Abstract
The histone fold is a structural element that facilitates heterodimerization, and histone fold heterodimers play crucial roles in gene regulation. Here, we investigated the nuclear import of two human histone fold pairs, which belong to the H2A/H2B family: CHRAC-15/CHRAC-17 and p12/CHRAC-17. Our results from in vitro nuclear import assays with permeabilized cells and in vivo cotransfection experiments reveal that importin 13 facilitates nuclear import of both histone fold heterodimers. Using glutathione S-transferase pulldown experiments, we provide evidence that heterodimers are required for efficient binding of importin 13 because the monomers alone do not significantly interact. Mutational analysis shows that stepwise substitution of basic amino acid residues conserved among the histone fold subunits leads to a progressive loss of importin 13 binding and nuclear accumulation of CHRAC-15/CHRAC-17 and p12/CHRAC-17. The distribution of basic amino acid residues among the histone fold subunits essential for nuclear uptake suggests that heterodimerization of the histone fold motif-containing proteins forms an importin 13-specific binding platform.
In eukaryotic cells, bidirectional exchange of macromolecules between the cytosolic and nuclear compartment occurs exclusively through the nuclear pore complexes (1, 2). Molecules larger than 40 kDa have to be actively transported via soluble transport receptors that bind specific amino acid residues or sequence elements within the cargo protein. Nuclear export signals are recognized by exportins (export karyopherins), whereas importins (import karyopherins) bind to nuclear localization signals (NLS)3 that can be further differentiated in classical and nonclassical types. Proteins bearing a classical NLS are imported by a heterodimer of importin α and β (3, 4). Proteins with a nonclassical NLS are directly recognized by one import receptor without the help of the importin α adapter molecule. In contrast to classical NLSs, the definition of a putative nonclassical NLS sequence is difficult because its length and the pattern of NLS-specific amino acids can vary strongly among proteins (5). Because of their structural flexibility, transport receptors can bind to very different signals, and some of them even function as import and export receptors. For instance, importin 13 recognizes the export substrate eIF1A and import cargoes such as hUbc9, RBM8 (6), paired type homeodomain transcriptions factors (7), the glucocorticoid receptor (8), and the actin-binding protein myopodin (9). In addition to the monomeric substrates, our recent studies have shown that importin 13 mediates nuclear import of the heterodimeric NF-YB/NF-YC complex of the transcriptional activator NF-Y (10). The subunits exhibit a histone fold motif (11, 12), the structural feature responsible for heterodimerization (13, 14).
In this study, we have analyzed the nuclear import of two related histone fold pairs, CHRAC-15/CHRAC-17 (CHRAC-15/17) and p12/CHRAC-17. The CHRAC-15/17 heterodimer is part of the chromatin accessibility complex (CHRAC) composed of SNF2H (sucrose nonfermenting protein 2 homolog) (15) and the regulatory subunit ACF1 (16-19). In humans, CHRAC represents one of five different SNF2H-containing chromatin remodeling complexes that use ATP to shift the position of nucleosomes on DNA, thus increasing the accessibility of sequence elements on DNA for regulatory proteins (for reviews see Refs. 20-23). In addition, chromatin remodelling factors are also involved in DNA and chromatin replication (24). The CHRAC-15/17 heterodimer presumably acts as an adapter between the remodeling complex and DNA, which results in an enhanced nucleosome sliding activity (17, 25).
The p12/CHRAC-17 heterodimer was identified as an integral component of DNA-polymerase ε (26). Polymerase ε is one of 14 DNA template-directed polymerases in humans responsible for DNA repair and DNA replication, and it is involved in cell cycle control (27, 28). In addition to the p12/CHRAC-17 complex, polymerase ε contains a 59-kDa subunit (p59) and the 261-kDa catalytic subunit (p261) (29).
Here, we show that both histone fold pairs CHRAC-15/17 and p12/CHRAC-17 are transported into the nucleus by importin 13. Importin 13 binds specifically to the dimerized histone fold subunits, whereas the monomeric subunits are neither bound efficiently nor imported. Mutational analysis reveals that basic amino acid residues conserved among the histone fold subunits are necessary for efficient nuclear uptake.
EXPERIMENTAL PROCEDURES
Cell Culture—HeLa P4-R5 MAGI (hereafter referred to as HeLa P4) cells (30) obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (catalog number 3580) were cultured in Dulbecco's modified Eagle's medium (Invitrogen). The medium was supplemented with 10% (v/v) fetal bovine serum (Biochrom), penicillin, streptomycin, and 2 mm glutamine.
Expression Constructs—The coding regions of the respective genes were amplified from plasmid DNA using specific primer pairs with appropriate restriction sites (for details see supplemental “Experimental Procedures”). All of the constructs were verified by DNA sequencing.
Site-directed Mutagenesis—To generate the nucleotide exchanges K47A, K92A, R69A, and R45A in p12; R25A, K70A, K47A, and R23A in CHRAC-15; K100A, R92A, K62A, and K86A in CHRAC-17; K18A, K63A, R40A, and R19A in NC2α and R101A, R102A, K95A, K64A, and K88A in NC2β site-directed mutagenesis was performed according to the QuikChange site-directed mutagenesis kit protocol (Stratagene). (For details see supplemental “Experimental Procedures.”)
Transfection Experiments—Transfection into HeLa P4 cells was performed with the Effectene™ Transfection Reagent (Qiagen) according to the manufacturer's instructions. The cells were fixed 24 h after transfection with 3% paraformaldehyde (w/v) in phosphate-buffered saline and analyzed directly by fluorescence microscopy. Inactivation of exportin 1 in transfected HeLa P4 cells was performed 24 h post-transfection with 10 ng/ml leptomycin B (Sigma-Aldrich) for up to 6 h.
For a semi-quantitative analysis, the cells were scored into three different categories: N>C (more reporter protein in the nucleus), N=C (equal distribution of reporter protein between nucleus and cytoplasm), and N<C (more reporter protein in the cytoplasm). Localization analysis of protein heterodimers was performed by measuring the fluorescence intensity in 20 cells using the software ImageJ (National Institutes of Health) followed by a calculation of the ratio between nuclear and cytoplasmic localization.
Recombinant Protein Expression and Purification—Epitope-tagged CHRAC-15/17 and p12/CHRAC-17 complexes were generated as follows: CHRAC-15 and CHRAC-17 as well as p12 and CHRAC-17 were coexpressed in Escherichia coli BL21 (DE3). The cultures were grown at 37 °C to an optical density of 0.8 at 600 nm. After shifting the temperature to 25 °C, bacterial protein expression was induced with 1 mm isopropyl-β-d-thiogalactopyranoside, and the cultures were grown for 3 h. The collected bacteria were resuspended in buffer A (20 mm Tris-HCl, pH 7.5, 300 mm NaCl, 5 mm MgCl2, and 5 mm β-mercaptoethanol) and lysed by sonication, and the recombinant complexes were purified on nickel nitrilotriacetic acid-agarose (Qiagen). For GST pulldown assays, a second purification step on glutathione-Sepharose 4B (GE Healthcare) followed.
The following proteins were expressed in E. coli BL21 (DE3) as indicated and subsequently purified on glutathione-Sepharose 4B according to the manufacturer's instructions: GST-CHRAC-15 and GST-CHRAC-17 at 25 °C for 3 h with 0.5 mm isopropyl-β-d-thiogalactopyranoside. The collected bacteria were resuspended in buffer B (50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 5 mm MgCl2, and 5 mm β-mercaptoethanol).
The following transport receptors were expressed in E. coli JM109 or TG1 as described in the literature indicated and were purified on nickel nitrilotriacetic acid-agarose (Qiagen) followed by gel filtration on Superdex 200 (GE Healthcare): Xenopus importin α1 (31), human importin β (32), Xenopus importin 7, human importin 5 (33), murine importin 9 (34), and human importin 13 (6). Expression and purification of NTF2 (32, 35), Ran, and RanQ69L (35) was performed as described.
Concentration Analysis of Recombinant Transport Receptors—After expression and purification of the transport receptors, the total protein concentration in the fractions was determined using the Quick Start Bradford Protein Assay Kit 3 (Bio-Rad). The percentage of full-length import receptor in these fractions was calculated using the program GelEval (Frogdance Software), comparing the band intensity of the full-length protein (verified by Western blotting) with the other bands. Based on these percentages, the protein concentration of the representative import receptor was calculated, and appropriate amounts were used for subsequent analysis.
GST Pulldown Assays—GST fusion proteins immobilized on glutathione-Sepharose 4B were used as affinity matrix for binding experiments. Appropriate amounts of affinity matrix were incubated for 3 h at 4 °C with 0.2 μm recombinant purified import receptors in buffer C (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 5 mm MgCl2, and 5 mm β-mercaptoethanol). The binding experiments were performed in the absence or presence of 2 μm RanQ69L(GTP). After washing the matrix-bound proteins were analyzed by SDS-PAGE followed by Coomassie staining.
In Vitro Nuclear Import Assays—Import reactions were performed as described previously (33) based on the method established by Adam et al. (36). Briefly, digitonin-permeabilized HeLa P4 cells were incubated at 37 °C for 30 min with 20 μl of a transport reaction mix consisting of 0.4 μm GST-CHRAC-15/His6-CHRAC-17 complex, either 10 μl of reticulocyte lysate (Promega) or 0.4 μm recombinant import receptor, and an energy-regenerating system (0.5 mm ATP, 0.5 mm GTP, 10 mm creatine phosphate, 50 μg/ml creatine kinase) in buffer D (20 mm HEPES-KOH, pH 7.4, 110 mm potassium acetate, 5 mm magnesium acetate, 0.5 mm EGTA, 2 mm dithiothreitol, 250 mm sucrose). Performing reconstitution experiments with recombinant import receptors a Ran mix (3 μm RanGDP, 0.5 μm NTF2) was added. The GST-CHRAC-15/His6-CHRAC-17 complex was detected by indirect immunofluorescence using an anti-GST polyclonal antibody (Santa Cruz). For quantification of nuclear import, 50-100 fluorescent cells were scored into the following categories: N>C (more reporter protein in the nucleus), N=C (equal distribution of reporter protein between nucleus and cytoplasm), and N<C (more reporter protein in the cytoplasm).
Immunoblotting—HeLa P4 cells were harvested, and the proteins were separated on a SDS-polyacrylamide gel and transferred electrophoretically to nitrocellulose. Uniform blotting was checked by staining the nitrocellulose with Ponceau S (Sigma). The blots were probed with a rabbit polyclonal anti-FLAG antibody (Sigma), polyclonal anti-green fluorescent protein antibody (Santa Cruz), a rabbit polyclonal anti-RFP antibody (MoBiTec), or anti-actin antibody (Sigma). The immunoreactive proteins were visualized using the chemiluminescence ECL plus detection system (Amersham Biosciences) after incubation with a horseradish peroxidase-conjugated goat anti-rabbit antibody (Sigma) at a 1:100,000 dilution.
RESULTS
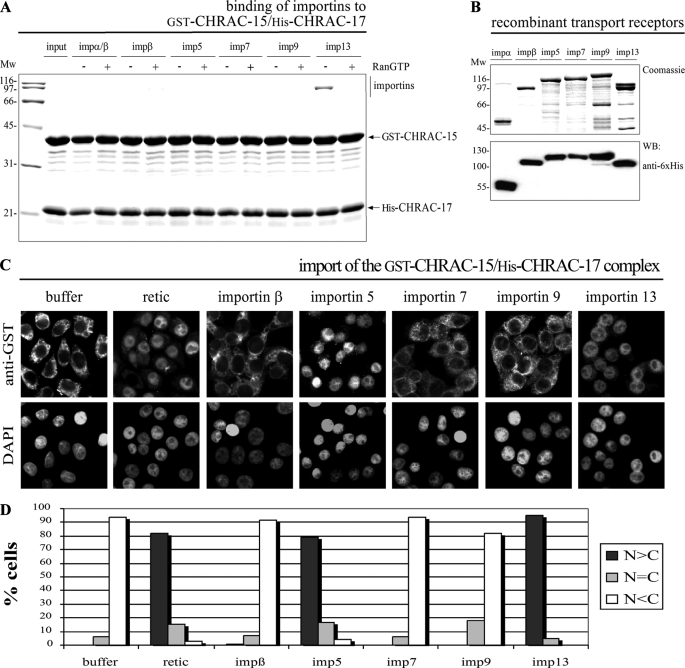
The CHRAC-15/17 Complex Is Recognized and Imported by Importin 13—As part of the human chromatin accessibility complex CHRAC (17), the CHRAC-15/17 complex fulfills its function in the nuclear compartment. This and the close relationship to the histone fold-containing NF-YB/NF-YC heterodimer, which is specifically imported into the nucleus by importin 13 (10), led to the question of whether the CHRAC-15/17 complex or histone fold heterodimers in general are recognized by importin 13. To address this question and to elucidate the potential role of importin 13, we first performed in vitro binding studies. For that purpose, the immobilized GST-CHRAC-15/His-CHRAC-17 complex was incubated with preassembled importin α/β, importin β, importin 5, importin 7, importin 9, and importin 13. Because dissociation of cargo proteins from the import receptors is controlled by RanGTP (37, 38), the binding studies were performed in the absence or presence of RanGTP. Importin 13 bound effectively and RanGTP sensitively to the CHRAC-15/17 complex, whereas binding of the other transport receptors to the heterodimer was not detected (Fig. 1A). To verify that importin 13 also mediates nuclear import of the CHRAC-15/17 complex, we next performed in vitro nuclear import assays (36) with the affinity-purified GST-CHRAC-15/His-CHRAC-17 complex (Fig. 1C). Incubation with reticulocyte lysate led to a strong accumulation of the CHRAC-15/17 complex in the nucleus, whereas in the absence of transport factors a strict cytoplasmic retention was observed. As suggested by the strong RanGTP-sensitive binding of importin 13, the CHRAC-15/17 complex was imported into the nuclei of permeabilized cells by importin 13 (for quantification see Fig. 1D). Surprisingly, importin 5 facilitated nuclear uptake of the heterodimer, although it had not bound to the CHRAC-15/17 dimer in the binding studies. Taken together, the data suggest that importin 13 is a primary nuclear transport receptor for the CHRAC-15/17 heterodimer.
FIGURE 1.
Accumulation of the CHRAC-15/17 heterodimer in the nucleus of permeabilized cells is mediated by importin 5 and importin 13. A, GST-CHRAC-15 and His-CHRAC-17 were coexpressed in E. coli and immobilized on glutathione-Sepharose. The CHRAC-15/17 complex was incubated with equal concentrations (∼0.2 μm) of recombinant purified importin α/β dimer, importin β, importin 5, importin 7, importin 9, or importin 13. Binding was performed in the absence (-) or presence (+) of RanGTP (Q69L mutant) to simulate cytoplasmic and nuclear conditions, respectively. Bound fractions were analyzed by SDS-PAGE followed by Coomassie staining. The CHRAC-15/17 heterodimer binds predominantly to importin 13 in a RanGTP-sensitive manner. Binding of other nuclear transport receptors was not detected. B, the transport receptors importin α, importin β, importin 5, importin 7, importin 9, and importin 13 used for in vitro studies were expressed in E. coli, analyzed by SDS-PAGE, and either stained with Coomassie or analyzed by Western blot using anti-His6 antibody. C, digitonin-permeabilized HeLa P4 cells were incubated with 0.4 μm purified GST-CHRAC-15/His-CHRAC-17 heterodimer, reticulocyte lysate (retic), or the indicated nuclear transport receptors (∼0.4 μm each), a RanGDP/NTF2 mix, and an energy regenerating system for 30 min at 37 °C. For a negative control, reticulocyte lysate was replaced by transport buffer (buffer). The cells were fixed, and the subcellular localization of the CHRAC-15/17 complex was visualized by indirect immunofluorescence (anti-GST). The DNA was counterstained using 4′,6-diamidino-2′-phenylindole. D, the mean distribution of the CHRAC-15/CHRAC-17 heterodimer was quantified for 50 cells/condition in the three categories: N>C, N=C, and N<C. Mw, molecular mass; imp, importin; WB, Western blot; DAPI, 4′,6′-diamino-2-phenylindole.
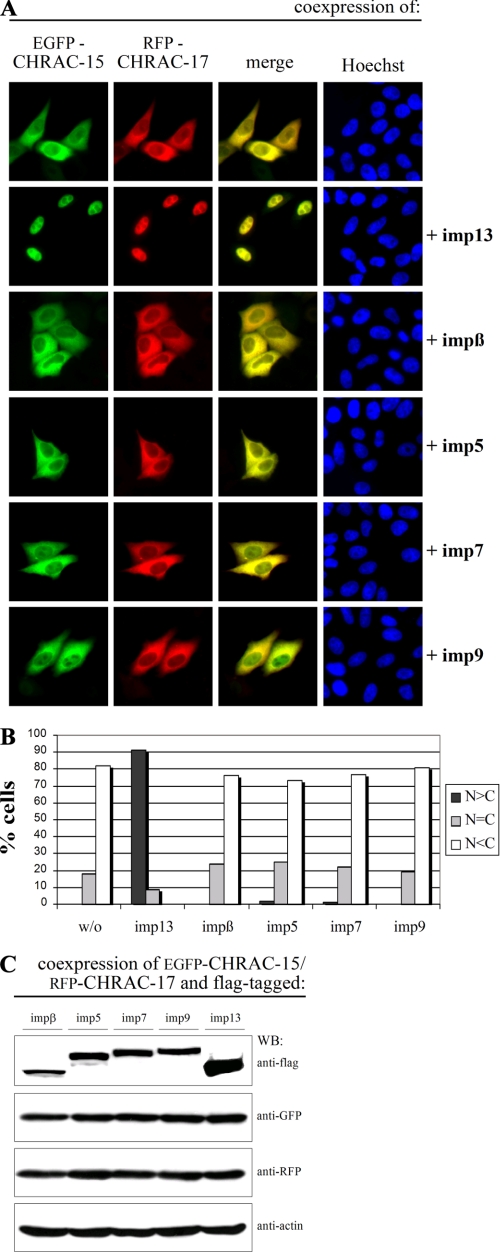
Nuclear Accumulation of Overexpressed CHRAC-15/17 Complex Requires Coexpression of Importin 13—To elucidate nuclear import of the CHRAC-15/17 heterodimer in vivo, we coexpressed EGFP-CHRAC-15 and RFP-CHRAC-17 in HeLa P4 cells. Despite the exclusive nuclear localization of endogenous CHRAC-15 and CHRAC-17 (data not shown) and the strong nuclear accumulation of the GST-CHRAC-15/His-CHRAC-17 complex in the presence of nuclear transport receptors (reticulocyte lysate) in vitro (Fig. 1C), the fluorescently labeled subunits remained exclusively in the cytoplasm of transfected cells (Fig. 2A, top row). The same result was observed when the fluorescent proteins were exchanged among the two CHRAC subunits (supplemental Fig. S1A) or when the position of the fluorescent tags was changed from the N terminus to the C terminus (data not shown). Thus, the position of the two fluorescent tags within the CHRAC complex has presumably no effect on the cytoplasmic retention of the CHRAC-15/17 heterodimer.
FIGURE 2.
Importin 13 mediates the nuclear import of the CHRAC-15/17 heterodimer in vivo. A, HeLa P4 cells were transiently transfected with plasmid DNA encoding EGFP-CHRAC-15, RFP-CHRAC-17, and FLAG-tagged importins as indicated. The subcellular localization of the green EGFP and the red RFP fusion proteins was determined by direct fluorescence 24 h post-transfection. The overlap is shown in yellow (merge). The DNA was counterstained with Hoechst. Coexpression of EGFP-CHRAC-15 and RFP-CHRAC-17 results in a colocalization in the cytoplasm of transfected cells (top row). The additional coexpression of FLAG-importin 13 leads to a nuclear accumulation of the CHRAC-15/17 complex. The additional coexpression of importin β, importin 5, importin 7, or importin 9 in contrast did not change the subcellular distribution of the EGFP-CHRAC-15/RFP-CHRAC-17 complex. B, the mean distribution of the EGFP-CHRAC-15/RFP-CHRAC-17 heterodimer with different coexpressed import factors was scored into the following three categories: N>C, N=C, and N<C. C, plasmid DNA coding for EGFP-CHRAC-15, RFP-CHRAC-17, and FLAG-tagged importin β, importin 5, importin 7, importin 9, or importin 13, respectively, were cotransfected in HeLa P4 cells. 24 h post-transfection the cells were harvested, and the import factors, CHRAC-17, and CHRAC-15 were analyzed by immunoblotting using anti-FLAG, anti-RFP, and anti-green fluorescent protein antibody. Anti-actin antibody was used to control equal loading. imp, importin; WB, Western blot; w/o, without importin.
Because cytoplasmic localization of proteins results not only from the absence of a NLS, but also from nuclear export, we examined a possible role of the export receptor Crm1/exportin 1 (39). For that purpose, HeLa P4 cells cotransfected with plasmid DNA encoding EGFP-CHRAC-15 and RFP-CHRAC-17 or RFP-CHRAC-15 and EGFP-CHRAC-17 were treated with leptomycin B, a specific inhibitor of exportin 1 (40). The cytoplasmic distribution of the fluorescently labeled CHRAC-15/17 complex was not affected by leptomycin B treatment (supplemental Fig. S2A), demonstrating that the CHRAC-15/17 dimer is not exported via exportin 1. Next, we examined whether inaccurate folding of the CHRAC-15/17 complex as a result of the overexpression was responsible for the cytoplasmic retention. For that, we coexpressed the HSP70 chaperone with fluorescently tagged CHRAC-15 and CHRAC-17 as shown in supplemental Fig. S2. The coexpression of HSP70, however, had no effect on the cytoplasmic localization of the CHRAC-15/17 complex (supplemental Fig. S2B). To investigate whether the CHRAC-15/17 dimer is imported in the nucleus via a piggy-back mechanism, the ATP-utilizing chromatin assembly and remodeling factor 1 (ACF1) was coexpressed with CHRAC-15 and CHRAC-17 fused to either EGFP or RFP, respectively. Overexpression of ACF1 did not change the cytoplasmic localization of the CHRAC-15/17 complex (supplemental Fig. S2C). Finally, the influence of additionally coexpressed transport receptors was tested by cotransfection of EGFP-CHRAC-15 and RFP-CHRAC-17 with FLAG-tagged importin 13, importin β, importin 5, importin 7, or importin 9 (Fig. 2A). The coexpression of exogenous importin 13 led to an exclusively nuclear distribution of the CHRAC-15/17 complex (for quantitative analysis see Fig. 2B). In contrast, the coexpression of importin β, importin 5, importin 7, and importin 9, although expressed to a slightly lower degree than importin 13 (Fig. 2C), did not lead to any nuclear accumulation of the CHRAC-15/17 dimer. The same results were obtained when the fluorescent subunits were exchanged among the two CHRAC subunits (supplemental Fig. S1A). In summary, the nuclear transport capacity for the CHRAC-15/17 complex was confirmed for importin 13. In addition, a functional significance for importin 5 in the nuclear import of CHRAC-15/17 was excluded, because importin 5 depletion by small interfering RNA did not affect the endogenous localization of CHRAC-17, and functionality of coexpressed FLAG-tagged importin 5 was verified via nuclear uptake of PGC7/Stella (supplemental Fig. S3), a known importin 5 cargo (41).
Full-length Importin 13 Is Required to Mediate Efficient Nuclear Import of the CHRAC-15/17 Complex—After confirming that importin 13 is the functional import receptor for the CHRAC-15/17 complex, we wanted to characterize the binding sites in importin 13. For that purpose, we generated different deletion constructs of importin 13 and tested their import capacity for the CHRAC-15/17 complex. According to the secondary structure prediction program PSIPRED (42, 43), importin 13 contains 38 α-helices. This is in line with the hypothesis that all transport receptors of the importin β family are comprised of 19 HEAT (huntingtin, elongation factor 3, protein phosphatase 2A, TOR1) repeats (44). Based on this prediction, we selectively deleted parts of importin 13 and coexpressed these truncated constructs with EGFP-CHRAC-15 and RFP-CHRAC-17. Among the different importin 13 deletions, amino acids 1-784 and 45-963 were still capable of facilitating nuclear accumulation of the CHRAC-15/17 complex (supplemental Fig. S4). All other importin 13 fragments did not transport CHRAC-15/17 into the nucleus. Because the importin 13 deletion that contains amino acids 45-963 lacks the putative RanGTP-binding site, which is essential for the dissociation of receptor-cargo complex, the last step of this particular nuclear transport process requires direct competition for either the cargo (CHRAC-15/17) or the receptor (importin 13).
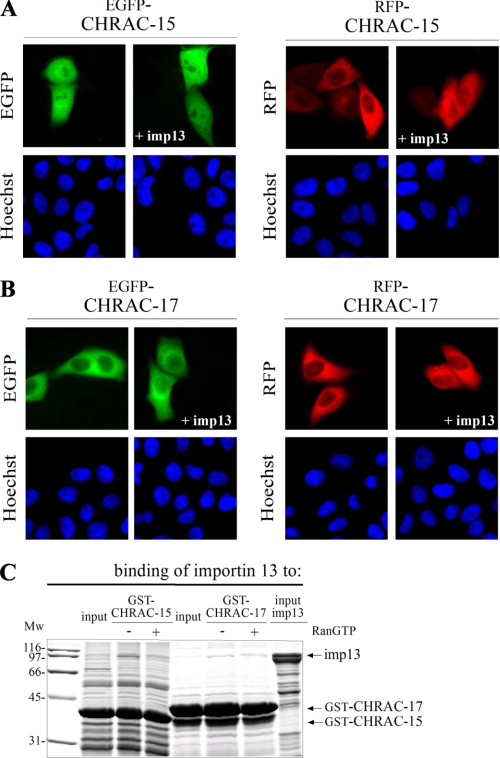
CHRAC-15 and CHRAC-17 Heterodimerize Prior to Nuclear Import, Which Is Not Altered by Subunit Phosphorylation—To analyze the influence of importin 13 on the subcellular localization of the individual CHRAC subunits, FLAG-tagged importin 13 was coexpressed separately with either fluorescently labeled CHRAC-15 or CHRAC-17 (Fig. 3, A and B). Whereas EGFP-CHRAC-15 was distributed homogenously within the cell, RFP-CHRAC-15 and the CHRAC-17 fusion proteins showed a strong cytoplasmic localization. In contrast to the CHRAC-15/17 dimer, the subcellular distribution of the monomeric histone fold subunits did not change upon coexpression of importin 13. Furthermore, nuclear accumulation of the individual subunits was not observed when other nuclear transport receptors were coexpressed (data not shown). Based on these data, it was not surprising that importin 13 bound only weakly to the individual GST-tagged subunits, CHRAC-15 or CHRAC-17 (Fig. 3C). These results demonstrate that the individual CHRAC subunits do not contain a NLS, and nuclear transport of CHRAC-15 and CHRAC-17 depends strictly on heterodimerization in the cytoplasm.
FIGURE 3.
Monomeric CHRAC-15 and CHRAC-17 are not imported by importin 13. The HeLa P4 cells were transiently transfected with plasmid DNA encoding CHRAC-15 (A) and CHRAC-17 (B) N-terminally fused either to EGFP or RFP. FLAG-tagged importin 13 was additionally coexpressed as indicated. The subcellular distribution of the transfected fusion proteins was determined by direct fluorescence 24 h post-transfection. The DNA was counterstained with Hoechst. Although EGFP-CHRAC-15 shows a homogenous distribution in transfected cells at steady state, RFP-CHRAC-15 and fluorescently labeled CHRAC-17 are localized in the cytoplasm. The coexpression of FLAG-importin 13 did not change the subcellular distribution of the fluorescently labeled CHRAC subunits. C, GST-tagged CHRAC-15 and CHRAC-17 were expressed in E. coli, immobilized on glutathione-Sepharose, and incubated with recombinant purified FLAG-tagged importin 13 in the absence (-) or presence (+) of RanGTP (Q69L mutant). The input of importin 13 represents 20% of the amount used in the assay. Bound fractions were analyzed by SDS-PAGE followed by Coomassie staining. Importin 13 does not bind efficiently to the monomeric CHRAC subunits. Mw, molecular mass; imp13, importin 13.
Gnad et al. (45) recently identified three in vivo phosphorylation sites in the sequence of CHRAC-15 (Ser124 and Ser131) and in CHRAC-17 (Ser122) by mass spectrometry-based proteomics. To elucidate the potential role of phosphorylation on the nuclear transport of the CHRAC-15/17 complex, we substituted the three serine residues for glutamate to mimic the effect of phosphorylation. However, substitution of these amino acids did not alter the subcellular distribution of the individual CHRAC-15 and CHRAC-17 subunits nor of the CHRAC-15/17 complex. Coexpression of importin 13 led to a strong nuclear accumulation of the mutated CHRAC-15(S124E,S131E)/CHRAC-17(S122E) complex (data not shown). In contrast to our results, Gnad et al. (45) found both phosphorylated subunits in the cytoplasmatic fraction.
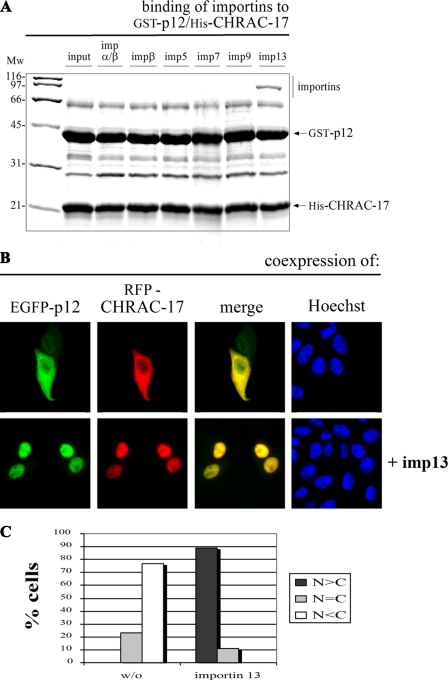
Importin 13 Mediates the Nuclear Import of the p12/CH-RAC-17 Complex—Because the CHRAC-15/17 heterodimer behaves like its histone fold relative NF-YB/NF-YC (10), we asked whether importin 13 also transports other histone fold heterodimers. To address this question, we analyzed the nuclear import of the p12/CHRAC-17 heterodimer of DNA-polymerase ε. In vitro binding experiments with the immobilized GST-p12/His-CHRAC-17 complex showed strong binding of importin 13 (Fig. 4A) comparable with the results observed for the GST-CHRAC-15/His-CHRAC-17 complex (see again Fig. 1A). Besides importin 13, none of the other tested transport receptors bound to the p12/CHRAC-17 heterodimer. Coexpression of EGFP-p12 and RFP-CHRAC-17 in HeLa P4 cells led to a cytoplasmic distribution that upon coexpression of importin 13 became predominantly nuclear (Fig. 4, B and C). The same results were obtained when the fluorescent proteins were exchanged among the two subunits (supplemental Fig. S1B). Coexpression of importin β, importin 5, importin 7, or importin 9 did not influence the cytoplasmic localization of the p12/CHRAC-17 complex (supplemental Fig. S1B). Taken together, these results demonstrate that importin 13 not only facilitates the import of CHRAC-15/CHRAC-17 but is in addition responsible for the nuclear accumulation of the p12/CHRAC-17 complex.
FIGURE 4.
Importin 13 also facilitates the nuclear accumulation of the p12/CHRAC-17 heterodimer. A, GST-p12 and His-CHRAC-17 were coexpressed in E. coli, immobilized on glutathione-Sepharose, and incubated with equal concentrations (0.2 μm) of recombinant purified importin α/β heterodimer, importin β, importin 5, importin 7, importin 9, or importin 13. Bound fractions were analyzed by SDS-PAGE followed by Coomassie staining. The histone fold heterodimer p12/CHRAC-17 binds exclusively to importin 13. B, HeLa P4 cells were transiently cotransfected with plasmid DNA coding for EGFP-p12 and RFP-CHRAC-17 (top row) and additional expression of FLAG-tagged importin 13 (bottom row). The localization of the EGFP-p12 and RFP-CHRAC-17 fusion proteins was determined 24 h post-transfection by direct fluorescence. Colocalization of p12 and CHRAC-17 is shown in yellow (merge). The DNA was counterstained with Hoechst. The largely cytoplasmic distribution of the p12/CHRAC-17 heterodimer was abolished in the presence of FLAG-importin 13, leading to a strong accumulation in the cell nucleus. C, for a semi-quantitative analysis, the mean distribution of the EGFP-p12/RFP-CHRAC-17 heterodimer with or without the additional coexpression of importin 13 was classified in the categories N>C, N=C, and N<C. Mw, molecular mass; imp, importin.
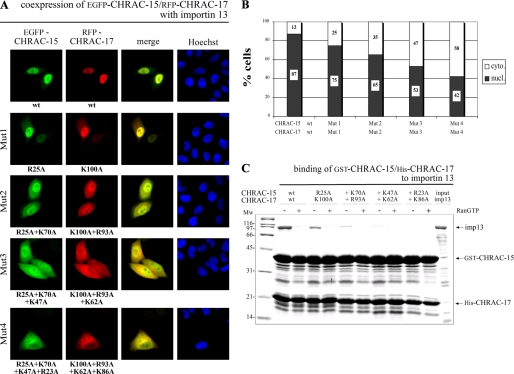
Basic Amino Acid Residues Conserved among the Histone Fold Pairs Are Necessary for Importin 13 Binding—Nonclassical NLSs are usually composed of long heterogeneous stretches of amino acids, making their prediction very difficult (1, 5). To identify sequence elements necessary for nuclear import of the CHRAC-15/17 complex, we looked for sequence similarities among four different histone fold pairs recognized by importin 13, including the NC2α/NC2β heterodimer of the negative cofactor NC2 (46). Because DNA-binding regions often overlap with NLSs (47), we aligned the related histone fold proteins and searched for conserved basic DNA binding residues not involved in subunit heterodimerization (supplemental Fig. S5, A and B). To elucidate the potential role of these residues for importin 13 binding, we gradually introduced point mutations, substituting up to four conserved basic amino acids against alanine (supplemental Fig. S5C). To determine the effect of the mutations on the subcellular distribution of the CHRAC-15/17 complex, we coexpressed mutated EGFP-CHRAC-15 and RFP-CHRAC-17 in the presence of FLAG-tagged importin 13. The progressive substitution of lysine or arginine residues led to an increased cytoplasmic localization of the CHRAC-15/17 dimer as compared with the wild type complex (Fig. 5A). Although single and double mutations in the CHRAC subunits still allowed nuclear uptake by importin 13, the ratio between nuclear and cytoplasmic distribution dropped to ∼50% and below when three or four basic amino acids, respectively, were substituted in each subunit. A quantification of the effect is shown in Fig. 5B. To further analyze the influence of basic amino acid substitution in CHRAC-15/17 on the binding capacity to importin 13, we performed in vitro binding assays with mutated GST-CHRAC-15/His-CHRAC-17 and recombinant importin 13 (Fig. 5C). The strong RanGTP-sensitive binding of importin 13 to the CHRAC-15/17 heterodimer was gradually reduced with increasing numbers of mutations and disappeared completely when four basic amino acids on each subunit were mutated. The necessity of basic amino acids in both subunits of the CHRAC-15/17 complex for binding to importin 13 explains the results from our in vivo transfection experiments (described above). It further led to the conclusion that positively charged amino acids of each histone fold subunit, CHRAC-15 and CHRAC-17, are part of the importin 13-binding platform.
FIGURE 5.
Basic amino acids in the CHRAC-15/17 subunits are necessary for importin 13 binding. A, HeLa P4 cells were transiently cotransfected with plasmid DNA coding for wt EGFP-CHRAC-15 and RFP-CHRAC-17 (top row) and fusion proteins carrying an increasing number of mutated basic amino acid residues as indicated (see also supplemental Fig. S5). FLAG-importin 13 was coexpressed in each approach. The subcellular localization of wild type and mutated CHRAC-15/17 complexes was determined by direct fluorescence 24 h post-transfection. The colocalization of RFP and EGFP fusion proteins is shown in yellow (merge). DNA was counterstained with Hoechst. The stepwise substitution of basic amino acids in CHRAC-15 and CHRAC-17 interferes with the importin 13-mediated nuclear transport and results in an increased cytoplasmic accumulation of the CHRAC-15/17 complex in transfected cells. B, for quantitative analysis, the fluorescence intensity of colocalized wild type and mutated CHRAC-15/17 complexes was measured in 20 cells using the ImageJ software (National Institutes of Health). The percentage of nuclear (nucl.) and cytoplasmic (cyto.) localization was calculated. Sites of mutations are indicated with Mut1, Mut2, Mut3, and Mut4 as in A. C, GST-CHRAC-15/His-CHRAC-17 with alanine substitutions of conserved basic amino acids were coexpressed in E. coli and immobilized on glutathione-Sepharose. Binding studies with the wt and the mutated heterodimers (described above) were performed using recombinant purified importin 13. 20% of importin 13 used in this assay are displayed as input. Each indicated amino acid substitution is additional compared with the preceding column. To imitate the RanGTP gradient between the cytoplasmic and the nuclear compartment, the binding was also performed in the presence (+) of RanGTP (Q69L mutant). After incubation, the bound fractions were displayed by SDS-PAGE and stained by Coomassie. The loss of positively charged amino acids in the CHRAC-15/17 complex results in a reduced binding of importin 13. Mw, molecular mass; imp13, importin 13.
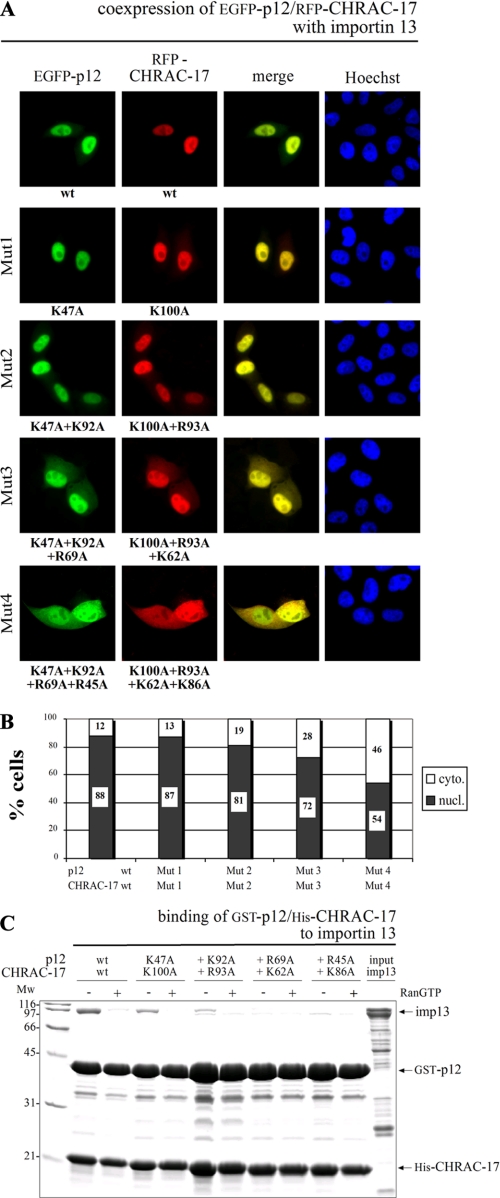
The Importin 13 Binding Platform Is Conserved between Different Histone Fold Heterodimers—To verify the necessity of positively charged amino acids conserved among the histone fold heterodimers for recognition of importin 13, we analyzed the nuclear transport of the p12/CHRAC-17 and NC2α/NC2β complexes deficient in conserved basic amino acids, analogous to previous transfection experiments with CHRAC-15/17 (Fig. 6 and supplemental Fig. S6). Although the substitution results in a less dramatic effect compared with mutated CHRAC-15/17, nuclear transport of p12/CHRAC-17 was reduced strongly if four basic amino acids on each subunit were mutated (Fig. 6A; see Fig. 6B for quantitative analysis). Additionally, we performed in vitro GST pulldown assays with mutated p12/CHRAC-17 complexes (Fig. 6C). The loss of basic amino acids in the p12/CHRAC-17 complex resulted in decreased importin 13 binding, similar to the observations made for the mutated CHRAC-15/CHRAC-17 complex (compare Figs. 6C and 5C). Because CHRAC-17 is part of both heterodimers, one could argue that the effect is caused solely by CHRAC-17. To test this, we performed transfection experiments with wild type RFP-CHRAC-17 in combination with either mutated EGFP-CHRAC-15 or mutated EGFP-p12 (supplemental Fig. S7). The loss of positively charged amino acids in CHRAC-15 and p12 alone led to a reduced nuclear accumulation of the corresponding heterodimers. Thus, the decline of nuclear transport does not exclusively result from mutations in CHRAC-17 but derives from the substitution of basic amino acid residues distributed among both histone fold subunits. In addition, as demonstrated for CHRAC-15/17 and p12/CHRAC-17, in vivo transfection assays with NC2α/NC2β complexes also led to a decreased nuclear uptake when progressively mutated at the predicted sites (supplemental Fig. S6). In conclusion, these data suggest that basic amino acids conserved among the histone fold heterodimers are necessary for binding of importin 13. Because of the necessity of positively charged amino acids, binding between the histone fold heterodimers and importin 13 is presumably mediated by electrostatic interactions.
FIGURE 6.
Basic amino acids conserved among the histone fold family are necessary for the interaction between importin 13 and the p12/CHRAC-17 heterodimer. A, HeLa P4 cells were transiently transfected with plasmid DNA encoding wt or stepwise mutated EGFP-p12 and RFP-CHRAC-17 carrying alanine residues in exchange for basic amino acids as indicated. In addition, plasmid DNA coding for FLAG-importin 13 was coexpressed. Subcellular localization of the p12/CHRAC-17 heterodimer was determined 24 h post-transfection by direct fluorescence. Colocalization is shown in yellow (merge), and the DNA was stained using Hoechst. Increasing the quantity of mutated basic residues in p12 and CHRAC-17 has an influence on the subcellular distribution of the heterodimer resulting in a cytoplasmic accumulation. B, the subcellular distribution of colocalized wt and mutated p12/CHRAC-17 complex was quantitatively analyzed using the program ImageJ (National Institutes of Health). The fluorescence intensity of 20 cotransfected cells was measured, and the ratio between nuclear (nucl.) and cytoplasmic (cyto.) localization was calculated. C, recombinant GST-p12/His-CHRAC-17 wt heterodimer and heterodimers with alanine substitutions of conserved basic amino acids were incubated with recombinant, purified His-importin 13. Binding was determined in the absence (-) or presence (+) of RanGTP (Q69L mutant). 20% of the applied importin 13 is represented as input. Bound fractions were analyzed by SDS-PAGE followed by Coomassie staining. Substitution of basic residues in p12 and CHRAC-17 leads to a decreasing binding capacity of the p12/CHRAC-17 complex to importin 13. Mw, molecular mass; imp13, importin 13.
Taken together, we have shown that nuclear accumulation of CHRAC-15/17 and p12/CHRAC-17 can be mediated by importin 13. Recognition of importin 13 depends on basic amino acid residues conserved among CHRAC-15/17, p12/CHRAC-17, and NC2α/NC2β.
DISCUSSION
In this study, we have analyzed the nuclear import of the heterodimeric CHRAC-15/17 and p12/CHRAC-17 protein complexes. We identified importin 13 as transport receptor that mediates nuclear import of these two histone fold pairs, both in vitro and in vivo. Thus, besides NF-YB/NF-YC (10) and NC2α/NC2β (46), CHRAC-15/17 and p12/CHRAC-17 represent two additional importin 13 cargoes that belong to the H2A/H2B family. Previous binding studies by Mingot et al. (6) using immobilized importin 13 as bait already identified CHRAC-17 (referred to as NF-YB-like) as potential import cargo, pointing toward a functional role of importin 13 for nuclear uptake of either CHRAC-17 alone or in complex. Our data indicate, however, that the individual histone fold subunits are not imported into the nucleus. The results of in vivo and in vitro experiments clearly demonstrate that only the dimerized subunits were efficiently bound and transported into the nucleus by importin 13.
Surprisingly, endogenous importin 13 alone was not able to translocate fluorescently labeled CHRAC-15/17 and p12/CHRAC-17 into the nucleus of transfected HeLa P4 cells. Only the additional coexpression of importin 13 led to the nuclear accumulation of these histone fold complexes. One explanation would be the partial masking of the NLS by the fused RFP and EGFP fluorescent proteins as shown by Wagstaff and Jans (48). However, exchanging the tags between the two histone fold subunits or the position of EGFP and RFP from the N to the C terminus did not alter the localization of the histone fold pairs. Another explanation for the necessity of exogenous importin 13 may be a comparatively low concentration of importin 13 in HeLa cells compared with other import factors. Hence, the HeLa cell content of importin 13 may just not suffice to support the transport of the strongly overexpressed histone fold complexes. As recently shown by Yang and Musser (49), higher concentrations of importin β lead to an increased import efficiency and an increased transit speed of signal-dependent cargoes across the nuclear envelope. Increasing the cellular concentration of importin 13 through additional coexpression most likely results in an advanced nuclear import of the histone fold heterodimers.
To characterize the binding sites in CHRAC-15/17, p12/CHRAC-17, and NC2α/NC2β for importin 13, we analyzed the nuclear import of histone fold pairs in which basic amino acids that are conserved among different histone fold pairs, but not involved in dimerization (50), were gradually mutated. Our results of in vitro binding studies show that these conserved basic residues are essential for efficient binding of importin 13. These results were further confirmed by in vivo cotransfection experiments. Substitution of three conserved positively charged amino acids on each subunit in CHRAC-15/17 and NC2α/NC2β and four on each subunit in p12/CHRAC-17 strongly reduced nuclear import in vivo.
The three-dimensional arrangement of the essential basic residues in CHRAC-15/17 based on the structure of the homologous Drosophila CHRAC-16/14 complex (51) and in NC2α/NC2β (50) shows that all of the basic residues except for Lys70 in CHRAC-15 and Lys63 in NC2α are clustered in close proximity to each other. This observation suggests that positively charged residues in the heterodimerized histone fold subunits form a region of positive electrostatic potential that permits favorable interactions with negatively charged amino acid residues of importin 13. Similarly, based on the three-dimensional structure of the Pax6 homeodomain (52) showing that two basic clusters essential for nuclear uptake of Pax6 localize close to one another, Ploski et al. (7) proposed that the homeodomain has a structural role of exposing the clusters in the proper position and orientation for importin 13 contact.
Sequence comparison showed that the positively charged residues essential for nuclear transport of the complexes studied here are not conserved in the histones H2A and H2B. Interestingly, the nuclear transport receptors that facilitate nuclear import of H2A/H2B and of the three related histone fold pairs differ. Nuclear import of H2A and H2B is mediated via multiple pathways but does not involve importin 13 (34, 53, 54). Thus, the H2A/H2B histone fold pair does not represent a bona fide importin 13-binding site. Additionally, the Ras activator Son of Sevenless, which contains an intramolecular histone fold pseudodimer of the H2A/H2B-type, is a cytoplasmic protein (55).
Regarding the binding mechanism of CHRAC-15/17 to importin 13, we propose the following model: Importin 13 simultaneously contacts both subunits, CHRAC-15 and CHRAC-17. This hypothesis is supported by our data showing that (i) nuclear uptake of histone fold subunits depends strictly on heterodimerization and (ii) the basic amino acids essential for nuclear accumulation are distributed on both histone fold subunits. This type of cooperativity between the individual histone fold subunits for nuclear import was also observed in situ for the nuclear accumulation of the p12 and CHRAC-17 homologs Dpb3 and Dpb4 in fission yeast. In that case, the repression of Dpb3 expression abrogated nuclear localization of Dpb4 (56).
Supplementary Material
Acknowledgments
We thank Patrick Varga-Weisz (The Babraham Institute, Cambridge, United Kingdom) for providing the plasmid DNA for CHRAC-17, CHRAC-15, and p12, the expression plasmid for hACF1, and the polyclonal antibody against human CHRAC-17; Dirk Görlich, José-Manuel Mingot, and Stefan Jäkel (Max-Planck-Institut für Biophysikalische Chemie, Göttingen, Germany) for providing the expression plasmids for the import factors as well as the antibody against importin 5; Ulrike Kutay (Institute for Biochemistry, Eidgenösische Technische Hochschule Zürich, Switzerland) for providing the expression plasmid for importin 9; Volker Haucke and Nadja Jung (Abteilung Membranbiochemie, Freie Universität Berlin, Germany) for providing the plasmid DNA of the pmRFP-N1 vector; and Ralf Dressel (Abteilung Zelluläre und Molekulare Immunologie, Georg-August-Universität Göttingen, Germany) for providing the plasmid DNA for HSP70. We acknowledge Kristine Henningfeld and Erik Meulmeesters for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 523: Protein und Membrantransport zwischen zellulären Kompartimenten).
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and supplemental Figs. S1-S7.
Footnotes
The abbreviations used are: NLS, nuclear localization signal(s); NC2, negative cofactor 2; CHRAC, chromatin accessibility complex; ACF, ATP-dependent chromatin assembly and remodeling factor; GST, glutathione S-transferase; EGFP, enhanced green fluorescent protein; RFP, red fluorescent protein; wt, wild type.
References
- 1.Strom, A. C., and Weis, K. (2001) Genome Biol. 2 REVIEWS3008 [DOI] [PMC free article] [PubMed]
- 2.Fahrenkrog, B., and Aebi, U. (2003) Nat. Rev. Mol. Cell Biol. 4 757-766 [DOI] [PubMed] [Google Scholar]
- 3.Nigg, E. A. (1997) Nature 386 779-787 [DOI] [PubMed] [Google Scholar]
- 4.Gorlich, D., and Mattaj, I. W. (1996) Science 271 1513-1518 [DOI] [PubMed] [Google Scholar]
- 5.Christophe, D., Christophe-Hobertus, C., and Pichon, B. (2000) Cell Signal 12 337-341 [DOI] [PubMed] [Google Scholar]
- 6.Mingot, J. M., Kostka, S., Kraft, R., Hartmann, E., and Gorlich, D. (2001) EMBO J. 20 3685-3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ploski, J. E., Shamsher, M. K., and Radu, A. (2004) Mol. Cell Biol. 24 4824-4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao, T., Lan, J., Presley, J. F., Sweezey, N. B., and Kaplan, F. (2004) Am. J. Respir. Cell Mol. Biol. 30 350-359 [DOI] [PubMed] [Google Scholar]
- 9.Liang, J., Ke, G., You, W., Peng, Z., Lan, J., Kalesse, M., Tartakoff, A. M., Kaplan, F., and Tao, T. (2008) Mol. Cell Biochem. 307 93-100 [DOI] [PubMed] [Google Scholar]
- 10.Kahle, J., Baake, M., Doenecke, D., and Albig, W. (2005) Mol. Cell Biol. 25 5339-5354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arents, G., and Moudrianakis, E. N. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 11170-11174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baxevanis, A. D., Arents, G., Moudrianakis, E. N., and Landsman, D. (1995) Nucleic Acids Res. 23 2685-2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goppelt, A., Stelzer, G., Lottspeich, F., and Meisterernst, M. (1996) EMBO J. 15 3105-3116 [PMC free article] [PubMed] [Google Scholar]
- 14.Maity, S. N., and de Crombrugghe, B. (1998) Trends Biochem. Sci. 23 174-178 [DOI] [PubMed] [Google Scholar]
- 15.Aihara, T., Miyoshi, Y., Koyama, K., Suzuki, M., Takahashi, E., Monden, M., and Nakamura, Y. (1998) Cytogenet Cell Genet. 81 191-193 [DOI] [PubMed] [Google Scholar]
- 16.Varga-Weisz, P. D., Wilm, M., Bonte, E., Dumas, K., Mann, M., and Becker, P. B. (1997) Nature 388 598-602 [DOI] [PubMed] [Google Scholar]
- 17.Poot, R. A., Dellaire, G., Hulsmann, B. B., Grimaldi, M. A., Corona, D. F., Becker, P. B., Bickmore, W. A., and Varga-Weisz, P. D. (2000) EMBO J. 19 3377-3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eberharter, A., Ferrari, S., Langst, G., Straub, T., Imhof, A., Varga-Weisz, P., Wilm, M., and Becker, P. B. (2001) EMBO J. 20 3781-3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito, T., Levenstein, M. E., Fyodorov, D. V., Kutach, A. K., Kobayashi, R., and Kadonaga, J. T. (1999) Genes Dev. 13 1529-1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsukiyama, T. (2002) Nat. Rev. Mol. Cell Biol. 3 422-429 [DOI] [PubMed] [Google Scholar]
- 21.Dirscherl, S. S., and Krebs, J. E. (2004) Biochem. Cell Biol. 82 482-489 [DOI] [PubMed] [Google Scholar]
- 22.Langst, G., and Becker, P. B. (2001) J. Cell Sci. 114 2561-2568 [DOI] [PubMed] [Google Scholar]
- 23.Eberharter, A., and Becker, P. B. (2004) J. Cell Sci. 117 3707-3711 [DOI] [PubMed] [Google Scholar]
- 24.Neves-Costa, A., and Varga-Weisz, P. (2006) Results Probl. Cell Differ. 41 91-107 [DOI] [PubMed] [Google Scholar]
- 25.Kukimoto, I., Elderkin, S., Grimaldi, M., Oelgeschlager, T., and Varga-Weisz, P. D. (2004) Mol. Cell 13 265-277 [DOI] [PubMed] [Google Scholar]
- 26.Li, Y., Pursell, Z. F., and Linn, S. (2000) J. Biol. Chem. 275 23247-23252 [DOI] [PubMed] [Google Scholar]
- 27.Nichols, A. F., and Sancar, A. (1992) Nucleic Acids Res. 20 2441-2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Syvaoja, J., and Linn, S. (1989) J. Biol. Chem. 264 2489-2497 [PubMed] [Google Scholar]
- 29.Li, Y., Asahara, H., Patel, V. S., Zhou, S., and Linn, S. (1997) J. Biol. Chem. 272 32337-32344 [DOI] [PubMed] [Google Scholar]
- 30.Charneau, P., Mirambeau, G., Roux, P., Paulous, S., Buc, H., and Clavel, F. (1994) J. Mol. Biol. 241 651-662 [DOI] [PubMed] [Google Scholar]
- 31.Gorlich, D., Prehn, S., Laskey, R. A., and Hartmann, E. (1994) Cell 79 767-778 [DOI] [PubMed] [Google Scholar]
- 32.Kutay, U., Izaurralde, E., Bischoff, F. R., Mattaj, I. W., and Gorlich, D. (1997) EMBO J. 16 1153-1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakel, S., and Gorlich, D. (1998) EMBO J. 17 4491-4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muhlhausser, P., Muller, E. C., Otto, A., and Kutay, U. (2001) EMBO Rep. 2 690-696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ribbeck, K., Lipowsky, G., Kent, H. M., Stewart, M., and Gorlich, D. (1998) EMBO J. 17 6587-6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adam, S. A., Marr, R. S., and Gerace, L. (1990) J. Cell Biol. 111 807-816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Izaurralde, E., Kutay, U., von Kobbe, C., Mattaj, I. W., and Gorlich, D. (1997) EMBO J. 16 6535-6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorlich, D., Pante, N., Kutay, U., Aebi, U., and Bischoff, F. R. (1996) EMBO J. 15 5584-5594 [PMC free article] [PubMed] [Google Scholar]
- 39.Fornerod, M., Ohno, M., Yoshida, M., and Mattaj, I. W. (1997) Cell 90 1051-1060 [DOI] [PubMed] [Google Scholar]
- 40.Nishi, K., Yoshida, M., Fujiwara, D., Nishikawa, M., Horinouchi, S., and Beppu, T. (1994) J. Biol. Chem. 269 6320-6324 [PubMed] [Google Scholar]
- 41.Nakamura, T., Arai, Y., Umehara, H., Masuhara, M., Kimura, T., Taniguchi, H., Sekimoto, T., Ikawa, M., Yoneda, Y., Okabe, M., Tanaka, S., Shiota, K., and Nakano, T. (2007) Nat. Cell Biol. 9 64-71 [DOI] [PubMed] [Google Scholar]
- 42.Jones, D. T. (1999) J. Mol. Biol. 292 195-202 [DOI] [PubMed] [Google Scholar]
- 43.McGuffin, L. J., Bryson, K., and Jones, D. T. (2000) Bioinformatics 16 404-405 [DOI] [PubMed] [Google Scholar]
- 44.Petosa, C., Schoehn, G., Askjaer, P., Bauer, U., Moulin, M., Steuerwald, U., Soler-Lopez, M., Baudin, F., Mattaj, I. W., and Muller, C. W. (2004) Mol. Cell 16 761-775 [DOI] [PubMed] [Google Scholar]
- 45.Gnad, F., Ren, S., Cox, J., Olsen, J. V., Macek, B., Oroshi, M., and Mann, M. (2007) Genome Biol. 8 R250.1-R250.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kahle, J., Piaia, E., Neimanis, S., Meisterernst, M., and Doenecke, D. (2009) J. Biol. Chem. 284 9382-9393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cokol, M., Nair, R., and Rost, B. (2000) EMBO Rep. 1 411-415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagstaff, K. M., and Jans, D. A. (2006) Anal. Biochem. 348 49-56 [DOI] [PubMed] [Google Scholar]
- 49.Yang, W., and Musser, S. M. (2006) J. Cell Biol. 174 951-961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamada, K., Shu, F., Chen, H., Malik, S., Stelzer, G., Roeder, R. G., Meisterernst, M., and Burley, S. K. (2001) Cell 106 71-81 [DOI] [PubMed] [Google Scholar]
- 51.Hartlepp, K. F., Fernandez-Tornero, C., Eberharter, A., Grune, T., Muller, C. W., and Becker, P. B. (2005) Mol. Cell Biol. 25 9886-9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, D. S., Guenther, B., Desplan, C., and Kuriyan, J. (1995) Cell 82 709-719 [DOI] [PubMed] [Google Scholar]
- 53.Baake, M., Bauerle, M., Doenecke, D., and Albig, W. (2001) Eur. J. Cell Biol. 80 669-677 [DOI] [PubMed] [Google Scholar]
- 54.Mosammaparast, N., Jackson, K. R., Guo, Y., Brame, C. J., Shabanowitz, J., Hunt, D. F., and Pemberton, L. F. (2001) J. Cell Biol. 153 251-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sondermann, H., Soisson, S. M., Bar-Sagi, D., and Kuriyan, J. (2003) Structure 11 1583-1593 [DOI] [PubMed] [Google Scholar]
- 56.Spiga, M. G., and D'Urso, G. (2004) Nucleic Acids Res. 32 4945-4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.