Abstract
Hepatoma-derived growth factor-related proteins (HRP) comprise a family of 6 members, which the biological functions are still largely unclear. Here we show that during embryogenesis HRP-3 is strongly expressed in the developing nervous system. At early stages of development HRP-3 is located in the cytoplasm and neurites of cortical neurons. Upon maturation HRP-3 relocalizes continuously to the nuclei and in the majority of neurons of adult mice it is located exclusively in the nucleus. This redistribution from neurites to nuclei is also found in embryonic cortical neurons maturing in cell culture. We show that HRP-3 is necessary for proper neurite outgrowth in primary cortical neurons. To identify possible mechanisms of how HRP-3 modulate neuritogenesis we isolated HRP-3 interaction partners and demonstrate that it binds tubulin through the N-terminal so called HATH region, which is strongly conserved among members of the HRP family. It promotes tubulin polymerization, stabilizes and bundles microtubules. This activity depends on the extranuclear localization of HRP-3. HRP-3 thus could play an important role during neuronal development by its modulation of the neuronal cytoskeleton.
Neuritogenesis is a key step in nervous system development in which neurons extend dendrites and axons and connect to different targets in and outside the nervous system. The proper regulation of this process is controlled by a number of extra- and intracellular molecules expressed by neurons themselves or non-neuronal cells in their surroundings. Multiple studies indicate that rearrangement of the neuronal cytoskeleton in response to extracellular signals is an important mechanism during neurite extension and pathfinding (1-3). Manipulation of the polymerization and depolymerization of microtubules has shown that regulation of microtubule assembly and maintenance is important for neuritogenesis (4). Microtubule dynamics are regulated by a huge number of regulatory proteins like tau or other microtubule-associated proteins (MAPs)4 (5). In addition, proteins like CRMP-2 that interact with tubulin dimers and accelerate the assembly of tubulin into microtubles have been shown to be involved in the regulation of neuronal polarity and neuritogenesis (6-10). Despite all advances, however, made in the understanding of the role of the cytoskeleton and its regulatory proteins during neuritic growth there are still many open questions regarding the regulation of these processes. Therefore identifying new molecules binding to and modulating the turnover of microtubules is of high interest for the understanding of how neurite outgrowth is regulated.
Hepatoma-derived growth factor (HDGF) is a protein that was purified from secretions of hepatoma cells by virtue of its growth factor activity. Subsequently 5 additional proteins were identified in which the 97 N-terminal amino acid residues show strong similarity to HDGF. Accordingly this family of proteins has been termed HDGF-related proteins (HRP) (11-13). HDGF has neurotrophic activity for hippocampal, spinal, and facial motor neurons (14, 15). So far, however, no receptor or signal transduction pathway involved has been identified for any of the HRPs.
Most HRPs are expressed in a variety of tissues. HRP-3, however, is the only family member in whose expression is restricted. It is only expressed in neurons and to a low extent in glial cells (16, 17). Like HDGF after transfection into human embryonic kidney cells HRP-3 exhibits proliferative activity (12). The strong and almost exclusive expression of HRP-3 in postmitotic neurons, however, suggests biological functions other than its growth factor activity (16).
In the present study we examine the expression and function of HRP-3 protein during mouse embryonic neuronal development. We demonstrate that the protein locates to the cytoplasm and neurites during early nervous system development, whereas most of HRP-3 can be found in the nucleus in adult neurons. We show that HRP-3 promotes neurite growth and suggest that this is due to the interaction of HRP-3 with the tubulin component of the neuronal cytoskeleton.
EXPERIMENTAL PROCEDURES
Immunolabeling of Tissue Sections—Embryos were removed from the uterus of a pregnant mouse under deep anesthesia and washed 3 times in prewarmed PBS each for 10 min, then fixed in 4% (w/v) paraformaldehyde. After fixation for 12 h, embryos were washed in PBS for several hours and dehydrated in a series of increasing concentrations of ethanol and xylol, then embedded in paraffin. Ten to 15-μm thick sagittal or coronal sections were cut using a Leica RM2155 microtome. For staining, the sections were deparaffinized and rehydrated in xylol and ethanol in decreasing concentration. After antigen retrieval steps, the endogenous peroxidase activity was blocked using 1% H2O2 in PBS for 10 min, washed once, and permeablized using 0.5% Triton X-100 for 10 min. Nonspecific binding sites were blocked by incubating the sections with 2% BSA for 2 h. Without further washing, the sections were incubated with the primary antibody diluted in the blocking solution (affinity purified rabbit anti-HRP-3, 25 μg/ml (17)) overnight at 4 °C. Afterward, the sections were washed for 3 × 5-min in PBS and incubated with biotinylated goat anti-rabbit antibody (diluted in the blocking solution; Dianova; 1:200) for 2 h and washed again as above. Bound antibodies were visualized using the ABC VectaStain kit and Histogreen as a chromogenic substrate according to the manufacturer's protocol (Vector Laboratories).
Cell Culture—Brains from 14.5-day-old NMRI mouse embryos were used. Cortices were dissected in ice-cold Hanks' balanced salt solution and incubated in 10 ml of 0.5% trypsin-EDTA for 10-12 min in a 37 °C water bath. The activity of trypsin was blocked using a mixture of trypsin inhibitor and 100 mg/ml BSA in H2O (in a ratio of 4:1) for 5 min at room temperature. The supernatant was carefully decanted and the cortices were triturated several times in plating medium (neurobasal medium + 2% B27, 0.5 mm glutamine, 25 μm l-glutamic acid, and 1 mm penicillin/streptomycin) using fire-polished Pasteur pipettes. Viability was assessed using trypan blue and 500-1000 neurons/mm2 were plated onto 12-mm glass coverslips coated overnight at 4 °C with 10 μg/ml poly-d-lysine (Sigma). Neurons were incubated at 37 °C with 5% CO2. For synthetic iRNA treatment 20 pmol of each siRNA (Ambion; siRNA1, GGTGATAGAGTAGAAGATG; siRNA2, GGAGTGAAATTTACTGGGT; siRNA3, GGGAGAAGGTGGAAATACT; negative control iRNA#1 catalog number AM4611) diluted in 50 μl of medium were mixed with 50 μl of Lipofectamine 2000 (diluted 1:50 in medium; Invitrogen) and incubated for 20 min at room temperature. The mixture was added to 250 μl of medium in a poly-d-lysine-coated well of a 24-well plate and mixed by gentle pipetting. 150,000 Neurons were added in an additional 250 μl of medium, shaken gently, and incubated for 2-3 days before analysis. To analyze the influence of HRP-3 reduction on neurite outgrowth, Cy3-labeled siRNAs were cotransfected to identify silenced neurons. Pictures were taken using a Zeiss Axiovert 100M microscope and the length of the longest neurite and the number of neurites arising from each cell were measured using the Zeiss axiovision software. Statistical analysis was performed using GraphPad Prism version 5.01. All experiments were performed at least three times and analyzed in a blinded fashion.
HEK293 cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, 0.5 mm glutamine, and 1 mm penicillin/streptomycin. Cells were split every 3 days and were used between passages 5 and 10 for analysis. To test the influence of HRP-3 on the acetylation of tubulin, different HRP-3 DNA constructs were transiently transfected into HEK cells in 6-well plates using Exgen (Fermentas) following the manufacturer's instructions. After 48 h cells were lysed using lysis buffer (50 mm Tris-HCl, pH 7.4, 50 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin, 1 mm phenylmethylsulfonyl fluoride). In parallel untransfected cells were treated with 10 μg/ml Nocodazol or 10 mm taxol for 2 h as negative and positive controls, respectively. Ten micrograms of protein of each condition was separated by SDS-PAGE and transferred onto nitrocellulose. Acetylated tubulin was detected using a specific antibody (1:4.000, Sigma). β-Actin was detected to control equal loading for every condition (1:2.000, Sigma). Bound primary antibodies were detected using fluorescently labeled antibody (goat antimouse Alexa 680, Molecular Probes) for use with an Odyssey far red scanner (LiCor).
B35 neuroblastma cells were maintained in Dulbecco's modified Eagle's medium/F-12 containing 5% fetal calf serum, 0.5 mm glutamine, and 1 mm penicillin/streptomycin. Cells were transfected using Exgen (Fermentas) following the manufacturer's instructions. To induce differentiation of B35 cells dibutyryl cyclic AMP (Bt2cAMP) was added to a final concentration of 1 mm for 3 days 24 h after transfection. To analyze the influence of HRP-3 on neuritic growth the different DNA constructs were cotransfected with a plasmid encoded for the green fluorescent protein to identify transfected cells. Fluorescent pictures were taken using a Zeiss axiovert microscope and the length of the longest neurites of at least 100 cells per transfected construct were measured using Zeiss axiovision software.
For immunocytochemistry, cells were fixed by incubation in 4% (w/v) paraformaldehyde + 10% sucrose for 10 min at 4 °C. After permeabilization with 0.5% Triton X-100 in PBS for 10 min and blocking in 2% BSA for 30 min, cells were incubated with an affinity-purified antibody against HRP-3 at a concentration of 2 μg/ml in blocking solution for 1 h at room temperature. For double staining, monoclonal antibodies against mouse microtubule-associated protein-2 (MAP-2, Sigma, 1:200), α-tubulin or acetylated tubulin (both Sigma, 1:200) were simultaneously incubated together with the anti-HRP-3 antibody. Bound antibodies were visualized using Cy3-conjugated goat anti-rabbit (1:500) or Cy2-conjugated goat anti-mouse (1:200) antibodies (Dianova, Hamburg, Germany). Cell nuclei were counterstained with 4,6-diamidino-2-phenylindole. Coverslips were mounted in Mowiol or polyvinyl alcohol fluorescent mounting medium with DABCO from Fluka and analyzed by epifluorescence microscopy on an Axiovert 100M instrument (Zeiss, Jena, Germany) or by laser scan microscopy using a Leica TCS SP2 instrument (Leica, Wetzlar, Germany).
Identification of HRP-3 Interaction Partners—Histidine-tagged proteins were produced in BL21 bacteria as described elsewhere (11). His-HRP-3 was immobilized on N-hydroxysuccinimide-activated columns (column volume 1 ml; Amersham Biosciences) as recommended by the manufacturer and washed three times with 10 column volumes of binding buffer (50 mm sodium phosphate, 300 mm NaCl, pH 7.4). A cytosolic newborn mouse brain extract was prepared as described before (16) and loaded onto a His-HRP-3 column. Nonspecifically bound proteins were washed out by 10 column volumes of binding buffer and then elution was performed using 5 column volumes of elution buffer (7 m urea, 1 m thiourea). After concentration by ultrafiltration (Centricon 10) proteins were analyzed by SDS-PAGE and Coomassie Brilliant Blue staining (Fermentas) followed by mass spectrometry (MALDI-TOF; STR Voyager, Applied Biosystems). For MALDI-TOF analysis stained proteins were cut from gels and subjected to in-gel digestion using sequencing grade trypsin (trypsin profile IGD Kit, Sigma) according to the manufacturer's instructions. Peptide masses obtained by mass spectrometry were used to search two different data bases (MS-fit and Mascot).
Cloning of HRP-3 Variants—Expression vectors coding for mutations in the nuclear localization signals (NLS) of HRP-3 were cloned via a two-step PCR approach using a pCDNA3 vector containing the HRP-3 coding sequence as a template. For mutation of the second NLS (NLS2) in a first step bases located 5′ of the NLS site were amplified using primers 5′-CTCGAGCTCAAGCTTGTCATGGCGCGTCCGCGG-3′ and 5′-TGCTGGACTAGTGTAGGACTTTTTCCGATTTGA-3′. The 480-bp PCR product was digested by HindIII/SpeI and cloned into a pBSK(-) vector. Bases located 3′ of the NLS were amplified using primers 5′-ACGCCGACTAGTAAGAAGTCTTCTAATCAGTCCCGGAACTCTCCA-3′ and 5′-CTAGCATTTAGGTGACACTATAG-3′. The 294-bp fragment was digested by SpeI and cloned into the pBSK(-) vector produced above. HRP-3 mutated in NLS1 was cloned using primers 5′-CTCGAGCTCAAGCTTGTCATGGCGCGTCCGCGG-3′ and 5′-GCTACGAATATTTGAGTTTCCAAACTTGTCTTTG-3′ for the 5′-part of the sequence (HindIII/SspI digest) and primers 5′-CAGCAAGCTTAGCGTGTTAACCAGAACGGATTTAATGAAGGAT-3′ and 5′-CTAGCATTTAGGTGACACTATAG-3′ (HpaI/NotI digest) for the 3′-part of the construct. Integrity of all constructs were confirmed by cycle sequencing. The vector coding for the double mutant (NLS1 + 2) was constructed by cutting a PstI fragment from the NLS2 mutant containing pBSK(-) vector (225 bp) and cloning it into the vector containing the NLS1 mutant. Expression vectors were constructed by subcloning into pCDNA3 vector containing the cytomegalovirus promoter for eukaryotic expression and a C-terminal signal coding for a StrepTag® peptide.
The C-terminal and N-terminal truncated versions of HRP-3 were cloned by PCR using the following primers, 5′-GTACGGATCCGCGCGTCCGCGGCCCCGCGAGTAC-3′ and 5′-TAGTAAGCTTTAAGTTTCTGAAGAGCTCTGTTG-3′ for the HATH region and 5′-AACAGAGGATCCGAAACTGAGGGAGAA-3′ and 5′-GCAGCCAAGCTTAGGTCCCTTCACCGG-3′ to amplify the non-HATH region of HRP-3. PCR products were digested by BamHI/HindIII and cloned in-frame to the N-terminal histidine tag into pQE80L vector coding for a N-terminal histidine tag (Qiagen).
Tubulin Overlay Assay—The tubulin overlay assay was performed as described by Kremer and co-workers (18). Briefly, 1 μg of recombinant wild type and truncated HRP-3 and 250 ng of tubulin were loaded on a 15% SDS-PAGE gel. After electrotransfer onto nitrocellulose membranes, free binding sites were blocked overnight at 4 °C in PIPES buffer (100 mm PIPES, pH 6.8, 1 mm MgCl2, 1 mm EGTA, 0.1% (v/v) Tween 20) containing 2% BSA (w/v). The next day the membranes were incubated 1 h at room temperature with 4 μg/ml tubulin dimers diluted in PIPES buffer and then washed 5 times for 10 min in buffer only. Protein interactions were stabilized with 0.1% formaldehyde (v/v) in PIPES buffer for 30 min at room temperature followed by incubation in 2% glycine (w/v) for 30 min in PIPES buffer. After three washes in 10 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.1% Tween 20 (TBS/T; 10 min each) bound tubulin was detected by incubation with an anti-α-tubulin antibody for 1 h at room temperature in TBS/T (Sigma, 1:2,000) and a fluorescently labeled antibody (goat anti-rabbit Alexa 680, Molecular Probes) for use with an Odyssey far red scanner (LiCor).
Microtubule Cosedimentation Assay—To obtain microtubules, purified tubulin (Cytoskeleton Inc.) was diluted in 50 μl of tubulin polymerizing buffer (100 mm Na-PIPES, pH 6.8, 1 mm MgCl2, 1 mm EGTA, 1 mm GTP, 20% tubulin glycerol buffer) to a concentration of 50 μm and kept on ice for 10 min. After incubation for 30 min at 37 °C, 20 μm taxol was added to stabilize microtubules and incubation was continued for a further 10 min at 37 °C. 5 μm Taxol-stabilized microtubules were mixed with different concentrations of recombinant proteins in 25 μl of tubulin polymerizing buffer and incubated for 30 min at 37 °C. After centrifugation at 100,000 × g (Beckman tabletop ultracentrifuge, rotor TLA110) for 30 min at 37 °C supernatants were collected and pellets were resuspended in 25 μl of SDS-PAGE sample buffer. Equal aliquots of supernatants and pellets were separated using 15% SDS-PAGE gels. After electrophoresis proteins were stained with Coomassie Brilliant Blue. Stained gels were scanned and analyzed using an Odyssey far red scanner.
Tubulin Assembly—Tubulin (15 μm) was polymerized with different concentrations (0, 0.5, 2.5, 5, 10, and 20 μm) of wild type HRP-3 in 20 μl of reassembly buffer (100 mm MOPS, pH 6.8, 0.1 mm EGTA, 0.5 mm MgCl2, 0.5 mm GTP) for 30 min at 37 °C. After centrifugation at 100,000 × g for 30 min at 37 °C pellets were resuspended in 20 μl of reassembly buffer and equal volumes of pellets and supernatants (5 μl) were loaded onto 12.5% SDS-PAGE gels. After electrophoresis proteins were stained with Coomassie Brilliant Blue. Stained gels were scanned and analyzed using an Odyssey far red scanner and the Odyssey software (LiCor).
Tubulin Stability—Stability against cold was investigated by incubating 2 μm polymerized microtubules in polymerization buffer for 30 min at 37 °C in the presence of 30 μm HRP-3, 30 μm GFP, and 20 μm taxol as negative and positive controls, respectively. After 2 additional minutes incubation on ice, samples were centrifuged at 100,000 × g for 30 min at 4 °C to collect the remaining microtubules. Stability against nocodazole treatment was tested by a modified method from Hasan and co-workers (19). 20 μm Tubulin was polymerized in 20 μl of polymerizing buffer in the presence or absence of 10 μm HRP-3. After incubation for 30 min at 37 °C, to half of each sample nocodazole was added to a final concentration of 50 μg/ml and all samples were incubated for another 10 min at 37 °C. After centrifugation for 30 min at 100,000 × g at 37 °C, pellets were resuspended in 20 μl of polymerizing buffer. For analysis equal volumes of supernatants and pellets were separated by SDS-PAGE and proteins were visualized by Coomassie Brilliant Blue staining. Stained gels were scanned and analyzed using an Odyssey far red scanner and Odyssey software (LiCor).
Chemical Cross-linking—His-HRP-3 or His-HDGF at a concentration of 10 μm were incubated for 30 min at room temperature in 50 μl of 50 mm phosphate buffer, pH 7.4, 150 mm NaCl with or without 1 mm disuccinimidyl suberate, the reaction was stopped by adding 50 μl of 2 × SDS-PAGE buffer. After incubation for 5 min at 95 °C, 15 and 2 μl of the reaction mixture were separated by 12.5% SDS-PAGE followed by Coomassie Brilliant Blue staining or immunoblotting using an anti-His tag antibody (Sigma, 1:2,000), respectively. Stained gels and immunoblots were scanned and analyzed using an Odyssey far red scanner and Odyssey software (LiCor).
Bundling of Microtubules—Rhodamine-labeled tubulin was polymerized together with unlabeled tubulin in a ratio of 1:4 for 40 min at 37 °C in polymerization buffer and stabilized with 10 μm taxol. Five hundred nanomolar of these microtubules were incubated in the presence of 2 μm HRP-3 or BSA and incubated for an additional 30 min at 37 °C. Samples were spotted on poly-l-lysine-coated coverslips for 30 min at 37 °C, washed with polymerization buffer, and mounted for analysis using fluorescence microscopy.
RESULTS
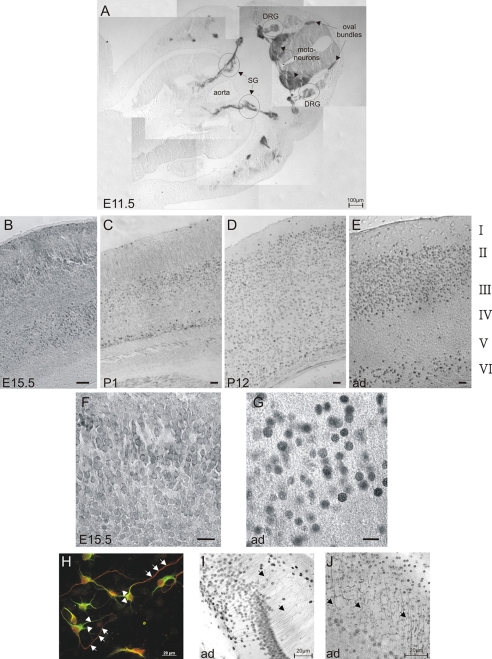
HRP-3 Is Expressed in Neuronal Extensions during Embryonic Development—HRP-3 is a member of the HDGF protein family that shows a temporally and spatially regulated expression during postnatal murine development (16). Highest expression is found around birth after which the expression decreases during the following postnatal weeks. Except for some minor expression in oligodendrocytes expression of HRP-3 is restricted to neurons (17). Because detailed data on the expression during prenatal development are not available, we investigated the embryonic HRP-3 expression pattern by using an HRP-3 specific affinity purified antibody on whole sections prepared from embryos at day 11.5 (E11.5) as well as transversal brain sections of embryos at E15.5. In addition we tested specimens from newborn (postnatal 1), 12-day-old (postnatal 12), as well as adult (ad) mice brains. At E11.5 HRP-3 is expressed in motoneurons, neurons of the dorsal root ganglia, and ganglia of the sympathetic system. Strong signals are also obtained in the oval bundle, the anterior neural root and the spinal nerve (Fig. 1A). At E15.5 nuclear as well as extranuclear HRP-3 immunoreactivity can be observed in neocortical neurons (Fig. 1B) that both are still present in neocortices of brains taken from newborn mice (Fig. 1C). This extranuclear immunoreactivity is remarkably reduced during postnatal development and can hardly be found in animals 12 days after birth (Fig. 1D) or in adults (Fig. 1E, see also Fig. 1, F and G)). Immunostainings on cultured cortical neurons using MAP-2 as a dendritic marker demonstrate that HRP-3 preferably locates to the axonal compartment of this type of cells but nevertheless is not excluded from dendrites (Fig. 1H). Localization to different kinds of neurites seems to be dependent on the neuronal cell type examined because, for example, hippocampal neurons show prominent axonal staining for HRP-3, whereas Purkinje cells of the cerebellum direct HRP-3 into their dendritic compartment (Fig. 1, I and J).
FIGURE 1.
HRP-3 is expressed in peripheral and central neurons during key stages of neuronal development. Various brain sections from mice of different embryonic days (E11.5 and E15.5), postnatal day 1 (P1) and 12 (P12) and adult mice (ad) were immunohistochemically stained using an affinity purified antiserum against mouse HRP-3. Bound antibodies were visualized by a biotinylated anti-rabbit antibody and streptavidin-coupled peroxidase. A, HRP-3 expression is prominent in dorsal root ganglia (DRG) as well as ganglia of the sympathetic system (SG) of an 11.5-day-old embryo. In addition, the area of motoneuron development and the oval bundles show strong HRP-3 immunoreactivity. B-E, pictures taken after HRP-3 immunostaining of brain slices taken from mice of different ages. In all cases the neocortical region above the hippocampal formation is shown. During embryonic and early postnatal development (E15.5 and P1) cortical neurons show extranuclear HRP-3 immunoreactivity (B and C), whereas neurons of later time points (P12 and adult) show a strong reduction of extranuclear HRP-3 signal (D and E). Numbers on the right label the different cortical layers of the adult mice. Bars in all figures are 40 μm. F and G, higher magnifications of cortical regions from E15.5 (F) and adult (G) mice brain demonstrate the localization change of HRP-3 during cortical development. Bars in both figures are 20 μm. H-J, cultured neocortical neurons were stained for MAP-2 (green) to label the dendritic compartment (arrowheads) and HRP-3 (red). The strongest signal for HRP-3 were found in neurites negative for MAP-2 representing axonal structures of these cells (arrows in H). Axonal staining can also be found in neurons of the hippocampal formation (arrows in I), whereas Purkinje cells in the cerebellum direct the protein to their dendritic compartment (arrows in J).
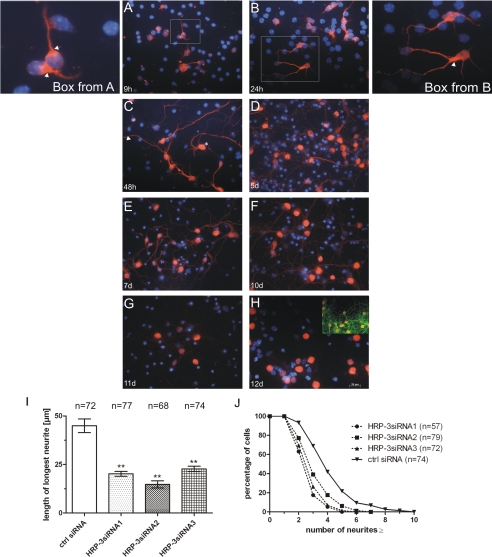
HRP-3 Expression in Cultured Embryonic Neurons—The expression and localization of HRP-3 was also examined during the maturation of a primary culture of mouse cortical neurons isolated from 14.5-day-old embryos. These cells were shown previously to contain substantial amounts of HRP-3 (16). After preparation the cells were stained with an HRP-3 specific antiserum. The earliest time point examined was 9 h after plating. At this time point neurites of cultured cells are strongly HRP-3 positive, whereas no or only little HRP-3 protein could be detected in the nuclear compartment (Fig. 2A). Interestingly, in many neurons HRP-3 staining seems to be polarized and most intense at the area of beginning neuritogenesis (arrows in box of Fig. 2, A and B). After 24 h of culture cells have already developed multiple extensions that all show HRP-3 reactivity (Fig. 2B). Still HRP-3 immunoreactivity in the nuclear compartment is comparatively low (see box from Fig. 2B). After 48 h most cells show both nuclear (arrow in Fig. 2C) as well as neurite staining for HRP-3. In some cells nuclear localization of HRP-3 is already a very pronounced (asterisk in Fig. 2C). This staining pattern is stable in culture for up to 10 days (see Fig. 2, D-F) but in the following days the extranuclear signal declines and after 12 days of culture, however, HRP-3 is exclusively localized in the nucleus (Fig. 2, G and H). Thus, the change of localization of HRP-3 during maturation of the nervous system from embryonic and adult mice, respectively, is also found in neurons maturing in cell culture.
FIGURE 2.
HRP-3 changes its localization during maturation of cultured neurons and is involved in neuritogenesis. Cortical neurons were prepared from E14.5 embryos and cultured for the time periods indicated in the figures. HRP-3 expression was visualized by immunocytochemistry using an HRP-3 specific affinity purified antiserum. Note that HRP-3 expression changes from an initially exclusively extranuclear localization (A) to an intermediate extranuclear and nuclear expression (B-F) to an exclusively nuclear localization in matured neurons (G and H)(blue, 4,6-diamidino-2-phenylindole). Box in H demonstrates the neuronal network established during culturing (visualized by MAP-2 staining in green). During early time points HRP-3 exhibits a polarized expression showing high expression at the base of an outgrowing neurite (arrows in boxes of A and B). I and J, neurons were treated with small interfering RNAs against HRP-3 before seeding. 48 h later treated neurons were analyzed for the length of their longest neurite (I) or the number of processes extending from each cell (J). Cells treated with three different siRNAs against HRP-3 showed a decreased rate of neurite growth as well as less extensions when compared with cells treated with a control (ctrl) siRNA. (Bars in E indicate the mean ± S.E., **, p < 0.01, n = number of cells analyzed.) Persons performing neurite measurements were blinded with respect to the identity of the samples.
These results point to a role of HRP-3 in neurite outgrowth during neuronal development. To examine the role of HRP-3 in neurite outgrowth, before seeding cortical neurons were treated with three different siRNAs targeting different areas of the coding sequence of HRP-3 mRNA. After plating the length of the longest neurite per cell was determined and the number of neurites produced by single neurons was counted. In cells treated with HRP-3 siRNAs the length of the longest neurites were clearly reduced (Fig. 2I). In addition, these cells produced significantly less neurites per cell than cells treated with a control siRNA (Fig. 2J).
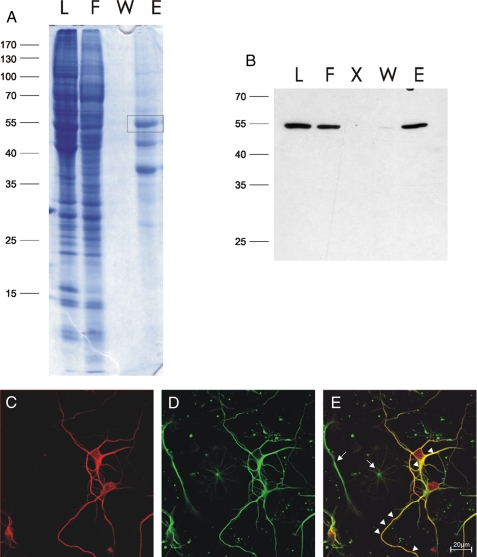
The HATH Domain of HRP-3 Binds to α-Tubulin and Microtubules—To investigate how HRP-3 might be involved in the regulation of neurite outgrowth during mouse embryonic development possible interaction partners were isolated by affinity chromatography. Recombinant His-HRP-3 expressed in bacteria was purified and coupled covalently to a Sepharose resin. Brain lysates from embryonic or newborn mice were passed over this His-HRP-3 affinity resin. Fig. 3A shows a typical elution pattern of this chromatography. After extensive washing the column bound proteins were eluted with 7 m urea, 1 m thiourea. Three main proteins eluted specifically from the HRP-3 column after stringent washing conditions. MALDI-TOF mass spectrometry of a tryptic digest identified one of the eluting proteins as α-tubulin (not shown). Western blots of the eluted fractions using specific antibodies against α-tubulin confirmed the results obtained by mass spectrometry (Fig. 3B). To corroborate a possible interaction between HRP-3 and α-tubulin we performed immunocytochemical analysis using specific antibodies against both proteins and laser scan microscopy. In primary cortical neurons α-tubulin and HRP-3 show a strong overlap in their localization in neuronal somata as well as in neurites (Fig. 3, C-E).
FIGURE 3.
HRP-3 interacts and colocalizes with tubulin in cortical mouse brain and neurons. A, HRP-3 interaction partners were isolated using immobilized bacterially expressed His-HRP-3 coupled to Sepharose beads as an affinity resin. After loading the column with soluble embryonic mouse brain proteins (L) and washing away unbound protein material (W), interacting proteins were eluted by an urea containing elution buffer (E). Lane F indicates the flow through. Eluted proteins were separated by one-dimensional SDS-gel electrophoresis and visualized by Coomassie Brilliant Blue. Possible interaction partners were identified by in-gel digestion with trypsin followed by MALDI-TOF mass spectrometry. The polypeptide boxed in A was identified to be α-tubulin. This was verified by probing all fractions of the affinity column with a mouse monoclonal antibody specific for α-tubulin (B). Costaining of cultured cortical neurons using affinity purified rabbit antibodies against HRP-3 (C, red) and the monoclonal α-tubulin antibody (D, green) demonstrates extensive colocalization of both proteins in neurites as well as somata of these cells (arrowheads in E). Arrows in E point to glial cells positive for tubulin but not stained by the antibody specific for HRP-3. X, free lane.
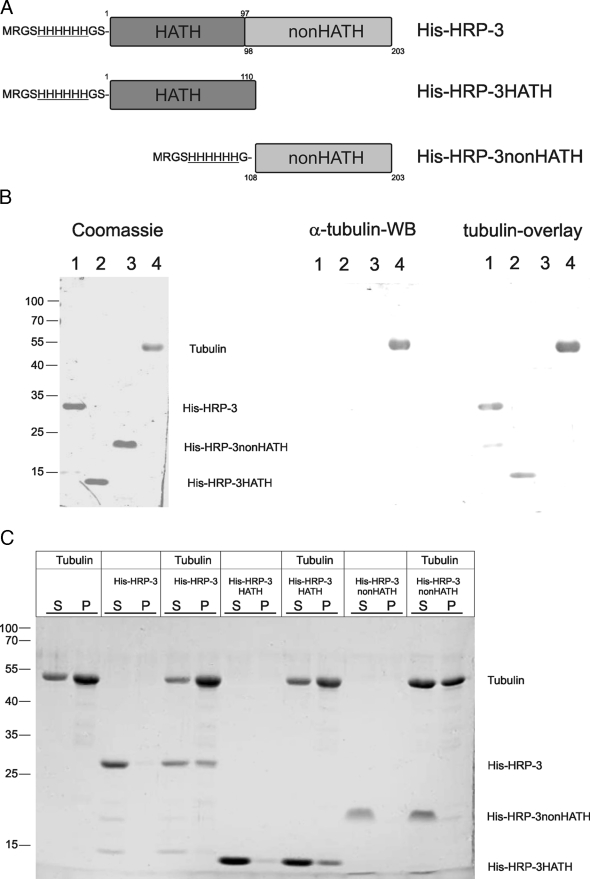
HRP-3 shows a modular structure with the N-terminal domain, called the HATH region, highly conserved among members of the HDGF family and a nonconserved C-terminal part also called non-HATH region. To examine which part is responsible for the interaction with α-tubulin we expressed full-length His-HRP-3 as well as two constructs encompassing only the N-terminal HATH region (His-HRP-3HATH) or the C-terminal non-HATH region of the protein (His-HRP-3non-HATH), respectively (Fig. 4A). These HRP-3 proteins were expressed in bacteria and purified via nickel chelating agarose. All three proteins as well as tubulin from bovine brain were subjected to SDS-PAGE (Fig. 4B, left panel), transferred onto a nitrocellulose membrane, and subjected to Western blot analysis with an α-tubulin specific antibody (Fig. 4B, middle panel). Whereas this antibody readily recognizes α-tubulin there is no cross-reaction with any of the His-HRP-3 proteins. In another Western blot experiment the membrane was incubated with purified tubulin dimers isolated from bovine brain and subsequently proteins on the membrane were cross-linked with formaldehyde. To examine whether tubulin bound to any of the HRP-3 proteins, after cross-linking the membranes were subjected to Western blot analysis with the antibody against α-tubulin (Fig. 4B, right panel). The results demonstrate that tubulin binds to the His-HRP-3 full-length protein as well as to the His-HRP-3HATH N-terminal domain but not to the His-HRP-3non-HATH C-terminal domain. Thus, the binding site of HRP-3 for tubulin is located in the HATH region.
FIGURE 4.
HRP-3 binds tubulin dimers as well as microtubules via its N terminus. A, His-HRP-3 full-length protein (lane 1 in B), its N-terminal domain (His-HRP-3HATH; lane 2 in B), or C-terminal domain (His-HRP-3nonHATH; lane 3 in B) were expressed in bacteria and purified using nickel-agarose via a N-terminal histidine tag. MRSGHHHHHHGS at the N terminus of each recombinant protein indicates the amino acid sequence containing the histidine tag (underlined) and the numbers indicate amino acid residues as they appear in wild type HRP-3. B, left panel, Coomassie Brilliant Blue staining of an SDS-PAGE proves the integrity and purity of the various recombinant HRP-3 proteins. Lane 4 shows tubulin as a control. Numbers on the left side indicate size of standards. B, middle panel, Western blot with an α-tubulin antibody of the Coomassie Brilliant Blue-stained gel shown on the left (α-tubulin-WB). B, right panel, the gel shown on the left was transferred onto nitrocellulose and incubated with tubulin dimers. After cross-linking of proteins with formaldehyde the membrane was subjected to Western blot (WB) analysis with a monoclonal antibody specific for α-tubulin (tubulin-overlay; asterisk labeling a HRP-3 fragment still binding tubulin). C, interaction of HRP-3 with preformed microtubules was analyzed by cosedimentation. His-HRP-3, His-HRP-3HATH, and His-HRP-3nonHATH were incubated with prepolymerized microtubules. Microtubules were collected by centrifugation and equal amounts of the supernatant (S) and the pellet (P) were processed by SDS-PAGE. Proteins were stained with Coomassie Brilliant Blue. A substantial amount of His-HRP-3 and His-HRP-3HATH cosediment with microtubules and therefore locate to the pellet fraction after centrifugation. His-HRP-3non-HATH does not cosediment with microtubules.
The dimeric form of tubulin can polymerize into microtubules. To examine whether HRP-3 interacts not only with tubulin dimers but also with microtubules we performed cosedimentation assays. Purified tubulin from bovine brain was polymerized into microtubules, which were stabilized by the addition of taxol. In contrast to tubulin dimers, microtubules can be sedimented by high speed 100,000 × g centrifugation. When recombinant His-HRP-3 was added to preformed microtubules and centrifuged at 100,000 × g, significant amounts of His-HRP-3 were found in the pellet. In contrast, in the absence of microtubules HRP-3 remained in the supernatant (Fig. 4C). Sedimentation experiments using the His-HRP-3HATH and His-HRP-3non-HATH domains of HRP-3 demonstrate that again the HATH region is responsible for the interaction with microtubules (Fig. 4C).
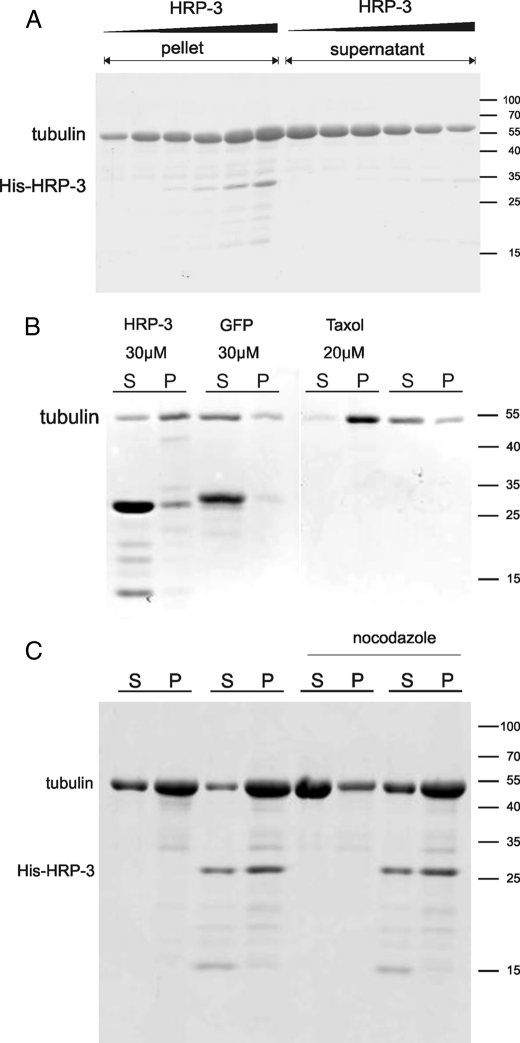
HRP-3 Promotes Tubulin Polymerization, Stabilizes and Bundles Microtubules—A number of proteins binding tubulin dimers influence tubulin dimer assembly into microtubules. To examine whether HRP-3 exhibits such an activity we incubated tubulin (15 μm) with increasing amounts of His-HRP-3 (0.5-20 μm) under conditions suboptimal for polymerization and analyzed the 100,000 × g pellet by SDS-PAGE and Coomassie Brilliant Blue staining (Fig. 5A). Without addition of His-HRP-3 the majority of tubulin remained in the supernatant and only 20% of total tubulin could be detected in the pellet fraction. With increasing concentrations of His-HRP-3 (0.5-20 μm) increasing amounts of tubulin were found in the pellet. Concomitantly the amount of tubulin in the supernatant decreased. A concentration of 20 μm His-HRP-3 caused 87% tubulin to polymerize into microtubules. These data demonstrate that HRP-3 promotes the polymerization of tubulin.
FIGURE 5.
HRP-3 accelerates the assembly of tubulin into polymers and stabilizes microtubules. A, 15 μm tubulin dimers were polymerized at 37 °C for 30 min in reassembly buffer in the presence of increasing amounts of His-HRP-3 (0.5-20 μm). Microtubules were collected by centrifugation and analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. The higher the HRP-3 concentration the more tubulin appears in the pellet indicating that the polymerization of tubulin into microtubules is more efficient. B, after polymerization microtubules were incubated in polymerization buffer alone, containing 20 μm taxol, 30 μm His-HRP-3 or His-GFP as a control protein of similar molecular weight for 30 min at 37 °C and an additional 2 min on ice. After incubation reactions were centrifuged and equal aliquots of the supernatant (S) and the pellet (P) were subjected to SDS-PAGE and Coomassie Brilliant Blue staining. Whereas His-GFP did not enhance the stability of microtubules against cold when compared with buffer alone 30 μm His-HRP-3 reached about 70% of the stabilization value of 20 μm taxol. C, prepolymerized microtubules were incubated with 10 μm HRP-3 for 30 min before adding nocodazole for an additional 10 min. Polymerized tubulin was collected by centrifugation and analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. HRP-3 protects the microtubules against the depolymerizing activity of nocodazole. Numbers on the right side indicate size of standards.
Because HRP-3 binds to microtubules we examined whether it can also stabilize already polymerized microtubules besides its ability to promote tubulin polymerization. Microtubules are unstable when incubated at low temperature. This instability can be overcome by stabilizing agents like taxol or some microtubule-associated proteins. To analyze whether His-HRP-3 has microtubule stabilizing activity the protein was incubated with preformed microtubules and centrifuged at 4 °C. Green fluorescent protein (His-GFP) as a protein with a molecular weight similar to HRP-3 was used as a negative control and taxol as a substance known to stabilize microtubules as a positive control, respectively. In the absence of any addition or in the presence of His-GFP, most tubulin is found in the supernatant as a sign of microtubule depolymerization, whereas in the presence of taxol most tubulin is found in the pellet indicating preservation of microtubules (see Fig. 5B). At 30 μm His-HRP-3 the majority of tubulin remained in the pellet reaching about 70% of the stabilizing activity of 20 μm taxol on pre-polymerized tubulin (Fig. 5B).
To confirm the microtubule stabilizing activity of His-HRP-3 we examined its ability to antagonize the activity of nocodazole, a substance known to destabilize microtubules. For this purpose 20 μm tubulin was polymerized in the presence or absence of 10 μm HRP-3. After polymerization 50 μg/ml nocodazole was added and the mixture incubated for an additional 10 min before centrifugation at 100,000 × g. Fig. 5C demonstrates that the addition of nocodazole causes a significant decrease in the amount of polymerized tubulin found in the pellet fraction. This effect of the drug is nearly completely abolished by the addition of His-HRP-3.
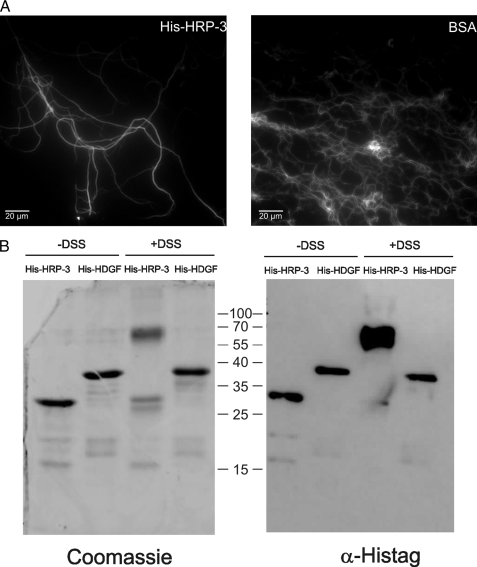
Certain stabilizing proteins also exhibit bundling activity for microtubules. We therefore tested the ability of HRP-3 to bundle by incubating microtubules containing fluorescently labeled tubulin with the protein or bovine serum albumin as a control. After incubation samples were mounted onto glass coverslips and analyzed using a fluorescence microscope. Fig. 6A demonstrates the bundling activity of HRP-3 when compared with incubation with BSA. The conserved HATH region has been shown to mediate dimerization of HDGF. Dimerization has been demonstrated also for a set of microtubule-associated proteins and can be involved in stabilization and bundling of this cytoskeletal component. We therefore tested the capacity of HRP-3 to dimerize via a cross-linking experiment. Therefore recombinant His-HRP-3 and His-HDGF for comparison were expressed in bacteria and purified via their histidine tags. Solutions of 10 μm of both full-length proteins in PBS were incubated for 30 min at room temperature with or without 1 mm disuccinimidyl suberate. After stopping the reaction via the addition of 2 × SDS-PAGE buffer and incubating at 95 °C, proteins were analyzed by Western blot using an antibody against the histidine tag. Whereas full-length HDGF showed no significant dimerization under these conditions HRP-3 migrated almost completely at the molecular weight of the dimer (Fig. 6B). This indicates a much stronger tendency of HRP-3 to dimerize than it is observed for HDGF. As mentioned before HRP-3 dimerization could be the prerequisite for the observed bundling activity of the protein.
FIGURE 6.
HRP-3 dimerizes and bundles microtubules. A, microtubules containing rhodamine-labeled tubulin were incubated with HRP-3 or BSA, mounted onto poly-l-lysine-coated coverslips, and analyzed via fluorescence microscopy. HRP-3 caused substantial amounts of the microtubules to assemble into long bundled fibers (A, left). In contrast no bundling was detected in the presence of BSA under these conditions (A, right). B, histidine-tagged HRP-3 and HDGF were expressed in bacteria and purified by nickel-nitrilotriacetic acid-agarose. Purified proteins were incubated with disuccinimidyl suberate (DSS) as a cross-linker and analyzed via SDS-PAGE and Coomassie Brilliant Blue staining or immunoblot against the N-terminal histidine tag. Whereas HDGF full-length protein did not dimerize under this condition, HRP-3 migrated at the molecular weight of the dimer after cross-linking. Numbers between the panels indicate size of standards.
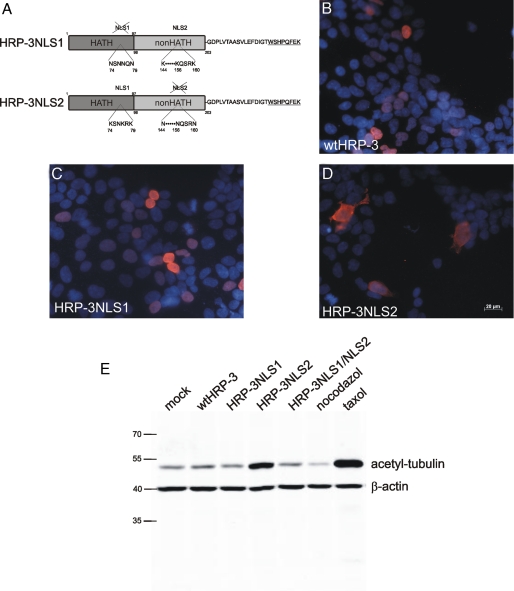
The NLS1 Region of HRP-3 Is Involved in Tubulin Interaction and Stabilization in Vivo—In vivo stabilization of microtubules results in an increased amount of acetylated tubulin. To investigate whether HRP-3 stabilizes microtubules also in vivo HRP-3 was transiently expressed in HEK293 cells and the cellular content of acetylated tubulin was determined by Western blot analysis using an antibody specifically binding acetylated tubulin. Expression of wild type HRP-3 (wtHRP-3) did not increase the amount of acetylated tubulin. It must be kept in mind, however, that in HEK293 cells HRP-3 localizes to the nucleus and has no access to cytoplasmic tubulin. HRP-3 has two putative NLS, one in the HATH region (NLS1) and the other one in the non-HATH region (NLS2), respectively (Fig. 7A). NLS1 and NLS2 were mutated independently (HRP-3NLS1 and HRP-3NLS2) and together (HRP-3NLS1/NLS2) and the respective HRP-3 proteins that were in addition fused to StrepTag at the C terminus (Fig. 7A, underlined) were expressed in HEK293 cells. Cells were immunocytochemically stained with an antibody detecting the StrepTag of the different HRP-3 proteins. Results show that wtHRP-3 and HRP-3NLS1 both localize to the nucleus, whereas HRP-3NLS2 largely localizes to the cytoplasma. It should be noted, however, that for unknown reasons in a considerable fraction of HEK293 cells HRP-3NLS2 still localized to the nucleus (Fig. 7, B-D).
FIGURE 7.
NLS1 mutation decreases the microtubule stabilization activity of HRP-3. A, schematic drawing of HRP-3 constructs used in the experiments shown in B to E, NLS1 and NLS2 indicate the two putative nuclear localization signals. In each case these were abolished by substitution of the lysines within these signals by asparagines. In addition, a StrepTag sequence (underlined) was fused to the C terminus. B-D, different HRP-3 constructs were expressed in HEK293 cells. The protein mutated in the first NLS sequence (HRP-3NLS-1, C) locates as the wild type protein (wtHRP-3, B) to the nuclear compartment of the cells, whereas HRP-3 mutated in the second (HRP-3NLS2, D) remains predominantly extranuclear. E, Western blot of cell lysates of transfected HEK293 cells using an antibody specific for acetylated tubulin. HEK293 cells were transfected with plasmids conferring wtHRP-3, HRP-3NLS-1, HRP-3NLS-2, and HRP-3NLS-1/NLS-2 expression. Actin served as a control protein. Cells treated with nocodazole or taxol were used as negative and positive controls, respectively. Only the overexpression of HRP-3NLS2 led to an increased signal for acetylated tubulin. Numbers on the left side indicate size of standards.
To corroborate that HRP-3 increases the amount of acetylated tubulin in vivo we analyzed protein extracts of cells transiently expressing wtHRP-3 or its NLS mutants by Western blot using an acetyl-tubulin specific antibody. The amount of acetylated tubulin was determined by densitometry using the Odyssey far red scanner and was related to the amount of β-actin in each lane (Fig. 7E). Taxol and nocodazole were used as positive and negative controls, respectively. Compared with mock transfected controls expression of wtHRP-3 and HRP-3NLS-1 did not increase the amount of acetylated tubulin. Expression of the HRP-3NLS-2, which localizes to the cytosol in most of the cells, yielded a substantial increase in the amount of acetylated tubulin. This proves that when present in the cytoplasm HRP-3 stabilizes microtubules also in vivo.
Interestingly, the HRP-3 mutated in both nuclear localization sequences (HRP-3NLS1/NLS2) did not increase the amount of acetylated tubulin despite its localization in the cytoplasm of HEK293 cells. This suggests that the NLS1 mutation localized in the N-terminal HATH region abolishes the microtubule stabilizing effect of HRP-3.
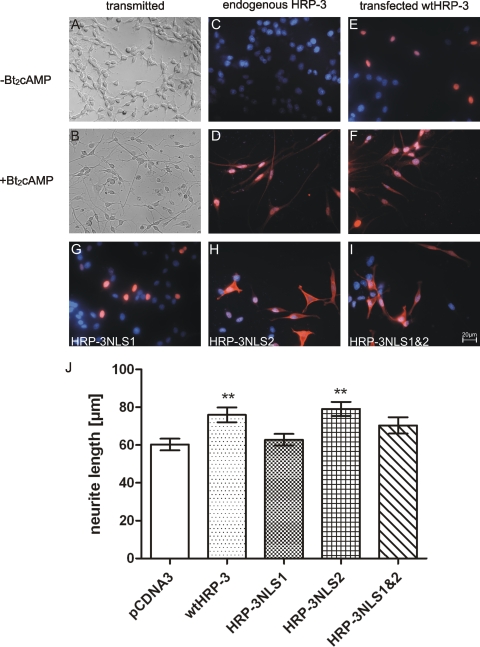
Expression of HRP-3 Lacking NLS1 Fails to Promote Neurite Outgrowth—To analyze the effects of mutations in the HRP-3 nuclear localization signals on neuronal differentiation we used a well transfectable inducible cell culture model for neurite outgrowth, B35 neuroblastoma cells (20). After incubation with dibutyryl cAMP these cells develop neurites but still grow as individual cells in culture (Fig. 8, A and B). Uninduced B35 cells expressed no detectable levels of HRP-3 but 3 days after induction of differentiation endogenous HRP-3 can be detected in the nuclear as well as extranuclear compartment of these cells (Fig. 8, C and D). Interestingly, HRP-3 protein expressed after transfection locates readily to the nuclear compartment before induction of differentiation (Fig. 8E), whereas the protein shows the same distribution as endogenous HRP-3 after treatment with dibutyryl cAMP (Fig. 8F). In addition to wtHRP-3 cells were transfected with plasmids conferring HRP-3NLS1, HRP3NLS2, or HRP-3NLS1/NLS2 expression. Immunocytochemistry performed on those cells show that before induction of differentiation HRP-3NLS1 as wtHRP-3 is predominantly located in the nucleus, whereas HRP-3NLS2 and HRP-3NLS1/NLS2 are mainly found in the cytoplasm. Therefore, as seen for HEK cells also in B35 neuroblastoma cells NLS2 is responsible for the nuclear localization of HRP-3. Twenty-four hours after transfection differentiation of transfected cells was induced by the addition of dibutyryl cAMP. Cells were cultured for another 72 h after which the length of neurites was determined. Neurites in B35 cells expressing the predominantly cytoplasmic HRP-3NLS2 were as long as those of cells expressing wtHRP-3. In contrast, expression of HRP-3 lacking NLS1 did not result in promotion of neurite development although the HRP-3NLS1/NLS2 protein is also largely cytoplasmic. These results support the idea that the impairment of tubulin stabilization caused by mutations in NLS1 results in decreased neurite growth and that the positive effect of HRP-3 on neurite length is due to its interaction with tubulin.
FIGURE 8.
NLS1 mutation abolishes the neurite outgrowth promoting activity of HRP-3. A and B, B35 neuroblastoma cells can be differentiated into a neuronal phenotype using dibutyryl cAMP (Bt2cAMP). C and D, endogenous HRP-3 in B35 cells was detected using an HRP-3 specific antiserum. Undifferentiated B35 cells do not express detectable levels of HRP-3 (C), whereas 3 days after induction with Bt2cAMP endogenous HRP-3 can be detected in both the nuclear as well as the extranuclear compartment (D). E and F, B35 cells were transfected with a DNA construct coding for wild type HRP-3 (wtHRP-3) with a C-terminal StrepTag (see also Fig. 7A). Detection of the protein using an anti-StrepTag antibody demonstrates an exclusively nuclear localization in undifferentiated B35 cells, which after induction of differentiation changes to a distribution similar to what is seen for the endogenous HRP-3 (compare F and D). G-I, cells transfected with DNA constructs coding for its NLS mutants were stained for the C-terminal StrepTag (see also Fig. 7A) to examine their intracellular localization demonstrating that NLS2 is responsible for the nuclear localization of the protein in undifferentiated cells (H and I). J, transfected cells were differentiated by addition of Bt2cAMP 1 day after transfection. Neurite length was measured after unspecific counterstaining of the cells and compared between mock transfected (pCDNA3) cells and cells expressing different HRP-3 protein variants. Cells expressing wtHRP-3 or HRP-3NLS2 extended significantly longer neurites than mock transfected cells. In contrast, cells expressing HRP-3NLS1 do not promote neurite growth in B35 cells. (Bars in J indicate the mean ± S.E., **, p < 0.01; n = 100 cells per sample).
DISCUSSION
So far little is known about biological functions of members of the HRP protein family. HDGF was the first member to be identified and several independent studies have shown that it has proliferative activity either when expressed in cells or when added externally as a recombinant protein. In line with this growth factor activity multiple studies correlate HDGF expression with increased malignancy of different types of tumors. So far, however, it is unclear how this proliferative activity is elicited nor has a receptor for any of the HRPs been identified. Various other functions have been ascribed to HRPs. Thus, HDGF may act as a transcriptional repressor, has been implicated in kidney development, and has neurotrophic activity on cultured neurons. None of these effects is understood on the molecular level. In contrast to other HRPs, which are expressed in various tissues, HRP-3 expression is restricted to neurons and to little extent also to oligodendroglia, suggesting rather specific functions in these cells. Our results suggest that HRP-3 is involved in neurite growth and exerts its effects through interaction with tubulin and microtubules. Thus, modulation of cellular HRP-3 content leads to changes in neuritogenesis. Reduction of HRP-3 levels by siRNA reduces neurite number and neurite length, whereas overexpression of the protein increases the number as well as the length of newly formed neurites. Moreover, HRP-3 is not equally distributed in developing neurons but clusters at the hillock of which the neurites emerge. Finally, the redistribution of HRP-3 from an extranuclear localization during development to a nuclear localization during maturation is highly suggestive for a role of HRP-3 during neuritogenesis. This neuritogenic effect is due to the interaction of HRP-3 with tubulin dimers and assembled microtubules. It promotes tubulin assembly into microtubules and stabilizes microtubules once they have formed. In addition, HRP-3 promotes bundling of microtubules. In particular for the microtubule stabilizing activity as measured by the extent of tubulin acetylation in transfected cells we provide evidence that HRP-3 acts on microtubules in vivo. This activity depends on the extranuclear localization of HRP-3. Furthermore, it is mediated by the N-terminal HATH region. An amino acid substitution within the NLS sequence located in the HATH region reduces the ability of the protein to stabilize microtubules and to support neuritogenesis in an inducible cell culture model. The importance of microtubule stabilization and tubulin acetylation for the differentiation of cortical neurons was also demonstrated by another recent approach (21). Besides the detected interaction with tubulin HRP-3 interacts directly or indirectly with a set of other proteins that have yet to be identified (Fig. 3). Therefore, there might be additional related and unrelated mechanisms on how the protein regulates neuritogenesis. These interactions and possible mechanisms must be one of the topics of HRP-3 research in the future.
Beside its neuritogenic effect HRP-3 may also play a role in the development of polarity. In very young neurons HRP-3 shows a polarized localization to the area of the beginning neuritogenesis. Interestingly cellular polarity and the initiation of neuritogenesis have been reported to be regulated by microtubules (for review, see Refs. 4 and 22). These processes are controlled by a set of microtubule stabilizing and destabilizing proteins. The localization of HRP-3 in young neurons and the fact that siRNA treatment of primary cortical neurons led to a reduced number of neurites formed by these cells point to an involvement of HRP-3 in the polarization processes. Uncovering this possible involvement is another major goal of future investigations on HRP-3.
HRP-3 is the first member of the HDGF protein family that is reported to interact with the cytoskeleton. Beside its homology to other family members in its N-terminal HATH region no substantial homologies of its amino acid sequence to other known MAPs can be found. As MAP2 and tau HRP-3 dimerizes and the HATH region, which binds tubulin, shows basic repeats similar to the region responsible for microtubule association in MAP2 and tau (23, 24). These basic repeats are also responsible for the interaction with glycosaminoglycans that were reported for tau as well as for members of the HDGF family (11, 25, 26). The interaction of tau with sulfated glycosaminoglycans was suggested to be the central event in development of neuropathology in the Alzheimer disease brain (27, 28). Interestingly, reducing the positive charge of such a stretch in the HATH region of HRP-3 (NLS1 region) reduces its positive effect on microtubule stabilization substantially. Leucine-rich acidic nuclear protein is another protein that like HRP-3 shuffles between the nucleus and cytosol and locates predominantly cytoplasmic during neuritogenesis. Interestingly, it was shown to modulate neurite outgrowth by interaction with a microtubule-associated protein (MAP1B) (29). Therefore shuffling to the cytoplasm and modulation of the tubulin cytoskeleton by nuclear proteins seems to be a more frequent mechanism during control of neurite outgrowth.
HRP-3 shows an interesting change of intracellular localization during development. After establishment of the neuronal network most of the protein is redirected into the nuclear compartment. For HDGF it was shown that its nuclear localization is a prerequisite for its proliferative activity. Because neurons are postmitotic nuclear HRP-3 in neurons must have another particular function (16, 30-32).
Other HDGF family members were shown to associate with chromatin pointing to a possible role of this protein family in nuclear transcription processes (33-36). HDGF has been shown to bind to a distinct DNA motif and function as a negative transcriptional regulator for different target genes. The N-terminal PWWP domain of HDGF, which is also found in HRP-3 is responsible for this interaction (36, 37). Transcriptional repression was also reported to be one of the nuclear activities of the leucine-rich acidic nuclear protein. In addition it inhibits histone acetylation by masking histones from histone acetyltransferase (38). Elongator, a protein complex also regulating histone acetylation in the nucleus have recently been reported to play an important role in the differentiation of cortical neurons by shuffling into the cytoplasm and regulating the acetylation of tubulin (21). It can be speculated that non-nuclear HRP-3 contributes to neuritogenesis and after this developmental phase has been completed is located to the nuclear compartment. Here it is possibly involved in the regulation of certain target genes, which may be of importance for maintaining the neuritic structure and connection of matured neurons. Its relocalization could be part of a pathway that signals the neuron the completion of neuritogenesis. This differentiation signal may change the transcriptional activity of various genes after entering the nuclear compartment. In addition, chromatin association has also been reported for tau (39) and processes like cell division and fate are regulated by microtubule-associated proteins in the nucleus, like for example, survivin (40). It will therefore be interesting in the future to isolate specific HRP-3 interaction partners using nuclear extracts from adult brain tissue or highly differentiated neuronal cell cultures. In addition, uncovering the mechanisms regulating the change in intracellular localization should be a main goal of HRP-3 research.
Taken together the properties of HRP-3 presented here make the protein an interesting candidate to be involved in cytoskeletal dynamics during developmental axon growth but also during dynamic reorganization processes of neurites. In addition, modulation of the cytoskeleton might also be an activity mediated by other members of the HDGF family because of the homology of the N-terminal HATH region. In this respect the extranuclear localization of HDGF found in different types of tumors might be interesting (41-43).
Acknowledgments
We thank J. Kappler, Institut für Biochemie und Molekularbiologie, for discussion and carefully reading of the manuscript.
Footnotes
The abbreviations used are: MAP, microtubule-associated protein; HDGF, hepatoma-derived growth factor; HRP, HDGF-related proteins; Bt2cAMP, dibutyryl cyclic AMP; NLS, nuclear localization signal; PBS, phosphate-buffered saline; BSA, bovine serum albumin; wt, wild type; siRNA, small interfering RNA; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; PIPES, 1,4-piperazinediethanesulfonic acid; MOPS, 4-morpholinepropanesulfonic acid; E, embryonic; GFP, green fluorescent protein.
References
- 1.Dent, E. W., and Gertler, F. B. (2003) Neuron 40, 209-227 [DOI] [PubMed] [Google Scholar]
- 2.Ledesma, M. D., and Dotti, C. G. (2003) Int. Rev. Cytol. 227, 183-219 [DOI] [PubMed] [Google Scholar]
- 3.Zhou, F. Q., and Snider, W. D. (2006) Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1575-1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dehmelt, L., and Halpain, S. (2004) J. Neurobiol. 58, 18-33 [DOI] [PubMed] [Google Scholar]
- 5.Avila, J., Dominguez, J., and Diaz-Nido, J. (1994) Int. J. Dev. Biol. 38, 13-25 [PubMed] [Google Scholar]
- 6.Cole, A. R., Knebel, A., Morrice, N. A., Robertson, L. A., Irving, A. J., Connolly, C. N., and Sutherland, C. (2004) J. Biol. Chem. 279, 50176-50180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arimura, N., Menager, C., Fukata, Y., and Kaibuchi, K. (2004) J. Neurobiol. 58, 34-47 [DOI] [PubMed] [Google Scholar]
- 8.Fukata, Y., Itoh, T. J., Kimura, T., Menager, C., Nishimura, T., Shiromizu, T., Watanabe, H., Inagaki, N., Iwamatsu, A., Hotani, H., and Kaibuchi, K. (2002) Nat. Cell Biol. 4, 583-591 [DOI] [PubMed] [Google Scholar]
- 9.Inagaki, N., Chihara, K., Arimura, N., Menager, C., Kawano, Y., Matsuo, N., Nishimura, T., Amano, M., and Kaibuchi, K. (2001) Nat. Neurosci. 4, 781-782 [DOI] [PubMed] [Google Scholar]
- 10.Gu, Y., and Ihara, Y. (2000) J. Biol. Chem. 275, 17917-17920 [DOI] [PubMed] [Google Scholar]
- 11.Dietz, F., Franken, S., Yoshida, K., Nakamura, H., Kappler, J., and Gieselmann, V. (2002) Biochem. J. 366, 491-500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikegame, K., Yamamoto, M., Kishima, Y., Enomoto, H., Yoshida, K., Suemura, M., Kishimoto, T., and Nakamura, H. (1999) Biochem. Biophys. Res. Commun. 266, 81-87 [DOI] [PubMed] [Google Scholar]
- 13.Izumoto, Y., Kuroda, T., Harada, H., Kishimoto, T., and Nakamura, H. (1997) Biochem. Biophys. Res. Commun. 238, 26-32 [DOI] [PubMed] [Google Scholar]
- 14.Marubuchi, S., Okuda, T., Tagawa, K., Enokido, Y., Horiuchi, D., Shimokawa, R., Tamura, T., Qi, M. L., Eishi, Y., Watabe, K., Shibata, M., Nakagawa, M., and Okazawa, H. (2006) J. Neurochem. 99, 70-83 [DOI] [PubMed] [Google Scholar]
- 15.Zhou, Z., Yamamoto, Y., Sugai, F., Yoshida, K., Kishima, Y., Sumi, H., Nakamura, H., and Sakoda, S. (2004) J. Biol. Chem. 279, 27320-27326 [DOI] [PubMed] [Google Scholar]
- 16.Abouzied, M. M., Baader, S. L., Dietz, F., Kappler, J., Gieselmann, V., and Franken, S. (2004) Biochem. J. 378, 169-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Tahir, H. M., Dietz, F., Dringen, R., Schwabe, K., Strenge, K., Kelm, S., Abouzied, M. M., Gieselmann, V., and Franken, S. (2006) BMC Neurosci. 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kremer, L., Dominguez, J. E., and Avila, J. (1988) Anal. Biochem. 175, 91-95 [DOI] [PubMed] [Google Scholar]
- 19.Hasan, M. R., Morishima, D., Tomita, K., Katsuki, M., and Kotani, S. (2005) FEBS J. 272, 822-831 [DOI] [PubMed] [Google Scholar]
- 20.Otey, C. A., Boukhelifa, M., and Maness, P. (2003) Methods Cell Biol. 71, 287-304 [DOI] [PubMed] [Google Scholar]
- 21.Creppe, C., Malinouskaya, L., Volvert, M. L., Gillard, M., Close, P., Malaise, O., Laguesse, S., Cornez, I., Rahmouni, S., Ormenese, S., Belachew, S., Malgrange, B., Chapelle, J. P., Siebenlist, U., Moonen, G., Chariot, A., and Nguyen, L. (2009) Cell 136, 551-564 [DOI] [PubMed] [Google Scholar]
- 22.Siegrist, S. E., and Doe, C. Q. (2007) Genes Dev. 21, 483-496 [DOI] [PubMed] [Google Scholar]
- 23.Sue, S. C., Chen, J. Y., Lee, S. C., Wu, W. G., and Huang, T. H. (2004) J. Mol. Biol. 343, 1365-1377 [DOI] [PubMed] [Google Scholar]
- 24.Sue, S.-C., Lee, W.-T., Tien, S.-C., Lee, S.-C., Yu, J.-G., Wu, W.-J., Wu, W.-g., and Huang, T.-h. (2007) J. Mol. Biol. 367, 456-472 [DOI] [PubMed] [Google Scholar]
- 25.Fatma, N., Singh, D. P., Shinohara, T., and Chylack, L. T., Jr. (2000) Investig. Ophthalmol. Vis. Sci. 41, 2648-2657 [PubMed] [Google Scholar]
- 26.Hasegawa, M., Crowther, R. A., Jakes, R., and Goedert, M. (1997) J. Biol. Chem. 272, 33118-33124 [DOI] [PubMed] [Google Scholar]
- 27.Goedert, M., Jakes, R., Spillantini, M. G., Hasegawa, M., Smith, M. J., and Crowther, R. A. (1996) Nature 383, 550-553 [DOI] [PubMed] [Google Scholar]
- 28.Paudel, H. K., and Li, W. (1999) J. Biol. Chem. 274, 8029-8038 [DOI] [PubMed] [Google Scholar]
- 29.Opal, P., Garcia, J. J., Propst, F., Matilla, A., Orr, H. T., and Zoghbi, H. Y. (2003) J. Biol. Chem. 278, 34691-34699 [DOI] [PubMed] [Google Scholar]
- 30.Everett, A. D., Stoops, T., and McNamara, C. A. (2001) J. Biol. Chem. 276, 37564-37568 [DOI] [PubMed] [Google Scholar]
- 31.Kishima, Y., Yamamoto, H., Izumoto, Y., Yoshida, K., Enomoto, H., Yamamoto, M., Kuroda, T., Ito, H., Yoshizaki, K., and Nakamura, H. (2002) J. Biol. Chem. 277, 10315-10322 [DOI] [PubMed] [Google Scholar]
- 32.Ortega-Paino, E., Fransson, J., Ek, S., and Borrebaeck, C. A. K. (2008) Blood 111, 1617-1624 [DOI] [PubMed] [Google Scholar]
- 33.Llano, M., Vanegas, M., Hutchins, N., Thompson, D., Delgado, S., and Poeschla, E. M. (2006) J. Mol. Biol. 360, 760-773 [DOI] [PubMed] [Google Scholar]
- 34.Lukasik, S. M., Cierpicki, T., Borloz, M., Grembecka, J., Everett, A., and Bushweller, J. H. (2006) Protein Sci. 15, 314-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turlure, F., Maertens, G., Rahman, S., Cherepanov, P., and Engelman, A. (2006) Nucleic Acids Res. 34, 1653-1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, J., and Everett, A. D. (2007) BMC Mol. Biol. 8, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, J., and Everett, A. D. (2009) J. Mol. Biol. 386, 938-950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kutney, S. N., Hong, R., Macfarlan, T., and Chakravarti, D. (2004) J. Biol. Chem. 279, 30850-30855 [DOI] [PubMed] [Google Scholar]
- 39.Hua, Q., He, R. Q., Haque, N., Qu, M. H., del Carmen Alonso, A., Grundke-Iqbal, I., and Iqbal, K. (2003) Cell Mol. Life Sci. 60, 413-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altieri, D. C. (2006) Curr. Opin. Cell Biol. 18, 609-615 [DOI] [PubMed] [Google Scholar]
- 41.Hu, T. H., Huang, C. C., Liu, L. F., Lin, P. R., Liu, S. Y., Chang, H. W., Changchien, C. S., Lee, C. M., Chuang, J. H., and Tai, M. H. (2003) Cancer 98, 1444-1456 [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto, S., Tomita, Y., Hoshida, Y., Takiguchi, S., Fujiwara, Y., Yasuda, T., Doki, Y., Yoshida, K., Aozasa, K., Nakamura, H., and Monden, M. (2006) Clin. Cancer Res. 12, 117-122 [DOI] [PubMed] [Google Scholar]
- 43.Yoshida, K., Tomita, Y., Okuda, Y., Yamamoto, S., Enomoto, H., Uyama, H., Ito, H., Hoshida, Y., Aozasa, K., Nagano, H., Sakon, M., Kawase, I., Monden, M., and Nakamura, H. (2006) Ann. Surg. Oncol. 13, 159-167 [DOI] [PubMed] [Google Scholar]