Abstract
The proper regulation of factors involved in mitosis is crucial to ensure normal cell division. Levels and activities of proteins are regulated in many ways, one of which is ubiquitin-mediated protein degradation. E3 ubiquitin ligases are involved in targeting specific substrates for degradation by facilitating their ubiquitination. In seeking to elucidate additional biological roles for Cul3 we performed a two-hybrid screen and identified Ctb9/KLHDC5 as a Cul3-interacting protein. Overexpression of Ctb9/KLHDC5 resulted in an increase in microtubule density as well as persistent microtubule bridges between post-mitotic cells. Conversely, down-regulation of Ctb9/KLHDC5 showed a pronounced reduction in microtubule density. Based on these observations, we examined the interactions between Cul3, Ctb9/KLHDC5, and the microtubule-severing protein, p60/katanin. Here we show that p60/katanin interacts with a complex consisting of Cul3 and Ctb9/KLHDC5, which results in ubiquitin laddering of p60/katanin. Also, Cul3-deficient cells or Ctb9/KLHDC5-deficient cells show an increase in p60/katanin levels, indicating that Cul3/Ctb9/KLHDC5 is required for efficient p60/katanin removal. We demonstrate a novel regulatory mechanism for p60/katanin that occurs at the level of targeted proteolysis to allow normal mitotic progression in mammalian cells.
Due to the irreversible nature of protein degradation, ubiquitin-mediated proteolysis is an effective method to sequentially order cell cycle events. During ubiquitin-mediated protein degradation, the E15 ubiquitin-activating enzyme forms an ATP-dependent thioester bond with a ubiquitin molecule. The activated ubiquitin is subsequently transferred to an E2 ubiquitin-conjugating enzyme, which can either directly attach ubiquitin onto its substrate or act in concert with an E3 ubiquitin ligase to achieve the same end (1, 2). The E3 serves a dual function in recruiting the E2 ubiquitin-conjugating enzyme to the substrate and in positioning the two in close proximity. Although attachment of a single ubiquitin molecule to a lysine side chain can serve as a cellular targeting or localization signal, multiple rounds of ubiquitination result in the formation of poly-ubiquitin chains added to substrate lysines, which can act as a signal to target the substrate for degradation by the 26 S proteasome (3). Cullins are the core of a major class of E3 ligase proteins. One well characterized E3 ligase of the cullin (Cul) family is the SCF complex, which is named for the essential core components Skp1-Cullin1-F-Box. Cul1 binds to the adaptor protein Skp1, which associates with an F-box protein to coordinate substrate recognition (4-10). Many F-box motif-containing proteins have been discovered, which function as adaptors to recruit specific substrates to the SCF complex (11, 12).
Cul3 is a related cullin family member that is involved in a variety of cellular processes. Cul3 is important in cell cycle regulation, targeting cyclin E for ubiquitination and degradation (13). In addition, we have shown that Cul3 activity is vital for the maintenance of quiescence in mammalian liver cells (14). Other substrates of mammalian Cul3 include the antioxidant transcription factor, Nrf2, Aurora B kinase, the GluR6 kainate receptor, Dishevelled, Daxx, RhoBTB2, Topoisomerase 1, and Ci (15-25).
A large family of proteins containing a BTB domain (Bric-abrac, Tramtrak, and Broad-complex), also known as POZ (Pox virus and zinc finger) (26-28) have been shown to interact with Cul3 (15-19, 21, 24, 29-32). Many of these BTB domain-containing proteins contain additional protein-protein interaction motifs, such as MATH domains, kelch (KLH) repeats, or zinc fingers (32). Several of these proteins have been identified as substrate-specific adaptors for Cul3.
Recent work suggests a possible trend in which proteins containing both BTB and kelch domains act as Cul3 adaptor proteins in mammalian systems. The human Keap1 KLH-BTB protein targets the transcription factor Nrf2 as well as PGAM5 for Cul3-mediated degradation, regulating gene expression in response to oxidative stress (16-18, 24, 33, 34). The substrate-specific adaptor KLHL12 binds to Cul3 via its BTB domain and, via kelch repeats, interacts with Dishevelled to negatively regulate the Wnt-B-catenin pathway (15). KLH-BTB 17, or actinfilin, acts as an adaptor to target the glutamate receptor subunit GluR6 for Cul3-based degradation (21). Most recently, Cul3 was shown to associate with KLHL9 and KLHL13 to target the Aurora B kinase for degradation (22). There are 183 known BTB domain-containing proteins in human cells, 49 of which also contain kelch domains (35), suggesting that a vast array of substrates may be targeted for degradation by the interaction of Cul3 with particular BTB proteins.
Regulation of progression through mitosis and cytokinesis is complex. A variety of spindle-associated proteins have been identified that are critical for the organization and function of the mitotic apparatus. Among these proteins are the kinesin family of motor proteins, dynein, NuMa, and γ-tubulin (36, 37). In addition, several proteins form a complex, the chromosome passenger complex, which is involved in coordinating chromosome segregation and cytokinesis (38). These proteins include Aurora B kinase that is found in a complex with survivin, borealin, and INCENP (39). During prophase, these proteins are localized along the length of chromosomes but become more concentrated at the inner centromere regions at the onset of prometaphase, and finally localize to the central spindle during anaphase. Aurora B is crucial in the kinetochore assembly pathway in mammalian cells (40) and is vital to establish and maintain the spindle checkpoint (41). Lastly, there is a window of time lasting approximately 1 h following anaphase during which the cell is competent to undergo cytokinesis, which also requires the APC complex (42, 43).
Studies in mammalian cells have revealed the importance of microtubules in the positioning and ingression of the cytokinesis furrow (44). An important microtubule-remodeling factor is katanin. Katanin is a microtubule-severing protein that consists of a catalytic p60 subunit and a non-catalytic subunit termed p80. p60 is a member of the AAA ATPase super family with microtubule-severing activity, whereas p80 serves to target katanin activity to the microtubules (45). The C-terminal portion of p60 shares homology with other AAA ATPase family members, and the N terminus mediates interaction with microtubules (46). p80 contains a C-terminal region required for dimerization with p60 that enhances activity, a central proline-rich region, and an N-terminal WD40 domain that may serve to negatively regulate p60 activity and to target katanin to the spindle pole (45, 47). It has been suggested that regulation of the ratio of p60 to p80 subunits may be one means of modifying katanin activity (48). Microtubule-associated proteins, such as Tau, protect the microtubule lattice from katanin-mediated severing (49). Phosphorylation of microtubule-associated proteins may promote their release from microtubules, making them more accessible to katanin (49).
An additional katanin, Mei-1, is the catalytic subunit of the katanin-like Mei-1/Mei-2 heterodimer (50). A genetic screen in Caenorhabditis elegans revealed a requirement for Mei-1 in spindle formation during oocyte meiosis. However, Mei-1 activity must cease before mitosis as loss of function mutants are unable to properly form the meiotic spindle, whereas gain of function mutants show Mei-1 persistence through mitosis, resulting in a small, misoriented mitotic spindle (51). Work in C. elegans suggests that microtubule severing by Mei-1 katanin increases the steady-state number of microtubules during both meiosis and mitosis (52) but increases the total polymer mass in meiotic spindles only (53). Genetic mutations in the microtubule-severing protein spastin show it to be responsible for hereditary spastic paraplegia, which is characterized by spasticity and weakness in the lower limbs (54). Spastin is involved in microtubule dynamics, and overexpression of spastin results in disassembly of microtubules, suggesting an underlying mechanism for hereditary spastic paraplegia pathogenesis (55). Recently, three microtubule-severing enzymes, katanin, spastin, and fidgetin, have been shown to contribute to moving chromosomes in Drosophila (56). Although katanin functions at the chromosomes during anaphase to promote microtubule plus end depolymerization, spastin and fidgetin are concentrated at the centrosome, where they contribute to the depolymerization of microtubule minus ends and flux.
The work described here indicates that a Cul3 complex is responsible for ubiquitin-mediated degradation of p60/katanin in human cells. We have identified KLHDC5 as a Cul3 substrate adaptor protein for p60/katanin and show that altered expression of Cul3 or KLHDC5 affects microtubule dynamics. The data presented here represent a novel means by which Cul3 can regulate cell division. Additionally, this work suggests a previously unrecognized mechanism of mitotic regulation, by control of p60/katanin abundance during mitosis in mammalian cells.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Screen—The Cul3 cDNA encoding amino acids 17-768 was subcloned in-frame to the LexA gene into the vector pLexA (Clontech). Yeast cells containing LexA-binding sites in the LEU2 promoter and carrying a plasmid containing a LexA-dependent promoter driving lacZ expression were transformed with the Cul3/LexA plasmid as well as a human testis cDNA library (Clontech). Transformed cells were grown under selection for the plasmids for 3 days and then harvested. The transformed library was titered and plated for two-hybrid interactions the next day. Greater than 2 × 106 clones were screened and >200 candidates were identified. Potential candidates were screened simultaneously for both leucine autotrophy and β-galactosidase activity. Positive clones were subsequently rechecked with an unrelated LexA-lamin bait for specificity. BTB domain-containing proteins identified as positives were further tested for interaction using Cul3ΔExon2 (amino acids 17-22 and 89-768 of Cul3) and Cul3Exon2 (amino acids 23-88 of Cul3) as baits.
Cell Culture, Transfection, and Transfection Plasmids—HeLa, HEK293, and NIH3T3 cells were grown under standard tissue-culture conditions. Transient transfection was accomplished using the standard calcium phosphate procedure (13), Lipofectamine2000 (Invitrogen), or Transmessenger (Qiagen).
Full-length clones of the BTB-containing proteins were obtained by PCR from a human embryonic brain library (Invitrogen). Clones were inserted into the Myc-CS2+ vector in-frame with the Myc tag. One milligram of each Myc-tagged BTB domain-containing plasmid was used for each 60-mm dish transfection. Ctb9/KLHDC5 BTB domain mutants and kelch domain mutants were created by mutagenesis PCR. p60/katanin and p80/katanin were cloned from a HeLa cDNA library (Stratagene) in-frame with the FLAG or Myc tag of the Myc-CS2+ or p3XFLAG-myc-CMV-24 (Sigma) vectors, respectively. 10 μg of each protein was used for transfections in a 60-mm cell culture dish. MG132 was added to cells at a final concentration of 1 μg/ml. ZM447493 (Tocris Bioscience) was used at 2 μm for 18-24 h.
siRNA oligonucleotides for Ctb9/KLHDC5 were designed against the following sequences: 1) AAUAGCCAGGAGAUCUCCGCU and 2) AAUAGCCAGGAGAUCUCCGCU in the Ctb9/KLHDC5 mRNA (Dharmacon Research, Inc.). RNA oligonucleotides for Cul3 siRNA were a “smart pool” from Dharmacon Research, Inc. siControl non-targeting siRNA #3 (Dharmacon) was used as a negative control. RNA oligonucleotides were targeted against the p60 sequence GCUUUGGAAAGAGAUAUAAUU (Thermo Scientific).
Antibodies—The cDNA encoding amino acids 1-287 of hCtb9/KLHDC5 was subcloned into the vector pET28 (Novagen). Protein expression, purification, and affinity-purified antibody production were performed as described (Singer, et al. 13): bacteria transformed with pET28-hCtb9 were induced with isopropyl 1-thio-β-d-galactopyranoside for 2 h and harvested with BugBuster (Novagen), and inclusion bodies were isolated. Inclusion bodies were then boiled in sample buffer and run on a preparative SDS gel. Protein bands were visualized using KCl staining (57) and excised, and Ctb9/KLHDC5 protein was then electroeluted. Purified protein was then injected into rabbits. To affinity-purify Ctb9/KLHDC5 antibodies, rabbit serum was incubated with a strip of polyvinylidene difluoride membrane-containing Ctb9/KLHDC5 protein, and Ctb9/KLHDC5-specific antibodies were eluted off the membrane using low pH glycine. Affinity-purified anti-Ctb9/KLHDC5 was used at a dilution of 1:200 on immunoblots and 1:5 for immunohistochemistry.
The following antibodies were used also used for immunoblots (IBs), immunoprecipitation (IP) and immunocytochemistry (IC): mouse anti-FLAG at 1:5,000 (IB) or 1:600 (IP) (Sigma), mouse anti-Myc at 1:2,000 (IB) or 1:600 (IP and IC) (Santa Cruz Biotechnology), horseradish peroxidase-conjugated donkey anti-rabbit and anti-mouse at 1:10,000 (IB) (Santa Cruz Biotechnology), rabbit anti-Cul3 at 1:200 (IB) or 1:20 for IC (13), mouse anti-α-tubulin at 1:1000 for IC (Sigma), goat anti-rabbit Alexafluor-568 or -488 and goat anti-mouse Alexafluor-568 at 1:500 for IC (Molecular Probes), mouse anti-HA at 1:1,000 (IB) (Covance), mouse anti-PanActin (NeoMarkers) at 1:1,000 (IB), and rabbit anti-p60 at 1:1,000 for IC (58).
Immunoprecipitation and immunoblot analyses were carried out using standard procedures. Transfected cells were lysed in radioimmune precipitation assay buffer with protease inhibitor mixture (Roche Applied Science). Protein A-conjugated Sepharose beads (Amersham Biosciences) were used in all IPs. All washes of beads were in radioimmune precipitation assay, and blots were done in 0.5% TNT (25 mm Tris, pH 7.5, 100 mm NaCl, 0.5% Triton).
Immunocytochemistry and Microscopy—Cells were grown on glass coverslips under standard tissue culture conditions. Cells were then either fixed for 10 min in methanol at -20 °C or for 30 min in 3.7% formaldehyde at room temperature, washed in PBS and then blocked for 1 h in 10% goat serum, 3% bovine serum albumin, 0.3% Triton X-100, 0.5 m NaCl in PBS (blocking buffer). Primary antibody was diluted in blocking buffer and added to fixed cells overnight. Coverslips were washed in PBS followed by addition of fluorescent secondary antibody for 1 h in blocking buffer without goat serum. Cells were counter-stained when appropriate with 20 mg/ml DAPI for 10 min. Coverslips were then mounted on slides with either a polyvinyl alcohol/DABCO mounting solution or Vectashield (Vector) followed by sealing with nail polish.
When using anti-α-tubulin, immunofluorescence was carried out as described (59). Cells were treated in 1% Triton X-100, 2 mm EGTA, 5 mm PIPES for 1 min, fixed in -20 °C methanol for 10 min, rehydrated for 5 min in PBS and then incubated in anti-α-tubulin (Sigma) in 0.1% bovine serum albumin/PBS overnight. Use of secondary antibodies, DAPI staining, and mounting was then carried out as described above. Fluorescence and light microscopy were performed on a Zeiss microscope using either a Photometrics Cool Snap HQ digital camera or a Photometrics Cool Snap CF camera.
For quantification, 100× images were used, and the data were obtained using MetaMorph software (Molecular Devices). A round or oval-shaped area (based on best overall fit to the cells) was drawn and then copied and pasted onto all cells analyzed. Therefore, all cells from each experiment were quantitated with an identical area. The average intensity of the area was determined for the particular color channel of interest by the software. Data were exported to Microsoft Excel for analysis. Statistical analysis was performed using the Student t distribution.
RESULTS
Cul3 Interacts with Proteins Containing a BTB Domain—Because components and organization of the Cul3 complex need further elucidation, we sought to identify proteins that interact with Cul3 by screening a yeast two-hybrid library constructed from human testes cDNA. This assay identified over 200 verified interacting proteins, which we refer to as Cullin three binding proteins, or Ctbs. Among the Ctbs, there were nine BTB domain-containing proteins (Fig. 1A). Two of these proteins also contained additional identifiable domains: Ctb9/KLHDC5 contained kelch domains (60), and Ctb75 contained a MATH (meprin and TRAF homology) domain (61). Seven of these proteins were novel, whereas two have been previously characterized. Ctb75, also known as Mel-26 (C. elegans), SPOP (mammalian), or HIB (Drosophila), has been shown to be a Cul3-interacting protein that mediates ubiquitination of Mei-1 in C. elegans (29, 30). In mammalian cells, Ctb75 (SPOP) has been shown to be a Cul3 substrate adaptor for two chromatin-remodeling proteins, BMI1 and MACROH2A (63), as well as the Daxx transcriptional repressor (19). In Drosophila, Ctb75 (HIB) interacts with Cul3 to mediate degradation of the transcription factor Ci (20). Ctb52 is also known as PDIP and has been shown to bind PCNA. The binding of PDIP to PCNA stimulates DNA synthesis (65, 66).
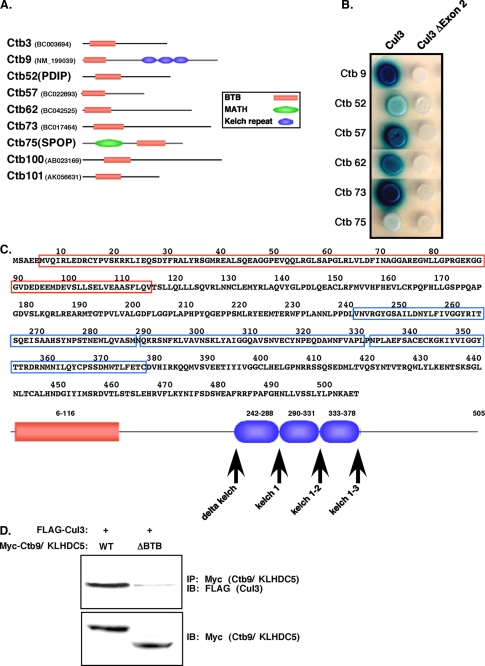
FIGURE 1.
Cul3 interacts with a variety of BTB domain-containing proteins. A, nine proteins containing a BTB domain were cloned from a yeast two-hybrid screen using Cul3 as bait. These Cul3-binding proteins (Ctbs) are depicted schematically with their conserved domains. Accession numbers are shown in parentheses. B, co-expression of several of the Ctbs with full-length Cul3 or Cul3Δexon 2 in yeast. Binding results in blue colonies. C, the amino acid sequence of Ctb9/KLHDC5 is shown, with the BTB domain highlighted in red, and kelch regions in blue. Below, is a schematic representation of Ctb9/KLHDC5, with arrows indicating the position of inserted stop codons. D, immunoprecipitation (IP) of Ctb9/KLHDC5 followed by immunoblot (IB) for Cul3 in HEK293 cells (top panel). Lower panel indicate expression levels of Ctb9/KLHDC5.
To verify the specificity of the two-hybrid interaction, the Cul3 bait plasmid and BTB domain-containing Ctb prey plasmids were reintroduced into yeast cells (Fig. 1B, left, selected Ctb interactions are shown). Binding of Cul3 to the Ctb results in transcription of β-galactosidase, allowing screening of the clones for β-galactosidase activity on X-gal-containing media. Ctb9/KLHDC5, Ctb57, Ctb62, and Ctb73 showed intense β-galactosidase activity (Fig. 1B, blue) indicating strong interactions with Cul3, whereas Ctb52 and Ctb75 showed weaker Cul3 interactions. To test for binding specificity, a Cul3 variant in which exon 2 (residues 23-88) has been deleted was used as a bait plasmid. Exon 2 has been shown to be responsible for interaction with the BTB domain (23, 29-32). This exon is occasionally spliced out and is a naturally occurring cDNA in cells from several adult mouse organs and in cultured cell lines (data not shown). Interestingly, exon 2 is identical in most Cul3 genes across multiple species, suggesting it has a critical role. As expected the Cul3 “bait” plasmid, lacking exon2 (Cul3Δexon2), was unable to interact with the Ctb proteins (Fig. 1B, right). In addition, a control prey plasmid containing Lamin A instead of Cul3 was tested against all the Ctbs and was found to have no β-galactosidase activity with any of the constructs (not shown).
Ctb9/KLHDC5 Contains Additional Protein Interaction Domains—We chose Ctb9/KLHDC5 for further study. We were interested in a more in-depth analysis of this protein for several reasons. We independently identified Ctb9/KLHDC5 as a Cul3-binding protein in a separate assay using a Cul3 tandem affinity purification-tagged construct and proteomics.6 In addition, others and we have identified other BTB/Kelch proteins as Cul3 substrate adaptors (15, 16, 21, 22). The Ctb9/KLHDC5 protein consists of 505 amino acids and, in addition to its BTB domain, contains three kelch repeat protein-protein interaction motifs (Fig. 1C). The BTB domain is located at the N-terminal end of the protein (red), whereas the kelch domains occur near the central region of the protein (blue), spanning amino acids 242-378 (Fig. 1C). Arrows indicate the positions of inserted stop codons in the constructs used in this study. We cloned the open reading frame for Ctb9/KLHDC5 in-frame with a myc tag and overexpressed it with Cul3. Immunoprecipitation of Ctb9/KLHDC5 followed by immunoblotting for Cul3 in human kidney 293 cells (HEK293) cells, confirmed that a physical interaction between Cul3 and Ctb9/KLHDC5 occurs in mammalian cells (Fig. 1D, left lane of the upper panel). Deletion of the BTB domain in Ctb9/KLHDC5 dramatically reduced its interaction with Cul3, verifying that the BTB domain was the site of Cul3 interaction (Fig. 1D, right lane).
Production of Antibodies against Ctb9/KLHDC5—We were interested in determining the in vivo expression patterns of Ctb9/KLHDC5 so we generated antibodies to Ctb9/KLHDC5 to allow analysis of endogenous Ctb9/KLHDC5 protein expression. Rabbit polyclonal antibodies were raised against the amino acids encoded by the first exon of Ctb9/KLHDC5 (amino acids 1-292). Antiserum was obtained from Ctb9/KLHDC5-injected rabbits, and anti-Ctb9/KLHDC5 antibodies were affinity-purified and tested for specificity by immunoblotting. Increasing total amount of protein from untransfected HEK293 cells resulted in the detection of a single band at the molecular weight corresponding to Ctb9/KLHDC5 that increased in intensity with increasing total protein (56.8 kDa, Fig. 2A). To verify the specificity of the purified antibodies, affinity-purified rabbit anti-Ctb9/KLHDC5 was epitope-blocked by prior incubation with purified Ctb9/KLHDC5 protein, which eliminated detection of transfected Ctb9/KLHDC5 by immunoblot (data not shown).
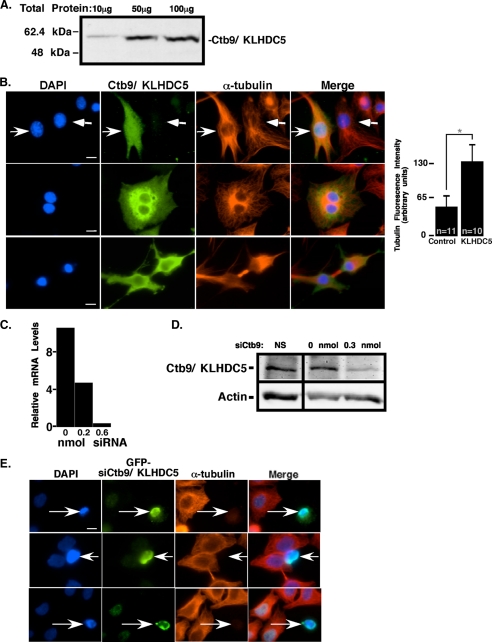
FIGURE 2.
Ctb9/KLHDC5 regulates microtubule dynamics in mammalian cells. A, affinity-purified anti-Ctb9/KLHDC5 antibodies were tested for specificity by immunoblot. The amount of lysate used is indicated above each lane, and protein size in kilodaltons is shown to the left. B, HeLa cells transfected with Myc-tagged Ctb9/KLHDC5 are stained with DAPI (blue), Myc (green), and α-tubulin (red) antibodies and visualized by immunofluorescence. Right facing arrows indicate cells that are overexpressing Ctb9/KLHDC5, and left facing arrows indicate cells that are not. Bar graph shows relative levels of α-tubulin in cells transfected with Ctb9/KLHDC5 versus cells transfected with a control vector. C, quantification of relative Ctb9/KLHDC5 mRNA levels as determined by quantitative real-time RT-PCR with addition of various amounts of siRNA targeted against Ctb9/KLHDC5. NS is a non-targeting siRNA used as a negative control. D, immunoblots for Ctb9/KLHDC5 in untreated HeLa cells compared with those treated with siCtb9. Actin blots below verify equal loading of lysates. E, HeLa cells co-transfected with 0.3 nmol of siCtb9/KLHDC5 and GFP stained with DAPI, GFP (green), and α-tubulin (red). Arrows indicate cells that received siRNA. Two independent siRNAs targeted against Ctb9/KLHDC5 were used. The upper two sets of panels depict cells treated with siRNA (1) and lower panels with siRNA (2). Control siRNA showed no change in tubulin staining (not shown). The size bar indicates 10 μm (*, p < 0.0001).
Over- and Under-expression of Ctb9/KLHDC5 Affects Microtubule Structure—To gain insight into the cellular function of Ctb9/KLHDC5, we transiently transfected Myc-tagged Ctb9/KLHDC5 into the human epithelial cervical carcinoma cell line, HeLa. The microtubule structures of the cells were visualized by immunofluorescence with an antibody against α-tubulin, and an antibody to Myc was used to identify transfected cells (Fig. 2). Cells overexpressing Ctb9/KLHDC5 (Fig. 2B, upper panel, right facing arrows) showed an increase in α-tubulin staining over non-transfected cells (Fig. 2B, upper panel, left facing arrows) (38% of 680 Ctb9/KLHDC5-transfected cells versus 15% of 100 control-transfected cells). This increase in α-tubulin staining was >2-fold when compared with staining in control cells (expressing empty vector) (Fig. 2B, bar graph). In addition 3.4% of cells overexpressing Ctb9/KLHDC5 (of the 680 cells examined) had persistent microtubule bridges between post-mitotic cells, suggesting mitosis was unable to occur normally (Fig. 2B, lower panels). Additionally, Ctb9/KLHDC5 overexpression frequently resulted in cells with multiple nuclei (Fig. 2B, center panels). These data suggest that overexpression of Ctb9/KLHDC5 stabilizes microtubules and may inhibit de-polymerization necessary for normal cell morphology and mitosis.
To examine the consequences of loss of endogenous Ctb9/KLHDC5 function, we reduced its expression by transient transfection of short interfering double-stranded RNA (siRNA) targeted against Ctb9/KLHDC5. To verify the effectiveness of the siRNA, HeLa cells transfected with Myc-tagged Ctb9/KLHDC5 were analyzed using quantitative real-time PCR to examine the impact of the siRNA oligonucleotides at the mRNA level (Fig. 2C). Increasing amounts of siRNA targeted against Ctb9/KLHDC5 effectively reduced transcript levels to a barely detectable level. Transfection with Ctb9/KLHDC5 siRNA also reduced endogenous Ctb9/KLHDC5 protein levels in HeLa cells as shown in Fig. 2D. Because a noticeable reduction in Ctb9/KLHDC5 protein levels was seen with an intermediate level of siRNA (0.3 nmol), this concentration was used for immunofluorescence experiments to avoid any potential deleterious effects due to higher concentrations of siRNA. Co-transfection of siRNA with green fluorescent protein (GFP) or the empty CS2+MT vector (not shown) was used to identify transfected cells for immunofluorescence (Fig. 2E, arrows indicate cells that received siRNA). Visualization of the microtubule structure in HeLa cells transfected with CS2+MT vector and 0.3 nmol of Ctb9/KLHDC5 siRNA revealed a dramatic loss of microtubule structure in 31.4% of 466 cells under expressing Ctb9/KLHDC5 compared with 7.4% of 94 cells treated with CS2+MT vector alone. Two independent siRNA constructs targeted against Ctb9/KLHDC5 were used, each with similar results. The total levels of tubulin protein (monomer) remained unchanged as assayed by immunoblot (data not shown). A non-targeting siRNA used as a negative control showed no change in microtubules (not shown). To confirm these results were not specific to a cancer-derived cell line, normal human embryonic lung fibroblasts were co-transfected with GFP and siRNA targeted against Ctb9/KLHDC5, revealing the same drastic reduction in the microtubule framework (not shown).
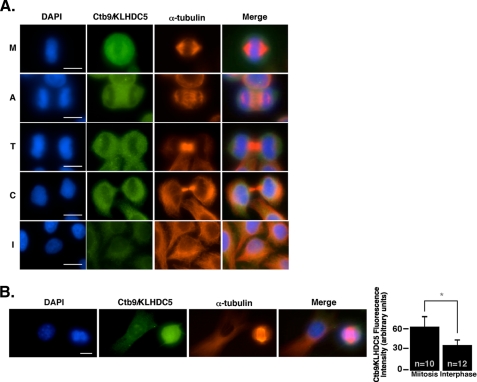
Endogenous Ctb9/KLHDC5 Is Expressed during Mitosis—To gain better insight into its function, we examined expression and localization of endogenous Ctb9/KLHDC5 protein by immunofluorescence. Untransfected HeLa cells synchronized by serum starvation and release were examined at various time points and stained with affinity-purified α-Ctb9/KLHDC5 antibody. This revealed very strong Ctb9/KLHDC5 expression in mitotic cells and diffuse staining in interphase cells (Fig. 3A). During metaphase (M), Ctb9/KLHDC5 is expressed throughout the cell, and is more widely dispersed than the microtubules (as seen in the upper set of panels). In anaphase cells (second set of panels, A), Ctb9/KLHDC5 is localized between the two sets of separated chromosomes as well as at the spindle poles. Finally, during telophase (T), Ctb9/KLHDC5 is again expressed in a circular pattern, surrounding the nuclei of the two daughter cells (middle panels). During cytokinesis (C), Ctb9/KLHDC5 expression appears absent from the midbody region as shown in the fourth set of panels. Interphase cells (I) are depicted in the lower set of panels. To confirm that similar Ctb9/KLHDC5 expression patterns occur in non-tumor-derived cells, we examined Ctb9/KLHDC5 expression in a mouse fibroblastic cell line, NIH3T3. Comparable to what was seen in the human cell line, Ctb9/KLHDC5 is expressed very highly during mitosis in NIH3T3 cells as indicated by immunofluorescence with only diffuse staining in interphase cells (Fig. 3B). Quantification of Ctb9/KLHDC5 fluorescence in mitotic versus interphase cells revealed the levels of Ctb9/KLHDC5 expression were nearly doubled during mitosis.
FIGURE 3.
Ctb9/KLHDC5 is expressed predominantly in mitotic cells. A, untransfected HeLa cells are immunostained with DAPI (blue), anti-Ctb9/KLHDC5 (green), and α-tubulin (red) during various mitotic stages to reveal the expression pattern for endogenous Ctb9/KLHDC5. Each stage of the cell cycle is indicated as follows: M (metaphase), A (anaphase), T (telophase), C (cytokinesis), and I (interphase). B, immunofluorescence showing endogenous Ctb9/KLHDC5 expression in NIH3T3 cells. The size bar indicates 10 μm. Quantification of Ctb9/KLHDC5 immunofluorescence staining intensity in mitotic versus interphase cells where n indicates the number of cells quantified (graph on right) (*, p = 0.0004).
The Microtubule-severing Protein p60/Katanin Interacts with Both Cul3 and Ctb9/KLHDC5 in Mammalian Cells—The observation that Ctb9/KLHDC5 is expressed in mitotic cells and that changing its expression levels dramatically changes microtubule networks, suggests that the normal function of Ctb9/KLHDC5 is to regulate microtubule dynamics throughout mitosis in mammalian cells. Additionally, the data showing that the gain and loss of Ctb9/KLHDC5 function affects microtubule structure in opposing directions led us to hypothesize that Ctb9/KLHDC5 may affect microtubule homeostasis through proteolytic regulation of a microtubule-severing protein by the mechanism of Cul3-mediated ubiquitin-dependent degradation.
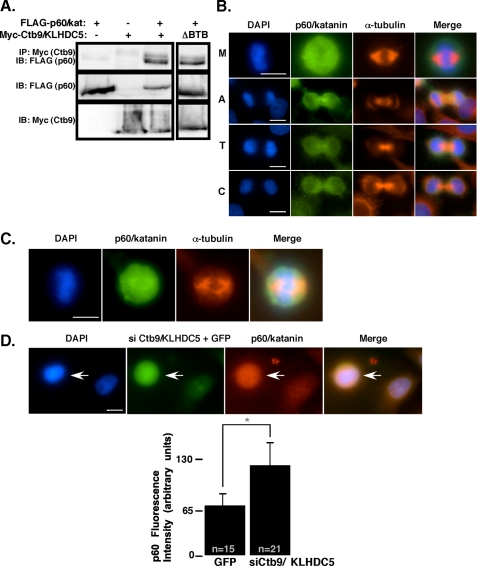
To determine if a microtubule-severing protein was the substrate of the Cul3/Ctb9/KLHDC5 E3 ligase, we cloned the mammalian p60/katanin homolog (KATNA1) and expressed it in HEK293 cells with or without co-expression of Ctb9/KLHDC5 and Cul3. Immunoprecipitation followed by immunoblot revealed binding between p60/katanin and Ctb9/KLHDC5 (Fig. 4A, left panel). This binding was not dependent on the BTB domain in Ctb9/KLHDC5 (Fig. 4A, right panel). Because p60/katanin appeared to be a potential candidate for regulation by the Cul3/Ctb9/KLHDC5 complex, we used an anti-p60 antibody (previously described (58)) to examine endogenous p60/katanin protein expression by immunofluorescence. Expression of endogenous p60/katanin in HeLa cells was detected primarily in mitotic cells, similar to the mitotic expression pattern seen for Ctb9/KLHDC5 (Fig. 4B). During metaphase, p60/katanin was expressed throughout the cell, extending beyond the area of microtubules (upper panel). The major differences seen in expression patterns of Ctb9/KLHDC5 and p60/katanin occur during anaphase and telophase in which the majority of the p60/katanin expression occurs in the midbody region, with some expression at the spindle poles (lower sets of panels). This is in contrast to Ctb9/KLHDC5 expression during telophase in which it is excluded from the midbody region (Fig. 3A). The same p60/katanin expression pattern was seen in human embryonic lung fibroblasts (Fig. 4C) and in NIH3T3 cells (supplemental Fig. S1). To more directly assess the effect of Ctb9/KLHDC5 on in vivo p60/katanin levels, Ctb9/KLHDC5 expression was knocked down with siCtb9/KLHDC5, and immunofluorescence was utilized to compare p60/katanin protein levels in transfected versus untransfected HeLa cells (Fig. 4D). p60/katanin levels were increased 2-fold in cells receiving siRNA targeted against Ctb9/KLHDC5 when compared with untransfected cells or cells transfected with GFP alone, providing strong evidence that Ctb9/KLHDC5 regulates p60/katanin levels in vivo.
FIGURE 4.
p60/katanin is also expressed in mitotic cells and associates with Ctb9/KLHDC5. A, HEK293 cells were transfected with FLAG-tagged p60/katanin, Myc-tagged Ctb9, or both FLAG-p60/katanin and Myc-Ctb9. Immunoprecipitation of Ctb9 (via the myc tag) followed by immunoblot for p60/katanin (FLAG) is shown in the upper panel to indicate protein interactions. On the right, a Ctb9 mutant construct lacking the BTB domain has been co-expressed with p60/katanin. The middle and lower panels show protein expression levels. B, expression of endogenous p60/katanin in untransfected HeLa cells stained with DAPI (blue), anti-p60/katanin (green), and α-tubulin (red). Each stage of the cell cycle is indicated as follows: M (metaphase), A (anaphase), T (telophase), and C (cytokinesis). C, immunofluorescence showing endogenous p60/katanin expression in human embryonic lung fibroblasts stained as in B. D, HeLa cells were co-transfected with siCtb9/KLHDC5 and GFP. The arrow indicates the cell that received siCtb9/KLHDC5. Cells were stained with DAPI, and p60/katanin levels were assessed by immunofluorescence (shown in red). The size bar indicates 10 μm. p60/katanin immunofluorescence levels were quantified for control cells versus siCtb9 treated cells where n represents the number of cells assessed for quantification (*, p < 0.0001).
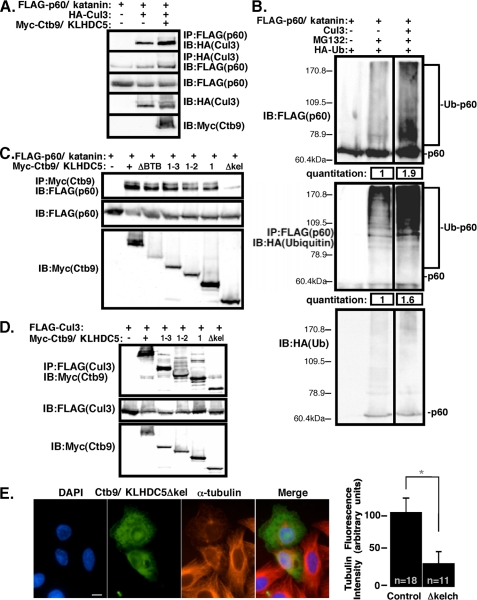
p60/Katanin Is Ubiquitinated by Cul3 in Cells—A suggested model, that Cul3 is the E3 ligase that ubiquitinates p60/katanin, was tested by co-expression in HEK293 cells. To determine whether p60/katanin was present in a complex with Cul3, HEK293 cells were co-transfected with p60/katanin and Cul3, with or without Ctb9/KLHDC5 (Fig. 5A). Immunoprecipitation of p60/katanin via the FLAG tag, followed by Cul3 immunoblot (upper panel) and Cul3 immunoprecipitation followed by immunoblot for p60/katanin (second panel) revealed an interaction between p60/katanin and Cul3 that was enhanced in the presence of Ctb9/KLHDC5. These data reinforce our hypothesis that Ctb9/KLHDC5 serves as a substrate recognition adaptor to recruit p60/katanin to the Cul3 complex. Cells were co-transfected with p60/katanin, HA-tagged ubiquitin, and Cul3, and levels of p60/katanin and ubiquitinated p60/katanin were determined by immunoblotting (Fig. 5B). Addition of the proteasome inhibitor MG132 resulted in ubiquitin laddering of p60/katanin when transfected alone showing that p60/katanin is ubiquitinated in vivo. When both MG132 treatment and Cul3 co-transfection were combined, it resulted in a 1.9-fold increase in overall p60/katanin protein levels (upper blot), and a 1.6-fold increase in ubiquitinated p60/katanin when compared with MG132 alone (lower blot). The upper panel depicts a p60/katanin blot, whereas the lower panel is a p60 (FLAG) IP followed by a ubiquitin (HA) blot to show the high molecular weight forms of p60/katanin are ubiquitinated p60. A breakdown of the quantification of the p60/katanin blot revealed that there is only a 1.2-fold increase in MG132-treated monomer p60/katanin with co-transfection of Cul3, but the higher molecular weight forms of p60/katanin increase by 2.3-fold in the presence of Cul3. These data show that the Cul3-based E3 ligase promotes poly-ubiquitination of p60/katanin and therefore acts as an E3 ligase to mediate p60/katanin ubiquitination-dependent degradation.
FIGURE 5.
Cul3 facilitates ubiquitination of p60/katanin in mammalian cells. A, HEK293 cells co-transfected with FLAG-p60/katanin and HA-Cul3, with or without Myc-Ctb9/KLHDC5. The upper panel shows immunoprecipitation for p60/katanin followed by Cul3 immunoblot. The second panel shows p60/katanin immunoblot following Cul3 immunoprecipitation. The lower three sets of panels indicate protein expression levels. B, upper panel: immunoblot for FLAG-tagged p60/katanin in lysates from HEK293 cells co-transfected with HA-tagged ubiquitin and Cul3. MG132 was added to the cells where indicated above. Lower panel: the same lysates immunoprecipitated for p60 (FLAG) and blotted for HA (ubiquitin) to show the high molecular weight forms are ubiquitinated p60/katanin. Quantification of ubiquitinated p60/katanin levels are shown at the bottom. C, lysates from transfected HEK293 cells were immunoprecipitated with anti-Ctb9/KLHDC5 (Myc) antibody, followed by immunoblot for p60/katanin (FLAG, upper panel). p60/katanin was co-transfected with various Ctb9/KLHDC5 mutants (depicted in Fig. 1) to determine the region of Ctb9/KLHDC5 important for the interaction with p60/katanin. The middle and lower panels show protein expression levels for each of the transfected proteins. D, in HEK293 cells, FLAG-tagged Cul3 was co-transfected with Myc-tagged Ctb9/KLHDC5 kelch domain mutants to determine whether the kelch regions of Ctb9/KLHDC5 are important for its interaction with Cul3. In the upper panel, immunoprecipitation for Cul3 (FLAG) was followed by immunoblot for Ctb9/KLHDC5 (Myc). The middle and lower panels show relative protein expression levels. E, HeLa cells transfected with a Ctb9/KLHDC5 Δkelch mutant construct are immunostained for DAPI (blue), Ctb9/KLHDC5 (green), and α-tubulin (red). The cell size bar indicates 10 μm. Quantification of α-tubulin immunofluorescence levels in control cells versus cells transfected with Ctb9/KLHDC5 Δkelch mutant are shown at the right where n indicates the number of cells quantified (*, p < 0.0001).
Ctb9/KLHDC5 Binds p60/Katanin via Its Kelch Domains—We have observed that immunoprecipitation followed by immunoblotting revealed that, although the BTB domain of Ctb9/KLHDC5 is necessary for interaction with Cul3, it is not needed for interaction with p60/katanin (Figs. 1D and 4A). Therefore, we hypothesized that interaction of Ctb9/KLHDC5 with p60/katanin may be mediated by one of the three kelch repeats in Ctb9/KLHDC5. We constructed four Myc-tagged Ctb9/KLHDC5 mutants containing stop codons as shown in Fig. 1C and examined the interactions between each Ctb9/KLHDC5 mutant and p60/katanin (Fig. 5C) and with Cul3 (Fig. 5D). We observed that, although the kelch domains of Ctb9/KLHDC5 are dispensable for binding with Cul3, at least one of the kelch domains is essential for its interaction with p60/katanin. Overexpression of the non-p60/katanin binding mutant Ctb9Δkelch in HeLa cells resulted in a vast reduction in microtubule density (Fig. 5E). This phenotype is comparable to siRNA knockdown of Ctb9/KLHDC5 and contrary to overexpression of full-length Ctb9/KLHDC5. It appears that the Ctb9Δkelch mutant acts as dominant negative protein, interfering with the function of endogenous Ctb9/KLHDC5.
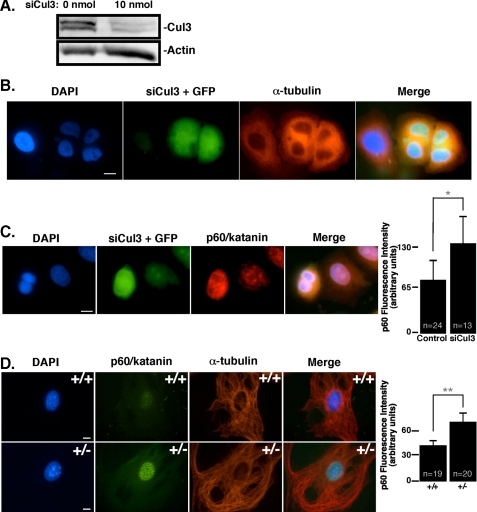
Cul3 Regulates the Abundance of p60/Katanin in Vivo, Which Is Vital for Faithful Progression of Mitosis—To determine if p60/katanin is an in vivo target for the Cul3/KLHDC5 ubiquitin ligase, HeLa cells were transiently transfected with siRNA targeted against Cul3. Immunoblots of lysates utilizing anti-Cul3 serum were used to verify the effectiveness of the Cul3 siRNA (Fig. 6A). Examination of endogenous Cul3 showed a dramatic decrease in Cul3 protein levels when cells were transfected with Cul3 siRNA (Fig. 6A, right lane) compared with untransfected controls (Fig. 6A, left lane). HeLa cells were co-transfected with Cul3 siRNA and GFP to identify transfected cells by immunofluorescence. Fixed cells were stained with DAPI and antibody to α-tubulin to visualize microtubules. HeLa cells under expressing Cul3 showed a dramatic increase in multinucleated cells (Fig. 6B). The presence of multinucleated cells is consistent with previous work (22), which attributed the failure of Cul3-defient cells to complete cytokinesis to the loss of regulation of Aurora B in these cells by the Cul3/KLHL9/KLHL13 complex. Our data also showed an occasional increase in Aurora B levels by immunofluorescence in Cul3-depleted cells (supplemental Fig. S2), but the results were inconsistent and some siRNA-treated cells showed no change in Aurora B expression compared with control cells (not shown).
FIGURE 6.
Reduction of Cul3 expression increases p60/katanin levels. A, HeLa cells were treated with siRNA targeted against Cul3. The upper panel shows endogenous Cul3 protein levels by immunoblot in untreated versus siRNA-treated HeLa cells, whereas the lower panel indicates actin levels. B, immunofluorescence of HeLa cells co-transfected with siCul3 RNA and GFP showing DNA and microtubules as visualized by DAPI and α-tubulin staining, respectively. C, Cul3 siRNA-treated HeLa cells are stained for p60/katanin (red). Expression of GFP indicates cells that received the siRNA. p60/katanin fluorescence levels in control versus siCul3-treated cells are quantified to the right where n equals the number of cells quantified. D, MEFs were stained by immunofluorescence for endogenous p60/katanin (shown in green) to compare expression levels in cells with contrasting levels of Cul3. Cul3+/+ MEFs are shown in the upper panel, whereas Cul3+/- MEFs (Cul3 hypomorphs) are shown below. Microtubules are stained in red, and DAPI staining is in blue. Quantification of p60/katanin immunofluorescence levels is shown to the right. n indicates the number of cells quantified. The size bar depicts 10 μm (*, p = 0.0004; **, p < 0.0001).
We wished to directly determine if p60 protein levels were also affected by loss of Cul3. To do so, cells were treated with Cul3 siRNA and stained for p60/katanin. Cells that received siRNA against Cul3 showed an increase in endogenous p60/katanin staining in interphase cells (Fig. 6C); nearly doubling the p60/katanin fluorescence intensity seen in control GFP-treated cells. These results further underscore the importance of Cul3 in targeting p60/katanin for ubiquitin-mediated degradation. We have previously shown that bona fide Cul3 substrates are more abundant in primary fibroblasts that express low levels of Cul3 (14, 21, 67). To determine if p60/katanin expression levels were also elevated in Cul3 hypomorphic cells, we examined p60/katanin protein levels by immunofluorescence in mouse embryonic fibroblasts (MEFs) (Fig. 6D). As expected, in comparison to wild-type MEFs (upper panel), Cul3+/- MEFs expressed greatly increased levels of p60/katanin (lower panels).
DISCUSSION
We identified nine proteins containing BTB domains that interact with Cul3, making them candidates for Cul3 substrate adaptor proteins. Further analysis of one of the nine proteins, Ctb9/KLHDC5, revealed its importance in the regulation of cellular microtubule structure. Overexpression of Ctb9/KLHDC5 resulted in an increased microtubule density, cytoplasmic bridges between cells, and bi-nucleated cells. In contrast, knockdown of Ctb9/KLHDC5 protein levels by double strand RNA or overexpression of a dominant negative mutant resulted in a dramatic reduction in the cellular microtubule framework. The phenotypes observed are consistent with regulation of the function of katanins or microtubule-severing proteins. Both Ctb9/KLHDC5 and p60/katanin were found to express predominantly during metaphase, anaphase, and telophase of mitotic cells. Although some staining was detectable in interphase cells, the staining was much less intense. During telophase Ctb9/KLHDC5 expression is lost in the midbody region, and p60/katanin expression is enhanced in that region. This observation is consistent with a role for Ctb9/KLHDC5 in the degradation of p60/katanin. In an area where Ctb9/KLHDC5 is gone, p60/katanin levels accumulate. Consistent with that data and providing mechanistic evidence, Ctb9/KLHDC5 forms a complex with p60/katanin, interacting with p60/katanin via one or more of its kelch domains, and with Cul3 through its BTB domain. Co-expression of Cul3 with p60/katanin results in ubiquitin laddering of p60/katanin.
Presumably, overexpression of Ctb9/KLHDC5 causes an increase in microtubule density, because more p60/katanin is targeted for ubiquitination and subsequent degradation than would be in cells expressing endogenous levels of Ctb9/KLHDC5. We hypothesize that, without proper regulation of p60/katanin microtubule-severing activity, cells that are in mitosis are unable to faithfully complete cell division by undergoing the necessary remodeling of their microtubule framework, resulting in the low percentages of cells with cytoplasmic bridges and bi-nucleated cells. Interphase cells that have excess Ctb9/KLHDC5 expression have reduced basal p60/katanin levels, which may prevent normal microtubule dynamics, by the failure of microtubules to be severed and restructured as they normally would. As expected, reduction of Ctb9/KLHDC5 expression by siRNA resulted in a dramatic decrease in the microtubule framework, because more p60/katanin is available for severing microtubules. We feel that the observed phenotypes are due to misregulation of p60/katanin, because we can reverse the phenotype of accumulation of cells containing excess microtubules by co-expression of both Ctb9/KLHDC5 and p60/katanin (supplemental Fig. S3). In addition, down-regulation of p60/katanin using siRNA phenocopies overexpression of Ctb9/KLHDC5 (supplemental Fig. S4).
When Cul3 expression is diminished by siRNA, mammalian cells are unable to complete cytokinesis, and binucleated cells accumulate (Fig. 6B and Ref. 22). This more complex phenotype compared with regulation of Ctb9/KLHDC5 expression is understandable, because Ctb9/KLHDC5 only targets a subset of the Cul3 substrates in cells. Thus, we would not expect the overall cellular phenotype resulting from reduced Cul3 expression to mirror what we see by manipulation of p60/katanin protein levels in cells, because changes in the expression of any of the diverse group of Cul3 substrates could result in a complex phenotype. In contrast, because Ctb9/KLHDC5 is the Cul3 substrate adaptor specific to p60/katanin, we expect changes in its expression to correlate closely with the affects of modifying p60 levels. Interestingly, Ctb9/KLHDC5 associates with other potential microtubule-severing proteins when assayed by overexpression, including human Mei-1, Skd-1, and Katna-27 and thus could serve as a Cul3 complex adaptor for these proteins as well. However, as mentioned above, we feel that the major substrate of the Cul3/Ctb9/KLHDC5 E3 ligase is p60/katanin.
The importance of microtubules for positioning and ingression of the cytokinesis furrow has previously been established (44). Analysis of cytokinesis in mammalian cells revealed that inhibition of the proteasome by the addition of MG132 drastically elongated the amount of time required for exit from cytokinesis (68). The anaphase-promoting complex or cyclosome (APC/C) has long been established as a major regulator of the cell cycle by targeting various cell cycle components such as cyclin A, cyclin B, securin, and CDC20 for ubiquitin-dependent proteasomal degradation (69). Although these observations may be attributable to the ubiquitin ligase activity of the APC/C, it is also feasible that Cul3 activity may be partly responsible for this phenomenon. Other studies as well as the work described here have begun to hint that Cul3 may be emerging as another major player in the ubiquitin-mediated regulation of cell division (14, 22).
Recent experiments have shown that RNA interference targeting Cul3 results in misaligned chromosomes and failure of HeLa cells to complete cytokinesis due to misorganization of the midzone and midbody regions (22). The BTB-domain-containing proteins KLHL9 and KLHL13 were shown to serve as Cul3 adaptors, which together regulate Aurora B localization on mitotic chromosomes. Although the authors attribute the observed mitotic defects to Aurora B activity, this study suggests these changes may be also due to misregulation of microtubule severing by p60/katanin and raises the possibility that multiple Cul3 substrates may prove important for faithful completion of mitosis. Although it is purely speculative, it is also possible that p60/katanin and Aurora B may have a more intimate involvement in addition to being targeted for ubiquitination by the same E3 ligase. Aurora A kinase, an Aurora B-related kinase, was shown to phosphorylate NDEL1 at mitotic entry, after which NDEL1 is rapidly degraded by ubiquitin-dependent proteolysis (62, 64). NDEL1 modulates recruitment of p60/katanin to the centrosome and regulates cytoplasmic dynein function in neurons (62). Treatment of HeLa cells with an Aurora B kinase inhibitor, ZM447493, resulted in an increase in p60/katanin protein levels (supplemental Fig. S5). It is tempting to speculate that Aurora B phosphorylation of p60/katanin may lead to its degradation. This is consistent with our hypothesis in that inhibition of Aurora B kinase activity may allow accumulation of unphosphorylated p60/katanin that is unable to be targeted for ubiquitination.
Reduction of Cul3 expression by the addition of double strand RNA caused an increased number of multinucleated cells, indicating a failure to complete mitosis in a normal fashion. p60/katanin protein levels were greatly elevated in cells having reduced Cul3 expression, further underscoring the importance of Cul3 in katanin regulation. Improper chromosome segregation leads to genetic instability that ultimately may result in tumorigenesis. We have previously shown that Cul3 is crucial for normal cell cycle regulation by targeting cyclin E for ubiquitin-mediated degradation (14). This study suggests an additional role of Cul3 in the regulation of cell cycle progression by its importance in promoting mitosis. Absence of Cul3 function may thus initiate tumorigenesis in a number of ways, both by promoting increased cyclin E protein stability and by contributing to genetic instability.
The data presented here suggest that Ctb9/KLHDC5 is essential for the completion of mitosis by regulating the abundance of the microtubule-severing katanin catalytic p60 subunit. How Cul3-Ctb9/KLHDC5 ubiquitination of p60/katanin is triggered is not known. We have narrowed down the region of binding to the first kelch domain on Ctb9/KLHDC5 but have not determined what amino acids on p60/katanin are essential for this interaction. Although there is some evidence that p60/katanin is phosphorylated, it is not clear if that is a trigger for recognition by the Cul3-based E3 ligase. It is intriguing to note that, when p60/katanin is immunoprecipitated by Ctb9/KLHDC5, it appears to be a doublet in which a higher molecular weight form of p60/katanin is enriched (Fig. 4A). We are tempted to speculate that the slower migrating band is a modified form that contains the signal for E3 ligase binding.
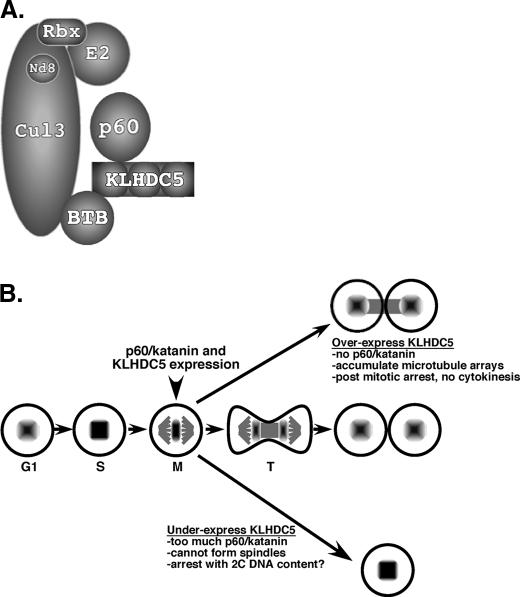
In conclusion we have identified p60/katanin as a novel Cul3 substrate, which is targeted for ubiquitin-mediated protein degradation following recognition by the substrate adaptor, Ctb9/KLHDC5 (Fig. 7A). The architecture of the complex consists of the adaptor (Ctb9/KLHDC5) bound to the N-terminal region of Cul3 carrying its cargo, p60/katanin. The C terminus is bound to an E2 ubiquitin-conjugating enzyme that has not been identified. Correct positioning of the substrate and E2 by the E3 complex mediates ubiquitin transfer. The severe phenotypes observed by interfering with this regulation (increased microtubule arrays, bi-nucleate cells) underscore the importance of p60/katanin degradation. However, these observations beg the question: what might the biological role for this regulation be? We suggest that timing of production, localization, and degradation of p60/katanin are crucial for properly completing mitosis (Fig. 7B). A major player in this process is composed of the production, activation, and possibly degradation of the Cul3 adaptor Ctb9/KLHDC5. Both proteins are expressed in early mitosis, but as mitosis proceeds Ctb9/KLHDC5 is excluded from the midbody and at the same time its substrate, p60/katanin, accumulates in the same location. This allows activity of the katanin to be high in the midbody region. We propose that overexpression of Ctb9/KLHDC5 results in too much degradation of p60/katanin, which results in an accumulation of microtubule arrays causing post mitotic arrest. In contrast, too little Ctb9/KLHDC5 results in an accumulation of p60/katanin that ultimately prevents microtubule formation causing cells to arrest before attempting mitosis.
FIGURE 7.
A model for Cul3-mediated ubiquitination of p60/katanin. A, Cul3 serves as a scaffold, binding the E2 ubiquitin conjugating enzyme and the essential ring finger protein Rbx1. KLHDC5 binds to Cul3 via its BTB domain, and to p60/katanin through its kelch repeats, and recruits p60/katanin for ubiquitination by the Cul3 E3 ligase. B, stages of the cell cycle that depend on p60/katanin degradation. Both p60/katanin and Ctb9/KLHDC5 are expressed early in mitosis. In telophase (T) Ctb9/KLHDC5 expression is limited by an unknown mechanism and excluded from the midbody allowing accumulation of p60/katanin in the area of the midbody. Overexpression of Ctb9/KLHDC5 results in too little p60/katanin and a buildup of microtubules, which prevents completion of mitosis. Under-expression of Ctb9/KLHDC5 results in too much p60/katanin, which prevents microtubules from forming, and therefore prevents progression of mitosis.
Supplementary Material
Acknowledgments
We thank Robert Sheaff for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant 7R01GM082940-0208 (to J. D. S.). This work was also supported by the Emerald Foundation (to J. D. S.), the Leukemia and Lymphoma Society (to J. D. S.), the Rhode Island Biomedical Research Infrastructure Network (National Center for Research Resources, NIH to J. D. S.), and the Center of Biological Research Excellence (National Center for Research Resources, NIH to J. D. S.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S5.
Footnotes
The abbreviations used are: E1, ubiquitin-activating enzyme; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; PIPES, 1,4-piperazinediethanesulfonic acid; Cul, cullin; BTB, Bric-a-brac, Tramtrak, and Broad-complex; siRNA, small interference RNA; IB, immunoblotting; IP, immunoprecipitation; IC, immunocytochemistry; PBS, phosphate-buffered saline; DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; MEF, mouse embryonic fibroblast; KLH, kelch; APC, anaphase-promoting complex.
W. Wimuttisuk and J. D. Singer, manuscript in preparation.
C. M. Cummings, C. A. Bentley, S. A. Perdue, P. W. Baas, and J. D. Singer, unpublished observations.
References
- 1.Ciechanover, A., Heller, H., Katz-Etzion, R., and Hershko, A. (1981) Proc. Natl. Acad. Sci. U. S. A. 78 761-765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hershko, A., Ciechanover, A., and Rose, I. A. (1981) J. Biol. Chem. 256 1525-1528 [PubMed] [Google Scholar]
- 3.Hershko, A. (1983) Cell 34 11-12 [DOI] [PubMed] [Google Scholar]
- 4.Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J. W., and Elledge, S. J. (1996) Cell 86 263-274 [DOI] [PubMed] [Google Scholar]
- 5.Lyapina, S. A., Correll, C. C., Kipreos, E. T., and Deshaies, R. J. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 7451-7456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulman, B. A., Carrano, A. C., Jeffrey, P. D., Bowen, Z., Kinnucan, E. R., Finnin, M. S., Elledge, S. J., Harper, J. W., Pagano, M., and Pavletich, N. P. (2000) Nature 408 381-386 [DOI] [PubMed] [Google Scholar]
- 7.Srayko, M., Buster, D. W., Bazirgan, O. A., McNally, F. J., and Mains, P. E. (2000) Genes Dev. 14 1072-1084 [PMC free article] [PubMed] [Google Scholar]
- 8.Yamanaka, A., Yada, M., Imaki, H., Koga, M., Ohshima, Y., and Nakayama, K. (2002) Curr. Biol. 12 267-275 [DOI] [PubMed] [Google Scholar]
- 9.Yan, Q., Kamura, T., Cai, Y., Jin, J., Ivan, M., Mushegian, A., Conaway, R. C., and Conaway, J. W. (2004) J. Biol. Chem. 279 43019-43026 [DOI] [PubMed] [Google Scholar]
- 10.Zheng, N., Schulman, B. A., Song, L., Miller, J. J., Jeffrey, P. D., Wang, P., Chu, C., Koepp, D. M., Elledge, S. J., Pagano, M., Conaway, R. C., Conaway, J. W., Harper, J. W., and Pavletich, N. P. (2002) Nature 416 703-709 [DOI] [PubMed] [Google Scholar]
- 11.Guardavaccaro, D., and Pagano, M. (2004) Oncogene 23 2037-2049 [DOI] [PubMed] [Google Scholar]
- 12.Willems, A. R., Schwab, M., and Tyers, M. (2004) Biochim. Biophys. Acta 1695 133-170 [DOI] [PubMed] [Google Scholar]
- 13.Singer, J. D., Gurian-West, M., Clurman, B., and Roberts, J. M. (1999) Genes Dev. 13 2375-2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McEvoy, J. D., Kossatz, U., Malek, N., and Singer, J. D. (2007) Mol. Cell. Biol. 27 3651-3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angers, S., Thorpe, C. J., Biechele, T. L., Goldenberg, S. J., Zheng, N., MacCoss, M. J., and Moon, R. T. (2006) Nat. Cell Biol. 8 348-357 [DOI] [PubMed] [Google Scholar]
- 16.Cullinan, S. B., Gordan, J. D., Jin, J., Harper, J. W., and Diehl, J. A. (2004) Mol. Cell. Biol. 24 8477-8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furukawa, M., and Xiong, Y. (2005) Mol. Cell. Biol. 25 162-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi, A., Kang, M. I., Okawa, H., Ohtsuji, M., Zenke, Y., Chiba, T., Igarashi, K., and Yamamoto, M. (2004) Mol. Cell. Biol. 24 7130-7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon, J. E., La, M., Oh, K. H., Oh, Y. M., Kim, G. R., Seol, J. H., Baek, S. H., Chiba, T., Tanaka, K., Bang, O. S., Joe, C. O., and Chung, C. H. (2006) J. Biol. Chem. 281 12664-12672 [DOI] [PubMed] [Google Scholar]
- 20.Ou, C.-Y., Lin, Y.-F., Chen, Y.-J., and Chien, C.-T. (2002) Gen. Dev. 16 2403-2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salinas, G. D., Blair, L. A., Needleman, L. A., Gonzales, J. D., Chen, Y., Li, M., Singer, J. D., and Marshall, J. (2006) J. Biol. Chem. 281 40164-40173 [DOI] [PubMed] [Google Scholar]
- 22.Sumara, I., Quadroni, M., Frei, C., Olma, M. H., Sumara, G., Ricci, R., and Peter, M. (2007) Dev. Cell 12 887-900 [DOI] [PubMed] [Google Scholar]
- 23.Wilkins, A., Ping, Q., and Carpenter, C. L. (2004) Genes Dev. 18 856-861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, D. D., Lo, S. C., Sun, Z., Habib, G. M., Lieberman, M. W., and Hannink, M. (2005) J. Biol. Chem. 280 30091-30099 [DOI] [PubMed] [Google Scholar]
- 25.Zhang, H. F., Tomida, A., Koshimizu, R., Ogiso, Y., Lei, S., and Tsuruo, T. (2004) Cancer Res. 64 1114-1121 [DOI] [PubMed] [Google Scholar]
- 26.Bardwell, V. J., and Treisman, R. (1994) Genes Dev. 8 1664-1677 [DOI] [PubMed] [Google Scholar]
- 27.Godt, D., Couderc, J. L., Cramton, S. E., and Laski, F. A. (1993) Development 119 799-812 [DOI] [PubMed] [Google Scholar]
- 28.Zollman, T. M., Austin, P. R., Jablonski, P. E., and Hovde, C. J. (1994) Protein Expr. Purif. 5 291-295 [DOI] [PubMed] [Google Scholar]
- 29.Xu, L., Yue, Y., Reboul, J., Vaglio, P., Shin, T.-H., Vidal, M., Elledge, S. J., and Harper, J. W. (2003) Nature 425 316-321 [DOI] [PubMed] [Google Scholar]
- 30.Pintard, L., Willis, J. H., Willems, A., Johnson, J. L., Srayko, M., Kurz, T., Glaser, S., Mains, P. E., Tyers, M., Bowerman, B., and Peter, M. (2003) Nature 425 311-316 [DOI] [PubMed] [Google Scholar]
- 31.Geyer, R., Wee, S., Anderson, S., Yates, J., and Wolf, D. A. (2003) Mol. Cell 12 783-790 [DOI] [PubMed] [Google Scholar]
- 32.Furukawa, M., He, Y. J., Borchers, C., and Xiong, Y. (2003) Nat. Cell Biol. 5 1001-1007 [DOI] [PubMed] [Google Scholar]
- 33.McMahon, M., Itoh, K., Yamamoto, M., and Hayes, J. D. (2003) J. Biol. Chem. 278 21592-21600 [DOI] [PubMed] [Google Scholar]
- 34.Lo, S. C., and Hannink, M. (2006) J. Biol. Chem. 281 37893-37903 [DOI] [PubMed] [Google Scholar]
- 35.Stogios, P. J., Downs, G. S., Jauhal, J. J., Nandra, S. K., and Prive, G. G. (2005) Genome Biol. 6 R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merdes, A., Ramyar, K., Vechio, J. D., and Cleveland, D. W. (1996) Cell 87 447-458 [DOI] [PubMed] [Google Scholar]
- 37.Scholey, J. M., Porter, M. E., Grissom, P. M., and McIntosh, J. R. (1985) Nature 318 483-486 [DOI] [PubMed] [Google Scholar]
- 38.Earnshaw, W. C. (2001) Curr. Biol. 11 R683. [DOI] [PubMed] [Google Scholar]
- 39.Vagnarelli, P., Morrison, C., Dodson, H., Sonoda, E., Takeda, S., and Earnshaw, W. C. (2004) EMBO Rep. 5 167-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murata-Hori, M., and Wang, Y. L. (2002) Curr. Biol. 12 894-899 [DOI] [PubMed] [Google Scholar]
- 41.Kallio, M. J., McCleland, M. L., Stukenberg, P. T., and Gorbsky, G. J. (2002) Curr. Biol. 12 900-905 [DOI] [PubMed] [Google Scholar]
- 42.Canman, J. C., Hoffman, D. B., and Salmon, E. D. (2000) Curr. Biol. 10 611-614 [DOI] [PubMed] [Google Scholar]
- 43.Martineau, S. N., Andreassen, P. R., and Margolis, R. L. (1995) J. Cell biol. 131 191-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wheatley, S. P., and Wang, Y. (1996) J. Cell Biol. 135 981-989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hartman, J. J., Mahr, J., McNally, K., Okawa, K., Iwamatsu, A., Thomas, S., Cheesman, S., Heuser, J., Vale, R. D., and McNally, F. J. (1998) Cell 93 277-287 [DOI] [PubMed] [Google Scholar]
- 46.Hartman, J. J., and Vale, R. D. (1999) Science 286 782-785 [DOI] [PubMed] [Google Scholar]
- 47.McNally, K. P., Bazirgan, O. A., and McNally, F. J. (2000) J. Cell Sci. 113 1623-1633 [DOI] [PubMed] [Google Scholar]
- 48.Yu, W., Solowska, J. M., Qiang, L., Karabay, A., Baird, D., and Baas, P. W. (2005) J. Neurosci. 25 5573-5583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baas, P. W., and Qiang, L. (2005) Trends Cell Biol. 15 183-187 [DOI] [PubMed] [Google Scholar]
- 50.Clark-Maguire, S., and Mains, P. E. (1994) Genetics 136 533-546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mains, P. E., Kemphues, K. J., Sprunger, S. A., Sulston, I. A., and Wood, W. B. (1990) Genetics 126 593-605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McNally, K., Audhya, A., Oegema, K., and McNally, F. J. (2006) J. Cell Biol. 175 881-891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srayko, M., O'Toole, E., T., Hyman, A. A., and Muller-Reichert, T. (2006) Curr. Biol. 16 1944-1949 [DOI] [PubMed] [Google Scholar]
- 54.Hazan, J., Fontaine, B., Bruyn, R. P., Lamy, C., van Deutekom, J. C., Rime, C. S., Durr, A., Melki, J., Lyon-Caen, O., Agid, Y., et al. (1994) Hum. Mol. Genet. 3 1569-1573 [DOI] [PubMed] [Google Scholar]
- 55.Errico, A., Ballabio, A., and Rugarli, E. I. (2002) Hum. Mol. Genet. 11 153-163 [DOI] [PubMed] [Google Scholar]
- 56.Zhang, D., Rogers, G. C., Buster, D. W., and Sharp, D. J. (2007) J. Cell Biol. 177 231-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hager, D. A., and Burgess, R. R. (1980) Anal. Biochem. 109 76-86 [DOI] [PubMed] [Google Scholar]
- 58.Karabay, A., Yu, W., Solowska, J. M., Baird, D. H., and Baas, P. W. (2004) J. Neurosci. 24 5778-5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Piperno, G., LeDizet, M., and Chang, X. J. (1987) J. Cell Biol. 104 289-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu, J., Gu, S., Wang, S., Dai, J., Ji, C., Jin, Y., Qian, J., Wang, L., Ye, X., Xie, Y., and Mao, Y. (2003) Mol. Biol. Rep. 30 239-242 [DOI] [PubMed] [Google Scholar]
- 61.Shimizu, C., Akazawa, C., Nakanishi, S., and Kageyama, R. (1995) Eur. J. Biochem. 229 239-248 [DOI] [PubMed] [Google Scholar]
- 62.Toyo-Oka, K., Sasaki, S., Yano, Y., Mori, D., Kobayashi, T., Toyoshima, Y. Y., Tokuoka, S. M., Ishii, S., Shimizu, T., Muramatsu, M., Hiraiwa, N., Yoshiki, A., Wynshaw-Boris, A., and Hirotsune, S. (2005) Hum. Mol. Genet. 14 3113-3128 [DOI] [PubMed] [Google Scholar]
- 63.Hernandez-Munoz, I., Lund, A. H., van der Stoop, P., Boutsma, E., Muijrers, I., Verhoeven, E., Nusinow, D. A., Panning, B., Marahrens, Y., and van Lohuizen, M. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 7635-7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mori, D., Yano, Y., Toyo-oka, K., Yoshida, N., Yamada, M., Muramatsu, M., Zhang, D., Saya, H., Toyoshima, Y. Y., Kinoshita, K., Wynshaw-Boris, A., and Hirotsune, S. (2007) Mol. Cell. Biol. 27 352-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He, H., Tan, C. K., Downey, K. M., and So, A. G. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 11979-11984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou, J., Hu, X., Xiong, X., Liu, X., Liu, Y., Ren, K., Jiang, T., Hu, X., and Zhang, J. (2005) J. Exp. Zool. Part A. Comp. Exp. Biol. 303 227-240 [DOI] [PubMed] [Google Scholar]
- 67.Watai, Y., Kobayashi, A., Nagase, H., Mizukami, M., McEvoy, J., Singer, J. D., Itoh, K., and Yamamoto, M. (2007) Genes Cells 12 1163-1178 [DOI] [PubMed] [Google Scholar]
- 68.Straight, A. F., Cheung, A., Limouze, J., Chen, I., Westwood, N. J., Sellers, J. R., and Mitchison, T. J. (2003) Science 299 1743-1747 [DOI] [PubMed] [Google Scholar]
- 69.Fry, A. M., and Yamano, H. (2006) Cell Cycle 5 1487-1491 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.