenous F-actin and SEPT9 were visualized for immunolabeling with fluorescent phalloidin and anti-SEPT9 antibodies. To define their location, nuclei were marked with DAPI. After deconvolution (Huygens Deconvolution Software; Scientific Volume Imaging), the confocal Z-stack images of cells were segmented, and fibers were localized in each cell. For each segmented cell, quantitative parameters (coordinates and area) were exported to an Excel file. For each segmented fiber, quantitative parameters (coordinates, area, and shape factor) were also exported to an Excel file, where fibers were analyzed per cell, and cells could subsequently be attributed to Class 1, 2, or 3. When comparing the Class frequencies for siRNA treatments, p values were accordingly obtained by chi square test.
RNA Interference—HeLa cells (0.8 × 105) were plated in 6-well plates (Techno Plastic Products) and transfected the following day using Oligofectamine (Invitrogen) following the manufacturer's instructions. Scramble sequence, custom designed from septin sequences ((sense) AUAAGCGACGUCCGCGUGGtt and (antisense) CCACGCGGACGUCGCUUAUtt), was applied as our control throughout experimentation (15). Custom scramble sequence, as well as pre-designed siRNAs for SEPT9 (ID#18228) and SEPT11 (ID#125139) were all from Ambion, and handled according to manufacturer's instructions. Cells were tested 72 h after siRNA transfection. To measure the efficiency of siRNA knockdown, the relative amounts of SEPT9 and SEPT11 proteins were quantified using GeneTools (SynGene), normalized to give 100% for the controls (average ± S.D. of triplicate experiments). Where mentioned, siRNA sequence for SEPT2 (ID#14709) (15), SEPT6 (ID#136764), and a second siRNA sequence for SEPT11 (ID#125138), was applied.
Scanning Electron Microscopy—For scanning electron microscopy analysis, non-infected siRNA-treated HeLa cells, and siRNA-treated HeLa cells incubated with InlB-coated beads, were prepared as described above, subsequently washed in phosphate-buffered saline, prefixed in 2.5% glutaraldehyde in 0.1 m cacodylate buffer for 30 min, and then rinsed in 0.2 m cacodylate buffer. After post-fixation in 1% osmium tetraoxide (in 0.2 m cacodylate buffer), samples were dehydrated in a series of ethanol concentrations. Specimens were critical point-dried using carbon dioxide, and then coated with gold and examined/photographed with a JEOL JSM-6700F scanning electron microscope.
Fluorescence Resonance Energy Transfer—As previously described by our laboratory (21), PI3K activation can be measured by the production of its lipid products phosphoinositide 3,4-bisphosphate and phosphoinositide 3,4,5-trisphosphate, which recruit the serine/threonine kinase Akt from the cytosol to the membrane, with the Akt pleckstrin homology (PH) domain specifically interacting with phosphoinositide 3,4-bisphosphate and phosphoinositide 3,4,5-trisphosphate (22, 23). The quantification of FRET between two coexpressed fluorescent chimeras of the Akt PH domain, YFP-AktPH and CFP-AktPH, can be used as readout of PI3K activity. Without stimulation, the fluorescent chimeras are too dispersed in the cytosol to undergo FRET. Upon PI3K activation, the fluorescent AktPH chimeras concentrate at the plasma membrane, consequently leading to an increase in the FRET signal.
siRNA-treated HeLa cells were cotransfected using jetPEI (PolyPlus Transfection) according to the manufacturer's instructions with plasmids encoding YFP-AktPH and CFP-AktPH. Details on FRET image acquisition, processing, and statistical analysis were introduced in Mostowy et al. (15). In brief, cells were automatically classified as not responsive to InlB stimulation if a maximum slope during FRET, i.e. a peak of signal intensity, could not be detected above background values during InlB stimulation. Final kinetics was obtained for 19 and 11 experiments, representing at least 3 biological replicates for each siRNA treatment, for control and SEPT11 siRNA-treated cells, respectively. To automatically detect whether a cell could be classified as having responded or not to InlB stimulation, we designed our own analysis software carried out using an Excel Macro. Our analysis software is based on these following criteria. From the original kinetic signals (i.e. the mean (EA + ED)/2 over the entire cell), a first median linear filter (formula: yi = (∑i-n/2,i+n/2 yi)/n, n = 3) was applied to smooth artifact arising from the acquisition of original signal. The slope of each filtered curve was subsequently calculated for each time point and was plotted as the processed “FRET response” (i.e. as in Fig. 4B). Starting from T0 (here defined as the time frame immediately following InlB stimulation, frame 16), Tmax was detected when the slope again reached a minimum, where minimum slope is based on a null second derivative function. Tmax-T0 was used to define the duration of FRET response (i.e.“Time of FRET,” where units are frames per 15 s). Between these two time points, the amplitude of the original signal (i.e. the mean (EA + ED)/2 over the entire cell) was calculated as a characteristic to qualify the response (i.e.“Amplitude of FRET”). As a final characteristic, the maximum slope was calculated between T0 and Tmax (i.e.“Maximum slope during FRET”; this value also served to determine if the FRET response of that cell was positive). In the case where Tmax could not be detected based on these criteria, such cells were then classified as not responding to InlB stimulation.
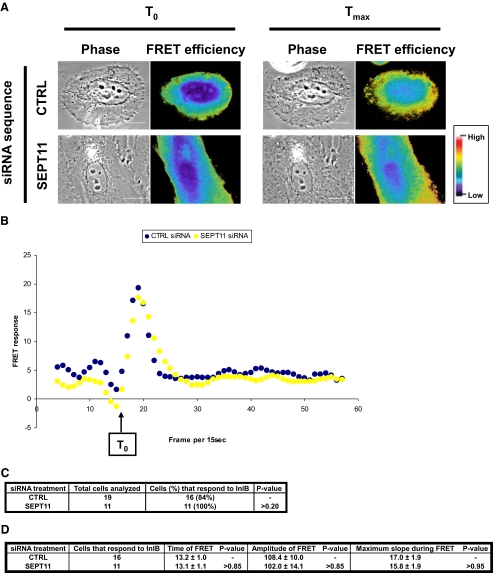
FIGURE 4.
SEPT11-depleted cells respond to InlB stimulation. A, representative FRET efficiency images for control (CTRL) and SEPT11-depleted HeLa cells expressing YFP-AktPH and CFP-AktPH stimulated with InlB. FRET efficiency at two time points are presented for each cell type: T0 (frame 16) and Tmax (frame corresponding to the maximum induction of FRET efficiency) after stimulation. Pseudocolor scale represents the range of FRET efficiency values from original signal ranging from blue (low) to red (high). Scale bars indicate 10 μm. Movies for the entire time course of InlB stimulation for these cells can be observed in the supplemental movies S1A and S1B. B, quantification of FRET response for septin-depleted cells. Processed FRET response was plotted against time (i.e. frame per 15 s), for the representative siRNA-treated cells depicted in Fig. 4A. Values from the control (CTRL) cell are plotted in black, and the SEPT11-depleted cell is in yellow. C, percentage of cells that respond to InlB treatment. Processed FRET response was determined for 11 or more experiments for each siRNA treatment. Cells were classified as not responding to InlB stimulation if the slope of processed FRET response could not be distinguished above baseline values. The percentage of SEPT11-depleted cells classified as not responding to InlB stimulation was statistically compared with expectations as derived from control cells by chi square test. D, summary of FRET characteristics for cells that respond to InlB treatment. Characteristics of the FRET response (time, amplitude, and maximum slope) were evaluated for control (CTRL)- and SEPT11-depleted cells that respond to InlB treatment, and p values were determined by Student's t test.
RESULTS
SEPT9 and SEPT11 Have Non-overlapping Roles in Filament Organization—We previously found that SEPT9 interacts with septins from other groups to form a complex, and in agreement with other reports (10, 24) identified SEPT11 as a binding partner of SEPT9 (15). In non-dividing, non-infected HeLa cells, both SEPT9 and SEPT11 colocalize with actin filaments, typically in the nuclear region and along the cell periphery, and rarely colocalize with microtubules (15, 24, 25).
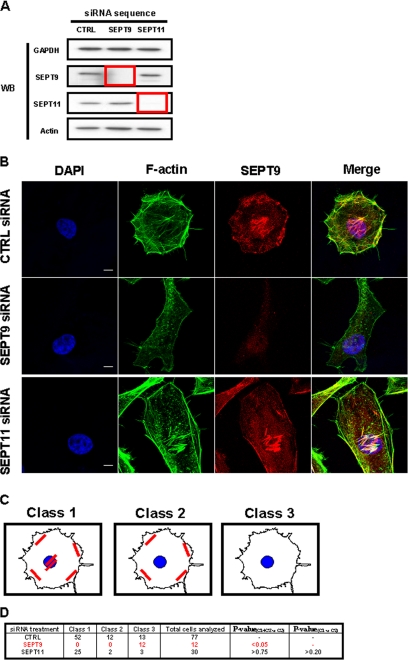
To establish the respective role of SEPT9 and SEPT11 in actin and septin filament formation, we treated HeLa cells, a cell line routinely used for siRNA depletion experiments, with siRNA against SEPT9 or SEPT11. siRNA depletion of each targeted septin protein was assessed following Western blot (Fig. 1A). Knockdown efficiency was quantified as 95 ± 2% for SEPT9 and 80 ± 13% for SEPT11. We then labeled siRNA-treated cells with fluorescein isothiocyanate-phalloidin and anti-SEPT9 antibody (Fig. 1B), and systematically analyzed actin and septin filament remodeling upon septin depletion. Confocal Z-stack images of siRNA-treated cells were then categorized by a septin filament tracking algorithm into three different pre-assigned classes of SEPT9 filament distribution (Fig. 1C): cells presenting filaments around the cell periphery and under the nucleus were designated as Class 1, cells with filaments only around the cell periphery, but not under the nucleus were designated as Class 2, and cells no longer presenting any filaments were designated as Class 3. The frequency of each filament class among septin-depleted cells was then statistically compared with values from control cells (Fig. 1D). As expected, SEPT9 filaments were absent from SEPT9-depleted cells. Actin filaments had also disappeared. In contrast, SEPT9 and actin filaments were not reduced in cells depleted for SEPT11 (p > 0.75), which retained their capacity to present both septin and actin filaments under the nucleus (p > 0.20). These results were also observed using a second siRNA sequence for SEPT11 (p > 0.15 and p > 0.30, respectively) (data not shown).
FIGURE 1.
The impact of septin depletion on HeLa cells. A, Western blot (WB) of HeLa cells transfected with siRNA against control (CTRL), SEPT9, or SEPT11. Cell lysates were separated by 10% SDS-PAGE before immunoblotting. The blots were probed with antibodies specific to GAPDH, SEPT9, SEPT11, and actin. GAPDH is shown as a loading control. The red box outlines depleted protein levels for targeted septins. B, septin depletions have different effects on cell shape and actin and septin filament distribution. Endogenous F-actin and SEPT9 were visualized by immunostaining with anti-F-actin (green) and anti-SEPT9 antibodies (red) for control (CTRL)-, SEPT9-, and SEPT11-depleted cells. Nuclei were marked with DAPI (blue). Scale bars indicate 10 μm. C, different classes of SEPT9 filament distribution. In siRNA-treated HeLa cells, septin and actin filaments were distributed into different patterns. Cells presenting filaments around the cell periphery and under the nucleus were designated as Class 1, cells with filaments only around the cell periphery, but not under the nucleus were designated as Class 2, and cells no longer presenting any filaments were designated as Class 3. D, different effects of septin inactivation on the organization of SEPT9 filaments. Confocal Z-stack images of siRNA-treated cells were categorized by a filament tracking algorithm into the three pre-assigned classes of filament distribution (Fig. 1C). The percentage of septin-depleted cells observed to still have filaments, i.e. % of cells in Class 1 and Class 2 versus % of cells in Class 3, was statistically compared with expectations as derived from control cells by chi square test (p value(C1+C2 versus C3)). To assess whether SEPT11-depleted cells retain SEPT9 filaments under the nucleus, the distribution of SEPT9 filaments in SEPT11-depleted cells, i.e. % of cells in Class 1 versus % of cells in Class 2, was statistically compared with expectations as derived from control cells by chi square test (p value(C1 versus C2)).
Thus our analysis, in contrast to previously published results for SEPT2, SEPT6, and SEPT7 (26, 27), showed that different septin inactivations have different effects on the organization of septin and actin filaments at least in HeLa cells. To further appreciate the impact of depleting different septin subunits, we have assessed the impact of SEPT2, SEPT9, and SEPT11 depletion on septin expression and filament formation (supplemental Fig. S1, A-C). Taken together these data suggest that SEPT11, unlike SEPT2 or SEPT9, is dispensable for septin and actin filament formation.
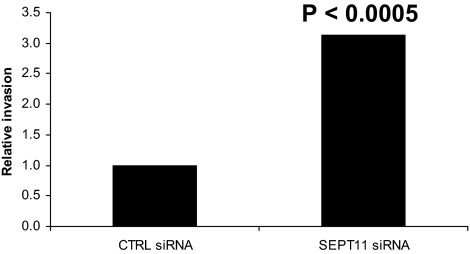
SEPT11 Inactivation Increases Listeria Invasion—Listeria invasion of non-phagocytic cells relies upon the polymerization and depolymerization of actin (13, 28) in concert also with the function of SEPT2 (15). To assess the functional consequence of SEPT11 depletion on Listeria invasion, we employed the classic gentamicin survival assay (16). In this assay, internalized bacteria are plated and counted 24 h after host cell lysis. Colony counts revealed that SEPT11 siRNA treatment increased Listeria invasion into HeLa cells (p < 0.0005) (Fig. 2), as opposed to results obtained for SEPT2 (15). Significantly increased Listeria invasion was also observed using a second siRNA sequence for SEPT11 (p < 0.01) (data not shown). Considering that SEPT2 depletion decreased Listeria entry (15), but SEPT11 depletion increased Listeria entry, these data strongly suggest that different septin inactivations can differentially modulate Listeria invasion.
FIGURE 2.
SEPT11 inactivation increases Listeria invasion. Gentamicin survival assays for L. monocytogenes EGD were performed in HeLa cells treated with control (CTRL) siRNA, or siRNA targeted against SEPT11. Graphs represent the relative number of intracellular bacteria found inside siRNA-treated cells after the survival assay, where CFU counts obtained from SEPT11-depleted cells were normalized to CTRL siRNA-treated cells. On the graph CTRL siRNA is figuratively presented as 1, and data represent the mean from n ≥ 9 experiments. Results were analyzed for statistical significance using the Student's t test.
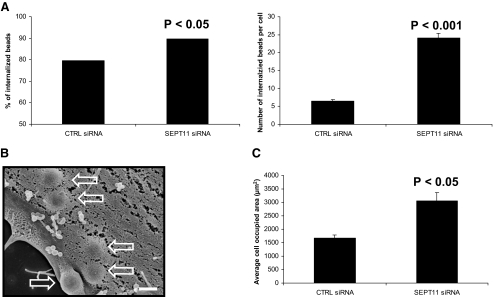
SEPT11 Inactivation Increases the Entry of InlB-coated Beads—Latex beads coated with purified recombinant InlB protein have been extensively used to study specifically the InlB-mediated entry of Listeria into HeLa cells (15, 19, 30, 31). To understand why L. monocytogenes invasion is increased upon SEPT11 depletion, we investigated particle uptake using 1-μm InlB-coated latex beads. siRNA-treated cells were incubated with InlB beads and were examined by a double immunofluorescence microscopy technique to distinguish intra-versus extracellular beads and provide a measure of InlB-induced phagocytosis (Fig. 3A). At 5-min postincubation, SEPT11-depleted cells presented an increased capacity to internalize beads compared with control cells (p < 0.05). On average, control cells internalized 6 beads per cell, whereas SEPT11-depleted cells internalized 24 beads per cell (p < 0.001).
FIGURE 3.
SEPT11 inactivation increases the entry of InlB-coated beads. A, quantification of InlB-induced phagocytosis for control (CTRL) and SEPT11-depleted HeLa cells. For internalization assays, beads were analyzed by immunofluorescence (see “Experimental Procedures”) for being extracellular or intracellular in at least 50 host cells counted for each of n ≥ 2 separate experiments per siRNA treatment. The left graph depicts the total % of internalized 1-μm InlB-coated beads at 5-min postincubation of siRNA-treated cells, calculated from the (total number of internalized beads)/(total number of cell-associated beads) × 100. Results were analyzed for statistical significance using the z-test for percentages. The right graph depicts the average number of internalized 1-μm InlB-coated beads per cell at 5-min postincubation of siRNA-treated cells. Graphed data represent this average value ± S.E., where results were analyzed for statistical significance using the two sample z-test. B, uptake of InlB beads by siRNA-treated cells. Control (CTRL) and SEPT11-depleted HeLa cells were incubated with 1-μm InlB beads for 5 min, and cells were processed for scanning electron microscopy. Depicted here is a representative images of InlB beads internalized for SEPT11-depleted cells. Hollow arrows indicate internalized beads. Magnification = 15,000 ×, where the scale bar indicates 1 μm. C, comparing the average surface areas of siRNA-treated cells. To quantify the surface area of siRNA-treated HeLa cells, confocal Z-stack images of cells were segmented using MetaMorph. Bar graphs depict the average cell occupied area (μm2) ± S.E. for control (CTRL)(n = 77)- and SEPT11 (n = 30)-depleted cells. The cell-occupied area for each septin-depleted cell type was then statistically compared with control cells by Student's t test.
Scanning electron microscopy was also used to examine the uptake of InlB beads (Fig. 3B). Of note, the membrane surface of SEPT11-depleted cells presented smoother membrane with a heterogeneous distribution of microvilli-like structures compared with control cells (supplemental Fig. S2A). At 5-min postincubation, the majority of beads were already internalized for control and SEPT11-depleted cells, but in agreement with the above immunofluorescence experiments, the number of internalized InlB beads per SEPT11-depleted cell was significantly higher than in control cells.
As previously reported for SEPT2-, SEPT6-, and SEPT7-depleted cells (26, 27), we found that HeLa cells become large and flat upon SEPT11 depletion (Fig. 1B). As a result, differences in the number of internalized InlB beads per siRNA-treated cell could reflect changes in cell size. To evaluate this, we measured the surface area of non-infected siRNA-treated HeLa cells after fixation (Fig. 3C and supplemental Fig. S2B). On average, the surface area for SEPT11-depleted cells was almost twice as large as control cells (p < 0.05). By comparison, SEPT2-depleted cells (15) occupied approximately the same surface area as control cells (p > 0.73) (data not shown). Thus, InlB invasion differences in SEPT11-depleted cells can in part be explained by differences in surface area. Taken together, these results definitively establish that SEPT11-depleted HeLa cells are competent to internalize Listeria and 1-μm InlB beads.
Signaling Events in Response to Met Stimulation by InlB Are Not Affected by the Depletion of SEPT11—Listeria enter HeLa cells via interactions between the cell surface receptor Met and the bacterial ligand InlB (32). Given the positive impact of SEPT11 depletion on particle entry, we hypothesized that signaling events induced upon InlB-Met interaction could be increased upon SEPT11 depletion. To test this, we used a FRET-based approach previously developed in our laboratory that focuses on phosphoinositide 3-kinase activation, a signaling pathway induced upon InlB-Met interaction (15, 21). The FRET assay is based on the fact that 3′-phosphoinositides produced by PI3K upon InlB-induced Met activation recruit the serine/threonine kinase Akt from the cytosol to the plasma membrane. Previous work with this assay revealed that SEPT2 has a critical role in these signaling events (15).
SEPT11-depleted cells and control cells were cotransfected with plasmids encoding YFP-AktPH and CFP-AktPH. The FRET stoichiometry measurement between YFP-AktPH and CFP-AktPH was then performed to determine FRET efficiency (Fig. 4, A and B, and supplemental movies S1A and S1B). As shown in Fig. 4, C and D, cells depleted of SEPT11 responded to InlB stimulation as well as control cells. Thus SEPT11, in contrast to SEPT2 (15), does not have an essential role in early signaling events in response to Met stimulation by InlB.
DISCUSSION
Septins are GTP-binding proteins that form filaments and are increasingly recognized as cytoskeleton components (5, 7). Although several studies have reported on septin function, little information is available for SEPT11. Using siRNA, we addressed the possible requirement for SEPT11 during Listeria infection. Our results show that SEPT11 is dispensable for InlB-mediated invasion, and surprisingly, its absence even leads to an increase in invasion.
A Dispensable Role for SEPT11 in Septin Filament Formation—SEPT11 can bind SEPT9 in vitro (10, 15, 24). In HeLa cells, SEPT9 depletion prevents septin filament formation and also actin filament formation. In contrast, SEPT11 depletion did not alter the presence and colocalization of SEPT9 and actin filaments (Fig. 1, B-D). That SEPT9, but not SEPT11, is required for septin filament formation strongly suggests non-overlapping functions for these different septins. Of note, neither SEPT9 nor SEPT11 depletion had any noticeable effect on microtubules (supplemental Fig. S3).
Septins Differentially Control the InlB-Met-mediated Entry of Listeria into Host Cells—To investigate how SEPT11 can regulate the internalization process, we performed our functional analysis on HeLa cells, because they allow the specific analysis of the InlB-Met entry pathway used by Listeria (28). Our present study together with previous work (15) reveals that septins differentially modulate the InlB-Met-mediated entry of L. monocytogenes, with an increase in SEPT11-depleted cells and a decrease in SEPT2-depleted cells. Moreover, FRET analysis confirmed that SEPT11, unlike SEPT2, is not required for the appropriate signaling cascade in response to Met stimulation by InlB. These data reveal a different effect of SEPT11 and SEPT2 depletion on the interaction of InlB with its host receptor Met, and consequently on bacterial entry.
How can one explain these results? Septin filaments colocalize in HeLa cells (data not shown), however we cannot exclude different localization between SEPT2 and SEPT11 at this stage. Using siRNA, we observe that SEPT11 filament formation relies on SEPT2 expression, but SEPT2 filaments continue to form in the absence of SEPT11 (supplemental Fig. S1, B and C). Structural data have shown that septins are ordered in the isolated complex as SEPT7-SEPT6-SEPT2-SEPT2-SEPT6-SEPT7 (9). As the central subunit in this septin complex, SEPT2 cannot be replaced in the conditions tested (15). Conversely, it has been proposed that members of the SEPT6 group are interchangeable, at least in the context of the SEPT2-SEPT6-SEPT7 complex (3, 12). Thus, some of the phenotypes we observe here in cells depleted for SEPT11 could be because the SEPT6 group members are partially redundant. Nevertheless, cells depleted for SEPT11 do not present all the control cell phenotypes. In particular they have an increased cell size and are more permissive to InlB-mediated invasion, suggesting that other SEPT6 group members cannot fully substitute for SEPT11. The extent to which SEPT6 group members share properties and/or functionally compete with each other is difficult to currently appreciate.
Why Are SEPT11-depleted Cells More Permissive to Listeria Invasion?—To explain differences in L. monocytogenes infection arising from the SEPT11 siRNA treatment, we analyzed phenotypes previously reported for septin-depleted cells. Of these, arrest in cytokinesis is a classic phenotype of septin depletion (1), itself typically assessed in mammalian cells by the increase of binucleated cells (25, 33-35). In agreement with these data, SEPT11 depletion resulted in an increase of binucleated cells (supplemental Fig. S4A). To determine if binucleation was influencing Listeria infection, we fixed and fluorescently labeled cells after gentamicin treatment to determine the frequency of infected cells that were binucleated. However, binucleated cells were as susceptible to infection as cells with a single nucleus (supplemental Fig. S4B).
The SEPT11 phenotype described here is different from the effects of global cytoskeleton disruption on bacterial entry. As well established (17, 18), inhibition of actin even by low concentrations of cytochalasin D or latrunculin B was found to have potent negative effects on bacterial invasion of HeLa cells (supplemental Fig. S5A). Depolymerization of microtubules by nocodazole, or stabilization of microtubules with paclitaxel, also resulted in a significant reduction in the number of invading Listeria (18) (supplemental Fig. S5B). We finally tested the impact of a novel drug, forchlorfeneuron, to specifically investigate septin function (29). Forchlorfeneuron treatment also had a negative impact on bacterial invasion (supplemental Fig. S5, C and D). As SEPT11 depletion increases entry, our study highlights a novel type of control for a cytoskeleton component.
As shown in Fig. 3C, SEPT11-depleted cells are larger than control cells, which in part may explain the higher permissivity. Nevertheless, the fact that SEPT11-depleted cells are more permissive suggests a role for SEPT11 in limiting entry into cells. Quite strikingly, preliminary work has revealed that SEPT6-depleted HeLa cells also are increased for Listeria invasion (p = 0.001) (supplemental Fig. S5E). These results strongly support that all SEPT6 group members play a similar role in restricting entry. Whether this is due to septin association with the plasma membrane or septin function in regulating cytoskeleton dynamics is currently under investigation.
CONCLUSIONS
On the basis of different recruitment we have previously distinguished septin from actin function during bacterial entry and identified SEPT2 as essential for bacterial entry (15). Here we demonstrate that SEPT11 depletion does not decrease, but surprisingly increases, the InlB-mediated entry of Listeria. Taken together, these data highlight the specific roles for two different septins, SEPT2 and SEPT11, during the InlB-mediated internalization process. These findings for the first time distinguish unique septin roles in regulating the efficiency of receptor-mediated bacterial invasion.
Supplementary Material
Acknowledgments
We thank Dr. Marc Lecuit and Dr. Elias Spiliotis for critical reading of the manuscript, Dr. Matteo Bonazzi for technical expertise, and members of the P. C. laboratory for helpful discussions. Thanks also to Dr. William Trimble (anti-SEPT2) and Dr. Makoto Kinoshita (anti-SEPT11) for antibodies.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S5 and movies S1A and S1B.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: siRNA, small interfering RNA; CFP, cyan fluorescent protein; DAPI, 4′, 6′-diamidino-2-phenylindole; DMEM, Dulbecco's modified Eagle's medium; FRET, fluorescence resonance energy transfer; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; InlB, internalin B; PH, pleckstrin homology; PI3K, phosphoinositide 3-kinase; SEPT, septin; YFP, yellow fluorescent protein.
References
- 1.Hartwell, L. H. (1971) Exp. Cell Res. 69 265-276 [DOI] [PubMed] [Google Scholar]
- 2.Versele, M., and Thorner, J. (2005) Trends Cell Biol. 15 414-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kinoshita, M. (2003) J. Biochem. (Tokyo) 134 491-496 [DOI] [PubMed] [Google Scholar]
- 4.Kinoshita, M. (2003) Genome Biol. 4 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weirich, C. S., Erzberger, J. P., and Barral, Y. (2008) Nat. Rev. Mol. Cell Biol. 9 478-489 [DOI] [PubMed] [Google Scholar]
- 6.Kinoshita, M. (2006) Curr. Opin. Cell Biol. 18 54-60 [DOI] [PubMed] [Google Scholar]
- 7.Spiliotis, E. T., and Nelson, W. J. (2006) J. Cell Sci. 119 4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spiliotis, E. T., and Nelson, W. J. (2008) in The Septins (Hall, P. A., Russell, H. S. E., and Pringle, J. R., eds) pp. 229-246, Wiley-Blackwell, Chippenham
- 9.Sirajuddin, M., Farkasovsky, M., Hauer, F., Kuhlmann, D., Macara, I. G., Weyand, M., Stark, H., and Wittinghofer, A. (2007) Nature 449 311-315 [DOI] [PubMed] [Google Scholar]
- 10.Nagata, K.-i., Asano, T., Nozawa, Y., and Inagaki, M. (2004) J. Biol. Chem. 279 55895-55904 [DOI] [PubMed] [Google Scholar]
- 11.Xie, Y., Vessey, J. P., Konecna, A., Dahm, R., Macchi, P., and Kiebler, M. A. (2007) Curr. Biol. 17 1746-1751 [DOI] [PubMed] [Google Scholar]
- 12.Ono, R., Ihara, M., Nakajima, H., Ozaki, K., Kataoka-Fujiwara, Y., Taki, T., Nagata, K.-i., Inagaki, M., Yoshida, N., Kitamura, T., Hayashi, Y., Kinoshita, M., and Nosaka, T. (2005) Mol. Cell Biol. 25 10965-10978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamon, M., Bierne, H., and Cossart, P. (2006) Nat. Rev. Micro. 4 423-434 [DOI] [PubMed] [Google Scholar]
- 14.Pizarro-Cerda, J., Jonquieres, R., Gouin, E., Vandekerckhove, J., Garin, J., and Cossart, P. (2002) Cell. Microbiol. 4 101-115 [DOI] [PubMed] [Google Scholar]
- 15.Mostowy, S., Nam Tham, T., Danckaert, A., Guadagnini, S. P., Boisson-Dupuis, S. P., Pizarro-Cerdá, J., and Cossart, P. (2009) PLoS ONE 4 e4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pizarro-Cerdá, J., Lecuit, M., and Cossart, P. (2002) in Molecular Cellular Microbiology (Sansonetti, P. J., and Zychlinsky, A., eds) Methods in Microbiology, Vol. 31, pp. 161-177, Academic Press, London [Google Scholar]
- 17.Ireton, K., Payrastre, B., Chap, H., Ogawa, W., Sakaue, H., Kasuga, M., and Cossart, P. (1996) Science 274 780-782 [DOI] [PubMed] [Google Scholar]
- 18.Kuhn, M. (1998) FEMS Microbiol. Lett. 160 87-90 [DOI] [PubMed] [Google Scholar]
- 19.Braun, L., Ohayon, H., and Cossart, P. (1998) Mol. Microbiol. 27 1077-1087 [DOI] [PubMed] [Google Scholar]
- 20.Braun, L., Dramsi, S., Dehoux, P., Bierne, H., Lindahl, G., and Cossart, P. (1997) Mol. Microbiol. 25 285-294 [DOI] [PubMed] [Google Scholar]
- 21.Seveau, S., Tham, T. N., Payrastre, B., Hoppe, A. D., Swanson, J. A., and Cossart, P. (2007) Cell. Microbiol. 9 790-803 [DOI] [PubMed] [Google Scholar]
- 22.Haugh, J. M., Codazzi, F., Teruel, M., and Meyer, T. (2000) J. Cell Biol. 151 1269-1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osaki, M., Oshimura, M., and Ito, H. (2004) Apoptosis 9 667-676 [DOI] [PubMed] [Google Scholar]
- 24.Nobuhiro, H., Koh-ichi, N., Aie, K., Takashi, S., Noriko, S., Yasuhisa, H., Shingo, M., and Masaki, I. (2004) FEBS Lett. 568 83-8815196925 [Google Scholar]
- 25.Surka, M. C., Tsang, C. W., and Trimble, W. S. (2002) Mol. Biol. Cell 13 3532-3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinoshita, M., Field, C. M., Coughlin, M. L., Straight, A. F., and Mitchison, T. J. (2002) Dev. Cell 3 791-802 [DOI] [PubMed] [Google Scholar]
- 27.Kremer, B. E., Adang, L. A., and Macara, I. G. (2007) Cell 130 837-850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cossart, P., Pizarro-Cerda, J., and Lecuit, M. (2003) Trends Cell Biol. 13 23-31 [DOI] [PubMed] [Google Scholar]
- 29.Hu, Q., Nelson, W. J., and Spiliotis, E. T. (2008) J. Biol. Chem. 283 29563-29571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lecuit, M., Ohayon, H., Braun, L., Mengaud, J., and Cossart, P. (1997) Infect. Immun. 65 5309-5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bierne, H., Gouin, E., Roux, P., Caroni, P., Yin, H. L., and Cossart, P. (2001) J. Cell Biol. 155 101-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen, Y., Naujokas, M., Park, M., and Ireton, K. (2000) Cell 103 501-510 [DOI] [PubMed] [Google Scholar]
- 33.Spiliotis, E. T., Kinoshita, M., and Nelson, W. J. (2005) Science 307 1781-1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagata, K.-i., Kawajiri, A., Matsui, S., Takagishi, M., Shiromizu, T., Saitoh, N., Izawa, I., Kiyono, T., Itoh, T. J., Hotani, H., and Inagaki, M. (2003) J. Biol. Chem. 278 18538-18543 [DOI] [PubMed] [Google Scholar]
- 35.Kouranti, I., Sachse, M., Arouche, N., Goud, B., and Echard, A. (2006) Curr. Biol. 16 1719-1725 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.