Abstract
Tetrahydrobiopterin (BH4) is an essential cofactor for aromatic acid hydroxylases, which control the levels of monoamine neurotransmitters. BH4 deficiency has been associated with many neuropsychological disorders. An inherited defect in BH4 biosynthesis is caused by the deficiency of sepiapterin reductase (SPR), which catalyzes the biosynthesis of BH4 from guanosine triphosphate at the terminal step. The human SPR gene has been mapped at the PARK3 locus, which is related to the onset of Parkinson disease. In this study, we report that mutant strains, lemon (lem) and its lethal allele lemon lethal (lem1) with yellow body coloration, of the silkworm Bombyx mori could be used as the first insect model for human SPR deficiency diseases. We demonstrated that mutations in the SPR gene (BmSpr) were responsible for the irregular body coloration of lem and leml. Moreover, biochemical analysis revealed that SPR activity in leml larvae was almost completely diminished, resulting in a lethal phenotype that the larvae cannot feed and that die immediately after the first ecdysis. Oral administration of BH4 and dopamine to leml larvae effectively increased their survival rates and feeding abilities. Our data demonstrate that BmSPR plays a crucial role in the generation of BH4, and monoamine neurotransmitters in silkworms and the lem (leml) mutant strains will be an invaluable resource to address many questions regarding SPR and BH4 deficiencies.
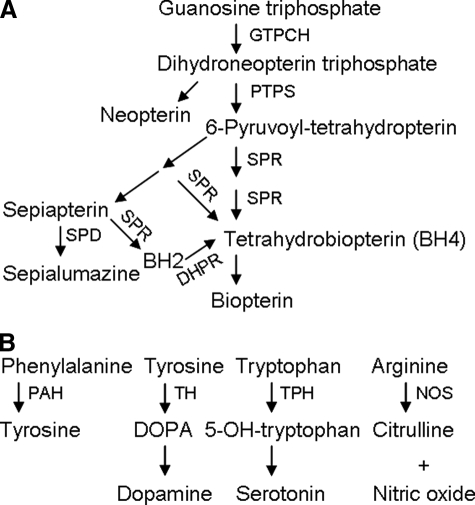
Tetrahydrobiopterin (BH4)2 is an essential cofactor for a number of enzymes, such as phenylalanine hydroxylase, tyrosine hydroxylase, tryptophan hydroxylase, and nitric-oxide synthase. These enzymes play an important role in the metabolism of aromatic amino acids and monoamine neurotransmitter biosynthesis (Fig. 1B) (1). Previous reports have shown that BH4 deficiency is associated with numerous metabolic syndromes and neuropsychological disorders (2, 3). BH4 is synthesized from GTP through a cascade of three enzymes (i.e. GTP cyclohydrolase I (EC 3.5.4.16), 6-pyruvoyl-tetrahydropterin (PTP) synthase (EC 4.6.1.10), and sepiapterin reductase (SPR; EC 1.1.1.153)) (Fig. 1A).
FIGURE 1.
A, the biosynthesis pathway of BH4 and pteridines from GTP. GTPCH, GTP cyclohydrolase; PTPS, PTP synthase; SPD, sepiapterin deaminase; DHPR, dihydrobiopterin reductase; BH2, dihydrobiopterin. B, physiological functions of BH4 as cofactors for aromatic acid hydroxylases and nitric-oxide synthase. TH and TPH, tyrosine hydroxylase and tryptophan hydroxylase, rate-limiting enzymes for the production of dopamine and serotonin, respectively. PAH, phenylalanine hydroxylase; NOS, nitric-oxide synthase.
SPR catalyzes the conversion of PTP to BH4 at the terminal step in the presence of reduced NADPH. It also catalyzes the reduction of sepiapterin (SP) to form BH4 by subsequent catalysis of dihydrobiopterin reductase (EC 1.6.99.7) (Fig. 1A). Mammalian SPRs have been a focus of study in recent years. The major symptoms of SPR deficiency are mental retardation, dystonia, spasticity, and movement disorder (1, 3). Diagnosis and therapy of SPR and/or BH4 deficiency-dependent genetic diseases, such as recessive DOPA-responsive dystonia and phenylketonuria, have been developed recently (3-5). Takazawa et al. (6) showed that the human SPR gene could be a causative gene for PARK3, the original reported pedigree of Parkinson disease (7). However, these findings are insufficient for developing treatments because patients from different families or regions exhibit distinct physiological and metabolic disorders due to SPR/BH4 deficiencies. Therefore, it is necessary to find or develop suitable models in other animals to obtain a better understanding of related human diseases. Recently, two groups generated Spr knock-out mice and concluded that these mice could be invaluable resources to address the issues regarding SPR/BH4 deficiencies (6, 8).
SPR has been previously purified from the silkworm Bombyx mori, and its activity in the fat body of normal larvae has been characterized (9, 10). However, the SPR gene (BmSpr) has not been identified yet in B. mori. lemon (lem) is a body color mutant of B. mori, which is regulated by a single recessive gene called lem (11). The lem silkworms display yellow body coloration during larval developmental stages, especially during molting, which is markedly different from that of wild-type strains (Fig. 2A). lemon lethal (leml) is a homozygous lethal allele of lem. The leml larvae grow normally in the first instar. After the first ecdysis, the leml larvae stop feeding, shake their heads frequently, and die within 3 days (Fig. 2, B and C) (12). lem (leml) was mapped onto the proximal end of the third linkage group of B. mori (11); however, the candidate gene itself remains unknown to date. Previous studies have shown that a large amount of yellow pteridines, SP, and sepialumazine accumulated in the integument of the lem larvae (13, 14). In addition, SPR activity was absent in the lem mutant silkworms (9). Lack of SPR activity accelerates the accumulation of SP and sepialumazine (Fig. 1A). Collectively, these results suggest that SPR was related to irregular body coloration in the lem (leml) mutant.
FIGURE 2.
A, larval body color comparison between normal (top, white) and lem (bottom, yellow) strains in the third molting. B, larval body color comparison between normal (top, white) and leml (bottom, bright yellow) strains at the beginning of the second instar. C, lethality of leml homologous larvae. They die from day 2 of the second instar, whereas +/+ or leml/+ larvae of the same strain grow normally. Scale bar, 5 mm.
In this study, we characterized and identified the mutations in BmSpr of lem and leml mutants. Linkage analysis showed that BmSpr is the candidate gene for the lem (leml) mutant. In addition, biochemical studies revealed that a decrease in BmSPR activity is responsible for the abnormal coloration, and a loss of BmSPR activity leads to the lethality of leml larvae. Furthermore, we succeeded in increasing the survival rates of the leml larvae by oral inoculation of BH4 and dopamine. Together, we conclude that BmSPR plays a crucial role in the generation of BH4 and monoamine neurotransmitters in B. mori, similar to that in mammals. The utility of the lem (leml) mutant as a potential insect model for human SPR deficiency has been discussed in this study.
EXPERIMENTAL PROCEDURES
Silkworm Strains—Five B. mori mutant lem strains (e36, l70, f40, r04, and b602) and one leml strain (a65) used in this study were obtained from Kyushu University (SilkwormBase; available on the World Wide Web). Normal strain 772 was obtained from the National Institute of Agrobiological Sciences. Normal strains of p50T and Sho-on were maintained in our laboratory. All larvae were fed fresh mulberry leaves under normal conditions (12 h light/12 h dark, 25 °C).
Genomic PCR and Reverse Transcription (RT)-PCR—Genomic DNA was prepared using a DNeasy blood and tissue kit (Qiagen). Genomic PCR was performed using TaKaRa Ex Taq (TaKaRa). Total RNA was extracted from whole body or tissues, as described previously (15). Expression profiles of BmSpr in different stages or tissues were analyzed by RT-PCR using a TaKaRa RNA PCR kit (TaKaRa). The PCR conditions were set up as recommended by the suppliers. The PCR primers used in the experiments are listed in Table 1 or available upon request.
TABLE 1.
Main primers used in this study
Artificial EcoRI sites are underlined. Attached His6 tag sequences are shown in boldface type.
| Name | Sequence (5′-3′) | Purpose |
|---|---|---|
| BmSprORF-fw | GGAGGACAAACCGGAGTCTG | RT-PCR and linkage analysis for lem |
| BmSprORF-re | CCCGCAAGGGGATACGAAGA | RT-PCR and linkage analysis for lem |
| BmSprMfw | CGTAACAAAGATCAGCTCGCCAT | Linkage analysis for leml |
| BmSprMre | CGAGCCTATCCTCTATCTCATC | Linkage analysis for leml |
| pETBmSPR-fw | CGGAATTCGATGGCTATGTCGTCTAGCATC | Expression in E. coli for three proteins |
| pETBmSPR-re1 | CGGAATTCTCAGTGGTGGTGGTGGTGGTGTTCGTCATCGAAATAGTCGAC | Expression in E. coli for normal and leml mutant type proteins |
| pETBmSPR-re2 | CGGAATTCTTAGTGATGGTGATGGTGATGGTCGACGTGCTCTGCCGGACT | Expression in E. coli for lem mutant type protein |
Cloning of B. mori Sepiapterin Reductase Gene—To identify whether the B. mori gene was homologous to the SPR gene, we screened expressed sequence tag data bases (16) and genome sequences (17, 18) using the BLAST program. By sequencing cDNA and genomic clones, we obtained the full-length BmSpr sequence that encodes a putative SPR and determined its genomic structure.
Protein Extract from Whole Body—Total proteins were extracted from each of the five individual pools at the beginning of the second larval instar. Whole bodies were homogenized in 10 mm phosphate-buffered saline (pH 7.0) containing a mixture of proteinase inhibitors (Roche Applied Science). The supernatant of the homogenates was collected by centrifugation (15,000 rpm, 4 °C, 20 min) and subjected to the PD-10 column (Amersham Biosciences) for desalting and buffer exchange with phosphate-buffered saline (pH 6.4; 200 mm). The resultant solution was concentrated using Amicon Ultra centrifugal filter devices (Millipore). The protein concentration was estimated using a Coomassie Plus protein assay reagent kit (Pierce) with bovine serum albumin as a standard.
Bacterial Expression and Purification of Recombinant BmSPR—The coding regions of the wild-type, lem mutant type, and leml mutant type BmSpr with a His6 tag sequence at the C terminus were amplified by PCR from the corresponding cDNA templates. The primers used are listed in Table 1. PCR products were digested with EcoRI and ligated into a pET24b vector (Novagen), resulting in three recombinant expression vectors, pET/BmSPR, pET/BmmtSPR, and pET/BmmtSPRl. They were transformed into Escherichia coli BL21 (DE3) competent cells. BmSPR expression was induced by 1 mm isopropyl-1-thio-β-d-galactopyranoside. The transformants were cultured overnight at 15 °C following isopropyl-1-thio-β-d-galactopyranoside induction. The cells were collected by centrifugation and suspended in a B-PER bacterial protein extraction reagent (Pierce) containing a mixture of proteinase inhibitors (Roche Applied Science), and the supernatant was collected by centrifugation. His-tagged BmSPRs were purified using a His GraviTrap nickel affinity column (GE Healthcare) according to the manufacturer's instructions. Finally, the eluate was desalted and concentrated. Protein concentration was determined as described above.
Immunoblot Analysis—Expression of recombinant BmSPRs were analyzed by immunoblot analysis using anti-His antibody (Qiagen), as described previously (19). After SDS-PAGE (20), the proteins were electrophoretically transferred to a polyvinylidene fluoride membrane (Immobilon-P; Millipore) using a blotting apparatus (Atto). The membrane was exposed to anti-His antibody (1:5000 dilution) and then to the secondary antibody, goat anti-mouse IgG-horseradish peroxidase conjugate (Zymed Laboratories Inc.) (1:5000 dilution). The blot was visualized using Western Lightning Chemiluminescence Reagent Plus (PerkinElmer Life Sciences) and a LAS 1000 imaging system (Fuji Film).
Enzyme Assay—SPR activity was assayed according to the method reported by Katoh (21) with slight modifications. Ten micrograms of protein from whole body or 0.1-0.5 μg of recombinant BmSPR was used in the assessment. The standard reaction mixture consisted of 100 μm NADPH (Sigma), 50 μm sepiapterin (Sigma), 100 mm potassium phosphate buffer (pH 6.4), and the enzyme in a final volume of 200 μl. The reaction was initiated by the addition of the enzyme and kept at 37 °C for 5-30 min. SPR activity was determined by measuring the rate of decrease in absorbance at 420 nm using a model 680 microplate reader (Bio-Rad). Reaction without the addition of the enzyme was used as a control. One unit of enzyme was defined as the amount of the enzyme/μg of protein that catalyzed the reduction of 1 nmol of sepiapterin/min (nmol/min/μg), using an extinction coefficient for sepiapterin of 1.04 × 104/mol/cm at 420 nm. The data were analyzed by one-way analysis of variance followed by Dunnett's test to localize the significant difference. A p value of less than 0.01 was considered significant.
To analyze the effect of pH on SPR activity, the final concentration of 20 mm Britton-Robinson buffer (pH 2.0-12.0) was used. To analyze the effect of temperature on SPR activity, each reaction mixture was kept at temperatures ranging from 20 to 90 °C for 5 min. The effect of the substrate concentration on SPR activity was analyzed by varying sepiapterin concentrations from 2.5 to 160 μm in the presence of 0.1 μg of protein and saturating amounts of 250 μm NADPH. The reaction was performed at 37 °C for 5 min. Kinetic parameters of maximal velocity (Vmax) and Michaelis constant (Km) were estimated using the double reciprocal (Lineweaver-Burk) plot (22). Inhibition of B. mori SPR by melatonin and N-acetylserotonin (Sigma) was investigated using 0.2 μg of protein at 37 °C for 10 min. Reactions with 1% ethanol were used as controls.
Linkage Analysis—Normal strain p50T, lem strain l70, and leml strain a65 were used in genetic linkage analysis. The F1 male moths of p50T and l70 were backcrossed with l70 females. Single nucleotide polymorphism at nt 786 in the BmSpr ORF of BC1 individuals was investigated by genomic PCR and direct sequencing of the PCR products (Table 2). The F1 moths of p50T and a65 (leml/+) were sibling-mated. Genotypes of F2 individuals randomly sampled from several broods were determined by a PCR marker in BmSpr ORF (Table 2). Genomic DNA was isolated using the Wizard SV 96 genomic DNA purification system (Promega), as described in the recommended protocol. The primers used are listed in Table 1. DNA sequences were determined using the ABI Big Dye Terminator Cycle Sequencing Ready Reaction kit version 3.1 (Applied Biosystems) and an ABI Prism 3130 genetic analyzer (Applied Biosystems).
TABLE 2.
Linkage analysis
|
lem
|
leml
|
|||||
|---|---|---|---|---|---|---|
| Phenotype (genotype) |
SNP in BmSpr ORF (nt 786)
|
Phenotype (genotype) |
PCR marker in BmSpr ORF
|
|||
| A/T | A/A | 325 bp only | 325 bp and 352 bp | 352 bp only | ||
| Normal (lem/+) | 50 | 0 | Normal (+/+) | 35 | 0 | 0 |
| Yellow (lem/lem) | 0 | 140 | Normal (leml/+) | 0 | 56 | 0 |
| Yellow and lethal (leml/leml) | 0 | 0 | 96 | |||
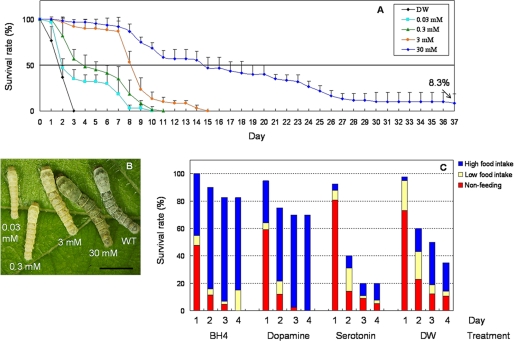
BH4 and Monoamine Feeding Experiment—To investigate the rescue effect of BH4 on the leml larvae, 10-fold diluted concentrations (0.03-30 mm) of BH4 (Wako) were orally supplied to newly hatched leml/leml larvae of the a65 strain. Fresh mulberry leaves were chopped and completely permeated in freshly prepared BH4 solutions. The leaves were dried in air and fed to the larvae every day. Survival rates were recorded from the beginning of second instar (day 0 in Fig. 6A). Oral administration of two monoamines, dopamine (Sigma) or serotonin (Wako), at a final concentration of 50 mm was performed against third instar leml larvae, which were survived by supplying 30 mm BH4 until the second ecdysis. Survival rate curves were recorded from day 1 to day 4. Feeding abilities of the living larvae were examined by counting their feces (more than 10 feces, high food intake; 1-10 feces, low food intake; no feces, nonfeeding). Distilled water-wetted leaves were used as a negative control.
FIGURE 6.
Rescue of the leml lethal larvae by BH4 and monoamine administration. A, survival rate curves by different doses of BH4. Eggs of a65 strain were randomly divided into five parts. Mulberry leaves wetted by various concentrations of BH4 were fed daily to newly hatched larvae. From the beginning of the second instar (day 0), 40 leml/leml larvae were grouped and used in each treatment. Survival rate curves were plotted daily throughout the larval stage. Distilled water (DW) was used as a negative control. The points indicate the mean ± S.D. (n = 3). B, the phenotypes of the leml larvae in each BH4 treatment on day 2 of the third instar. WT, wild type. Scale bar, 1 cm. C, survival rates and feeding abilities of leml larvae with oral administration of dopamine and serotonin. Final concentration of monoamines was 50 mm. Forty leml/leml larvae at the beginning of third instar survived by 30 mm BH4 from first instar were used in each group. Survival rate curves were recorded from day 1 to day 4. Feeding abilities of the living larvae were examined by counting their feces: more than 10 feces, high food intake; 1-10 feces, low food intake; no feces, nonfeeding. BH4 (30 mm) and distilled water were used as control reagents.
RESULTS
Characterization of BmSpr Gene in Normal and Mutant Strains—Previous studies showed that SPR deficiency is involved in abnormal body coloration of the lem (leml) mutant (9, 13, 14). To examine whether the B. mori SPR gene (BmSpr) corresponds to lem (leml), we first determined a full-length cDNA sequence of BmSpr using expressed sequence tag data bases (16). The sequence is located on Bm_scaf63, which is a newly integrated scaffold near the end of chromosome 3 (KAIKObase; available on the World Wide Web).
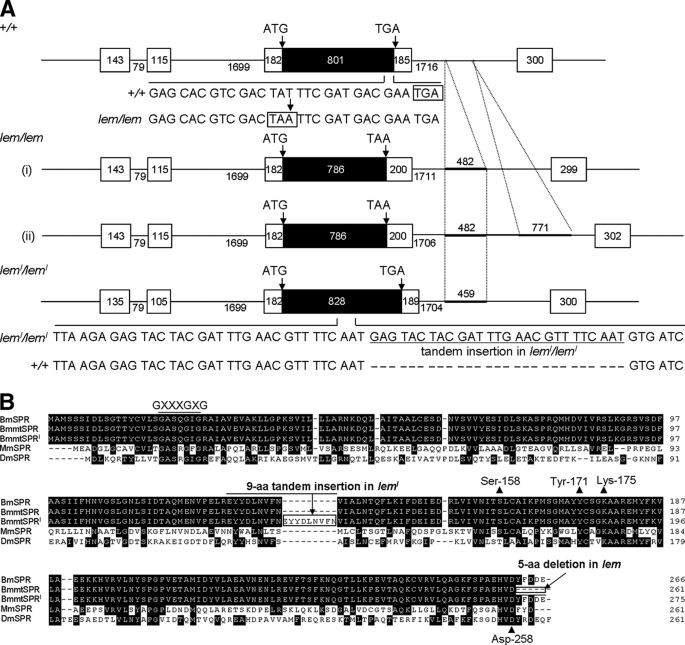
We determined the gene structures of BmSpr in normal and mutant strains by genomic primer walking and identified the mutations in BmSpr open reading frames (ORFs) of the mutants (Fig. 3A). BmSpr comprised an ORF of 798 bp, which encodes a 266-aa protein with a predicted molecular mass of 29.2 kDa (Fig. 3B). In the BmSpr ORF of five lem strains, a single nucleotide mutation was identified at the same position (T786A), which formed an abnormal forward stop codon and resulted in a 5-aa deletion (-YFDDE) at the C terminus. In the leml strain, 27 nt were inserted in tandem in the middle of the BmSpr ORF, which resulted in an addition of 9 aa (-EYYDLNVFN-) (Fig. 3, A and B). Moreover, one or two genomic insertions were found in the third intron of the mutant BmSpr (Fig. 3A).
FIGURE 3.
Schematic representation of the BmSpr gene structure and alignment of amino acid sequences. A, the top gene structure is the normal type (strains p50T and Sho-on). The middle two are structures in lem strains (i, strains l70, e36, r04, and b602; ii, strain f40), and the bottom structure is in leml strain (a65). The boxes indicate exons in black for the coding region and white for the noncoding region. Numerals show the length of exons or introns. Start and stop codons are indicated by arrows. The single nucleotide substitution (T → A) at the end of the BmSpr ORF in lem strains led to an abnormal forward stop codon (TAA, boxed). Tandem insertion of 27 nt into BmSpr ORF is underlined, which caused a 9-aa increase in the leml strain. Genomic insertions found in the third intron of mutant types of BmSpr are indicated as thick bars. B, three types of B. mori SPRs were aligned with mouse SPR (Mus musculus; GenBank™ accession number Q64105) and Drosophila SPR (Drosophila melanogaster; GenBank™ accession number NP_727265) using the ClustalX program. Amino acid residues conserved among more than four SPR sequences are highlighted. Five amino acid deletions in lem (-YFDDE) and 9-aa tandem insertions in leml (-EYYDLNVFN-) are aligned and boxed. Important residues for mammalian SPR activity, such as GXXXGXG (Rossmann fold, for NADPH binding), Ser158-Tyr171-XXXK175 triad (catalytic site), and Asp-258 (an aspartate anchor, for pterin substrate) are also shown.
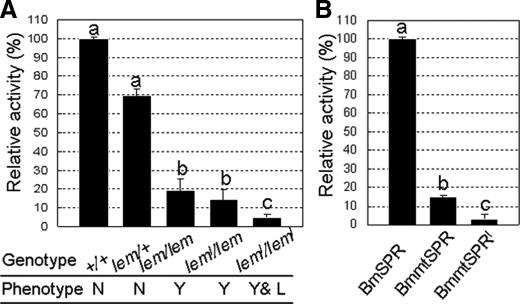
Enzyme Activity of BmSPR—We investigated the SPR activities in different genotypic larvae at the beginning of the second instar (Fig. 4A). When compared with the SPR activities in normal phenotypes, the activities in lem mutant silkworms were significantly low, and almost no activity was detected in the leml mutant silkworms. To investigate the enzymatic properties of BmSPR and compare the activities between normal and mutant proteins, we produced the three recombinant proteins using a bacterial expression system. Expression and purification of recombinant BmSPRs were analyzed by SDS-PAGE and immunoblot analysis (Fig. 5, A and B). Our analyses showed that BmSPR was a typical NADPH-dependent enzyme (Fig. 5C), which exhibited suitable enzymatic parameters to the substrate of SP with Km of 28.3 μm and Vmax of 14.5 nmol/min/μg (Table 3). Increase in activity was observed at pH 4-6 with pH 5 being an optimal condition (data not shown). The most suitable reaction temperature for BmSPR was 50 °C (data not shown). Moreover, two potent inhibitors of mammalian SPRs, melatonin and N-acetylserotonin, significantly inhibited BmSPR activity, with IC50 of 100 and 200 μm, respectively (data not shown). Comparison among the three proteins showed that the enzyme activity of BmmtSPR (lem type) and BmmtSPRl (leml type) was 15 and 3%, respectively, as compared with that of the normal BmSPR (Fig. 4B). The values were consistent with SPR activities measured in the second instar larvae of the normal and lem and leml mutants (Fig. 4A). These data show that reduced SPR activity correlates with abnormal coloration and mortality in the mutant strains.
FIGURE 4.
Enzyme assays for BmSPR. A, comparison of SPR activity among different genotypes. Total proteins were extracted from whole bodies of five individuals at the beginning of the second larval instar. Ten micrograms of proteins were used. The reaction was carried out as a standard enzyme assay at 37 °C for 30 min. Corresponding phenotypes are shown under each genotype. N, normal; Y, yellow; L, lethal. B, comparison of SPR activity among the three types of recombinant BmSPR proteins. Recombinant proteins (0.5 μg) were added to the reaction mixture. The reaction was carried out at 37 °C for 20 min. The bars indicate the mean ± S.D. (n = 3). Values indicated using different letters are significantly different (p < 0.01).
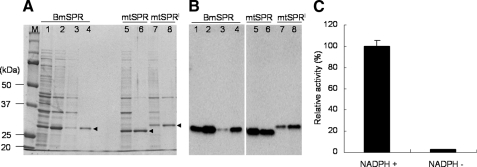
FIGURE 5.
Expression of recombinant BmSPR proteins and enzymatic property of normal BmSPR. Three recombinant proteins, normal type BmSPR, and two mutant types, BmmtSPR (lem) and BmmtSPRl (leml), were expressed using a bacterial expression system and purified using nickel chromatography columns. Expression and purification of recombinant BmSPRs were analyzed by SDS-PAGE (A) and immunoblot analysis (B). Lane 1, total cell proteins before isopropyl-1-thio-β-d-galactopyranoside induction; lanes 2, 5, and 7, total cell proteins induced by 1 mm isopropyl-1-thio-β-d-galactopyranoside; lane 3, proteins not binding to the nickel column; lanes 4, 6, and 8, eluted proteins using a buffer containing 20 mm sodium phosphate, 500 mm NaCl, and 200 mm imidazole (pH 7.4). M, protein standard. C, effect of cofactor NADPH on BmSPR activity. The reaction was carried out as a standard enzyme assay using 0.5 μg of enzyme with (+) or without (-) 100 μm NADPH. The reaction was carried out at 37 °C for 30 min. The data indicate the mean ± S.D. (n = 3).
TABLE 3.
Comparison of enzymatic properties of SPRs from B. mori and other organisms
Linkage Analysis—To determine the consistency between the lem or leml phenotype and the BmSpr genotype, we performed linkage analysis between the normal and lem or leml using p50T, l70, and a65 strains, respectively. We backcrossed F1 male moths of the normal and lem with lem females. Single nucleotide polymorphism of a total of 190 individuals from (lem/lem × lem/+) was sequenced in BmSpr ORF at nt 786 (Table 2). Moreover, we analyzed the genotypes of 187 F2 individuals of the normal and leml by a PCR marker in BmSpr ORF (Table 2). The results showed that phenotypes of all of the individuals were identical with their genotypes (i.e. no recombination between BmSpr and lem or leml was detected among their progenies). Based on these results, we concluded that the candidate gene BmSpr corresponds to lem (leml).
Therapeutic Effects of BH4 and Monoamine Administration—The leml larvae, in which SPR activity was almost completely diminished, do not eat and die immediately after the first ecdysis. To verify whether SPR deficiency-induced lack of BH4 is responsible for the lethality of the leml mutant, we performed BH4 administration experiments by oral inoculation. We found that BH4 administration effectively improved the growth and development of the leml larvae in a dose-dependent manner (Fig. 6, A and B). Larvae fed with lower concentrations of BH4 ate fewer mulberry leaves and developed more slowly than those fed with 30 mm BH4, which showed body size similar to that of the wild type (Fig. 6B). Although a majority of the larvae fed with 30 mm BH4 died during the larval stage, about 8% individuals grew normally with a 7-11-day-longer larval period and successfully accomplished the morphological transition from larvae to pupae (Fig. 6A). Furthermore, we performed oral administration experiments with two monoamines, dopamine and serotonin. The results clearly showed that, similar to BH4, dopamine administration effectively increased the survival rates of leml larvae, because this treatment drastically improved their feeding abilities (Fig. 6C). In contrast, serotonin administration did not show any positive effects on survival rates and feeding behavior of the leml larvae (Fig. 6C). Taken together, these results suggest that BH4 deficiency results in the lethal phenotype observed in the leml mutant, which is mainly due to the lack of dopamine.
DISCUSSION
In this paper, we report the identification and characterization of BmSpr and conclude that BmSpr corresponds to the yellow body color mutant lem (leml) of the silkworm B. mori. Using genetic and biochemical approaches, we demonstrated that the mutations in BmSpr significantly reduced SPR activity both in the lem (leml) larvae and in mutant BmSPR proteins. Moreover, oral administration of BH4 and dopamine successfully increased the survival rates of the leml larvae, suggesting that BH4 deficiency induced by loss of BmSPR activity leads to the lethality of leml larvae, probably due to the lack of dopamine. Therefore, we propose that the lem (leml) mutant can be regarded as a useful insect model for human SPR-deficient diseases.
Various pteridine derivates synthesized from GTP are the primary components in insect body coloration (Fig. 1A). The larval body color in B. mori is determined by the concentrations of melanin in the cuticle and of xanthommatin, sepialumazine, sepiapterin, and uric acid in the epidermis (23-25). Synthesis of pteridine from GTP is possible when a number of enzymes, including SPR (EC 1.1.1.153) and sepiapterin deaminase (EC 3.5.4.24) (Fig. 1A), cooperate. Our data indicated that the 5-aa deletion at the end of BmSPR in lem and the 9-aa insertion in the middle of BmSPR in leml caused a marked decrease in SPR activity, although important motifs for SPR activity are highly conserved in lem and leml (Figs. 3B and 4) (26, 27). These data proved that mutations in BmSpr are responsible for the abnormal accumulation of yellow pteridines in the integuments of lem (leml) (13, 14).
Park et al. (28) proposed that in the case of complete SPR defect, BH4 biosynthesis from PTP could be compensated by carbonyl and/or aldose reductases in humans. However, Bonafe et al. (1) showed that the compensation might be true for some peripheral tissues but not for the brain. Similarly, in the silkworm, Iino et al. (29, 30) discovered two carbonyl enzymes in the fat body and integument, which could reduce PTP to form BH4. However, the BH4 forming activity in the lem mutant was 10-fold lower than in the normal strain (29). These studies indicate that the salvage pathways cannot provide sufficient BH4, unlike the BH4 biosynthesis pathway catalyzed by SPR. Biochemical analysis showed that BmSPR exhibited enzymatic properties more similar to those of mammalian SPRs than to Drosophila SPR (Table 3) (21, 31, 32), which was consistent with a previous report (10). Collectively, the biosynthetic pathway of BH4 and the enzymatic properties of SPR are similar between Bombyx and mammals, suggesting that the silkworm is a suitable animal for studying human SPR/BH4 deficiency.
In mammals, BH4 exhibits various physiological functions, such as acting as a cofactor for aromatic hydroxylases and nitric-oxide synthase. Therefore, appropriate levels of BH4 are necessary for the metabolism of phenylalanine and the production of monoamine neurotransmitters (Fig. 1B) (8). In general, patients with BH4 deficiency present progressive neuronal deterioration, convulsions, abnormal movements, and difficulty in swallowing (2). Abnormal symptoms observed in the leml larvae, such as head shaking and feeding inability after the first ecdysis, are strikingly similar. Oral inoculation of BH4 effectively improved the feeding ability of the leml larvae and enabled them to grow normally through the larval developmental stage (Fig. 6, A and B), suggesting that loss of BmSPR activity reduced BH4 to a lethal level in the leml larvae. Further, we observed that oral inoculation of dopamine also effectively improved the survival rate and feeding ability of leml larvae (Fig. 6C). These results demonstrate that SPR deficiency-induced lack of dopamine results in the abnormal behavior observed in the leml larvae.
SPR deficiency in patients was frequently misdiagnosed as dihydrobiopterin reductase deficiency (3). Although diagnosis and therapy of SPR/BH4 deficiencies have advanced in recent years (1, 3-6), it is necessary to develop appropriate animal models to obtain better understanding the complexity of these diseases. Several invertebrates, such as Caenorhabditis elegans and Drosophila, have been used as animal models for human diseases (33-35). Together with these invertebrate animal models, the silkworm larvae have a number of advantages as an animal model; they are genetically tractable, easily maintained in laboratories throughout the year using artificial diets, and can be reared on a large scale at low cost. Moreover, the advantage of the large body size of silkworm larvae, which makes handling easier when injecting drugs and microorganisms but can be a practical problem in small animals, has made B. mori a useful model for infection with human pathogenic microorganisms (36-38). Based on the present results, we propose the lem (leml) mutant as the first insect model for human SPR deficiency. Recent studies have shown that Spr-null mice display greatly decreased amounts of BH4, severe monoamine deficiencies, and growth retardation. Also, a majority of mice died within 1-2 months (6, 8). Similar to the rescue effects of BH4 and dopamine feeding on the leml larvae, oral administration of BH4 and neurotransmitters completely rescued dwarfism and phenylalanine metabolism (6, 8). Together with the present results, we hope that the silkworm lem (leml) mutant can provide useful information for clinical diagnosis, for therapy options, and for screening therapeutic agents or new drug candidates for human SPR deficiency.
Acknowledgments
Most materials (silkworms, related DNA clones, and their information) were provided by the National Bioresource Project of the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) AB465548-AB465551.
This work was supported by a Grant-in-aid for Scientific Research 17018007 (to T. S.) from the Ministry of Education, Culture, Sports, Science, and Technology, the Professional Program for Agricultural Bioinformatics from Japan Science and Technology Agency, and Japan Society for the Promotion of Science Postdoctoral Fellowship for Foreign Researchers Grant P07427 (to Y. M.).
Footnotes
The abbreviations used are: BH4, tetrahydrobiopterin; PTP, 6-pyruvoyl-tetrahydropterin; SP, sepiapterin; SPR, sepiapterin reductase; ORF, open reading frame; RT, reverse transcription; nt, nucleotide(s); aa, amino acid(s).
References
- 1.Bonafe, L., Thony, B., Penzien, J. M., Czarnecki, B., and Blau, N. (2001) Am. J. Hum. Genet. 69, 269-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blau, N., Barnes, I., and Dhondt, J. N. (1996) J. Inherit. Metab. Dis. 19, 8-14 [DOI] [PubMed] [Google Scholar]
- 3.Blau, N., Bonafe, L., and Thony, B. (2001) Mol. Genet. Metab. 74, 172-185 [DOI] [PubMed] [Google Scholar]
- 4.Abeling, N. G., Duran, M., Bakker, H. D., Stroomer, L., Thöny, B., Blau, N., Booij, J., and Poll-The, B. T. (2006) Mol. Genet. Metab. 89, 116-120 [DOI] [PubMed] [Google Scholar]
- 5.Friedman, J., Hyland, K., Blau, N., and MacCollin, M. (2006) Neurology 67, 2032-2035 [DOI] [PubMed] [Google Scholar]
- 6.Takazawa, C., Fujimoto, K., Homma, D., Sumi-Ichinose, C., Nomura, T., Ichinose, H., and Katoh, S. (2008) Biochem. Biophys. Res. Commun. 367, 787-792 [DOI] [PubMed] [Google Scholar]
- 7.Wszolek, Z. K., Cordes, M., Calne, D. B., Muenter, M. D., Cordes, I., and Pfeifer, R. F. (1993) Nervenarzt 64, 331-335 [PubMed] [Google Scholar]
- 8.Yang, S., Lee, Y. J., Kim, J.-M., Park, S., Peris, J., Laipis, P., Park, Y. S., Chung, J. H., and Oh, S. P. (2006) Am. J. Hum. Genet. 78, 575-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsubara, M., Tsusué, M., and Akino, M. (1963) Nature 199, 908-909 [DOI] [PubMed] [Google Scholar]
- 10.Iino, T., Dohke, K., and Tsusué, M. (1992) Zool. Sci. 9, 119-125 [Google Scholar]
- 11.Banno, Y., Fujii, H., Kawaguchi, Y., and Yamamoto, K. (2005) A Guide to the Silkworm Mutants 2005: Gene Name and Gene, Institute of Genetic Resources, Kyushu University, Fukuoka, Japan
- 12.Tsujita, M. (1955) Jpn. J. Genet. 30, 107-117 [Google Scholar]
- 13.Tsusué, M., and Akino, M. (1965) Zool. Mag. 74, 91-94 [Google Scholar]
- 14.Goto, M., Konishi, M., Sugiura, K., and Tsusué, M. (1966) Bull. Chem. Soc. Japan 39, 929-932 [Google Scholar]
- 15.Meng, Y., Omuro, N., Funaguma, S., Daimon, T., Kawaoka, S., Katsuma, S., and Shimada, T. (2008) Arch. Insect Biochem. Physiol. 67, 9-19 [DOI] [PubMed] [Google Scholar]
- 16.Mita, K., Morimyo, M., Okano, K., Koike, Y., Nohata, J., Kawasaki, H., Kadono-Okuda, K., Yamamoto, K., Suzuki, M. G., Shimada, T., Goldsmith, M. R., and Maeda, S. (2003) Proc. Natl. Acad. Sci. U. S. A. 100, 14121-14126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mita, K., Kasahara, M., Sasaki, S., Nagayasu, Y., Yamada, T., Kanamori, H., Namiki, N., Kitagawa, M., Yamashita, H., Yasukochi, Y., Kadono-Okuda, K., Yamamoto, K., Ajimura, M., Ravikumar, G., Shimomura, M., Nagamura, Y., Shin, I. T., Abe, H., Shimada, T., Morishita, S., and Sasaki, T. (2004) DNA Res. 11, 27-35 [DOI] [PubMed] [Google Scholar]
- 18.Xia, Q., Zhou, Z., Lu, C., Cheng, D., Dai, F., Li, B., Zhao, P., Zha, X., Cheng, T., Chai, C., Pan, G., Xu, J., Liu, C., Lin, Y., Qian, J., Hou, Y., Wu, Z., Li, G., Pan, M., Li, C., Shen, Y., Lan, X., Yuan, L., Li, T., Xu, H., Yang, G., Wan, Y., Zhu, Y., Yu, M., Shen, W., Wu, D., Xiang, Z., Yu, J., Wang, J., Li, R., Shi, J., Li, H., Li, G., Su, J., Wang, X., Li, G., Zhang, Z., Wu, Q., Li, J., Zhang, Q., Wei, N., Xu, J., Sun, H., Dong, L., Liu, D., Zhao, S., Zhao, X., Meng, Q., Lan, F., Huang, X., Li, Y., Fang, L., Changfeng Li, D., Sun, Y., Zhang, Z., Yang, Z., Huang, Y., Xi, Y., Qi, Q., He, D., Huang, H., Zhang, X., Wang, Z., Li, W., Cao, Y., Yu, Y., Yu, H., Li, J., Ye, J., Chen, H., Zhou, Y., Liu, B., Wang, J., Ye, J., Ji, H., Li, S., Ni, P., Zhang, J., Zhang, Y., Zheng, H., Mao, B., Wang, W., Ye, C., Li, S., Wang, J., Wong, G. K.-S., and Yang, H. (2004) Science 306, 1937-194015591204 [Google Scholar]
- 19.Daimon, T., Taguchi, T., Meng, Y., Katsuma, S., Mita, K., and Shimada, T. (2008) J. Biol. Chem. 283, 15271-15279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. (1970) Nature 227, 680-685 [DOI] [PubMed] [Google Scholar]
- 21.Katoh, S. (1971) Arch Biochem. Biophys. 146, 202-214 [DOI] [PubMed] [Google Scholar]
- 22.Lineweaver, H., and Burk, D. (1934) J. Am. Chem. Soc. 56, 658-666 [Google Scholar]
- 23.Mazda, T., Tsusué, M., and Sakate, S. (1980) Insect Biochem. 10, 357-362 [Google Scholar]
- 24.Ohashi, M., Tsusué, M., Yoshitake, N., Sakate, S., and Kiguchi, K. (1983) J. Seric. Sci. Jpn. 52, 498-504 [Google Scholar]
- 25.Kato, T., Sawada, H., Yamamoto, T., Mase, K., and Nakagoshi, M. (2006) Pigment Cell Res. 19, 337-345 [DOI] [PubMed] [Google Scholar]
- 26.Auerbach, G., Herrmann, A., Gütlich, M., Fischer, M., Jacob, U., Bacher, A., and Huber, R. (1997) EMBO J. 16, 7219-7230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujimoto, K., Hara, M., Yamada, H., Sakurai, M., Inaba, A., Tomonura, A., and Katoh, S. (2001) Chem. Biol. Interact. 130, 825-832 [DOI] [PubMed] [Google Scholar]
- 28.Park, Y. S., Heizmann, C. W., Wermuth, B., Levine, R. A., Steinerstauch, J., Guzman, J., and Blau, N. (1991) Biochem. Biophys. Res. Commun. 175, 738-744 [DOI] [PubMed] [Google Scholar]
- 29.Iino, T., Sawada, H., Tsusué, M., and Takikawa, S. (1996) Biochim. Biophys. Acta 1297, 191-199 [DOI] [PubMed] [Google Scholar]
- 30.Iino, T., Takikawa, S. I., Yamamoto, T., and Sawada, H. (2000) Arch. Biochem. Biophys. 373, 442-446 [DOI] [PubMed] [Google Scholar]
- 31.Ruiz-Vazquez, P., Silva, F. J., and Ferre, J. (1996) Comp. Biochem. Physiol. B 113, 131-136 [DOI] [PubMed] [Google Scholar]
- 32.Sueoka, T., and Katoh, S. (1982) Biochim. Biophys. Acta 717, 265-271 [DOI] [PubMed] [Google Scholar]
- 33.Mahajan-Miklos, S., Tan, M. W., Rahme, L. G., and Ausubel, F. M. (1999) Cell 96, 47-56 [DOI] [PubMed] [Google Scholar]
- 34.Wu, Y., Bolduc, F. V., Bell, K., Tully, T., Fang, Y., Sehgal, A., and Fischer, J. A. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 12399-12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, Z., Wang, X., Yu, Y., Li, X., Wang, T., Jiang, H., Ren, Q., Jiao, Y., Sawa, A., Moran, T., Ross, C. A., Montell, C., and Smith, W. W. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 2693-2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaito, C., Akimitsu, N., Watanabe, H., and Sekimizu, K. (2002) Microb. Pathog. 32, 183-190 [DOI] [PubMed] [Google Scholar]
- 37.Hamamoto, H., et al. (2004) Antimicrob. Agents Chemother. 48, 774-779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orihara, Y., Hamamoto, H., Kasuga, H., Shimada, T., Kawaguchi, Y., and Sekimizu, K. (2008) J. Gen. Virol. 89, 188-194 [DOI] [PubMed] [Google Scholar]