Abstract
Decorin, an archetypal member of the small leucine-rich proteoglycan gene family, regulates collagen fibrillogenesis and cell growth. To further explore its biological function, we examined the role of Decorin during zebrafish development. Zebrafish Decorin is a chondroitin sulfate proteoglycan that exhibits a high degree of conservation with its mammalian counterpart and displays a unique spatiotemporal expression pattern. Morpholino-mediated knockdown of zebrafish decorin identified a developmental role during medial-lateral convergence and anterior-posterior extension of the body plan, as well as in craniofacial cartilage formation. decorin morphants displayed a pronounced shortening of the head-to-tail axis as well as compression, flattening, and extension of the jaw cartilages. The morphant phenotype was efficiently rescued by zebrafish decorin mRNA. Unexpectedly, microinjection of excess zebrafish decorin mRNA or proteoglycan/protein core into one-cell stage embryos caused cyclopia. The morphant and overexpression phenotype represent a convergent extension defect. Our results indicate a central function for Decorin during early embryogenesis.
Proteoglycan-enriched extracellular matrices provide powerful messages to the cells via signaling events that vary from storing growth factors and morphogens to modulating their bioactivity and interactions with their cognate receptors (1). Decorin belongs to the family of the small leucine-rich proteoglycans (SLRPs)3 (2-4). The Decorin protein core directly binds type I collagen, a key biological interaction that controls the pace and extent of collagen fibril formation both in vitro and in vivo (5). The attached glycosaminoglycan (GAG) chain also contributes by coordinating the proper spacing between the fibrils (6). The structural requirement of Decorin during these events was clearly manifested by the decorin-null mice. Gene targeting of murine decorin resulted in irregular collagen fibril morphology associated with fragility of the skin (7). In addition to various collagen types, Decorin binds to Zn2+, fibrin, fibronectin, C1q, thrombospondin, transforming growth factor β, LRP1, and EGFR (8-17). Decorin is also involved in the pathogenesis of renal diseases (18-20), angiogenesis (21), wound healing (22), myocardial infarction (23), lung mechanics (24), tooth development (25), and bone marrow stromal cell biology (26). The function of Decorin through the EGFR has been extensively linked to the pathobiology of cancer (27-33).
Notably, double mutant mice lacking both decorin and the tumor suppressor p53 die early as a consequence of aggressive lymphomas (34), suggesting that Decorin is permissive for tumorigenesis. In line with this hypothesis, a recent study utilizing decorin-null animals, which were backcrossed into a different genetic background, has shown that the lack of decorin favors spontaneous occurrence of intestinal tumors in ∼30% of the cases, and this tumor burden and frequency were exacerbated by subjecting the mutant mice to a high risk diet (35).
In this study, to further explore the functions of Decorin, we utilized the zebrafish, Danio rerio, as a model organism. We identified zebrafish Decorin as a chondroitin sulfate proteoglycan that maintains a significant degree of conservation with the mammalian counterpart. Focusing on embryonic development, we defined the developmental expression profile of zebrafish decorin and applied a morpholino knockdown approach to block endogenous Decorin expression. The decorin morphants displayed a range of phenotypes characterized by progressive shortening of the body axis associated with abnormal craniofacial cartilage development. Zebrafish decorin mRNA was capable of rescuing the morphant phenotype. Interestingly, we found that an excess of zebrafish decorin mRNA induced cyclopia. Both the morphant and overexpressing phenotypes represent defects in embryonic convergent extension, the coordinated movement of embryonic cells in the anterior-posterior and medial-lateral directions (36). Convergent extension cell movements are required for proper establishment of the vertebrate body plan and are largely mediated by the noncanonical Wnt pathway (planar cell polarity pathway) converging on RhoA and Rac as downstream effectors of cell movement (37-40). Collectively, our findings indicate a novel and crucial role for Decorin function during establishment of the embryonic body plan.
EXPERIMENTAL PROCEDURES
Cloning, Analysis, Synthesis, and Purification of Zebrafish decorin mRNA and Protein—Zebrafish decorin was PCR-amplified from a zebrafish cDNA template with the following primer pair, 5′-GGCGCGCCTTATGAAATCGGCCTGTCTCTCCCTG-3′ and 5′-CTCGAGCTTCTTCCTGTAGTTGCCGAGCT-3′ (Operon). The zebrafish decorin coding sequence was cloned into pCRII:TOPO (Invitrogen). Zebrafish decorin was then subcloned into the AsCI and XhoI sites of the pCEP-Pu-Hulk vector. pCEP-Pu-Hulk-zDcn was transfected by Lipofectamine (Invitrogen) into human embryonic kidney cells (293-EBNA) for the synthesis and purification of recombinant zebrafish Decorin as described previously (41, 42).
Chondroitinase ABC Digestion—Recombinant zebrafish Decorin (∼0.5 μg) was subjected to chondroitinase ABC digestion (6.5 milliunits, Sigma) for 5 h at 37 °C in a buffer containing 0.1 m Tris base, 30 mm sodium acetate, pH 8.0. Human recombinant Decorin (0.2 μg) was used as positive control for the digestion reaction. Samples were then separated on an 8% SDS-PAGE and detected by immunoblotting with a monoclonal anti-His antibody (Qiagen).
Functional Assays with Zebrafish Decorin—For EGFR analysis, HeLa cells were plated on a 12-well plate (Nunc) and grown to subconfluency followed by serum starvation overnight. The next day cells were incubated with zebrafish Decorin (30 μg/ml) for 2 h, washed twice with Dulbecco's PBS, followed by extraction with RIPA buffer. Lysates were then separated on an 8% SDS-PAGE and subjected to immunoblotting with polyclonal anti-EGFR (Santa Cruz Biotechnology) and anti-phospho-EGFR-Tyr-1068 (Cell Signaling). β-Actin was used as loading control. For matrix binding analysis, glass slides were coated with collagen type I, fibronectin, or laminin 111 (BD Biosciences). Slides were incubated overnight at 37 °C with conditioned media from 293-EBNA cells, as negative control, or 293-EBNA cells expressing zebrafish decorin. The next day, wells were washed three times with Dulbecco's PBS and subjected to standard immunostaining with monoclonal anti-His (Qiagen), followed by secondary anti-mouse rhodamine-conjugated antibody (Santa Cruz Biotechnology). Fluorescence images were acquired using an Olympus BX51 microscope driven by SPOT advanced version 4.0.9 imaging software (Diagnostic Instruments, Inc.). The fluorescence intensity (pixels) representing zebrafish Decorin bound to the matrix components was obtained using the histogram function of Adobe Photoshop CS2®. For tumor cell proliferation and apoptosis assays, HeLa cells were seeded in 96-well plates and grown in full serum medium for 48 h in the presence or absence of zDcn (30 μg/ml). Media were changed after 24 h. Proliferation was measured by incubation with CellTiter 96® AQueous One Solution (Promega) at day 0 and after 24 and 48 h of zDcn treatment. Apoptosis was measured using the Caspase-Glo® 3/7 Assay (Promega). Both kits were used following the manufacturer's instructions. Proliferation activity was measured at 490 nm, and luminosity was measured to detect apoptosis.
Zebrafish Decorin Glycosaminoglycan Analysis—Recombinant zebrafish Decorin glycosaminoglycan chains (GAGs) were released from purified proteoglycan samples following treatment with 1 m NaBH4 in 0.5 n NaOH at 37 °C. The released GAGs were precipitated from the solution with cold ethanol and resuspended in water. Selected samples were then treated with either chondroitinase ABC or nitrous acid, pH 1.5. Samples were applied to cellulose acetate membranes (Super Sepraphore, Pall Corp.) along with hyaluronic acid/chondroitin sulfate mixed standards, and electrophoresis was performed in 0.1 m KH2PO4, pH 2.0, for 3 h at 35 mA. Following electrophoresis, GAGs were visualized by staining with 0.2% Alcian blue.
Zebrafish Embryos, decorin Morpholino Design, and Microinjection—Wild-type zebrafish embryos were cultured at ∼28 °C according to common procedures in phenylthiourea-supplemented embryo medium to prevent pigmentation. All embryos were housed in the zebrafish facility at Thomas Jefferson University according to the guidelines put forth by IACUC. Morpholino antisense oligonucleotides (Gene Tools, LLC) were designed to target/block the 5′-untranslated region/translation start (Dcn-MOSTART) or to target/block a splice junction (Dcn-MOSPLICE) within zebrafish decorin. decorin morpholino sequences were as follows: Dcn-MOSTART GACAGGCCGATTTCATGTTGCTGAC and Dcn-MOSPLICE GCAGACCTGGGCATTTTGACACAGA. The nonspecific standard morpholino was used as a control (Control-MO). The standard control morpholino sequence was as follows: Control-MO CCTCTTACCTCAGTTACAATTTATA. Nonspecific morpholino off-target effects were eliminated by co-knockdown with p53-MO as described previously (43). The p53 morpholino sequence was as follows: p53-MO GCGCCATTGCTTTGCAAGAATTG. Morpholino (≤20 ng per embryo) was microinjected into 1-cell stage zebrafish embryos according to common practice (44). Phenotype observations and overall gross morphology were visualized with either a Leica MZFIII stereo microscope and photographed with an Axiocam camera (Carl Zeiss, Inc.) and AxioVision software version 3.0.6.1 or a Leica MZ16FA stereo microscope equipped with a DFC500 camera and Application Suite version 2.5.0.r1 (Leica). Embryos were imaged in either 4% methylcellulose, PBS plus Tween 20 (0.1%) or embryo medium, and anesthetized with Tricaine when necessary. The morphant phenotype was defined by comparison with matched control embryos. All embryos were classified by 2 dpf as normal and described by 180° full extension of the head-to-tail axis; mild was described by body curvature associated with a nearly full extension of the tail without straightening; moderate was described by tail extension just past the yolk ball margin; and severe was described by abnormal short bodies associated with a tail that did not extend past the yolk ball.
Reverse Transcription-PCR—Zebrafish total RNA (n = 51 embryos/group for developmental analysis and n = 26 embryos/group for morpholino splice junction blocking) verification was isolated according to the TRIzol method (Invitrogen). For reverse transcription, ∼5 μg of total RNA was annealed with oligo(dT) primer (Roche Applied Science) at 70 °C for 5 min followed by the addition of 5× Moloney murine leukemia virus buffer (Thermo Fisher Scientific), 10 mm deoxynucleotide triphosphates (Thermo Fisher Scientific), RNasin (Promega), Moloney murine leukemia virus reverse transcriptase (Thermo Fisher Scientific), and incubation at 42 °C for 1 h. Reverse transcription reactions were heated at 90 °C for 10 min followed by incubation at 4 °C for 2 min before usage, or reactions were kept at 4 °C for prolonged cDNA storage. decorin PCRs contained cDNA, 10 mm deoxynucleotide triphosphates (Thermo Fisher Scientific), Taq polymerase (Fisher), 10 pmol/μl zebrafish decorin forward primer GCTTTTGCTGATCTGAAGAGGGTCT, and 10 pmol/μl zebrafish decorin reverse primer CTGCGGTCACTTTGGTGATCTTGTT (Operon). PCRs were analyzed on 2.5-4% agarose gel electrophoresis.
Zebrafish decorin Riboprobe Generation and Whole Mount in Situ Hybridization—For decorin riboprobe generation, zebrafish decorin sense/antisense riboprobes were synthesized by in vitro transcription from the TOPO:zebrafish decorin plasmid with Sp6 (Thermo Fisher Scientific) or T7 (Promega) RNA polymerase and were digoxigenin-labeled via digoxigenin-UTP (Roche Applied Science) incorporation during the in vitro transcription reaction. For ISH, RNA localization/detection with sense/antisense riboprobes was performed on groups of 10-20 embryos essentially as described previously (45). All embryos were photographed on an Axioplan2 microscope with an Axiocam camera and Axiovision software version 3.0.6.1.
Immunohistochemistry—For frozen sections, decorin ISH, RNA, or protein injected and uninjected embryos were fixed in 4% paraformaldehyde (Thermo Fisher Scientific) in PBS overnight. Embryos were washed in fix buffer (4% sucrose, 0.1 mm CaCl2, 16 mm NaH2PO4, 4 mm Na2HPO4; Fisher) three times for 5 min each. Embryos were immersed in 30% sucrose followed by embedding in OCT (Miles) at -20 °C. Frozen sections were prepared using a Thermo Shandon cryostat. All sections were placed on glass slides and stored at -80 °C. For immunohistochemistry, sections were immersed in ice-cold acetone for 5 min followed by immersion in 5% FBS (Sigma) for 1 h at room temperature. Sections were incubated with primary antibody, rabbit anti-human Decorin (a kind gift from Larry W. Fisher) at a 1:50 dilution in 1% FBS for 2 h at room temperature. Sections were washed in 1% FBS three times for 5 min each, followed by incubation with secondary antibody, goat anti-rabbit rhodamine (Santa Cruz Biotechnology), at a 1:300 dilution in 1% FBS for 2 h at room temperature. Sections were washed in 1% FBS three times for 5 min each. Sections were incubated with 4′,6-diamidino-2-phenylindole (Sigma) for 1 min followed by washing in 1% FBS for 5 min. Decorin-injected zebrafish sections incubated with the secondary antibody alone served as control. All sections were photographed on an Olympus BX 51 with a SPOT camera.
Alcian Blue Staining of the Zebrafish Cartilage—Five-day embryos were fixed overnight at 4 °C in 4% paraformaldehyde. Embryos were cleared in PBS containing Tween 20 (0.2%), followed by cartilage staining in 0.1% Alcian blue (Chroma-Gesellschaft) according to the procedure described previously (43) with the following modifications: 4 h of staining in sterilefiltered Alcian blue at 24 °C, followed by dehydration in ethanol series, washing in PBST, and incubation at 37 °C with 0.05% trypsin, 2.21 mm EDTA in Hanks' balanced salt solution (Cell-gro) for 2 h to digest excess tissue for clear visualization.
decorin RNA-based Rescue, Overexpression, and Protein Microinjection—The zebrafish decorin coding sequence was subcloned into the SpeI and BglII sites of pT3TS vector for the synthesis of in vitro transcribed mRNA via T3 polymerase (Ambion). For decorin RNA-based rescue experiments, zebrafish decorin RNA was microinjected alone or in combination with Dcn-MOSPLICE at the 1-cell stage. For Decorin protein-based experiments, human Decorin was microinjected into 1-cell stage zebrafish embryos. 0-4 h post-injection, injected and matched uninjected embryos were collected for immunoblot analysis. Total embryo protein lysates were collected by syringe homogenization in RIPA buffer over ice. Extracts were centrifuged at ∼7000 rpm at 4 °C for 10 min to remove insoluble material. Sample supernatants were separated by SDS-PAGE and subjected to immunoblotting with goat anti-human Decorin (Calbiochem) followed by rabbit anti-goat horseradish peroxidase (Calbiochem). Immunoblot densitometry was analyzed via Scion Image 4.0.4 Beta (Scion Corp.). For zebrafish embryo load control, a portion of the gel was stained with the colloidal blue staining kit (Invitrogen, LC6025).
RESULTS
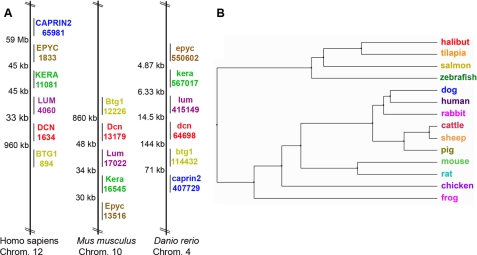
Characterization and Analysis of Zebrafish decorin—Our analysis of the zebrafish genome revealed the presence of one single gene encoding decorin (Gene ID: 64698). Analysis of the available data base indicated that the zebrafish decorin gene was physically located on chromosome 4 and maintained conserved synteny with human chromosome 12 and mouse chromosome 10 decorin genes (Fig. 1A). Interestingly, the zebrafish decorin gene clustered in a syntenic manner with related SLRP family members, including epiphycan, keratocan, and lumican. The zebrafish decorin mRNA sequence (∼1.1 kb) is composed of seven exons interspersed by six introns and encodes a protein of ∼373 amino acids (supplemental Fig. S1, A and B), with a predicted molecular mass of ∼38 kDa (mature protein). Comparative analysis of Decorin amino acid sequences from 14 different species highlighted common ancestry and fish species evolutionary relationships compared with mammalian, amphibian, and avian counterparts (Fig. 1B). Amino acid alignment of zebrafish, mouse, and human Decorin indicated that zebrafish Decorin was ∼67% identical and ∼78% homologous to the human counterpart (supplemental Fig. S1A). Zebrafish Decorin maintains classic SLRP architecture characterized by a central region of ten leucine-rich repeats flanked by N- and C-terminal Cys-rich regions.
FIGURE 1.
Chromosomal mapping and phylogenetic analysis of zebrafish decorin. A, zebrafish decorin exhibits conserved synteny. The zebrafish decorin gene (dcn) clusters in a syntenic manner with related SLRP family members, including epiphycan (epyc), keratocan (kera), and lumican (lum), as well as btg1 and caprin2, comparable with mouse and human. Species names are followed by the chromosome (Chrom.) number of interest; colored gene symbols are matched with the corresponding Entrez Gene ID, and distances between genes are displayed in kilo- or mega-base pairs. B, comparative phylogenetic analysis of Decorin among 14 different species. The tree was constructed on the basis of percent identity at the amino acid level.
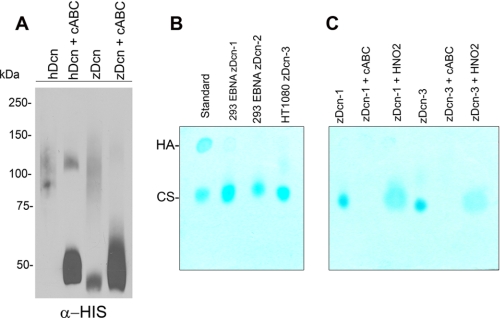
We cloned the entire zebrafish decorin by RT-PCR into pCRII:TOPO and subcloned the coding sequence into pCEP-Pu/HULK for the synthesis and purification of His-tagged recombinant zebrafish Decorin by 293-EBNA or HT1080 cells. Our initial characterization, based solely on amino acid sequence, predicted that zebrafish Decorin was a heparan sulfate (HS) proteoglycan. Interestingly, we found that zebrafish Decorin harbors one N-terminal SGD motif flanked by additional residues, which conform to the predicted HS attachment sites (46, 47), and which is strikingly similar to one of the known HS attachment sites found in human perlecan (supplemental Fig. S1C). Human and mouse Decorin harbor one N-terminal SG motif that serves as the attachment site for one chondroitin sulfate GAG chain. Despite the sequence-based prediction, we found that recombinant zebrafish Decorin was sensitive to chondroitinase ABC (cABC) digestion (Fig. 2A) indicating that zebrafish Decorin is a chondroitin sulfate proteoglycan. Further GAG analysis from conditioned media of 293-EBNA or HT1080 clones producing zebrafish Decorin indicated that the GAG chains of zebrafish Decorin were composed of chondroitin sulfate and were sensitive to chondroitinase ABC but not HNO2 (Fig. 2, B and C). Notably, zebrafish Decorin was capable of binding various matrix components and down-regulating EGFR levels, indicating that several biological functions of Decorin are well conserved (supplemental Fig. S2, A and B). Additionally, zebrafish Decorin was capable of inhibiting tumor cell proliferation and increasing tumor cell apoptosis (supplemental Fig. S2, C and D).
FIGURE 2.
Characterization and analysis of zebrafish Decorin. A, immunoblotting of zebrafish Decorin purified from human 293-EBNA cells. Notice that zebrafish Decorin is fully sensitive to cABC digestion (3rd and 4th lanes). Human Decorin proteoglycan (1st and 2nd lanes) serves as a positive control. B, glycosaminoglycan analysis of the conditioned medium of 293-EBNA (2nd and 3rd lanes) or HT1080 (4th lane) clones producing zebrafish Decorin indicates chondroitin sulfate comprises the GAG chain of zebrafish Decorin. 1st lane serves as a standard, hyaluronic acid (HA) and chondroitin sulfate (CS). C, GAG analysis from conditioned medium of 293-EBNA and HT1080 synthesizing zebrafish Decorin clones reveals sensitivity to cABC but not to HNO2. 1st and 4th lanes, untreated media; 2nd and 5th lanes, cABC-treated media; and 3rd and 6th lanes, HNO2-treated media. Numbers correspond to the clone labels as used in B.
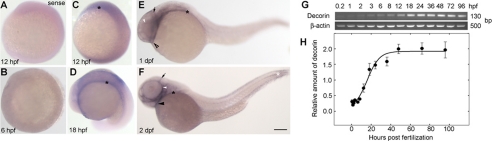
To examine the spatial and temporal expression pattern of decorin mRNA throughout zebrafish embryonic development, we performed whole mount ISH and reverse transcription-PCR across various stages of zebrafish development (Fig. 3). By ISH, decorin mRNA could not be detected earlier than the 6-somite stage, reflecting the low expression levels as detected by RT-PCR (Fig. 3B, shield stage, 6 hpf). Whole mount ISH using a digoxigenin-labeled decorin antisense riboprobe was capable of localizing decorin mRNA at the 6-somite stage (12 hpf) in regions of the developing head and tail (Fig. 3C). At the 20-somite stage (18 hpf) decorin mRNA was expressed throughout the lateral plate mesoderm (*) all along the trunk of the embryo (Fig. 3D). The expression level in the lateral plate mesoderm was decreased over time, with lower levels detected at the prime-5 stage (1 dpf) and hatching stage (2 dpf) (Fig. 3, E and F, respectively).
FIGURE 3.
Spatiotemporal analysis of zebrafish decorin developmental expression profile. A represents ISH with a decorin sense riboprobe. B-F, whole mount in situ hybridization with a digoxigenin-labeled decorin antisense riboprobe for the localization of decorin mRNA. decorin mRNA cannot be detected at the shield stage (6 hpf; B), but expression can be detected at 12 hpf (6 somites) in regions of the developing head and tail (*, C). At 18 hpf (18 somites), decorin mRNA is localized to the lateral plate mesoderm along the trunk (D). E and F, by 1 and 2 dpf, decorin expression can also be detected in regions of the developing fin (white *), head mesenchyme (black arrow), heart (black arrowhead), lateral plate mesoderm (*), and otic capsule (white arrowhead). G, zebrafish decorin expression analysis by RT-PCR. 1st to 13th lanes correspond to template cDNA derived from 0.2 to 96 hpf zebrafish embryos. Positive detection of decorin expression yields a 130-bp PCR product. PCR amplification of β-actin serves as a load control for the assay. H, quantification of zebrafish decorin expression level relative to β-actin derived from G. All PCR products were analyzed by AGE. A-E, bar, 315 μm; F, 500 μm.
The mesoderm gives rise to many kinds of tissue, including muscle, blood, cartilage, bone, and connective tissue, suggesting Decorin may contribute to the development of these tissues. The expression of decorin mRNA could also be detected at 1 and 2 dpf throughout the head mesenchyme (black arrow), otic capsule (white arrowhead), heart (black arrowhead), and developing fin (white *) (Fig. 3, E and F). Moreover, the expression pattern in these regions remained the same at 5 dpf (data not shown).
By using decorin-specific primers, we were able to detect the predicted 130-bp PCR product in cDNA derived from as early as 0.2 hpf to 4 dpf zebrafish embryos (Fig. 3, G and H). Analysis of decorin expression levels relative to β-actin over time suggested that very low decorin mRNA levels were present in the embryos at cleavage stage (0.2-2 hpf) (Fig. 3, G and H). Our findings indicate that low amounts of maternal decorin mRNA were deposited in the embryos because the onset of zygotic transcription does not initiate until at least 3-3.5 hpf. Low level mRNA was present in the embryos at blastula stage (3 hpf) and gastrula stage (shield stage, 6 hpf; 75% epiboly stage, 8 hpf). Higher levels of decorin mRNA could be detected at the 6-somite stage (12 hpf) (Fig. 3, G and H). The levels of decorin mRNA were higher at the 20-somite stage (18 hpf), prime-5 stage (1 dpf), prime-25 stage (36 hpf), and hatching stage (2 dpf), respectively (Fig. 3H). We found the levels of decorin mRNA to remain constant from 2 to 4 dpf (Fig. 3H). These results correspond to the decorin expression data available on the zebrafish developmental profile web resource (supplemental Fig. S3) (48).
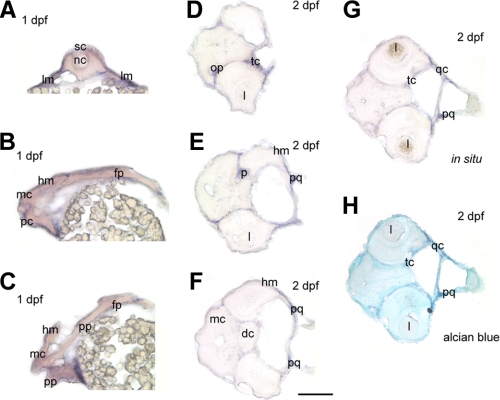
Cross-sections through 1 dpf zebrafish embryos further revealed decorin expression within the lateral plate mesoderm along both sides of the trunk (Fig. 4A) and in the head mesenchyme and heart region (Fig. 4B and C). Our expression profile in the zebrafish is consistent with the previously characterized murine decorin also detected in the heart of mouse embryos (49). Cross-sections through 2 dpf embryos also localized expression within the olfactory placode, trabecular cartilage, palatoquadrate, and quadrate cartilages (Fig. 4, D-G). decorin expression could also be detected in a region corresponding to the possible pituitary gland (Fig. 4E), in line with the murine expression profile (49). In addition, high levels of decorin expression were also detected in the parachondral cartilage and pharyngeal arches (data not shown). These results reveal decorin expression that is largely localized throughout the developing cartilage. Moreover, after cryosection of whole mount ISH embryos, the sections were counterstained with Alcian blue. The positive regions of decorin in situ hybridization colocalized with Alcian blue-stained regions (Fig. 4H) thereby supporting the cartilage localization.
FIGURE 4.
Cryosections of 1 and 2 dpf zebrafish embryos. All sections were obtained from whole mount embryos that have undergone decorin in situ hybridization. A, cross-section through a 1 dpf embryo at the trunk level high-lighting decorin expression in the lateral plate mesoderm (lm). B and C, longitudinal sections of a 1 dpf embryo through the head region indicating decorin expression in the developing head mesenchyme (hm) and heart region. D-G, cross-sections through 2 dpf embryos at the head region, localizes decorin expression within the olfactory placode (op), trabecular cartilage (tc), placode (p), palatoquadrate (pq), quadrate cartilage (qc), and head mesenchyme (hm). H, cross-section from G was counterstained with Alcian blue correlating staining of the cartilage with regions of decorin positive expression by ISH. Spinal cord, sc; notochord, nc; mesencephalon, mc; prosencephalon, pc; placordal plate, pp; floor plate, fp; lens, l; diencephalon, dc. Bar, 250 μm.
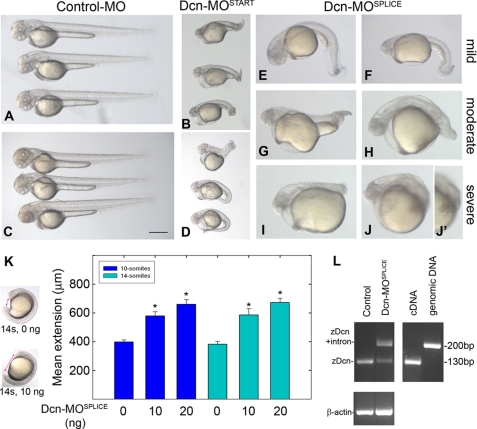
Consequences of decorin Knockdown, Abnormal Convergent Extension—To examine the role of Decorin during development, we applied a morpholino-based approach to knockdown zebrafish decorin. We designed two morpholinos of nonoverlapping sequence, one targeting and blocking the Decorin translation start (Dcn-MOSTART) and one targeting and blocking the splice junction between exons 4 and 5 of zebrafish decorin (Dcn-MOSPLICE). Both morpholinos induced a similar “truncated body” phenotype characterized by progressive shortening of the trunk and tail (Fig. 5, A-J′). By 2 dpf, all decorin morphants could be classified into the following three groups: (i) mild, described by body curvature associated with a nearly full extension of the tail without straightening (Fig. 5, E and F); (ii) moderate, described by tail extension just past the yolk ball margin (Fig. 5, G and H); and (iii) severe, described by abnormal short bodies associated with a tail that did not extend past the yolk ball (Fig. 5, I-J′). The mean morphant phenotype range could be represented by 23% mild, 25% moderate, 52% severe (n = 140, 2dpf). The shortened body axis phenotype suggested a possible defect in convergent extension.
FIGURE 5.
Morpholino-mediated knockdown of zebrafish decorin. A-J′, two morpholinos of nonoverlapping sequence, both targeting zebrafish decorin, induced a similar truncated body phenotype (B, translation start blocker Dcn-MOSTART, and E-J′, splice junction blocker Dcn-MOSPLICE). A represents microinjection with a nonspecific control-MO. C and D, decorin co-knockdown with p53-MO maintains the Dcn-MOSTART morphant phenotype (B) and eliminates morpholino off-target effects. C represents microinjection with p53-MO alone. E-J′, decorin morphants display a range of truncated body phenotypes, classified as mild (E and F), moderate (G and H), or severe (I-J′). K, measurement of the head-to-tail axis (arrows, left panel) and summary of the convergent extension defect (n = 78, 66, 43, 46, 34, 44 embryos each; *, p < 0.0001). L, RT-PCR verification of the splice junction blocking intron retaining effect of Dcn-MOSPLICE. Notice the presence of incorrectly spliced upper band (220 bp) in Dcn-MOSPLICE as well as the correctly spliced lower band (130 bp), suggesting partial knockdown. All lanes represent zebrafish decorin PCR using the following templates: uninjected embryo cDNA (n = 26, 1st lane), Dcn-MOSPLICE embryo cDNA (n = 26, 2nd lane), control cDNA (3rd lane), and genomic DNA (4th lane). PCR amplification of β-actin serves as a load control (lower panel). All PCR products were analyzed by agarose gel electrophoresis. All embryos including those used in RT-PCR are 2 dpf. Bar, ∼400 μm.
To examine this possibility, we measured the distance between the head and tail at earlier time points during which convergent extension cell movements establish the embryo body plan (Fig. 5K). Overall, we observed a significant increase in morphant head-to-tail distance as compared with matched control embryos (Fig. 5K), supporting a defect in convergent extension when low levels of endogenous Decorin are present.
To establish specificity of morpholino-targeted effects, we utilized two approaches. First, we used a nonspecific morpholino as a control in our experiments, and this did not induce any observable phenotype (Fig. 5A). Second, to eliminate nonspecific off-target morpholino effects, we performed co-knockdown experiments with p53-MO as described previously (43). decorin co-knockdown with p53-MO maintained the decorin morphant truncated body phenotype (Fig. 5, B and D). Knockdown of p53 alone by microinjection of p53-MO did not result in any observable phenotype consistent with previous reports that rule out a developmental requirement for p53 (Fig. 5C).
We verified the effects of the splice-blocking Dcn-MOSPLICE by RT-PCR. Total RNA extracted from a pool of 26 2-dpf control or morphant embryos was reverse-transcribed for the synthesis of cDNA. We used the cDNA template for PCR amplification of the morpholino-targeting splice junction boundary. Amplification of this region indicated intron retention as demonstrated by the presence of the upper band of ∼220 bp in the morphant embryos compared with the correctly spliced controls, lower band of ∼130 bp (Fig. 5L). We used zebrafish cDNA and genomic DNA as controls for decorin PCR and β-actin as a control for template loads (Fig. 5L). The presence of a smaller amount of the correctly spliced band in the morphant embryos suggests a partial knockdown effect by Dcn-MOSPLICE.
Consequences of decorin Knockdown, Abnormal Craniofacial Development—A closer examination of the decorin morphant phenotype revealed a possible mild craniofacial defect characterized by abnormal jaw appearance and a possible slight reduction in head size. Because our ISH revealed significant positive expression in cartilage, we hypothesized that decorin knockdown may influence cartilage development. To this end, we stained the decorin morphant embryos with Alcian blue to visualize cartilage development.
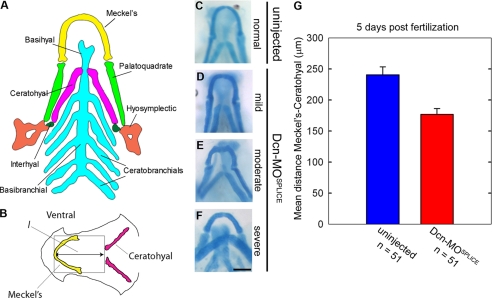
Zebrafish cartilage is derived from neural crest cells, and the first cartilaginous structures to develop include the Meckel's and hyosymplectic followed by interhyal, all contributing to the future supportive structures of the jaw (50). Fig. 6A represents a schematic dorsal view of the cartilage anatomy in 5-dpf embryos. The major framework of the future jaw (Fig. 6A) includes the outer u-shaped Meckel's, left and right palatoquadrate, and the hyosymplectic cartilages. The inner cartilages are composed of a basibranchial running down the midline accompanied by 5 pairs of ceratobranchials on each side. The ceratohyal and interhyal cartilages lie between the outer and inner structures. Because the Meckel's, ceratohyal, and palatoquadrate cartilages are clearly visible as distinct structures by 5 dpf, we performed our comparative analysis of craniofacial cartilage development at this time.
FIGURE 6.
decorin morphants display compression of the craniofacial cartilages. A, 5 dpf ventral view of zebrafish cranial cartilage anatomy. B, measurements of Dcn-MOSPLICE craniofacial cartilage compression was estimated by l, the distance between the peak of the Meckel's and ceratohyal cartilage. C-F, 5 dpf Dcn-MOSPLICE embryos display a range of cartilage compression that can be classified as mild (D), moderate (E), or severe (F). C serves as a control for comparison. G, graphical summary of the measurements of l comparing control and morphant embryos, ± S.E. Bar, ∼100 μm.
Zebrafish control cartilage exhibited an elongated Meckel's and ceratohyal, forming an arrow within an arrow conformation with anterior projection (Fig. 6, B and C). In contrast the Dcn-MOSPLICE morphants displayed compression of the cartilage (Fig. 6D), described by a narrowing between the Meckel's and ceratohyal cartilages, or flattening/reversal of the ceratohyal, described by a triangle or diamond shape conformation (Fig. 6, E and F, respectively). The Meckel's, ceratohyal, and palatoquadrate were abnormally short, whereas the ceratobranchials appeared reduced or absent. As well as direction of growth and shortening, the structure of the morphant Meckel's, ceratohyal, and palatoquadrate was frequently malformed, often exhibiting bends or kinks.
To provide a quantification of these observations, we measured the distance between the peak of the Meckel's and ceratohyal (l) as an estimate of compression to quantify these differences (Fig. 6G). We found that morphants with a reversed ceratohyal had an l value similar to that of the controls (281 versus 302 μm, respectively), whereas the morphants with anterior projecting (216 μm) or flattened ceratohyal (233 μm) had l values significantly lower than the controls (n = 51 each group). A similar phenotype was found in Dcn-MOSTART (data not shown).
Notably, we found a similar effect even in the absence of visible defects in convergent extension or edema, supporting a role for decorin during normal cartilage development. Therefore, the cartilage phenotype does not appear to be a secondary effect caused by pressure from pericardial edema or a raised yolk sac mechanically damaging cartilage development. We conclude that decorin knockdown causes defects in craniofacial cartilage development characterized by severe structural malformations and misdirected projections of the cartilage.
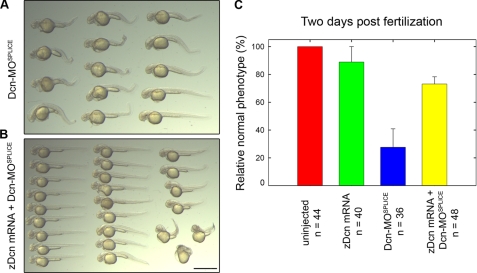
Zebrafish decorin mRNA Can Rescue the decorin Morphant Phenotype—Morpholino target specificity can be verified via rescue experiments with the target mRNA. To assess our decorin morpholino, design we attempted to rescue decorin knockdown with in vitro transcribed zebrafish decorin mRNA. We found that co-injection of Dcn-MOSPLICE along with zebrafish decorin mRNA was capable of rescuing the decorin morphant phenotype (Fig. 7).
FIGURE 7.
Zebrafish decorin mRNA can rescue Dcn-MOSPLICE. A, overview of the Dcn-MOSPLICE truncated body phenotype. B, overview, coinjection of zebrafish decorin mRNA and Dcn-MOsplice enhances survival and rescues the truncated body phenotype. C, 2dpf summary of zebrafish decorin mRNA-based rescue. Bar, ∼1.5 mm.
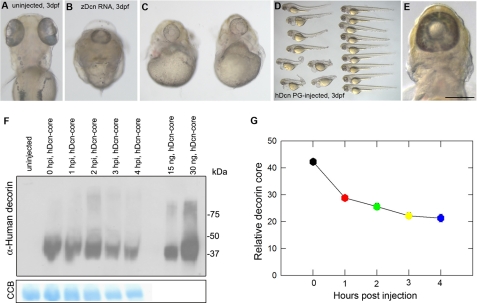
Microinjections of Excess Zebrafish Decorin mRNA or Human decorin Cause Defective Convergent Extension and Cyclopia—Injection of zebrafish decorin alone at the rescue dose did not result in any significant phenotype (data not shown). However, to our surprise, we found that microinjection of an excess amount of zebrafish decorin mRNA was capable of inducing a rather striking phenotype in ∼30% of injected embryos. These embryos exhibited a severely truncated body lacking a trunk region but maintaining a visible head and tail. By 3 dpf, and the onset of eye pigmentation, we found that these embryos had one single eye located centrally on the head and positioned above the heart (Fig. 8, A-C). The cyclopia and abnormal body plans are also characteristic defects of convergent extension cell movements.
FIGURE 8.
Microinjection of excess decorin mRNA or protein disrupts convergent extension and causes cyclopia. A-C, 3 dpf zebrafish decorin mRNA-injected embryos exhibit defects in convergent extension and display cyclopia (dorsal view in B; frontal view in C). A represents a dorsal view of a 3-dpf uninjected control embryo for comparison. D and E, overview of n = 17 3-dpf embryos injected with human Decorin proteoglycan. Twenty four percent of protein-injected embryos display visible convergent extension defects (lower left region of D), some associated with cyclopia (E). F, microinjected human Decorin core can be detected by immunoblot analysis with goat anti-human Decorin antibody (top) 0-4 h post-injection. Colloidal Coomassie Blue (CCB) staining (bottom) serves as a load control. Decorin recovery over time (n = 6 experiments) can be summarized as in G. Bar, ∼140 μm.
Next, we tested whether a similar phenotype could be evoked by excess amounts of human Decorin proteoglycan/protein core, which we have purified from 293-EBNA cells and routinely tested in our laboratory in functional assays both in vitro and in mouse models of tumorigenesis (21, 30-32, 41, 51). Zebrafish embryos injected with an excess of human Decorin displayed a similar shortened body phenotype associated with cyclopia (Fig. 8, D and E), further corroborating the microinjection data obtained with zebrafish mRNA.
To verify the presence and examine the turnover rate of Decorin protein post-microinjection, we performed immunoblot analysis on pools of injected embryos over time (Fig. 8F). We collected embryos every hour starting immediately post-injection (0 time point) up to 4 h. We utilized gel staining with colloidal blue as a load control. Using an antibody that recognizes Decorin, we detected the injected protein across all time points with ∼50% of the injected material still present even 4 h later (Fig. 8G). Using immunohistochemistry, we detected the injected Decorin pericellularly in frozen sections of 4-hpf embryos (supplemental Fig. S4). Our results indicate a relatively long half-life for the microinjected Decorin protein core and corroborate the results obtained with microinjections of capped zebrafish decorin mRNA.
DISCUSSION
Our analysis found that zebrafish decorin maintains a significantly high degree of conservation with the mammalian counterparts. Zebrafish decorin maintains synteny and includes seven exons versus eight as found in human or mouse species. The exon organization still maintains the standard SLRP design at the amino acid level. The zebrafish Decorin protein is composed of 373 amino acids arranged by 10 internal leucine-rich repeats flanked by N- and C-terminal cysteine-rich regions. Interestingly the N-terminal region does not maintain the classic SG glycosaminoglycan attachment site for chondroitin sulfate as found in mouse and human Decorin. Zebrafish Decorin harbors one SGD motif, flanked by additional residues that actually conform to the classic HS attachment sites as found in perlecan. Despite our observation at the amino acid level, chondroitinase ABC sensitivity and subsequent GAG analysis, both based upon recombinant zebrafish Decorin, revealed that zebrafish Decorin is a chondroitin sulfate proteoglycan. Although our analysis of in vitro synthesized recombinant zebrafish Decorin is indeed a chondroitin sulfate proteoglycan, we cannot rule out the possibility of in vivo attachment of HS chains.
Our application of the zebrafish, Danio rerio, has led to the identification of a requirement for periecan during developmental angiogenesis (45). Such a role had not previously been defined by classical mouse Hspg2 knock-out approaches (52, 53), possibly because of compensatory mechanisms. In a similar context the decorin null mice mainly display skin fragility manifested at the ultra-structural level by abnormal collagen fibrillogenesis (7). To further explore the function of Decorin, we generated the first decorin “knockdown” embryos. The decorin morphant phenotype could be characterized by 2 dpf as a progressive shortening of the embryo body head-to-tail axis. Examination of early embryonic head-to-tail distance during somitogenesis (<24 hpf) supported our observations at later time points. Combined, the decorin morphant phenotype represents a convergent extension defect and indicates for the first time a role for Decorin during these events. The morphant craniofacial cartilage defect and cyclopia observed in overexpressing embryos also represent common features of convergent extension defects (37, 38, 54). The craniofacial cartilage defect may be independent of convergent extension because milder embryos, without visible convergent extension defects, still exhibited the phenotype. Convergent extension cell movements define the proper establishment of the embryo anterior-posterior, mediallateral axis (36). Convergent extension is largely mediated by the noncanonical Wnt pathway converging on downstream effectors RhoA and Rac (36). decorin knockdown phenocopies several zebrafish mutants or morphants of noncanonical Wnt signaling pathway components. For example, mutation of the noncanonical Wnts, wnt5/pipetail or wnt11/silberblick, display defects in convergent extension associated with cyclopia (39, 55, 56). Interestingly, decorin-deficient mice develop tumors in the intestine with a concurrent up-regulation of β-catenin levels (35), a known downstream effector of the Wnt pathway. The exact role of Decorin function during convergent extension cell movements, possibly via the noncanonical Wnt pathway, is the current focus of our investigation.
In this study, we have applied a protein-based approach within the context of Decorin and Perlecan, as reported previously (45). However, we find a largely biased preponderance throughout the current literature for classical mRNA approaches with very few reports on protein-based experiments. To this end, we would like to establish the feasibility of protein-based methods in zebrafish. First, we observed similar phenotypic consequences upon microinjection of decorin RNA or protein. Second, we verified the presence of the injected protein by whole mount immunohistochemistry. Additionally, we could immunologically detect the presence of apparently intact Decorin protein core within the embryo lysate even 4 h post-injection. We consider the protein as a valid alternative to the RNase-sensitive nature of in vitro transcribed mRNA. Finally, we suggest that under certain instances protein injection would bypass the need for assumed translation of the injected mRNA. Together our protein-based methods represent an equal or perhaps better approach compared with classical RNA-based methods.
In conclusion, we report for the first time a complete molecular analysis and characterization of zebrafish decorin. Our study highlights convergent extension and craniofacial cartilage development as two novel aspects of Decorin function. Finally, we substantiate protein-based zebrafish experiments as opposed to classical RNA-based methods.
Supplementary Material
Acknowledgments
We thank A. McQuillan for expert technical assistance and for the artistic contribution, A. Lee for technical assistance with cloning of zebrafish decorin, and M. Hammerschmidt for valuable advice.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 CA39481, RO1 CA47282, and RO1 CA120975 (to R. V. I.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S4.
Footnotes
The abbreviations used are: SLRPs, small leucine-rich proteoglycans; GAG, glycosaminoglycan; EGFR, epidermal growth factor receptor; HS, heparan sulfate; cABC, chondroitinase ABC; ISH, in situ hybridization; FBS, fetal bovine serum; PBS, phosphate-buffered saline; dpf, days post-fertilization; hpf, hours post-fertilization; RT, reverse transcription; MO, morpholino.
References
- 1.Ramirez, F., and Rifkin, D. B. (2003) Matrix Biol. 22 101-107 [DOI] [PubMed] [Google Scholar]
- 2.Ameye, L., and Young, M. F. (2002) Glycobiology 12 R107-R116 [DOI] [PubMed] [Google Scholar]
- 3.Iozzo, R. V. (1999) J. Biol. Chem. 274 18843-18846 [DOI] [PubMed] [Google Scholar]
- 4.Schaefer, L., and Iozzo, R. V. (2008) J. Biol. Chem. 283 2135-2139 [Google Scholar]
- 5.Reed, C. C., and Iozzo, R. V. (2002) Glycoconj. J. 19 249-255 [DOI] [PubMed] [Google Scholar]
- 6.Rühland, C., Schönherr, E., Robenek, H., Hansen, U., Iozzo, R. V., Bruckner, P., and Seidler, D. G. (2007) FEBS J. 274 4246-4255 [DOI] [PubMed] [Google Scholar]
- 7.Danielson, K. G., Baribault, H., Holmes, D. F., Graham, H., Kadler, K. E., and Iozzo, R. V. (1997) J. Cell Biol. 136 729-743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang, V. W. C., LaBrenz, S. R., Rosenberg, L. C., McQuillan, D., and Höök, M. (1999) J. Biol. Chem. 274 12454-12460 [DOI] [PubMed] [Google Scholar]
- 9.Dugan, T. A., Yang, V. W. C., McQuillan, D. J., and Höök, M. (2006) J. Biol. Chem. 281 38208-38216 [DOI] [PubMed] [Google Scholar]
- 10.Schmidt, G., Robenek, H., Harrach, B., Glössl, J., Nolte, V., Hörmann, H., Richter, H., and Kresse, H. (1987) J. Cell Biol. 104 1683-1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krumdieck, R., Höök, M., Rosenberg, L. C., and Volanakis, J. E. (1992) J. Immunol. 149 3695-3701 [PubMed] [Google Scholar]
- 12.Winnemöller, M., Schön, P., Vischer, P., and Kresse, H. (1992) Eur. J. Cell Biol. 59 47-55 [PubMed] [Google Scholar]
- 13.Hildebrand, A., Romaris, M., Rasmussen, L. M., Heinegård, D., Twardzik, D. R., Border, W. A., and Ruoslahti, E. (1994) Biochem. J. 302 527-534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferdous, Z., Wei, V. M., Iozzo, R. V., Höök, M., and Grande-Allen, K. J. (2007) J. Biol. Chem. 282 35887-35898 [DOI] [PubMed] [Google Scholar]
- 15.Iozzo, R. V., Moscatello, D., McQuillan, D. J., and Eichstetter, I. (1999) J. Biol. Chem. 274 4489-4492 [DOI] [PubMed] [Google Scholar]
- 16.Santra, M., Reed, C. C., and Iozzo, R. V. (2002) J. Biol. Chem. 277 35671-35681 [DOI] [PubMed] [Google Scholar]
- 17.Brandan, E., Cabello-Verrugio, C., and Vial, C. (2008) Matrix Biol. 27 700-708 [DOI] [PubMed] [Google Scholar]
- 18.Schaefer, L., Macakova, K., Raslik, I., Micegova, M., Gröne, H.-J., Schönherr, E., Robenek, H., Echtermeyer, F. G., Grässel, S., Bruckner, P., Schaefer, R. M., Iozzo, R. V., and Kresse, H. (2002) Am. J. Pathol. 160 1181-1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaefer, L., Mihalik, D., Babelova, A., Krzyzankova, M., Grone, H. J., Iozzo, R. V., Young, M. F., Seidler, D. G., Lin, G., Reinhardt, D., and Schaefer, R. M. (2004) Am. J. Pathol. 165 383-396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams, K. J., Qiu, G., Usui, H. K., Dunn, S. R., McCue, P., Bottinger, E., Iozzo, R. V., and Sharma, K. (2007) Am. J. Pathol. 171 1441-1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant, D. S., Yenisey, C., Rose, R. W., Tootell, M., Santra, M., and Iozzo, R. V. (2002) Oncogene 21 4765-4777 [DOI] [PubMed] [Google Scholar]
- 22.Järveläinen, H., Puolakkainen, P., Pakkanen, S., Brown, E. L., Höök, M., Iozzo, R. V., Sage, H., and Wight, T. N. (2006) Wound Repair Regen. 14 443-452 [DOI] [PubMed] [Google Scholar]
- 23.Weis, S. M., Zimmerman, S. D., Shah, M., Covell, J. W., Omens, J. H., Ross, J., Jr., Dalton, N., Jones, Y., Reed, C. C., Iozzo, R. V., and McCulloch, A. D. (2005) Matrix Biol. 24 313-324 [DOI] [PubMed] [Google Scholar]
- 24.Fust, A., LeBellego, F., Iozzo, R. V., Roughley, P. J., and Ludwig, M. S. (2005) Am. J. Physiol. 288 L159-L166 [DOI] [PubMed] [Google Scholar]
- 25.Goldberg, M., Septier, D., Rapoport, O., Iozzo, R. V., Young, M. F., and Ameye, L. G. (2005) Calcif. Tissue Int. 77 297-310 [DOI] [PubMed] [Google Scholar]
- 26.Bi, Y., Stueltens, C. H., Kilts, T., Wadhwa, S., Iozzo, R. V., Robey, P. G., Chen, X.-D., and Young, M. F. (2005) J. Biol. Chem. 280 30481-30489 [DOI] [PubMed] [Google Scholar]
- 27.Moscatello, D. K., Santra, M., Mann, D. M., McQuillan, D. J., Wong, A. J., and Iozzo, R. V. (1998) J. Clin. Investig. 101 406-412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Csordás, G., Santra, M., Reed, C. C., Eichstetter, I., McQuillan, D. J., Gross, D., Nugent, M. A., Hajnóczky, G., and Iozzo, R. V. (2000) J. Biol. Chem. 275 32879-32887 [DOI] [PubMed] [Google Scholar]
- 29.Santra, M., Eichstetter, I., and Iozzo, R. V. (2000) J. Biol. Chem. 275 35153-35161 [DOI] [PubMed] [Google Scholar]
- 30.Reed, C. C., Waterhouse, A., Kirby, S., Kay, P., Owens, R. A., McQuillan, D. J., and Iozzo, R. V. (2005) Oncogene 24 1104-1110 [DOI] [PubMed] [Google Scholar]
- 31.Seidler, D. G., Goldoni, S., Agnew, C., Cardi, C., Thakur, M. L., Owens, R. A., McQuillan, D. J., and Iozzo, R. V. (2006) J. Biol. Chem. 281 26408-26418 [DOI] [PubMed] [Google Scholar]
- 32.Goldoni, S., Seidler, D. G., Heath, J., Fassan, M., Baffa, R., Thakur, M. L., Owens, R. A., McQuillan, D. J., and Iozzo, R. V. (2008) Am. J. Pathol. 173 844-855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldoni, S., and Iozzo, R. V. (2008) Int. J. Cancer 123 2473-2479 [DOI] [PubMed] [Google Scholar]
- 34.Iozzo, R. V., Chakrani, F., Perrotti, D., McQuillan, D. J., Skorski, T., Calabretta, B., and Eichstetter, I. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 3092-3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bi, X., Tong, C., Dokendorff, A., Banroft, L., Gallagher, L., Guzman-Hartman, G., Iozzo, R. V., Augenlicht, L. H., and Yang, W. (2008) Carcinogenesis 29 1435-1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tada, M., Concha, M. L., and Heisenberg, C.-P. (2002) Semin. Cell Dev. Biol. 13 251-260 [DOI] [PubMed] [Google Scholar]
- 37.Topczewski, J., Sepich, D. S., Myers, D. C., Walker, C., Amores, A., Lele, Z., Hammerschmidt, M., Postlethwait, J., and Solnica-Krezel, L. (2001) Dev. Cell 1 251-264 [DOI] [PubMed] [Google Scholar]
- 38.Marlow, F., Zwartkruis, F., Malicki, J., Neuhauss, S. C. F., Abbas, L., Weaver, M., Driever, W., and Solnica-Krezel, L. (1998) Dev. Biol. 203 382-399 [DOI] [PubMed] [Google Scholar]
- 39.Heisenberg, C.-P., Tada, M., Rauch, G.-J., Saúde, L., Concha, M. L., Geisler, R., Stemple, D. L., Smith, J. C., and Wilson, S. W. (2000) Nature 405 76-81 [DOI] [PubMed] [Google Scholar]
- 40.Marlow, F., Topczewski, J., Sepich, D., and Solnica-Krezel, L. (2002) Curr. Biol. 12 876-884 [DOI] [PubMed] [Google Scholar]
- 41.Zhu, J.-X., Goldoni, S., Bix, G., Owens, R. A., McQuillan, D., Reed, C. C., and Iozzo, R. V. (2005) J. Biol. Chem. 280 32468-32479 [DOI] [PubMed] [Google Scholar]
- 42.Goldoni, S., Owens, R. T., McQuillan, D. J., Shriver, Z., Sasisekharan, R., Birk, D. E., Campbell, S., and Iozzo, R. V. (2004) J. Biol. Chem. 279 6606-6612 [DOI] [PubMed] [Google Scholar]
- 43.Robu, M. E., Larson, J. D., Nasevicius, A., Beiraghi, S., Brenner, C., Farber, S. A., and Ekker, S. C. (2007) PLoS Genet. 3 e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nasevicius, A., and Ekker, S. C. (2000) Nat. Genet. 26 216-220 [DOI] [PubMed] [Google Scholar]
- 45.Zoeller, J. J., McQuillan, A., Whitelock, J., Ho, S.-Y., and Iozzo, R. V. (2008) J. Cell Biol. 181 381-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bishop, J. R., Schuksz, M., and Esko, J. D. (2007) Nature 446 1030-1037 [DOI] [PubMed] [Google Scholar]
- 47.Wang, H., Julenius, K., Hryhorenko, J., and Hagen, F. K. (2007) J. Biol. Chem. 282 14586-14597 [DOI] [PubMed] [Google Scholar]
- 48.Ouyang, M., Garnett, A. T., Han, T. M., Hama, K., Lee, A., Deng, Y., Lee, N., Liu, H.-Y., Amacher, S. L., Farber, S. A., and Ho, S.-Y. (2008) Gene Expr. Patterns 8 171-180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scholzen, T., Solursh, M., Suzuki, S., Reiter, R., Morgan, J. L., Buchberg, A. M., Siracusa, L. D., and Iozzo, R. V. (1994) J. Biol. Chem. 269 28270-28281 [PubMed] [Google Scholar]
- 50.Neuhauss, S. C. F., Solnica-Krezel, L., Schier, A. F., Zwartkruis, F., Stemple, D. L., Malicki, J., Abdelilah, S., Stainier, D. Y. R., and Driever, W. (1996) Development (Camb.) 123 357-367 [DOI] [PubMed] [Google Scholar]
- 51.Reed, C. C., Gauldie, J., and Iozzo, R. V. (2002) Oncogene 21 3688-3695 [DOI] [PubMed] [Google Scholar]
- 52.Costell, M., Gustafsson, E., Aszódi, A., Mörgelin, M., Bloch, W., Hunziker, E., Addicks, K., Timpl, R., and Fässler, R. (1999) J. Cell Biol. 147 1109-1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arikawa-Hirasawa, E., Watanabe, E., Takami, H., Hassell, J. R., and Yamada, Y. (1999) Nat. Genet. 23 354-358 [DOI] [PubMed] [Google Scholar]
- 54.Matsui, T., Raya, A., Kawakami, Y., Callol-Massot, C., Capdevilla, J., Rodríguez-Esteban, C., and Belmonte, J. C. I. (2005) Genes Dev. 19 164-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hammerschmidt, M., Pelegri, F., Mullins, M. C., Kane, D. A., Brand, M., van Eeden, F. J. M., Furutani-Seiki, M., Granato, M., Haffter, P., Heisenber, C.-P., Jiang, Y.-J., Kelsh, R. N., Odenthatl, J., Warga, R. M., and Nüsslein-Volhard, C. (1996) Development (Camb.) 123 143-151 [DOI] [PubMed] [Google Scholar]
- 56.Kilian, B., Mansukoski, H., Barbosa, F. C., Ulrich, F., Tada, M., and Heisenberg, C.-P. (2003) Mech. Dev. 120 467-476 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.