Abstract
Parkinson disease (PD) is characterized by the presence of ubiquitylated inclusions and the death of dopaminergic neurons. Seven in absentia homolog (SIAH) is a ubiquitin-ligase that ubiquitylates α-synuclein and synphilin-1 and is present in Lewy bodies of PD patients. Understanding the mechanisms that regulate the ubiquitylation of PD-related proteins might shed light on the events involved in the formation of Lewy bodies and death of neurons. We show in this study that the recently described synphilin-1 isoform, synphilin-1A, interacts in vitro and in vivo with the ubiquitin-protein isopeptide ligase SIAH and regulates its activity toward α-synuclein and synphilin-1. SIAH promotes limited ubiquitylation of synphilin-1A that does not lead to its degradation by the proteasome. SIAH also increases the formation of synphilin-1A inclusions in the presence of proteasome inhibitors, supporting the participation of ubiquitylated synphilin-1A in the formation of Lewy body-like inclusions. Synphilin-1A/SIAH inclusions recruit PD-related proteins, such as α-synuclein, synphilin-1, Parkin, PINK1, and UCH-L1. We found that synphilin-1A robustly increases the steady-state levels of SIAH by decreasing its auto-ubiquitylation and degradation. In addition, synphilin-1A blocks the ubiquitylation and degradation of the SIAH substrates synphilin-1 and deleted in colon cancer protein. Furthermore, synphilin-1A strongly decreases the monoubiquitylation of α-synuclein by SIAH and the formation of α-synuclein inclusions, supporting a role for monoubiquitylation in α-synuclein inclusion formation. Our results suggest a novel function for synphilin-1A as a regulator of SIAH activity and formation of Lewy body-like inclusions.
Parkinson disease (PD)3 is characterized by progressive degeneration of dopaminergic neurons in the substantia nigra and the presence of Lewy bodies in surviving neurons (1). The majority of PD cases are sporadic, but mutations in different genes have been found responsible for familial PD (1, 2). α-Synuclein plays a crucial role in the disease. It is mutated in some familial forms of PD and represents a major component of Lewy bodies in sporadic PD as well (3, 4).
Dysfunction of the ubiquitin-proteasome system has been proposed to play a role in PD as the proteasome activity is decreased in the substantia nigra of PD patients (5). Highlighting the role of the ubiquitin-proteasome system in PD is the finding that proteins mutated in the disease, such as Parkin and UCH-L1, are also components of this system (6, 7). Furthermore, different PD-related proteins, including α-synuclein, synphilin isoforms, UCH-L1 and PINK1, were shown to be ubiquitylated and to accumulate into ubiquitylated inclusions (8-14).
Synphilin-1 interacts in vivo with α-synuclein, and their coexpression promotes the formation of Lewy body-like inclusions (15). Synphilin-1 is present in Lewy bodies of PD and Diffuse Lewy Body disease (16-19). Synphilin-1 was shown to be ubiquitylated by different E3 ubiquitin-ligases, including Parkin, Dorfin, and SIAH (8, 20-22). Inability of the proteasome to degrade polyubiquitylated synphilin-1 leads to robust formation of inclusions (8). Moreover, ubiquitylation of synphilin-1 is essential for its aggregation, as inactive SIAH mutants do not elicit formation of synphilin-1 inclusions (8). Thus, we raised the possibility that ubiquitylation of synphilin-1 and other PD-related proteins might represent an important step for Lewy body formation (13, 23, 24).
In addition to ubiquitylated synphilin-1, SIAH interacts with and monoubiquitylates α-synuclein at lysines found to be ubiquitylated in α-synuclein purified from Lewy bodies (8, 14). Monoubiquitylation of α-synuclein leads to its aggregation and inclusion formation, which are toxic to dopaminergic cells (14). The presence of SIAH in Lewy bodies of PD patients (8) indicates that it might represent an additional component of the ubiquitin-proteasome system involved in the disease.
We recently identified a novel synphilin-1 isoform, synphilin-1A, that is expressed in the brain of different α-synucleinopathies and is a neurotoxic and aggregation-prone protein (25). The importance of synphilin-1A in PD is suggested by its ability to interact with α-synuclein and synphilin-1, and to accumulate in insoluble brain fractions of patients with Diffuse Lewy Body disease (25). In this study, we sought to investigate the interaction between SIAH and synphilin-1A. We present evidence that synphilin-1A interacts with SIAH. We found that SIAH ubiquitylates synphilin-1A and increases the formation of synphilin-1A inclusions, but it does not promote synphilin-1A degradation. On the other hand, synphilin-1A decreases SIAH E3 ubiquitin-ligase activity as well as toxicity. Synphilin-1A also decreases the monoubiquitylation of α-synuclein promoted by SIAH as well as the formation of α-synuclein inclusions. Our data indicate that synphilin-1A is a regulator of SIAH activities, with implications for regulation of α-synuclein monoubiquitylation and aggregation.
EXPERIMENTAL PROCEDURES
Materials—Ubiquitin aldehyde, purified ubiquitin-activating enzyme, UbcH5b, lactacystin, and 3-MA were purchased from Sigma.
Cell Culture and Transfections—HEK293 and SH-SY5Y cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum in a 5% CO2 atmosphere. Cells were transiently transfected with N-terminal tagged pRK5 and pFLAG-CMV-2 plasmids utilizing Lipofectamine 2000 (Invitrogen) and processed after 36 h.
For experiments using siRNA, HEK293 cells were transfected with 100 nm siRNAs using Lipofectamine 2000. After 48 h, cells were again transfected with 100 nm siRNAs and processed after 36 h. siRNAs to synphilin-1A (sense siRNA strand, 5′-GCUGAUUGAAGAGCAUCUG-3′, and antisense siRNA strand, 5′-CAGAUGCUCUUCAAUCAGC-3′; sense siRNA strand, 5′-CCUCUAACAUGCUGAUUGA-3′, and antisense siRNA strand, 5′-UCAAUCAGCAUGUUAGAGG-3′) as well as control siRNA were obtained from Sigma.
Western Blot Analysis—Samples were homogenized as described (8). Blots were probed with antibodies mouse anti-HA (Covance), mouse anti-Myc (Sigma), mouse anti-α-synuclein (BD Biosciences), rabbit anti-FLAG (Sigma), rabbit anti-Myc, rabbit anti-HA, goat anti-SIAH-1 (N-15), goat anti-SIAH-2 (N-14), goat anti-SIAH-1/2 (H-18), and mouse anti-actin (Santa Cruz Biotechnology). Purified anti-synphilin-1A and anti-SIAH-1 (990) antibodies were generated as described before (8, 25).
In Vitro Binding Assays—Extracts of transfected HEK293 cells were incubated with 5 μg/ml GST fusion proteins for 2 h at 4 °C in buffer containing 50 mm Tris-HCl, pH 7.4, 140 mm NaCl, 1% Triton X-100, 0.1% SDS, 30 μm MG132 and protease inhibitors (Complete; Roche Applied Science). Beads were washed in the same buffer. Bound proteins were detected by Western blot.
Co-immunoprecipitation Assays—Transfected HEK293 cells were lysed in buffer containing 50 mm Tris, pH 7.4, 140 mm NaCl, 1% Triton X-100, 0.1% SDS, 30 μm MG132, and protease inhibitor mixture (Complete; Roche Applied Science). Cell extracts were clarified by centrifugation at 13,000 × g for 5 min, and the supernatant was incubated with anti-HA for 4 h as described (8). Immunoprecipitates were washed with lysis buffer containing 500 mm NaCl and 1% Chaps and detected by Western blot.
Endogenous Co-immunoprecipitation Assays—Rat brains were homogenized in buffer containing 50 mm Tris, pH 7.4, 140 mm NaCl, 1% Chaps, 30 μm MG132, and protease inhibitor mixture (Complete, Roche Applied Science). Brain homogenates were clarified by centrifugation at 13,000 × g for 5 min. Antibodies to SIAH-1 or SIAH-2 (N-15 and N-14, respectively) (Santa Cruz Biotechnology) were coupled to protein G beads (8) and incubated for 7 h with brain homogenate (2 mg/ml). Immunoprecipitates were washed with lysis buffer containing 500 mm NaCl and 1% Chaps and detected by Western blot using rabbit anti-synphilin-1A (25).
In Vitro Ubiquitylation Assays—For the in vitro ubiquitylation assays, synphilin-1A, synphilin-1, and SIAH were translated using TnT wheat germ in vitro translation kit (Promega) using [35S]methionine (Amersham Biosciences). His-α-synuclein was purified from bacteria using TALON beads according to the manufacturers instructions (BD Biosciences). In vitro translated proteins or recombinant α-synuclein was incubated in reaction medium containing 40 mm Tris, pH 7.6, 5 mm MgCl2, 2 mm dithiothreitol, 1 mm ATPγS, 7.5 μg of ubiquitin, 1 μm ubiquitin aldehyde, 100 ng of E1, and 200 ng of UbcH5b, in the presence or absence of 500 ng of SIAH-2. Reactions were incubated at 37 °C for 1 h, terminated by the addition of SDS sample buffer, and resolved on SDS-polyacrylamide gels. Ubiquitylated [35S]synphilin-1A, [35S]synphilin-1, and [35S]SIAH-1 were determined by PhosphorImager analysis. Ubiquitylated α-synuclein was determined by Western blot using α-synuclein antibody.
In Vivo Ubiquitylation Assays—Transfected HEK293 or SH-SY5Y cells were incubated in the absence or in the presence of 10 μm lactacystin for 12 h. Transfected cells were then resuspended in buffer containing 50 mm Tris, pH 7.4, 140 mm NaCl, 1% SDS and boiled for 5 min. Cell extracts were sonicated, diluted 10-fold with buffer containing 50 mm Tris, pH 7.4, 140 mm NaCl, 1% Triton X-100, 30 μm MG132, and protease inhibitor mixture (Complete, Roche Applied Science), and then incubated for 16 h with anti-HA beads (Sigma). Immunoprecipitates were washed with lysis buffer containing 500 mm NaCl and detected by Western blot.
Pulse-Chase Experiments—Transfected HEK293 cells were washed, incubated with methionine/cysteine-free medium for 1 h, pulsed with methionine/cysteine-free medium containing 100 μCi of [35S]methionine/cysteine (PerkinElmer Life Sciences) for 3 h, and subsequently chased in normal medium for the times specified. Cells were harvested, and HA-SIAH-1 immunoprecipitation was carried out as described above for the in vivo ubiquitylation assays. Immunoprecipitates were resolved on 10% SDS-polyacrylamide gels and the amount of 35S-labeled SIAH-1 was quantified by PhosphorImager analysis.
Immunocytochemistry Assays—Transfected HEK293 and SH-SY5Y cells were incubated with or without 10 μm lactacystin, 10 mm NH4Cl, and 10 mm 3-MA for 12 h, fixed with 4% paraformaldehyde for 15 min, and blocked in phosphate-buffered saline containing 0.2% Triton X-100 and 5% normal goat serum. Cells were labeled with anti-HA (Covance) and anti-Myc antibodies (Santa Cruz Biotechnology) as described (8). Immunolabeling was detected using fluorescein isothiocyanate- and Cy3-labeled secondary antibodies (The Jackson Laboratories). Statistics of the number of inclusion-containing cells were analyzed by Student's t test.
Cell Death Analysis—Transfected HEK293 and SH-SY5Y cells were transfected and processed for immunocytochemistry as described above. Before mounting, nuclei were stained with Hoechst 33342. Cell death was determined by counting the number of transfected cells that were positive for nuclear condensation and fragmentation. Statistics of cell death was done by Student's t test.
Primary Neuronal Cultures—Embryonic day 18 primary cortical cultures were prepared from Sprague-Dawley rats as described (26). The animals were killed by decapitation according to the protocol approved by the committee for animal experimentation at the Technion-Israel Institute of Technology. Neurons were maintained in neurobasal medium supplemented with B27 (Invitrogen) and 0.5 mm l-glutamine (Biological Industries, Beit-Haemek, Israel). After 2 weeks of culture, neurons were fixed for immunocytochemical studies as described above for cell lines.
RESULTS
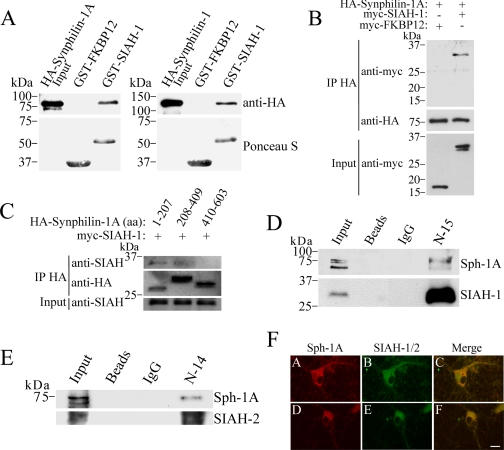
Synphilin-1A Interacts in Vitro and in Vivo with SIAH and Co-localizes in Neurons—We have previously identified a novel synphilin-1 isoform, called synphilin-1A, that is present in Lewy bodies and is toxic to neurons (25). We now investigated if synphilin-1A interacts with SIAH, which was shown to ubiquitylate synphilin-1 (8). To determine whether they interact in vitro, GST pulldown experiments were carried out by incubating GST-SIAH-1 fusion protein with the extract of HEK293 cells transfected with HA-synphilin-1A (Fig. 1A, left panel). HA-Synphilin-1A specifically interacted with GST-SIAH-1 but not with the control protein GST-FKBP12 (Fig. 1A, left panel). The in vitro interaction of synphilin-1A and SIAH-1 was similar to that observed between synphilin-1 and SIAH-1 (Fig. 1A, right panel).
FIGURE 1.
SIAH interacts with synphilin-1A in vitro and in vivo. A, left panel, extract of HEK293 cells transfected with HA-synphilin-1A was incubated with the indicated GST fusion proteins. Binding was analyzed by using an anti-HA antibody. Right panel, extract of HEK293 cells transfected with HA-synphilin-1 was incubated with the indicated GST fusion proteins. Binding was analyzed using anti-HA antibody. The total amount of GST fusion proteins used in the experiments was determined by staining the membranes with Ponceau S (lower panels). B, SIAH-1 co-immunoprecipitates with synphilin-1A from co-transfected HEK293 cells. HA-synphilin-1A was immunoprecipitated (IP) from extracts of HEK293 cells using an anti-HA antibody. Co-immunoprecipitation was determined by Western blot using an anti-Myc antibody. C, mapping the SIAH binding region in synphilin-1A. HA-synphilin-1A fragments were immunoprecipitated from extracts of co-transfected HEK293 cells using an anti-HA antibody. The co-immunoprecipitation of SIAH-1 with synphilin-1A fragments was determined by using the anti-SIAH antibody 990. The detection of immunoprecipitated synphilin-1A fragments was done using an anti-HA antibody. aa, amino acids. D, synphilin-1A (Sph-1A) co-immunoprecipitates with endogenous SIAH-1. SIAH-1 was immunoprecipitated from rat brain homogenate using the anti-SIAH-1 antibody (N-15), and detection of co-immunoprecipitation was carried out using anti-synphilin-1A antibody (25). The detection of immunoprecipitated-SIAH-1 was done using the anti-SIAH-1 antibody (990) (8). E, synphilin-1A also co-immunoprecipitates with endogenous SIAH-2. SIAH-2 was immunoprecipitated from rat brain homogenate using the anti-SIAH-2 antibody (N-14), and detection of co-immunoprecipitation was carried out using anti-synphilin-1A antibody. The detection of immunoprecipitated-SIAH-2 was done using the anti-SIAH-1/2 antibody (H-18). F, rat cortical neurons grown for 14 days in vitro were immunolabeled against synphilin-1A (panels A and D), SIAH-1 with N-15 antibody (panel B), and SIAH-1/2 with H-18 antibody (panel E). The co-localization of endogenous synphilin-1A and SIAH in the cytoplasm of neurons, including cell bodies and neuronal processes, is observed in the merge pictures (panels C and F). Scale bar, 20 μm.
To test whether synphilin-1A interacts with SIAH in cells, we also carried out co-immunoprecipitation experiments using HEK293 cells co-transfected with HA-synphilin-1A and myc-SIAH-1. We found that SIAH-1, but not the control protein FKBP12, interacts with synphilin-1A (Fig. 1B). In addition, SIAH-2 also co-immunoprecipitated with synphilin-1A in cells (data not shown).
To map the SIAH-binding domain of synphilin-1A, HEK293 cells were co-transfected with different HA-synphilin-1A constructs and myc-SIAH-1. By carrying out co-immunoprecipitation experiments, we identified that SIAH co-immunoprecipitated with the first third as well as the second third of synphilin-1A (Fig. 1C). The last third of synphilin-1A did not interact with SIAH. Thus, the first two-thirds of synphilin-1A seem to be responsible for the interaction with SIAH-1.
In addition, synphilin-1A specifically co-immunoprecipitated with SIAH-1 from rat brain tissue (Fig. 1D) but not with control beads alone or coupled with IgG, indicating that these two proteins interact in vivo. Synphilin-1A also co-immunoprecipitated with SIAH-2 from rat brain (Fig. 1E), indicating that, similar to SIAH-1, SIAH-2 interacts in vivo with synphilin-1A.
Using antibodies that we previously characterized as specific to synphilin-1A (Fig. 1F, panels A and D), SIAH-1 (panel B), and SIAH-1/2 (panel E) (8, 14, 25), we found in immunocytochemistry experiments of rat primary neuronal cultures that endogenous synphilin-1A co-localizes with endogenous SIAH proteins in neuronal cytosol, including cell bodies and processes, as determined by confocal laser microscopy (Fig. 1F, panels C and F).
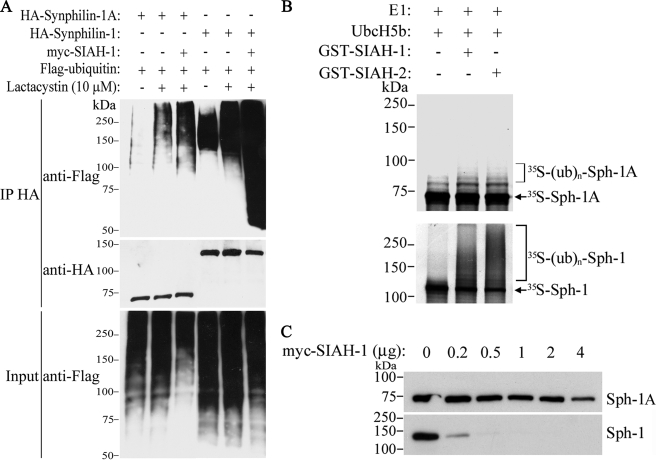
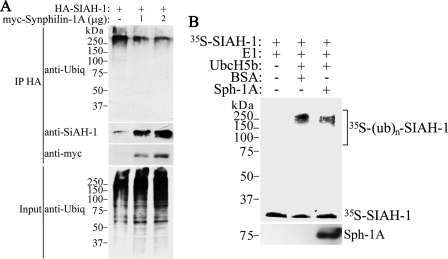
Characterization of Synphilin-1A Ubiquitylation and Inclusion Formation—We next investigated whether SIAH ubiquitylates synphilin-1A. For this, in vivo ubiquitylation experiments were carried out by co-transfecting HEK293 cells with HA-synphilin-1A, FLAG-ubiquitin, in the absence or in the presence of myc-SIAH-1. In the presence of lactacystin, SIAH-1 promoted only a slight increase in synphilin-1A ubiquitylation (Fig. 2A, 3rd lane of upper panel). By contrast, SIAH-1 promoted a robust increase in synphilin-1 ubiquitylation (Fig. 2A, 6th lane of upper panel) (8). We also carried out in vitro ubiquitylation experiments where SIAH-1 and -2 promoted only a slight ubiquitylation of synphilin-1A (Fig. 2B, upper panel), although they lead to robust ubiquitylation of synphilin-1 (Fig. 2B, lower panel) (8).
FIGURE 2.
SIAH promotes slight ubiquitylation and degradation of synphilin-1A. A, HEK293 cells were co-transfected with HA-synphilin-1A, FLAG-ubiquitin, in the absence or in the presence of myc-SIAH-1. As control, co-transfections were carried out with synphilin-1 instead of synphilin-1A. Cells were incubated 12 h with 10 μm lactacystin, and HA-synphilin-1A and HA-synphilin-1 were immunoprecipitated (IP) with anti-HA antibody. Ubiquitylation of synphilin-1A and synphilin-1 was detected by Western blot using anti-FLAG antibody. The middle panel shows the levels of immunoprecipitated synphilin-1A and synphilin-1 using anti-HA antibody. The lower panel shows the total ubiquitylation levels of co-transfected cells with anti-FLAG antibody. B, in vitro translated 35S-synphilin-1A was incubated with recombinant SIAH-1 or SIAH-2, UbcH5b, ubiquitin, and the other purified components of the ubiquitin system. The levels of 35S-synphilin-1A in vitro ubiquitylation were determined by PhosphorImager analysis (upper panel). As control, in vitro translated 35S-synphilin-1 was incubated with recombinant SIAH-1 or SIAH-2 under the same conditions as for 35S-synphilin-1A, and the levels of 35S-synphilin-1 ubiquitylation were determined by PhosphorImager analysis (lower panel). E1, ubiquitin-activating enzyme. C, effect of SIAH-1 on synphilin-1A (Sph-1A) steady-state levels. HEK293 cells were transfected with HA-synphilin-1A and myc-SIAH-1. HA-synphilin-1A and HA-synphilin-1 from total cell lysates were detected by Western blot using anti-HA antibody.
To determine whether SIAH promotes the degradation of synphilin-1A, HEK293 cells were co-transfected with HA-synphilin-1A and increasing amounts of myc-SIAH-1, and the steady-state levels of synphilin-1A were analyzed. No major decrease in the steady-state of synphilin-1A was observed even in the presence of large amounts of SIAH-1 (Fig. 2C, upper panel), suggesting that SIAH-1 does not promote significant degradation of synphilin-1A. By contrast, SIAH-1 promoted a sharp decrease in synphilin-1 steady state levels (Fig. 2C, lower panel) (8). Similar results were obtained when SIAH-2 was used instead SIAH-1 (data not shown). Thus, contrasting with synphilin-1, synphilin-1A seems to be a poor SIAH substrate.
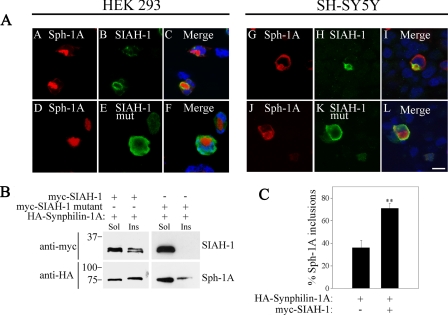
We have shown that overexpression of synphilin-1A into cultured cells promotes its accumulation into inclusions (25). To determine whether SIAH can be recruited to synphilin-1A inclusions, HEK293 cells were co-transfected with HA-synphilin-1A and myc-SIAH-1 or myc-SIAH-1 C55A,C59H,C72S mutant devoid of catalytic activity (8). By carrying out immunocytochemistry experiments, we found that only catalytically active SIAH-1 is recruited to synphilin-1A inclusions (Fig. 3A, left panels). In addition, SIAH-1 co-localized with synphilin-1A inclusions also in the dopaminergic cells SH-SY5Y (Fig. 3A, right panels), indicating that the recruitment of SIAH to synphilin-1A inclusions is not specific to a single cell type. Similar to SIAH-1, SIAH-2 was also recruited to synphilin-1A inclusions (data not shown), further supporting the interaction between SIAH proteins and synphilin-1A. Finally, the co-localization of SIAH-1 and SIAH-2 with synphilin-1A inclusions was observed either in the absence or in the presence of the proteasome inhibitor lactacystin (Fig. 3A and data not shown). Furthermore, no recruitment of inactive SIAH-1 mutant to synphilin-1A inclusions was observed (Fig. 3A, left and right lower panels).
FIGURE 3.
SIAH is recruited to synphilin-1A inclusions. A, immunofluorescence of transfected HEK293 (left panels) and SH-SY5Y (right panels) cells reveals the co-localization of myc-SIAH-1 but not myc-SIAH-1 catalytically inactive mutant (C55A,C59H,C72S) with HA-synphilin-1A inclusions. Immunocytochemistry was carried out with anti-HA (red) and anti-Myc antibody (green). Nuclei were revealed with TOPRO-3. Scale bar, 25 μm. B, synphilin-1A (Sph-1A) recruits wild-type SIAH-1 but not the catalytically inactive mutant to the Triton X-100-insoluble fraction. HEK293 cells transfected with HA-synphilin-1A and myc-SIAH-1 or myc-SIAH-1 inactive mutant were lysed, and cell lysates were divided into Triton-soluble (Sol) and Triton-insoluble (Ins) fractions. The distribution of SIAH-1 (wild-type and inactive mutant) between soluble and insoluble fractions was determined using anti-Myc antibody (upper panels). The distribution of synphilin-1A between the same fractions was determined using anti-HA antibody. C, quantification of synphilin-1A inclusion formation in SH-SY5Y cells transfected with HA-synphilin-1A, in the absence or in the presence of myc-SIAH-1. Cells were incubated 12 h with 10 μm lactacystin prior to the immunocytochemistry assay. Error bars represent standard error of five independent experiments. **, significantly different from control at p < 0.01.
To confirm the recruitment of SIAH to synphilin-1A inclusions, we investigated the effect of synphilin-1A on the distribution of SIAH-1 into soluble and insoluble fractions by Western blot assays. HEK293 cells were co-transfected with HA-synphilin-1A and myc-SIAH-1 (wild-type or mutant devoid of catalytic activity), and cell homogenates were separated into fractions soluble and insoluble to 1% Triton X-100. In the presence of synphilin-1A, SIAH-1 distributed between soluble and insoluble fractions, whereas SIAH-1 devoid of catalytic activity was present only in the soluble fraction (Fig. 3B). Therefore, we confirmed by biochemical means that insoluble synphilin-1A is able to recruit only active SIAH-1.
We have previously shown that ubiquitylation by SIAH in the presence of proteasome inhibition promotes about 5-fold increase in the formation of synphilin-1 inclusions (8). In the presence of lactacystin, SIAH-1 also increased the formation of synphilin-1A inclusions (Fig. 3C). However, this increase in synphilin-1A inclusions promoted by SIAH-1 was smaller than that previously observed for synphilin-1, as SIAH-1 leads to less than a 2-fold increase in synphilin-1A inclusions (Fig. 3C). This result is compatible with the small ubiquitylation extent of synphilin-1A promoted by SIAH, if compared with that observed with synphilin-1 (Fig. 2A).
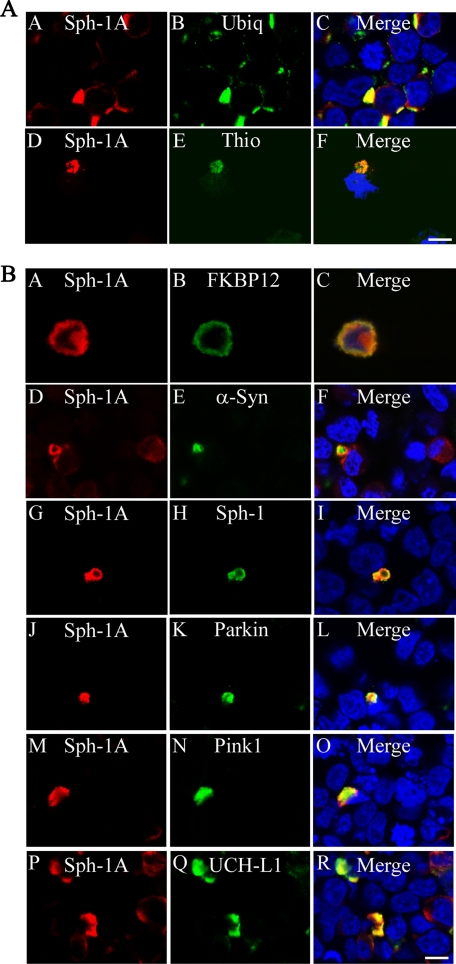
We next characterized synphilin-1A/SIAH inclusions. In immunocytochemistry experiments, synphilin-1A/SIAH inclusions were thioflavine S- and ubiquitin-positive (Fig. 4A). Also, synphilin-1A/SIAH inclusions were able to recruit PD-related proteins, such as α-synuclein, synphilin-1, Parkin, PINK1, and UCH-L1 (Fig. 4B), indicating that SIAH does not compete with the binding of PD-related proteins to synphilin-1A inclusions. These results were observed for synphilin-1A/SIAH inclusions either in the absence (Fig. 4, A and B) or in the presence of lactacystin (data not shown). No significant recruitment of the control protein FKBP12 to synphilin-1A/SIAH inclusions was observed, suggesting that, together with SIAH, aggregated synphilin-1A may work as anchor for PD proteins to form Lewy bodies.
FIGURE 4.
Co-localization of PD-related proteins with synphilin-1A/SIAH inclusions. A, SH-SY5Y cells were co-transfected with HA-synphilin-1A (Sph-1A), untagged SIAH-1, and FLAG-ubiquitin (Ubiq). Immunocytochemistry was carried out with anti-HA (red) and anti-FLAG or thioflavine (Thio) S (14) (green). Nuclei were stained with TOPRO-3. Scale bar, 25 μm. B, SH-SY5Y cells were co-transfected with HA-synphilin-1A, untagged SIAH-1, and myc-FKBP12, myc-α-synuclein (α-Syn), myc-synphilin-1, myc-Parkin, myc-PINK1, or myc-UCH-L1. Immunocytochemistry was carried out with anti-HA (red) and anti-Myc (green). Nuclei were stained with TOPRO-3. Scale bar, 25 μm. The figures are representative of 3-6 independent experiments.
Synphilin-1A Decreases SIAH Auto-ubiquitylation and Degradation—Because synphilin-1A strongly binds to SIAH in vivo and in vitro, but is a very poor ubiquitylation substrate, we wondered whether synphilin-1A could in fact affect SIAH functions. To determine whether synphilin-1A could change SIAH ubiquitin-ligase activity, the effect of synphilin-1A on SIAH auto-ubiquitylation was investigated. For this, HEK293 cells were co-transfected with HA-SIAH-1 and increasing amounts of myc-synphilin-1A; SIAH was immunoprecipitated with an anti-HA antibody, and the levels of SIAH auto-ubiquitylation were determined using an anti-ubiquitin antibody. We found that increasing amounts of synphilin-1A significantly decreased SIAH-1 auto-ubiquitylation (Fig. 5A). In vitro ubiquitylation experiments were also carried out by incubating in vitro translated SIAH with purified components of the ubiquitin system in the absence or in the presence of recombinant synphilin-1A. Similar to the in vivo ubiquitylation experiments, addition of synphilin-1A decreased the in vitro auto-ubiquitylation of SIAH (Fig. 5B).
FIGURE 5.
Synphilin-1A decreases the ubiquitylation of SIAH-1. A, HEK293 cells were transfected with HA-SIAH-1 and increasing amounts of myc-synphilin-1A. Cells were incubated 12 h with 10 μm lactacystin. HA-SIAH-1 was immunoprecipitated (IP) with anti-HA antibody, and ubiquitylated SIAH-1 was detected by Western blot using anti-ubiquitin (Ubiq) antibody. The amount of immunoprecipitated SIAH-1 was determined using anti-SIAH-1 antibody N-15 (2nd panel). The levels of synphilin-1A (Sph-1A) were determined with anti-Myc antibody (3rd panel). B, in vitro translated SIAH-1 was incubated with UbcH5b, ubiquitin, and the other purified components of the ubiquitin system in the absence or in the presence of recombinant synphilin-1A. The levels of SIAH-1 ubiquitylation were determined by PhosphorImager analysis. The amount of synphilin-1A added was determined by Western blot using anti-synphilin-1A antibody (lower panel). BSA, bovine serum albumin.
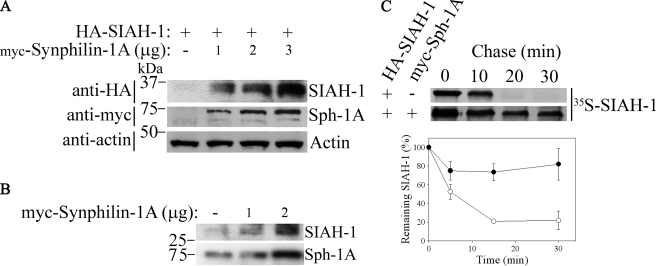
Because auto-ubiquitylation of SIAH is required for its degradation (27), we next examined if synphilin-1A leads to decreased degradation of SIAH. HEK293 cells were co-transfected with HA-SIAH-1 and myc-synphilin-1A, and the steady-state levels of SIAH-1 were analyzed. We found that increasing amounts of synphilin-1A significantly increased the steady-state levels of SIAH-1 (Fig. 6A). In experiments where only synphilin-1A was transfected, increasing amounts of synphilin-1A were able to significantly raise the steady-state levels of endogenous SIAH-1 protein (Fig. 6B). In addition, pulse-chase experiments revealed that SIAH-1 half-life was significantly prolonged in the presence of synphilin-1A (Fig. 6C), indicating that the increase of SIAH-1 steady-state levels promoted by synphilin-1A is because of decreased degradation and not to changes in transcription. Taken together, our data indicate that synphilin-1A decreases the auto-ubiquitylation and degradation of SIAH-1.
FIGURE 6.
Synphilin-1A inhibits the degradation of SIAH-1. A, HEK293 cells were co-transfected with HA-SIAH-1 and increasing amounts of myc-synphilin-1A. HA-SIAH-1 steady-state levels from total cell lysates were detected by Western blot with anti-HA antibody. The expression levels of synphilin-1A were determined using anti-Myc antibody. Loading control was monitored with anti-actin antibody (lower panel). B, HEK293 cells were transfected with increasing amounts of myc-synphilin-1A. Steady-state levels of endogenous SIAH-1 from total cell lysates were determined using anti-SIAH-1 antibody (N-15). The expression levels of synphilin-1A were determined using anti-Myc antibody. C, synphilin-1A (Sph-1A) increases the half-life of SIAH-1. Transfected HEK293 cells were chased for the indicated time points, and HA-SIAH-1 was immunoprecipitated using anti-HA antibody. Immunoprecipitates were analyzed by SDS-PAGE and autoradiography (upper panel). Graph shows the quantification of remaining SIAH-1 at indicated time points. Error bars represent standard error of three independent experiments (lower panel).
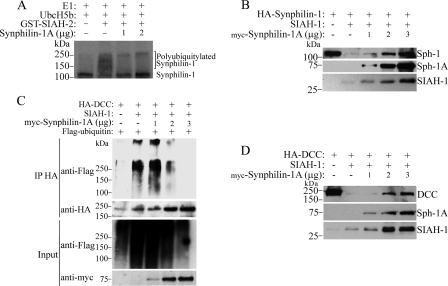
Synphilin-1A Decreases SIAH Ligase Activity and Cell Toxicity—To determine whether synphilin-1A also changes SIAH ability to ubiquitylate its substrates, we carried out ubiquitylation of synphilin-1 and DCC promoted by SIAH in the absence and in the presence of synphilin-1A. Increasing amounts of synphilin-1A significantly decreased the ability of SIAH-1 to ubiquitylate synphilin-1 as well as DCC (Fig. 7, A and C). Also, increasing amounts of synphilin-1A prevented the degradation of both synphilin-1 and DCC promoted by SIAH (Fig. 7, B and D). Therefore, in addition to decreasing the auto-catalytic activity of SIAHs, synphilin-1A decreases the ability of SIAH to ubiquitylate and to degrade its substrates.
FIGURE 7.
Synphilin-1A decreases the ubiquitylation and degradation of the SIAH-1 substrates synphilin-1 and DCC. A, in vitro translated synphilin-1 was incubated with increasing amounts of synphilin-1A, UbcH5b, ubiquitin, and the other purified components of the ubiquitin system, in the absence or in the presence of SIAH-1. The levels of synphilin-1 ubiquitylation were determined by PhosphorImager analysis. E1, ubiquitin-activating enzyme. B, HEK293 cells were transfected with HA-synphilin-1, untagged SIAH-1, and increasing amounts of synphilin-1A. HA-synphilin-1 from total cell lysates was detected by Western blot using anti-HA antibody (upper panel). The expression levels of synphilin-1A (Sph-1A) and SIAH-1 were determined with anti-Myc and anti-SIAH-1 (N-15) antibodies, respectively. C, HEK293 cells were transfected with HA-DCC and increasing amounts of myc-synphilin-1A. Cells were incubated 12 h with 10 μm lactacystin. HA-DCC was immunoprecipitated with anti-HA antibody, and ubiquitylated DCC was detected by Western blot using anti-FLAG antibody. The amount of immunoprecipitated DCC was determined using anti-HA antibody (2nd panel). The levels of synphilin-1A were determined with anti-Myc antibody (4th panel). D, HEK293 cells were transfected with HA-DCC, untagged SIAH-1, and increasing amounts of synphilin-1A. HA-DCC from total cell lysates was detected by Western blot using anti-HA antibody (upper panel). The expression levels of synphilin-1A and SIAH-1 were determined with anti-Myc and anti-SIAH-1 (N-15) antibodies, respectively.
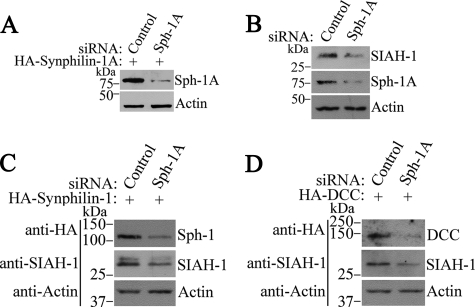
To demonstrate the relevance of synphilin-1A in the metabolism of endogenous proteins, we analyzed SIAH activity in the presence of siRNA to synphilin-1A. Because we found low levels of synphilin-1A expression in HEK293 (data not shown), all the siRNA experiments were carried out in SH-SY5Y cells. First, we generated a pair of siRNAs that efficiently reduce synphilin-1A expression (Fig. 8A). We next investigated the ability of synphilin-1A siRNA to interfere with SIAH activity. As steady-state levels of SIAH were shown to correlate with its ligase activity (27), we investigated the effect of synphilin-1A on the steady-state levels of SIAH. We found that siRNA-mediated synphilin-1A knockdown promoted a significant reduction in SIAH steady-state levels (Fig. 8B), suggesting that decreased synphilin-1A expression leads to improvement of SIAH ligase activity with increased auto-ubiquitylation and degradation. Also, siRNA to synphilin-1A decreased the steady state of the SIAH substrates synphilin-1 and DCC (Fig. 8, C and D), indicating that the absence of synphilin-1A increases the activity of SIAH toward its substrates as well. Taken together, our data suggest that the decrease in synphilin-1A expression increases the activity of endogenous SIAH as well as its ability to degrade substrates, confirming that endogenous synphilin-1A negatively modulates the ligase activity of SIAH.
FIGURE 8.
Reduction of synphilin-1A expression decreases the steady-state levels of SIAH-1 and its substrates synphilin-1 and DCC. A, SH-SY5Y cells were transfected with HA-synphilin-1A, in the presence of siRNA control or siRNA to synphilin-1A (Sph-1A). HA-synphilin-1A from total cell lysates was detected by Western blot using anti-HA antibody (upper panel). B, SH-SY5Y cells were transfected with siRNA control or siRNA to synphilin-1A. The endogenous levels of SIAH-1 from total cell lysates were detected by Western blot using the 990 anti-SIAH-1 antibody (8) (upper panel). Middle panel shows the levels of endogenous synphilin-1A with anti-synphilin-1A antibody. C, SH-SY5Y cells were transfected with HA-synphilin-1, in the presence of siRNA control or siRNA to synphilin-1A. HA-synphilin-1 from total cell lysates was detected by Western blot using anti-HA antibody (upper panel). Middle panel shows the levels of endogenous SIAH-1 with 990 anti-SIAH-1 antibody. D, SH-SY5Y cells were transfected with HA-DCC, in the presence of siRNA control or siRNA to synphilin-1A. HA-DCC from total cell lysates was detected by Western blot using anti-HA antibody (upper panel). Middle panel shows the levels of endogenous SIAH-1 with 990 anti-SIAH-1 antibody. Lower panels in A-D show the loading control of protein extracts determined by incubating the membranes with anti-actin antibody.
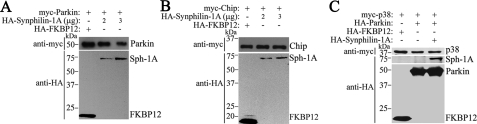
Next, we examined whether the inhibitory activity of synphilin-1A is specific for SIAH-1. For this, we investigated the ability of synphilin-1A to interfere with the ligase activity of two other E3 ubiquitin-ligases, Parkin and CHIP (carboxyl terminus of Hsc70-interacting protein). Because the autoubiquitylation of Parkin and CHIP were shown to reflect their catalytic activity (28-30), we measured the ability of synphilin-1A to modulate the steady-state levels of Parkin and CHIP. We found that synphilin-1A did not promote the accumulation of Parkin or CHIP as it did for SIAH (Fig. 9, A and B), indicating that synphilin-1A does not interfere with the activities of Parkin and CHIP. Also, synphilin-1A did not promote the accumulation of one of the Parkin bona fide substrates p38 (31, 32) (Fig. 9C), confirming that synphilin-1A does not inhibit Parkin ligase activity. Therefore, these findings indicate that synphilin-1A specifically modulates SIAH activity.
FIGURE 9.
Synphilin-1A does not interfere with the steady-state levels of Parkin and its substrate p38 as well as with CHIP. A, HEK293 cells were transfected with myc-Parkin and increasing amounts of synphilin-1A (Sph-1A) or HA-FKBP12. myc-Parkin from total cell lysates was detected by Western blot using anti-Myc antibody (upper panel). The expression levels of synphilin-1A and FKBP12 were determined with anti-HA. B, HEK293 cells were transfected with myc-CHIP and increasing amounts of synphilin-1A or HA-FKBP12. myc-CHIP from total cell lysates was detected by Western blot using anti-Myc antibody (upper panel). The expression levels of synphilin-1A and FKBP12 were determined with anti-HA. C, HEK293 cells were transfected with myc-p38, HA-Parkin, and HA-FKBP12 in the absence or in the presence of HA-synphilin-1A. myc-p38 from total cell lysates was detected by Western blot using anti-Myc antibody (upper panel). The expression levels of Parkin, synphilin-1A, and FKBP12 were determined with anti-HA.
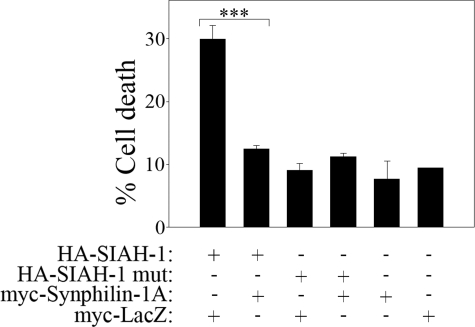
SIAH has been shown to cause cell death (33). To investigate if synphilin-1A can also decrease the toxicity promoted by SIAH, we transfected HEK293 cells with SIAH, in the absence or in the presence of synphilin-1A, and measured cell toxicity by determining the amount of nuclear condensation/fragmentation by Hoechst 33342. We found that synphilin-1A significantly decreased the cell death promoted by SIAH (Fig. 10). Different from wild-type SIAH, the ligase-inactive mutant of SIAH did not promote cell death when compared with vector alone (Fig. 10). In addition, cell death promoted by synphilin-1A, in the absence or in the presence of ligase-inactive SIAH, was similar to that observed with vector alone, indicating that synphilin-1A, alone or combined with ligase-inactive SIAH, is not toxic to HEK293 cells (Fig. 10). Therefore, synphilin-1A modulates both SIAH ligase activity (Figs. 5, 6, 7) and its toxicity (Fig. 10), suggesting that the ligase and the toxic activity of SIAH are interconnected.
FIGURE 10.
Synphilin-1A decreases the toxicity of SIAH. HEK293 cells were transfected with HA-SIAH-1, HA-SIAH-1 C55A,H59A,C72S (ligase-inactive mutant (mut)) (8), myc-synphilin-1A, and myc-LacZ. Immunocytochemistry was carried out using anti-HA and anti-Myc antibodies, and cell death was determined by quantifying the nuclear fragmentation and condensation of transfected HEK293 cells with Hoechst 33342. Error bars represent standard error of 4-7 independent experiments. ***, significantly different from vehicle control at p < 0.001.
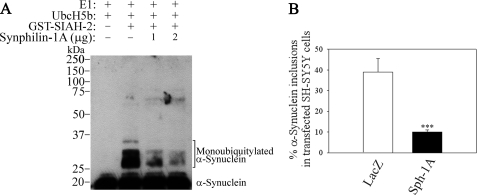
Synphilin-1A Decreases the Monoubiquitylation of α-Synuclein by SIAH and Its Inclusion Formation—We next investigated the effect of synphilin-1A on the ability of SIAH to ubiquitylate substrates that are not the target for degradation, such as α-synuclein (8, 14). Addition of increasing amounts of synphilin-1A decreased the monoubiquitylation of α-synuclein (Fig. 11A). In addition, no significant change in α-synuclein steady-state levels was observed in the presence of SIAH alone or combined with increasing amounts of synphilin-1A (data not shown).
FIGURE 11.
Synphilin-1A decreases the monoubiquitylation of α-synuclein promoted by SIAH and the formation of monoubiquitylated α-synuclein inclusions. A, His-α-synuclein was incubated with increasing amounts of synphilin-1A (Sph-1A), UbcH5b, and the other components of the ubiquitin system, in the absence or in the presence of recombinant SIAH-2. His-α-synuclein ubiquitylation was determined by Western blot using an antibody to α-synuclein. E1, ubiquitin-activating enzyme. B, SH-SY5Y cells were transfected with HA-α-synuclein, untagged SIAH-2, in the presence of myc-synphilin-1A or LacZ Myc. Cells were incubated for 12 h with a combination of 10 μm lactacystin, 10 mm NH4Cl, and 10 mm 3-MA. Immunocytochemistry was carried out using anti-HA and anti-Myc antibodies, and the percent of α-synuclein inclusions was quantified in transfected SH-SY5Y cells. Error bars represent standard error of five independent experiments. ***, significantly different from vehicle control at p < 0.001.
Because α-synuclein monoubiquitylation may affect inclusion formation in dopaminergic cells (14), we investigated the effect of synphilin-1A in the formation of α-synuclein inclusions. For this, SH-SY5Y cells were transfected with α-synuclein and SIAH-2, in the absence or in the presence of synphilin-1A. To promote the accumulation of monoubiquitylated α-synuclein, transfected cells were incubated with proteasome, lysosome, and autophagy inhibitors, and the amounts of monoubiquitylated α-synuclein inclusions were determined by immunocytochemistry. In accordance with its ability to inactivate SIAH ligase activity, synphilin-1A significantly decreased the formation of monoubiquitylated α-synuclein inclusions promoted by SIAH (Fig. 11B).
DISCUSSION
We have previously identified an isoform of synphilin-1 we called synphilin-1A that is prone to aggregation and that accumulates in the brain of Diffuse Lewy Body disease (25). Also, synphilin-1A interacts with both α-synuclein and synphilin-1, suggesting a possible role in PD. In this study, we described the interaction between synphilin-1A and the E3 ubiquitin-ligase SIAH. We found that synphilin-1A is ubiquitylated by SIAH but that this ubiquitylation is not enough to promote synphilin-1A degradation by the proteasome. As a poorly degraded substrate, synphilin-1A inactivates SIAH, inhibiting SIAH ligase and toxic activities. Accumulation of ubiquitylated synphilin-1A leads to increased formation of synphilin-1A inclusions, which recruits SIAH. Synphilin-1A decreases the auto-ubiquitylation of SIAH and its ligase activity toward other substrates, including synphilin-1, DCC, and α-synuclein. In parallel to a decrease in synphilin-1 and DCC ubiquitylation, synphilin-1A reduced their degradation by the proteasome. The ability of synphilin-1A to interfere with SIAH activity was also observed for endogenous proteins as siRNA mediated synphilin-1A knockdown significantly decreased the steady-state of endogenous SIAH and its substrates, synphilin-1 and DCC. Furthermore, the inhibitory activity of synphilin-1A is specific for SIAH because synphilin-1A did not promote the accumulation of other E3 ubiquitin ligases, such as Parkin and CHIP. Moreover, synphilin-1A decreased α-synuclein monoubiquitylation and the formation of α-synuclein inclusions, supporting a role of α-synuclein monoubiquitylation in inclusion body formation.
SIAH has been shown to interact with its substrates by binding to the consensus VXP (34). Synphilin-1A is devoid of VXP consensus, but it interacts with endogenous SIAH, as observed in co-immunoprecipitation experiments with rat brains. Supporting an interaction of SIAH and synphilin-1A inaVXP-independent manner is the finding that some SIAH substrates, such as synaptophysin, do not contain the VXP motif (34). Also, mutations in the VXP consensus of some substrates, such as Kid, do not change the interaction with SIAH (34), suggesting that some substrates of SIAH may interact in a VXP-independent manner.
Further supporting a specific interaction of synphilin-1A and SIAH, we found in immunocytochemistry experiments of primary neuronal cultures that both endogenous proteins co-localize in the cytosol of neurons, including cell bodies and processes. In agreement, both proteins were previously shown to be widely expressed in the brain and to be present at the presynapse (25, 35, 36). In addition, SIAH interacts with and promotes the degradation of different synaptic proteins, including the group 1 metabotropic glutamate receptor and the synaptic vesicle protein synaptophysin (36, 37). Moreover, endogenous SIAH was shown to be present and to co-localize with synaptophysin in neuritic processes of neuronally differentiated PC12 cells (37).
The inability of SIAH to promote the degradation of synphilin-1A is not unique as several substrates of SIAH, such as PHYL and Vav, are not targeted for degradation by the proteasome (38). Nevertheless, the small extent of synphilin-1A ubiquitylation promoted by SIAH increased the formation of synphilin-1A inclusions in the presence of proteasome inhibitors. Therefore, synphilin-1A/SIAH interaction promotes the formation of synphilin-1A inclusions rather than the degradation of synphilin-1A. Because both SIAH and synphilin-1A are present in Lewy bodies, ubiquitylation of synphilin-1A might be important for the formation of these inclusions.
SIAH ligase activity has been shown previously to be inhibited by the protein Disabled-1, but it was not clear whether it constitutes SIAH substrate (27). Synphilin-1A works as a “suicide” substrate as it inhibits both the autoubiquitylation and the ability of SIAH to ubiquitylate the substrates α-synuclein, synphilin-1, and DCC. The ability of synphilin-1A to inhibit both auto-ubiquitylation and ubiquitylation of SIAH toward substrates indicates that these activities of SIAH are interconnected. Further supporting the correlation between auto-ubiquitylation and ubiquitylation of substrates is the finding that phosphorylation by Cdk5 inhibits Parkin auto-ubiquitylation as well as ubiquitylation of its substrates p38 and synphilin-1 (29). SIAH is a pro-apoptotic protein that seems to promote cell death by activation of glyceraldehyde-3-phosphate dehydrogenase and JNK pathways (33, 39-41). In an attempt to dissect the connection between the ligase activity and toxicity of SIAH, we also investigated the effect of synphilin-1A on the ability of SIAH to promote cell death. We could not investigate the effect of synphilin-1A on the toxicity of SIAH in primary neuronal cultures because synphilin-1A is per se toxic in neurons (25). To avoid the toxic component of synphilin-1A, we chose to carry out the cell toxicity experiments in HEK293 cells, where we found that synphilin-1A exerts no significant direct toxicity. We found that, in addition to decreasing the ligase activity of SIAH, synphilin-1A decreased the ability of SIAH to promote cell death in HEK293 cells. Therefore, we propose that the toxicity of SIAH correlates with its ability to ubiquitylate substrates involved in triggering cell death. This observation is in accordance with the finding that activation of JNK requires the E3 ligase activity of SIAH (41).
Although synphilin-1A is toxic to primary cultured neurons from rat brain (25), we now found that it does not promote toxicity in HEK293 (Fig. 10) or SH-SY5Y cells (data not shown). The lack of toxicity in these cell types allowed us to investigate the role of synphilin-1A in SIAH toxicity and accumulation of monoubiquitylated α-synuclein inclusions. By inhibiting SIAH, synphilin-1A led to a significant decrease in the formation of monoubiquitylated α-synuclein inclusions in SH-SY5Y cells (Fig. 11B). On the other hand, our attempts to determine changes in α-synuclein inclusion formation by synphilin-1A in primary neuronal cultures were not successful because of synphilin-1A neurotoxicity.
Even though SH-SY5Y cells have a neuronal origin, the ability of synphilin-1A to promote cell death seems to be restricted to primary neurons. Differences in protein toxicity depending on the cell type have been previously reported by several studies. Overexpression of wild-type α-synuclein was shown to be toxic to neurons (42), but several studies have found that wild-type α-synuclein is not toxic to different types of cultured cells (43-46). Therefore, we speculate that dissimilarities between the cellular machinery of primary neuronal cultures and cultured cell lines could account for the difference in synphilin-1A toxicity. Moreover, this indicates that synphilin-1A toxicity in neurons may be mediated by still unidentified cellular pathways. Alternatively, aggregation of synphilin-1A, as observed in patients with Diffuse Lewy Body disease (25), could lead to less available synphilin-1A to neutralize SIAH toxicity in vivo. Future studies are necessary to determine the cellular pathways involved in the toxicity of synphilin-1A in neurons.
Similar to α-synuclein, synphilin-1 has been shown to play a synaptic role. Synphilin-1 is a presynaptic protein (47) and promotes the release of neurotransmitters in a cell model (22). Because synphilin-1A increases the steady-state levels of synphilin-1 (Fig. 7B), it is possible that synphilin-1A may modulate the synaptic function of synphilin-1 by increasing its concentration in the cell. Furthermore, by changing the steady-state levels of DCC (Fig. 7D), synphilin-1A may modulate axonal outgrowth (48). Likewise, synphilin-1A may affect hypoxic response by changing the degradation of HIF1α, a process controlled by SIAH through PHD proteins (49). Thus, synphilin-1A may be linked to different physiological events, such as axonal growth, neuronal activity, and hypoxic response.
In summary, we report that synphilin-1A interacts in vivo and inhibits the endogenous catalytic activity of SIAH, promoting accumulation of SIAH itself as well as substrates that it normally degrades, such as synphilin-1 and DCC. Our results also shed light on the mechanisms regulating α-synuclein monoubiquitylation, which has been implicated in α-synuclein aggregation (14). By interfering with the activity of SIAH, synphilin-1A also leads to decreased monoubiquitylation of α-synuclein and its aggregation into inclusions. Because monoubiquitylated α-synuclein inclusions are toxic to cells (14, 50), the ability of synphilin-1A to decrease the formation of α-synuclein inclusions indicates that synphilin-1A represents a novel therapeutic target to PD.
Acknowledgments
We thank Dr. K. L. Lim (National Neuroscience Institute, Singapore) for providing SH-SY5Y cells, Dr. O. Corti and Prof. A. Brice (INSERM, France) for the p38 cDNA, and Prof. M. Tessier-Lavigne (Genentech, San Francisco) for the DCC cDNA.
This work was supported by Israel Academy of Sciences, Ministry of Health/Ezvonot, The B. Rappaport Foundation, A. J. Berman Neurological Research Fund, Brauda Foundation Parkinson Disease Research Fund, M. Rojek Memorial Fund for Lewy Body Research, and the Technion Research funds (to S. E.).
Footnotes
The abbreviations used are: PD, Parkinson disease; E3, ubiquitin-protein isopeptide ligase; HA, hemagglutinin; SIAH, seven in absentia homolog; siRNA, short interfering RNA; GST, glutathione S-transferase; ATPγS, adenosine 5′-O-(thiotriphosphate); Chaps, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; JNK, c-Jun N-terminal kinase; DCC, deleted in colon cancer.
References
- 1.Moore, D. J., West, A. B., Dawson, V. L., and Dawson, T. M. (2005) Annu. Rev. Neurosci. 28, 57-87 [DOI] [PubMed] [Google Scholar]
- 2.Hardy, J., Cai, H., Cookson, M. R., Gwinn-Hardy, K., and Singleton, A. (2006) Ann. Neurol. 60, 389-398 [DOI] [PubMed] [Google Scholar]
- 3.Spillantini, M. G., Crowther, R. A., Jakes, R., Hasegawa, M., and Goedert, M. (1998) Proc. Natl. Acad. Sci. U. S. A. 95, 6469-6473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee, V. M., and Trojanowski, J. Q. (2006) Neuron 52, 33-38 [DOI] [PubMed] [Google Scholar]
- 5.McNaught, K. S., and Jenner, P. (2001) Neurosci. Lett. 297, 191-194 [DOI] [PubMed] [Google Scholar]
- 6.Kitada, T., Asakawa, S., Hattori, N., Matsumine, H., Yamamura, Y., Minoshima, S., Yokochi, M., Mizuno, Y., and Shimizu, N. (1998) Nature 392, 605-608 [DOI] [PubMed] [Google Scholar]
- 7.Leroy, E., Boyer, R., Auburger, G., Leube, B., Ulm, G., Mezey, E., Harta, G., Brownstein, M. J., Jonnalagada, S., Chernova, T., Dehejia, A., Lavedan, C., Gasser, T., Steinbach, P. J., Wilkinson, K. D., and Polymeropoulos, M. H. (1998) Nature 395, 451-452 [DOI] [PubMed] [Google Scholar]
- 8.Liani, E., Eyal, A., Avraham, E., Shemer, R., Szargel, R., Berg, D., Bornemann, A., Riess, O., Ross, C. A., Rott, R., and Engelender, S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101, 5500-5505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Junn, E., Lee, S. S., Suhr, U. T., and Mouradian, M. M. (2002) J. Biol. Chem. 277, 47870-47877 [DOI] [PubMed] [Google Scholar]
- 10.Ardley, H. C., Scott, G. B., Rose, S. A., Tan, N. G., and Robinson, P. A. (2004) J. Neurochem. 90, 379-391 [DOI] [PubMed] [Google Scholar]
- 11.Muqit, M. M., Abou-Sleiman, P. M., Saurin, A. T., Harvey, K., Gandhi, S., Deas, E., Eaton, S., Payne Smith, M. D., Venner, K., Matilla, A., Healy, D. G., Gilks, W. P., Lees, A. J., Holton, J., Revesz, T., Parker, P. J., Harvey, R. J., Wood, N. W., and Latchman, D. S. (2006) J. Neurochem. 98, 156-169 [DOI] [PubMed] [Google Scholar]
- 12.Avraham, E., Szargel, R., Eyal, A., Rott, R., and Engelender, S. (2005) J. Biol. Chem. 280, 42877-42886 [DOI] [PubMed] [Google Scholar]
- 13.Eyal, A., and Engelender, S. (2006) Cell Cycle 5, 2082-2086 [DOI] [PubMed] [Google Scholar]
- 14.Rott, R., Szargel, R., Haskin, J., Shani, V., Shainskaya, A., Manov, I., Liani, E., Avraham, E., and Engelender, S. (2008) J. Biol. Chem. 283, 3316-3328 [DOI] [PubMed] [Google Scholar]
- 15.Engelender, S., Kaminsky, Z., Guo, X., Sharp, A. H., Amaravi, R. K., Kleiderlein, J. J., Margolis, R. L., Troncoso, J. C., Lanahan, A. A., Worley, P. F., Dawson, V. L., Dawson, T. M., and Ross, C. A. (1999) Nat. Genet. 22, 110-114 [DOI] [PubMed] [Google Scholar]
- 16.Wakabayashi, K., Engelender, S., Yoshimoto, M., Tsuji, S., Ross, C. A., and Takahashi, H. (2000) Ann. Neurol. 47, 521-523 [PubMed] [Google Scholar]
- 17.Wakabayashi, K., Engelender, S., Tanaka, Y., Yoshimoto, M., Mori, F., Tsuji, S., Ross, C. A., and Takahashi, H. (2002) Acta Neuropathol. 103, 209-214 [DOI] [PubMed] [Google Scholar]
- 18.Bandopadhyay, R., Kingsbury, A. E., Muqit, M. M., Harvey, K., Reid, A. R., Kilford, L., Engelender, S., Schlossmacher, M. G., Wood, N. W., Latchman, D. S., Harvey, R. J., and Lees, A. J. (2005) Neurobiol. Dis. 20, 401-411 [DOI] [PubMed] [Google Scholar]
- 19.Murray, I. J., Medford, M. A., Guan, H. P., Rueter, S. M., Trojanowski, J. Q., and Lee, V. M. (2003) Acta Neuropathol. 105, 177-184 [DOI] [PubMed] [Google Scholar]
- 20.Chung, K. K., Zhang, Y., Lim, K. L., Tanaka, Y., Huang, H., Gao, J., Ross, C. A., Dawson, V. L., and Dawson, T. M. (2001) Nat. Med. 7, 1144-1150 [DOI] [PubMed] [Google Scholar]
- 21.Ito, T., Niwa, J., Hishikawa, N., Ishigaki, S., Doyu, M., and Sobue, G. (2003) J. Biol. Chem. 278, 29106-29114 [DOI] [PubMed] [Google Scholar]
- 22.Nagano, Y., Yamashita, H., Takahashi, T., Kishida, S., Nakamura, T., Iseki, E., Hattori, N., Mizuno, Y., Kikuchi, A., and Matsumoto, M. (2003) J. Biol. Chem. 278, 51504-51514 [DOI] [PubMed] [Google Scholar]
- 23.Szargel, R., Rott, R., and Engelender, S. (2008) Cell. Mol. Life Sci. 65, 80-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engelender, S. (2008) Autophagy 4, 372-374 [DOI] [PubMed] [Google Scholar]
- 25.Eyal, A., Szargel, R., Avraham, E., Liani, E., Haskin, J., Rott, R., and Engelender, S. (2006) Proc. Natl. Acad. Sci. U. S. A. 103, 5917-5922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kartvelishvily, E., Shleper, M., Balan, L., Dumin, E., and Wolosker, H. (2006) J. Biol. Chem. 281, 14151-14162 [DOI] [PubMed] [Google Scholar]
- 27.Park, T. J., Hamanaka, H., Ohshima, T., Watanabe, N., Mikoshiba, K., and Nukina, N. (2003) Biochem. Biophys. Res. Commun. 302, 671-678 [DOI] [PubMed] [Google Scholar]
- 28.Sriram, S. R., Li, X., Ko, H. S., Chung, K. K., Wong, E., Lim, K. L., Dawson, V. L., and Dawson, T. M. (2005) Hum. Mol. Genet. 14, 2571-2586 [DOI] [PubMed] [Google Scholar]
- 29.Avraham, E., Rott, R., Liani, E., Szargel, R., and Engelender, S. (2007) J. Biol. Chem. 282, 12842-12850 [DOI] [PubMed] [Google Scholar]
- 30.Xu, Z., Kohli, E., Devlin, K. I., Bold, M., Nix, J. C., and Misra, S. (2008) BMC Struct. Biol. 8, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corti, O., Hampe, C., Koutnikova, H., Darios, F., Jacquier, S., Prigent, A., Robinson, J. C., Pradier, L., Ruberg, M., Mirande, M., Hirsch, E., Rooney, T., Fournier, A., and Brice, A. (2003) Hum. Mol. Genet. 12, 1427-1437 [DOI] [PubMed] [Google Scholar]
- 32.Ko, H. S., von Coelln, R., Sriram, S. R., Kim, S. W., Chung, K. K., Pletnikova, O., Troncoso, J., Johnson, B., Saffary, R., Goh, E. L., Song, H., Park, B. J., Kim, M. J., Kim, S., Dawson, V. L., and Dawson, T. M. (2005) J. Neurosci. 25, 7968-7978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roperch, J. P., Lethrone, F., Prieur, S., Piouffre, L., Israeli, D., Tuynder, M., Nemani, M., Pasturaud, P., Gendron, M. C., Dausset, J., Oren, M., Amson, R. B., and Telerman, A. (1999) Proc. Natl. Acad. Sci. U. S. A. 96, 8070-8073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.House, C. M., Frew, I. J., Huang, H. L., Wiche, G., Traficante, N., Nice, E., Catimel, B., and Bowtell, D. D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100, 3101-3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu, G., Chung, Y. L., Glover, T., Valentine, V., Look, A. T., and Fearon, E. R. (1997) Genomics 46, 103-111 [DOI] [PubMed] [Google Scholar]
- 36.Moriyoshi, K., Iijima, K., Fujii, H., Ito, H., Cho, Y., and Nakanishi, S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101, 8614-8619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wheeler, T. C., Chin, L. S., Li, Y., Roudabush, F. L., and Li, L. (2002) J. Biol. Chem. 277, 10273-10282 [DOI] [PubMed] [Google Scholar]
- 38.Polekhina, G., House, C. M., Traficante, N., Mackay, J. P., Relaix, F., Sassoon, D. A., Parker, M. W., and Bowtell, D. D. (2002) Nat. Struct. Biol. 9, 68-75 [DOI] [PubMed] [Google Scholar]
- 39.Nemani, M., Linares-Cruz, G., Bruzzoni-Giovanelli, H., Roperch, J. P., Tuynder, M., Bougueleret, L., Cherif, D., Medhioub, M., Pasturaud, P., Alvaro, V., der Sarkissan, H., Cazes, L., Le Paslier, D., Le Gall, I., Israeli, D., Dausset, J., Sigaux, F., Chumakov, I., Oren, M., Calvo, F., Amson, R. B., Cohen, D., and Telerman, A. (1996) Proc. Natl. Acad. Sci. U. S. A. 93, 9039-9042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hara, M. R., Agrawal, N., Kim, S. F., Cascio, M. B., Fujimuro, M., Ozeki, Y., Takahashi, M., Cheah, J. H., Tankou, S. K., Hester, L. D., Ferris, C. D., Hayward, S. D., Snyder, S. H., and Sawa, A. (2005) Nat. Cell Biol. 7, 665-674 [DOI] [PubMed] [Google Scholar]
- 41.Xu, Z., Sproul, A., Wang, W., Kukekov, N., and Greene, L. A. (2006) J. Biol. Chem. 281, 303-312 [DOI] [PubMed] [Google Scholar]
- 42.Xu, J., Kao, S. Y., Lee, F. J., Song, W., Jin, L. W., and Yankner, B. A. (2002) Nat. Med. 8, 600-606 [DOI] [PubMed] [Google Scholar]
- 43.Tanaka, Y., Engelender, S., Igarashi, S., Rao, R. K., Wanner, T., Tanzi, R. E., Sawa, A., Dawson, V., Dawson, T. M., and Ross, C. A. (2001) Hum. Mol. Genet. 10, 919-926 [DOI] [PubMed] [Google Scholar]
- 44.Colapinto, M., Mila, S., Giraudo, S., Stefanazzi, P., Molteni, M., Rossetti, C., Bergamasco, B., Lopiano, L., and Fasano, M. (2006) Biochem. Biophys. Res. Commun. 349, 1294-1300 [DOI] [PubMed] [Google Scholar]
- 45.Qian, J. J., Cheng, Y. B., Yang, Y. P., Mao, C. J., Qin, Z. H., Li, K., and Liu, C. F. (2008) Neurosci. Lett. 435, 142-146 [DOI] [PubMed] [Google Scholar]
- 46.da Costa, C. A., Ancolio, K., and Checler, F. (2000) J. Biol. Chem. 275, 24065-24069 [DOI] [PubMed] [Google Scholar]
- 47.Ribeiro, C. S., Carneiro, K., Ross, C. A., Menezes, J. R., and Engelender, S. (2002) J. Biol. Chem. 277, 23927-23933 [DOI] [PubMed] [Google Scholar]
- 48.Hu, G., Zhang, S., Vidal, M., Baer, J. L., Xu, T., and Fearon, E. R. (1997) Genes Dev. 11, 2701-2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakayama, K., Frew, I. J., Hagensen, M., Skals, M., Habelhah, H., Bhoumik, A., Kadoya, T., Erdjument-Bromage, H., Tempst, P., Frappell, P. B., Bowtell, D. D., and Ronai, Z. (2004) Cell 117, 941-952 [DOI] [PubMed] [Google Scholar]
- 50.Lee, J. T., Wheeler, T. C., Li, L., and Chin, L. S. (2008) Hum. Mol. Genet. 17, 906-917 [DOI] [PubMed] [Google Scholar]