Abstract
The cytokine transforming growth factor-β (TGF-β) is an important negative regulator of adaptive immunity1–3. TGF-β is secreted by cells as an inactive precursor that must be activated to exert biological effects4, but the mechanisms that regulate TGF-β activation and function in the immune system are poorly understood. Here we show that conditional loss of the TGF-β-activating integrin αvβ8 on leukocytes causes severe inflammatory bowel disease and age-related autoimmunity in mice. This autoimmune phenol-type is largely due to lack of αvβ8 on dendritic cells, as mice lacking αvβ8 principally on dendritic cells develop identical immunological abnormalities as mice lacking αvβ8 on all leukocytes, whereas mice lacking αvβ8 on T cells alone are phenotypically normal. We further show that dendritic cells lacking αvβ8 fail to induce regulatory T cells (TR cells) in vitro, an effect that depends on TGF-β activity. Furthermore, mice lacking αvβ8 on dendritic cells have reduced proportions of TR cells in colonic tissue. These results suggest that αvβ8-mediated TGF-β activation by dendritic cells is essential for preventing immune dysfunction that results in inflammatory bowel disease and autoimmunity, effects that are due, at least in part, to the ability of αvβ8 on dendritic cells to induce and/or maintain tissue TR cells.
The critical pathways for activation of latent TGF-β at specific sites in vivo remain controversial. One important mechanism for activation of two of the three mammalian TGF-β isoforms (TGF-β1 and TGF-β3) is through interaction with members of the integrin recaptor family5–7. The in vivo importance of TGF-β activation by integrins has been recently demonstrated in mice with a mutated integrin-binding motif in TGF-β1 (ref. 8). These mice completely phenocopy Tgfb1−/− mice, which die from multi-organ inflammatory disease1, showing that integrin-mediated TGF-β activation is vital to maintain immune homeostasis. However, the integrin(s) responsible for TGF-β activation in the immune system in vivo, where these integrins are expressed, and the mechanisms by which integrin-mediated TGF-β activation maintains immune homeostasis remain a mystery.
The integrin αvβ6 has previously been shown to activate TGF-β (ref. 6), but this integrin is restricted to a subset of epithelial cells and is not expressed on immune cells. Furthermore, the inflammatory phenotype of mice lacking αvβ6 is mild compared to mice lacking TGF-β1 or TGF-β3 (ref. 5). The αvβ8 integrin can also activate TGF-β (ref. 7). Mice completely lacking αvβ8 die before or shortly after birth from defects in brain vascular development9, so tissue-specific deletion was required to determine whether αvβ8-mediated TGF-β activation has a function in regulating adaptive immunity. Polymerase chain reaction with reverse transcription (RT–PCR) analysis revealed that β8 was expressed in total splenocytes, CD4+ T cells and dendritic cells (Supplementary Fig. 1a) but was present at very low or undetectable levels in macrophages, CD8+ T cells, natural killer cells and B cells. To assess the in vivosignificance of αvβ8 expression in the immune system, we produced mice with conditional loss of β8 on leukocytes by crossing mice homozygous for a floxed β8 (Itgb8) allele10 with mice expressing Cre recombinase under the control of the Vav1 promoter11 (hereafter called (Vav1-cre)Itgb8fl/fl). Quantitative (q)RT–PCR revealed efficient knockdown (>98%) of Itgb8 messenger RNA expression in CD4+ T cells and dendritic cells from (Vav1-cre)Itgb8fl/fl mice (Supplementary Fig. 1b).
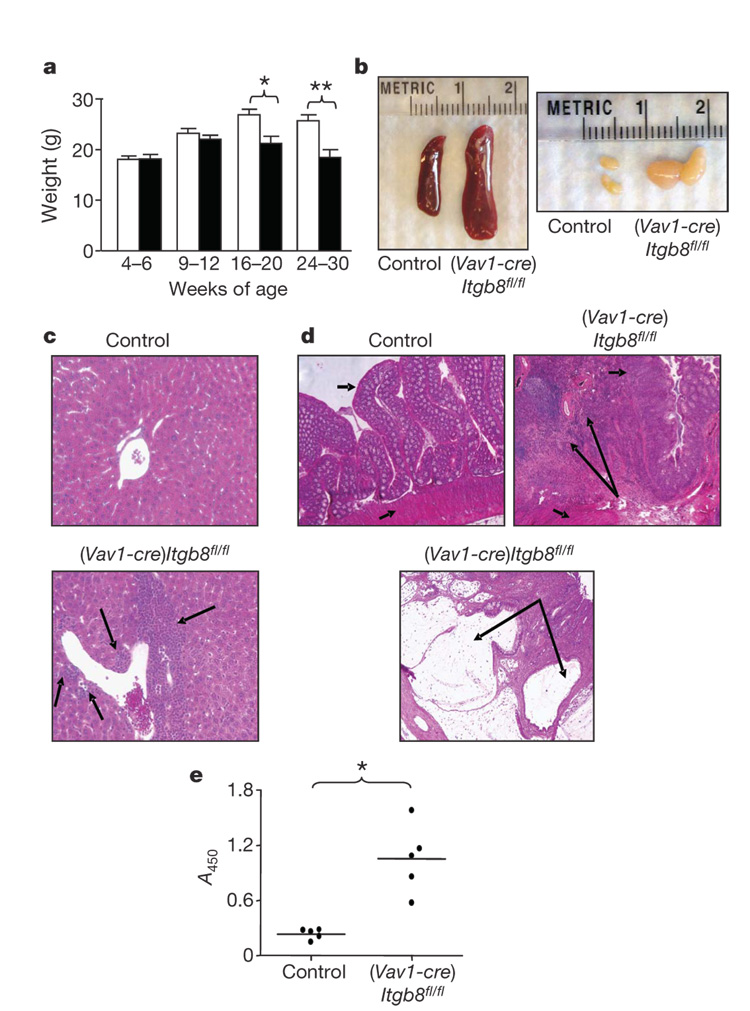
(Vav1-cre)Itgb8fl/fl mice were phenotypically normal until 4–5 months of age, when they began to develop a progressive wasting disorder (Fig. 1a). (Vav1-cre)Itgb8fl/fl mice also developed spleno-megaly, massive enlargement of mesenteric lymph nodes (Fig. 1b) and accumulations of mononuclear cells adjacent to portal triads of the liver (Fig. 1c). By 10 months of age, all surviving (Vav1-cre)Itgb8fl/fl mice developed severe colitis, characterized by cellular infiltration of the colonic wall with eosinophils and plasma cells, and formation of colonic cysts (Fig. 1d). These mice also developed high levels of auto-antibodies directed against double-stranded DNA and ribonuclear proteins (Fig. 1e). These findings are remarkably similar to phenotypes described for mice with a partial loss of TGF-β signalling in T cells as a result of expression of a dominant negative TGF-β receptor12, and for mice lacking the key TGF-β signalling protein Smad4 in T cells13. Mice lacking Smad4 in T cells also had increased tumorigenesis13, a finding we did not observe.
Figure 1. (Vav1-cre)Itgb8fl/fl mice develop age-related autoimmunity.
a, Weight loss in control and (Vav1-cre)Itgb8fl/fl mice (white, control; black, (Vav1-cre)Itgb8fl/fl; n=7 per group; asterisk, P=0.011; double asterisk, P=0.0026). Error bars represent s.e.m. b, Lymphoid organs of 5-month-old mice. c, Haematoxylin- and eosin-stained sections of livers (10 months, original magnification ×200). Arrows show cellular infiltrates. d, Haematoxylin- and eosin-stained colon sections (9 months, original magnification ×50). Short arrows, epithelium (top arrow) and smooth muscle (bottom arrow); large arrows, cellular infiltrates (top panel) and large cyst (bottom panel). e, ELISA for anti-dsDNA and anti-ribonuclear protein (9–12 months, n=5, P=0.0013).
PCR of stool samples revealed the presence of the common intestinal bacteria Helicobacter hepaticus and Helicobacter ganmani from all control and experimental mice tested. These organisms, endemic in our facility and in over 85% of mouse research colonies worldwide 14,15, are not pathogenic in most strains of mice, but have been reported to cause colitis and hepatic inflammation in some immune-suppressed strains16. We therefore cannot exclude the possibility that the presence of these organisms, or other unmeasured microbial flora, contribute to the severity of colonic and/or hepatic pathology in (Vav1-cre)Itgb8fl/fl mice. Such a response could be relevant to inflammatory bowel diseases in humans, where abnormal responses to normally non-pathogenic intestinal flora have been suggested to have a role17.
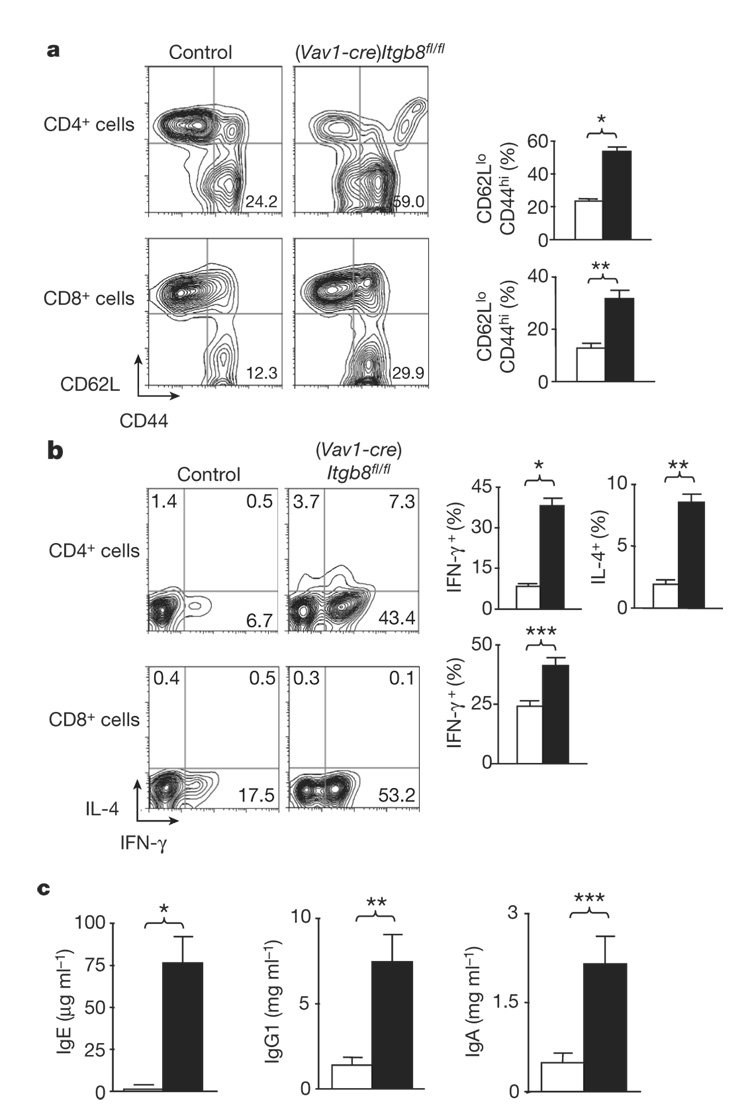
Mice with impaired T-cell responsiveness to TGF-β were also shown to have increased numbers of activated peripheral T cells, increased circulating levels of IgA and IgG1, and increased numbers of T cells that produce interleukin-4 (IL-4) and/or interferon-γ (IFN-γ)12. (Vav1-cre)Itgb8fl/fl mice (4–6 months old) also had enhanced numbers of activated/memory CD4+ and CD8+ T cells (Fig. 2a), and increased numbers of CD4+ T cells producing IFN-γ and IL-4, and CD8+ cells producing IFN-γ (Fig. 2b and Supplementary Fig. 2). (Vav1-cre)Itgb8fl/fl mice also had increased levels of circulating IgE, IgG1 and IgA (Fig. 2c), whereas levels of IgM, IgG2a, IgG2b and IgG3 were not significantly different (Supplementary Fig. 3). Again, these features were virtually identical to those described in mice with a partial loss of TGF-β signalling in T cells, suggesting that αvβ8 on leukocytes has an important role in activating TGF-β for presentation to T cells. This phenotype is not due to the mixed genetic background analysed in these initial experiments, because mice bred five generations to pure C57BL/6 background have a similar immune phenotype and also develop colitis (Supplementary Fig. 4).
Figure 2. (Vav1-cre)Itgb8fl/fl mice develop enhanced numbers of activated/memory T cells expressing IL-4 and IFN-γ, and increased serum IgE, IgG1 and IgA levels.
a, Activated/memory T cells from spleen (CD62LlowCD44high, 4–6-month-old mice) were analysed by flow cytometry. Representative flow cytometry plots and plotted mean values are shown (white, control; black, (Vav1-cre)Itgb8fl/fl; n=14; asterisk, P=2.7×10−11; double asterisk, P=4.9×10−5). b, IL-4 and IFN-γ levels in T cells from spleen were analysed by intracellular flow cytometry. Representative flow cytometry plots and plotted mean values are shown (white, control; black, (Vav1-cre)Itgb8fl/fl; n=9; asterisk, P=1.8×10−8; double asterisk, P=1.8×10−7; triple asterisk, P=0.00048). c, ELISA for IgE, IgG1 and IgA levels in sera (4–6-month-old mice; white, control; black, (Vav1-cre)Itgb8fl/fl; n=4; asterisk, P=0.0028; double asterisk, P=0.011; triple asterisk, P=0.016). All error bars represent s.e.m.
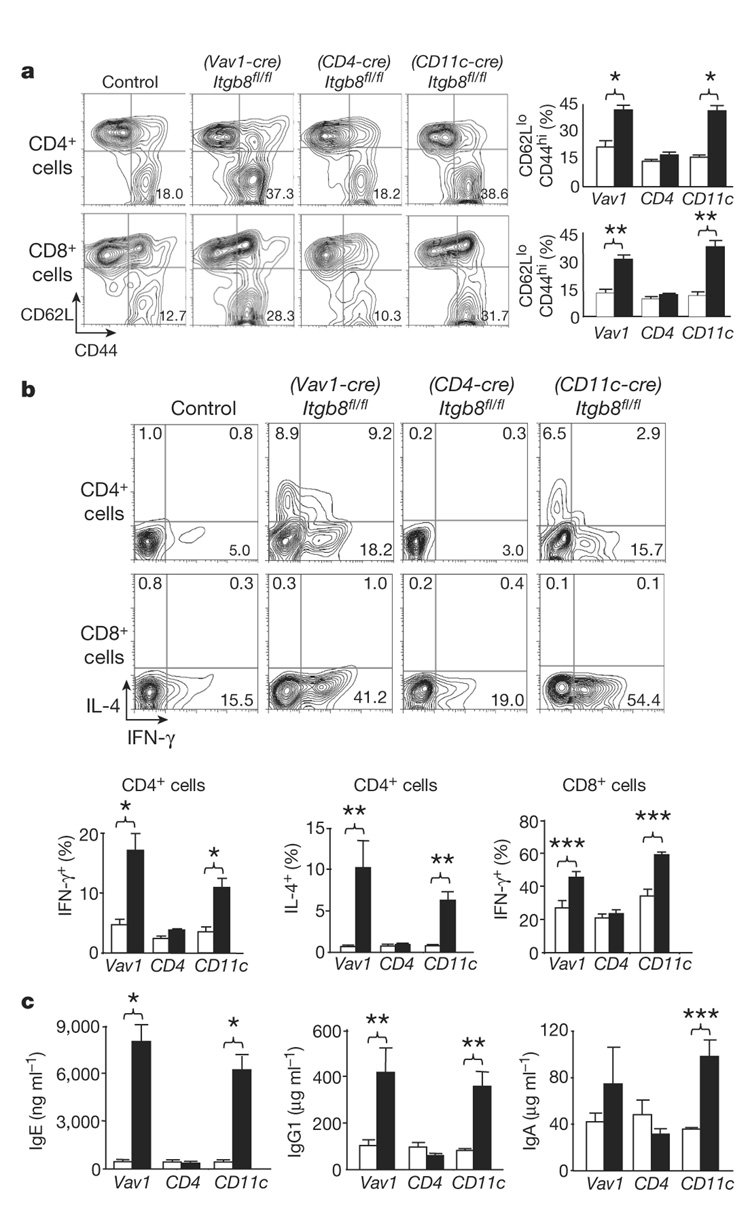
We next assessed whether loss of αvβ8 on either dendritic cells or T cells was responsible for the in vivo phenotype observed in (Vav1-cre)Itgb8fl/fl mice by crossing mice homozygous for the Itgb8-floxed allele with mice expressing Cre recombinase principally in T cells (CD4-Cre)18 or dendritic cells (CD11c-Cre)19. By qRT–PCR we saw a greater than 99% reduction in Itgb8 mRNA levels in CD4+ T cells from Itgb8fl/fl mice expressing CD4-Cre ((CD4-cre)Itgb8fl/fl), with a modest reduction in dendritic cells (Supplementary Fig. 5a). Greater than 99% reduction in Itgb8 mRNA levels was found in dendritic cells from Itgb8fl/fl mice expressing CD11c-Cre ((CD11c-cre)Itgb8fl/fl), with a modest reduction in CD4+ T cells (Supplementary Fig. 5b). Therefore, (CD4-cre)Itgb8fl/fl mice completely lacked β8 expression in CD4+ T cells, but only partially in dendritic cells, whereas (CD11c-cre) Itgb8fl/fl mice completely lacked β8 expression in dendritic cells, but only partially in CD4+ T cells.
By 4–6 weeks of age, (Vav1-cre)Itgb8fl/fl mice already had increased numbers of activated/memory T cells, increased T-cell production of IFN-γ and/or IL-4, and increased circulating levels of IgE, IgG1 and IgA (Fig. 3). In contrast, (CD4-cre)Itgb8fl/fl mice were indistinguishable from control mice, and showed no signs of illness up to at least 14 months of age. (CD11c-cre)Itgb8fl/fl mice show an identical immunological phenotype to mice lacking β8 integrin on all leukocytes, with significantly increased numbers of activated/memory T cells (Fig. 3a), increased T cells expressing IFN-γ and/or IL-4 (Fig. 3b), and increases in serum IgE, IgG1 and IgA levels (Fig. 3c). Older (CD11c-cre)Itgb8fl/fl mice (6–7 months) manifested an identical phenotype to age-matched mice lacking β8 on all leukocytes, with significant weight loss, enlarged spleen and mesenteric lymph nodes, infiltrates in portal triads of the liver, and development of colonic inflammation (Supplementary Fig. 6). These results suggest that expression of the αvβ8 integrin on dendritic cells is vital for the negative regulation of adaptive immunity. Loss of αvβ8 on T cells is not by itself sufficient to cause any aspects of this phenotype. However, because CD11c-Cre did cause a modest reduction in β8 expression in CD4+ T cells, we cannot exclude a minor contribution from partial loss of αvβ8 on these cells.
Figure 3. Mice lacking integrin β8 on dendritic cells develop an identical immune phenotype to mice lacking β8integrin on all leukocytes.
a, Activated/memory T cells from spleen (CD62LlowCD44high, 4–6-week-old mice) were analysed by flow cytometry. Representative flow cytometry plots and plotted mean values are shown (white, control; black, Itgb8 conditional knockout; n=6 per group; asterisk, P<8.2×10−4; double asterisk, P<1.4×10−4). b, IL-4 and IFN-γ levels in T cells from spleen were analysed by intracellular flow cytometry. Representative flow cytometry plots and plotted mean values are shown (white, control; black, Itgb8 conditional knockout; n=6 per group; asterisk, P<0.0063; double asterisk, P=0.0055; triple asterisk, P=0.0045). c, ELISA for IgE, IgG1 and IgA levels in sera (4–6-week-old mice; white, control; black, Itgb8 conditional knockout; n=6; asterisk, P<0.0018; double asterisk, P<0.016; triple asterisk, P=0.0015). All error bars represent s.e.m.
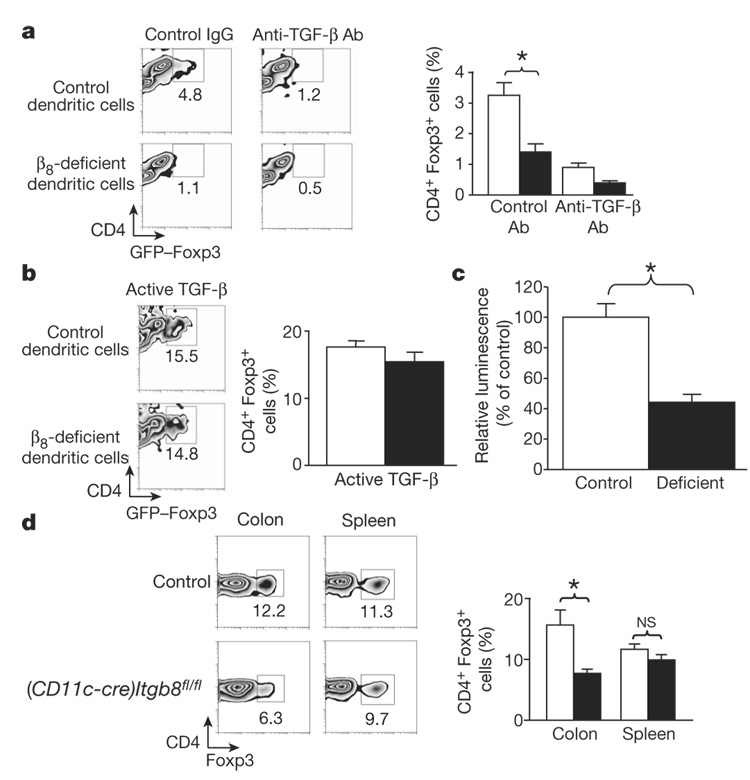
Mice lacking TR cells also develop severe autoimmunity20, and recent work has shown an important role for TGF-β in the biology of TR cells21–24. We therefore examined the ability of control or β8-deficient dendritic cells to induce TR cells from CD4+ T cells in an in vitro T-cell activation assay. Both control and β8-deficient dendritic cells showed similar populations of myeloid (CD11chighCD8−), lymphoid (CD11chighCD8+) and plasmacytoid (CD11clowB220+) dendritic cells (Supplementary Fig. 7a), with comparable expression of the activation markers CD86 and major histocompatibility complex (MHC) class II (Supplementary Fig. 7b), indicating that lack of β8 integrin does not affect the development or maturation of dendritic cells. In the presence of control dendritic cells approximately 3% of CD4+ T cells converted to TR cells (determined by expression of the TR-cell-specific reporter gene Foxp3-Gfp25) and induction was inhibited by an anti-TGF-β antibody (Fig. 4a). However, in the presence of β8-deficient dendritic cells, TR cell induction was markedly reduced (Fig. 4a). Addition of active TGF-β to cultures containing either control or β8-deficient dendritic cells resulted in a similar induction of TR cells (Fig. 4b). Co-culture with TGF-β-luciferase reporter cells also demonstrated decreased TGF-β activity generated by β8-deficient dendritic cells compared to control dendritic cells (Fig. 4c).
Figure 4. β8-deficient dendritic cells fail to induce TR cells in vitro, and (CD11c-cre)Itgb8fl/fl mice have reduced proportions of TR cells in colonic tissue.
a, b, Induction of TR cells (CD4+GFP–Foxp3+) by control or β8-deficient dendritic cells in the presence of control or anti-TGF-β antibody (a) or active TGF- β (b). Representative flow cytometry plots and mean data plots (expressed as percentage of CD4+ cells that expressed GFP–Foxp3) are shown (white, control dendritic cells; black, β8-deficient dendritic cells; n=5; asterisk, P=0.013). c, TGF-β activation by control or β8-deficient dendritic cells, detected using mink lung reporter cells30 (white, control dendritic cells; black, β8-deficient dendritic cells; n=6; asterisk, P=0.0006). d, TR cell proportions present in spleen or colonic lamina propria. Representative flow cytometry plots and mean data plots (expressed as percentage of CD4+ cells that expressed GFP–Foxp3) are shown (white, control; black, (CD11c-cre)Itgb8fl/fl; n=6; asterisk, P=0.012). NS, not significant. All error bars represent s.e.m.
To determine whether there was a defect in TR cell induction by β8-deficient dendritic cells in vivo, we analysed CD4+Foxp3+ TR cells in the spleen or colon of young (8–14 weeks) control or (CD11c-cre) Itgb8fl/fl mice. No difference in the proportion of TR cells was observed in the spleens, but there was a greater than 50% reduction in the proportion of TR cells recovered from the colonic lamina propria of (CD11c-cre)Itgb8fl/fl mice (Fig. 4d). We cannot rule out the possibility that CD4+ effector cell expansion contributes to the observed reduction in the fractional number of TR cells in the colon. However, taken together, our in vitro and in vivo results suggest that activation of TGF-β by αvβ8 on dendritic cells is important for the induction of TR cells in peripheral tissues such as the colon, and that a defect in TR cell induction and/or maintenance could contribute to the autoimmunity and colitis in (CD11c-cre)Itgb8fl/fl mice.
Because the phenotype we observed is markedly similar to that of mice with impaired TGF-β signalling in T cells, and αvβ8 on dendritic cells activates TGF-β, our results suggest a key negative regulatory role for presentation of active TGF-β from αvβ8 on dendritic cells to T cells. Our findings further suggest that the aberrant immune homeostassis we observed may be explained by the known role of TGF-β in induction of Foxp3 expression21 and resultant differentiation of T cells to TR cells. However, whereas TR cells in the colon were decreased in (CD11c-cre)Itgb8fl/fl mice, numbers of TR cells in the spleen were normal. These results may be explained by a difference in the mechanisms underlying the generation of natural/innate TR cells that are the primary contributor to TR cells in the spleen, lymph nodes and circulation, and adaptive TR cells that are critical for maintaining peripheral homeostasis and preventing diseases such as colitis26,27. In addition to TR cell abnormalities, the observed autoimmune phenotype could be partially explained by a role for dendritic cell αvβ8-mediated TGF-β activation in the direct regulation of T-cell function, as it is known that TGF-β can also directly suppress effector T cells28.
As described earlier, mutation of the RGD integrin-binding site in TGF-β1 results in a more severe autoimmune phenotype reminiscent of loss of TGF-β1 (ref. 8). Therefore, it is likely that activation of TGF-β by other integrins or cell types is also important for the main-tenance of immune homeostasis. For example, the αvβ6 integrin on epithelial cells or the αvβ8 integrin on resident tissue cells might also contribute to TR cell induction and immune suppression at specific sites. Notably, in the absence of the αvβ8 integrin on dendritic cells, mice demonstrate widespread evidence of immune activation, autoimmunity and colitis, suggesting that genetic or acquired abnormalities in αvβ8 might contribute to the development of inflammatory bowel disease or other autoimmune disorders.
METHODS SUMMARY
Mice
Mice expressing a floxed allele of Itgb8 have been described10, and were maintained on a mixed CD1, FVB and C57BL/6 background. Vav1-Cre mice were a gift from D. Kioussis11. CD4-Cre mice18 were purchased from Taconic. For some experiments, alleles were bred for five generations to C57BL/6 mice. CD11c-Cre mouse generation is described in a separate manuscript19. Foxp3–GFP mice were a gift from A. Rudensky25. All mice were maintained under pathogen-free conditions, according to institutional guidelines.
Cell purification and flow cytometry
Cells were purified using magnetic beads (Miltenyi) or by flow cytometry. Flow cytometry data analysis was performed using FloJo software (Treestar).
Quantitative RT–PCR
RNA was isolated using an RNeasy mini kit (Qiagen), and reverse transcribed using Superscript II RT (Invitrogen). Quantitative PCR was performed using a Sybr Green qRT–PCR kit (Invitrogen). Primers used are detailed in the Methods.
Cytokine production assays
Splenocytes (5×106 ml−1) were incubated with 50 ng ml−1 PMA (Sigma) and 1 µM ionomycin (Calbiochem). For intracellular flow cytometry, cells were cultured for 2 h followed by addition of 2 µM monensin (eBioscience) and cultured for another 2 h. For ELISA analysis, cells were cultured for 24 h and supernatants harvested.
Regulatory T-cell induction assay
Briefly, non-TR CD4+ cells (CD4+GFP– Foxp3− T cells from GFP–Foxp3 mice25) were incubated with CD11c+ dendritic cells in the presence of anti-CD3 antibody and either control antibody, anti-TGF-β antibody or active TGF for 3 days. TR cell induction was measured by flow cytometry analysis of CD4+GFP–Foxp3+ cells. See Methods for full details of TR cell induction and TGF-β activation assays.
Colon lamina propria cell preparation
Colonic lamina propria lymphocytes were isolated as described29, with slight modification (see Methods).
Statistical analysis
All statistics were generated using a Student’s t-test. All error bars in figures represent s.e.m.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Supplementary Information is linked to the online version of the paper at http://www.nature.com/nature.
Acknowledgements
We thank D. Kioussis for providing the Vav1-Cre mice and A. Rudensky for providing theGFP–Foxp3 mice. This work was supported by grants from the National Heart, Lung and Blood Institute (to D.S.), the National Institute of Allergy and Infectious Diseases (to J.A.B and B.R.) and funds from the Sandler Program for Asthma Research (to B.R.). M.A.T. was the recipient of an American Lung Association Research Fellowship.
Footnotes
Author Information Reprints and permissions information is available at http://www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Shull MM, et al. Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marie JC, Liggitt D, Rudensky AY. Cellular mechanisms of fatal early-onset autoimmunity in mice with the T cell-specific targeting of transforming growth factor-β receptor. Immunity. 2006;25:441–454. doi: 10.1016/j.immuni.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Li MO, Sanjabi S, Flavell RA. Transforming growth factor-β controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 2006;25:455–471. doi: 10.1016/j.immuni.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFβ activation. J. Cell Sci. 2003;116:217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- 5.Huang XZ, et al. Inactivation of the integrin β6 subunit gene reveals a role of epithelial integrins in regulating inflammation in the lung and skin. J. Cell Biol. 1996;133:921–928. doi: 10.1083/jcb.133.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munger JS, et al. The integrin αvβ6 binds and activates latent TGFβ1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 1999;96:319–328. doi: 10.1016/s0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- 7.Mu D, et al. The integrin αvβ8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-β1. J. Cell Biol. 2002;157:493–507. doi: 10.1083/jcb.200109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z, et al. Absence of integrin-mediated TGFβ1 activation in vivo recapitulates the phenotype of TGFβ1-null mice. J. Cell Biol. 2007;176:787–793. doi: 10.1083/jcb.200611044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J, et al. β8 integrins are required for vascular morphogenesis in mouse embryos. Development. 2002;129:2891–2903. doi: 10.1242/dev.129.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Proctor JM, Zang K, Wang D, Wang R, Reichardt LF. Vascular development of the brain requires β8 integrin expression in the neuroepithelium. J. Neurosci. 2005;25:9940–9948. doi: 10.1523/JNEUROSCI.3467-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Boer J, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 12.Gorelik L, Flavell RA. Abrogation of TGFβ signaling in T cells leads to spontaneous T cell differentiation and autoimmune disease. Immunity. 2000;12:171–181. doi: 10.1016/s1074-7613(00)80170-3. [DOI] [PubMed] [Google Scholar]
- 13.Kim BG, et al. Smad4 signalling in T cells is required for suppression of gastrointestinal cancer. Nature. 2006;441:1015–1019. doi: 10.1038/nature04846. [DOI] [PubMed] [Google Scholar]
- 14.Bohr UR, et al. Prevalence and spread of enterohepatic Helicobacter species in mice reared in a specific-pathogen-free animal facility. J. Clin. Microbiol. 2006;44:738–742. doi: 10.1128/JCM.44.3.738-742.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor NS, Xu S, Nambiar P, Dewhirst FE, Fox JG. Enterohepatic Helicobacter species are prevalent in mice obtained from commercial and academic institutions in Asia, Europe, and North America. J. Clin. Microbiol. 2007;45:2166–2172. doi: 10.1128/JCM.00137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whary MT, Fox JG. Detection, eradication, and research implications of Helicobacter infections in laboratory rodents. Lab Anim. (NY) 2006;35:25–36. doi: 10.1038/laban0706-25. [DOI] [PubMed] [Google Scholar]
- 17.Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J. Clin. Invest. 2007;117:514–521. doi: 10.1172/JCI30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 19.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8− dendritic cells in the spleen. J. Exp. Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakaguchi S, et al. Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol. Rev. 2006;212:8–27. doi: 10.1111/j.0105-2896.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- 21.Chen W, et al. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J. Exp. Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura K, et al. TGF-β1 plays an important role in the mechanism of CD4+CD25+ regulatory T cell activity in both humans and mice. J. Immunol. 2004;172:834–842. doi: 10.4049/jimmunol.172.2.834. [DOI] [PubMed] [Google Scholar]
- 23.Fahlen L, et al. T cells that cannot respond to TGF-β escape control by CD4+CD25+ regulatory T cells. J. Exp. Med. 2005;201:737–746. doi: 10.1084/jem.20040685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marie JC, Letterio JJ, Gavin M, Rudensky AY. TGF-β1 maintains suppressor function and Foxp3 expression in CD4+CD25+ regulatory T cells. J. Exp. Med. 2005;201:1061–1067. doi: 10.1084/jem.20042276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fontenot JD, et al. Regulatory T cell lineage specification by the forkhead transcription factor Foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 26.Bluestone JA, Abbas AK. Natural versus adaptive regulatory T cells. Nature Rev. Immunol. 2003;3:253–257. doi: 10.1038/nri1032. [DOI] [PubMed] [Google Scholar]
- 27.Bluestone JA, Tang Q. How do CD4+CD25+ regulatory T cells control autoimmunity? Curr. Opin. Immunol. 2005;17:638–642. doi: 10.1016/j.coi.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-β regulation of immune responses. Annu. Rev. Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 29.Lefrancois L, Lycke N. Isolation of mouse small intestine intraepithelial lymphocytes, Peyer’s patch, and lamina propria cells. Curr. Protocols Immunol. 2001 doi: 10.1002/0471142735.im0319s17. Unit 3.19 doi:10.1002/0471142735.im0319s17. [DOI] [PubMed] [Google Scholar]
- 30.Abe M, et al. An assay for transforming growth factor-β using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal. Biochem. 1994;216:276–284. doi: 10.1006/abio.1994.1042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at http://www.nature.com/nature.