Abstract
The ability of mammals to identify and distinguish among many thousands of different odorants suggests a combinatorial use of odorant receptors, with each receptor detecting multiple odorants and each odorant interacting with multiple receptors. Numerous receptors may be devoted to the sampling of particularly important regions of odor space. Here we explore the similarities and differences in the molecular receptive ranges of four mouse odorant receptors (MOR23-1, MOR31-4, MOR32-11 and MOR40-4), which have previously been identified as receptors for aliphatic carboxylic acids. Each receptor was expressed in Xenopus oocytes, along with Gαolf and the cystic fibrosis transmembrane regulator to allow electrophysiological assay of receptor responses. We find that even though these receptors are relatively unrelated, there is extensive overlap among their receptive ranges. That is, these receptors sample a similar region of odor space. However, the receptive range of each receptor is unique. Thus, these receptors contribute to the depth of coverage of this small region of odor space. Such a group of receptors with overlapping, but distinct receptive ranges, may participate in making fine distinctions among complex mixtures of closely related odorant compounds.
Keywords: olfactory receptors, Xenopus oocytes, electrophysiology
Introduction
The mammalian olfactory system can detect and distinguish among thousands of diverse chemical structures. To accomplish this immense ligand recognition task, mammals employ a large family of odorant receptors (ORs) (Buck and Axel, 1991). Mammalian ORs are rhodopsin-like G-protein coupled receptors (GPCRs), located on the cilia of olfactory sensory neurons (OSNs). Odorant ligand recognition by ORs initiates a signal transduction cascade that ultimately results in depolarization of the OSNs (Mombaerts, 2004; Reed, 2004). ORs constitute the largest gene family in the mammalian genome and, based on sequence similarity, can be grouped into two broad classes and further subdivided into numerous subfamilies (Young et al., 2002; Zhang and Firestein, 2002; Godfrey et al., 2004). While ORs are identified by several sequence motifs, these receptors are very diverse. The greatest variability occurs within the ligand binding transmembrane region, likely accounting for the high diversity in ligand specificity (Buck and Axel, 1991).
While the OR family is large, it is exceeded by many orders of magnitude by the number of detectable odorants. The vast range of volatile compounds in the air around us can be termed odor space. To effectively sample odor space, the olfactory system is thought to use ORs combinatorially, with each odorant being recognized by an array of ORs and each OR recognizing an array of odorants (Malnic et al., 1999). The array of odorants recognized by an individual OR is termed the molecular receptive range (MRR) of the OR (Araneda et al., 2000). The collective MRRs of the entire OR family would then provide a coverage of odor space relevant to a particular species (Zhang and Firestein, 2002; Godfrey et al., 2004). However, the breadth and depth of odor space coverage may not be uniform. Broad (but shallow) coverage of less important regions of odor space may be provided by ORs with unique and non-overlapping MRRs. Highly detailed coverage of more important regions of odor space may be provided by ORs with extensively overlapping MRRs. For example, a wide variety of ORs have been shown to respond to aliphatic ligands such as octanoic acid and nonanoic acid (Malnic et al., 1999; Saito et al., 2004).
Understanding how ORs are deployed to sample odor space requires detailed characterization of the MRRs of more ORs. Unfortunately, progress in characterizing ORs has been slow due to difficulties in functionally expressing ORs in heterologous systems (McClintock and Sammeta, 2003; Lu et al., 2004). However, the Xenopus oocyte expression system has been used to functionally express several ORs from various species (Speca et al., 1999; Wetzel et al., 1999; Wetzel et al., 2001; Katada et al., 2003). Our group has recently used Xenopus oocytes and robotic electrophysiology to develop a robust system for functional characterization of a wide variety of mammalian ORs (Abaffy et al., 2006; Abaffy et al., 2007). Here we use this assay system to explore the MRRs of four MORs, from different OR subfamilies, that have been previously reported to respond to the aliphatic carboxylic acids.
Experimental Procedures
Materials
Xenopus laevis frogs were purchased from Nasco (Fort Atkinson, WI). The care and use of X. laevis frogs in this study was approved by the University of Miami Animal Research Committee and meet the guidelines of the National Institutes of Health. RNA transcription kits were from Ambion (Austin, TX). Collagenase B was from Boehringer-Mannheim (Indianapolis, IN). All other compounds and all odorants were from Sigma–Aldrich (St. Louis, MO).
Expression Constructs
We refer to MORs using the nomenclature of Zhang and Firestein (2002). MORs used in this study are: MOR23-1, MOR31-4, MOR32-11, MOR 40-4 (S83), MOR174-9 (mOR-EG) and MOR267-13 (MOR23). Constructs containing the MOR23-1, MOR31-4, MOR32-11, MOR174-9, MOR267-13 and the mouse accessory protein RTP1, each in the pCI expression vector (Promega), were generously provided by Dr. Hiroaki Matsunami (Saito et al., 2004). The coding region of MOR40-4 was amplified by PCR from mouse genomic DNA (BD Biosciences/Clontech), subcloned into the pCI vector and confirmed by sequencing. All OR constructs used in this study contain an N-terminal extension consisting of the N-terminal 20 amino acid residues of human rhodopsin. This “rhodopsin-tag” is thought to aid in surface expression of mammalian ORs in heterologous cells (Krautwurst et al., 1998; Saito et al., 2004; Abaffy et al., 2006). The construct containing human Gαolf was purchased from the UMR cDNA Resource Center. The human CFTR construct was kindly provided by Dr. Ian Dickerson (University of Rochester). cRNA encoding each protein was generated using mMessage mMachine kits (Ambion, Austin, TX).
Preparation of oocytes and cRNA injection
Oocytes were surgically removed from mature Xenopus laevis frogs (Nasco, Fort Atkinson, WI). Follicle cells were removed by treatment with Collagenase B (Boehringer Mannhem, Indianapolis, IN) for 2 hours at room temperature. For determination of MOR receptive ranges, oocytes were injected with cRNA in 23 nl of water. cRNA quantities injected per oocyte: MORs, 25 ng; Gαolf, 10 ng, CFTR, 1 ng. In addition, expression of MOR23-1 required inclusion of 10 ng RTP1 cRNA (Abaffy et al., 2006). For dose-response experiments, oocytes were often injected with a lower total amount of cRNA (in the same ratio as above), in order to reduce the current amplitudes in response to high concentrations of odorant. Oocytes were incubated at 18°C in Barth's saline (in mM: 88 NaCl, 1 KCl, 2.4 NaHCO3, 0.3 CaNO3, 0.41 CaCl2, 0.82 MgSO4, 15 HEPES, pH 7.5 and 12 μg/ml tetracycline) for 2-4 days prior to electrophysiological recording.
Electrophysiology and Data Analysis
Odorant induced Cl− currents, resulting from cAMP mediated activation of the co-expressed CFTR reporter channel (Uezono et al., 1993), were measured 2-4 days after cRNA injection using two-electrode voltage clamp in an automated parallel electrophysiology system (OpusExpress 6000A, Molecular Devices). Micropipettes were filled with 3M KCl and had resistances of 0.2-2.0 MΩ. The holding potential was −70 mV. Current responses, filtered (4-pole, Bessel, low pass) at 20 Hz (−3db) and sampled at 100 Hz, were captured and stored using OpusXpress 1.1 software (Molecular Devices). Initial analysis was done using Clampfit 9.1 software (Molecular Devices). Oocytes were perfused with ND96 (in mM: 96 NaCl, 2 KCl, 1 CaCl2, 1 MgCl2, 5 HEPES, pH 7.5). Odorants were stored under argon and high concentration (1M) stock solutions of each odorant were prepared in DMSO. Each odorant, diluted in ND96, was applied for 15 sec (Abaffy et al., 2006). IBMX (1 mM) was used to activate the CFTR in a receptor independent manner. This occurs both through the inhibition of phosphodiesterase and consequent increase in cAMP concentration, and through a direct action on the CFTR (Schultz et al., 1999). The CFTR can be directly activated by a wide variety of structures (Ma et al., 2002). Thus, to guard against false positives, all compounds (at all concentrations) used in our studies were tested with oocytes expressing Gαolf and CFTR, but no odorant receptors (termed “R-” oocytes). Oocytes are generally stable under voltage clamp for one to several hours, providing a useful platform for ligand screening. The oocytes used to screen MOR23-1 (recordings from one of these oocytes are shown in Fig. 2) were unusually stable, providing 8 hours of recordings. All four recordings in Figure 2 were obtained from the same oocyte (one of 8 oocytes in the screen).
Figure 2. A ligand screen for MOR23-1.
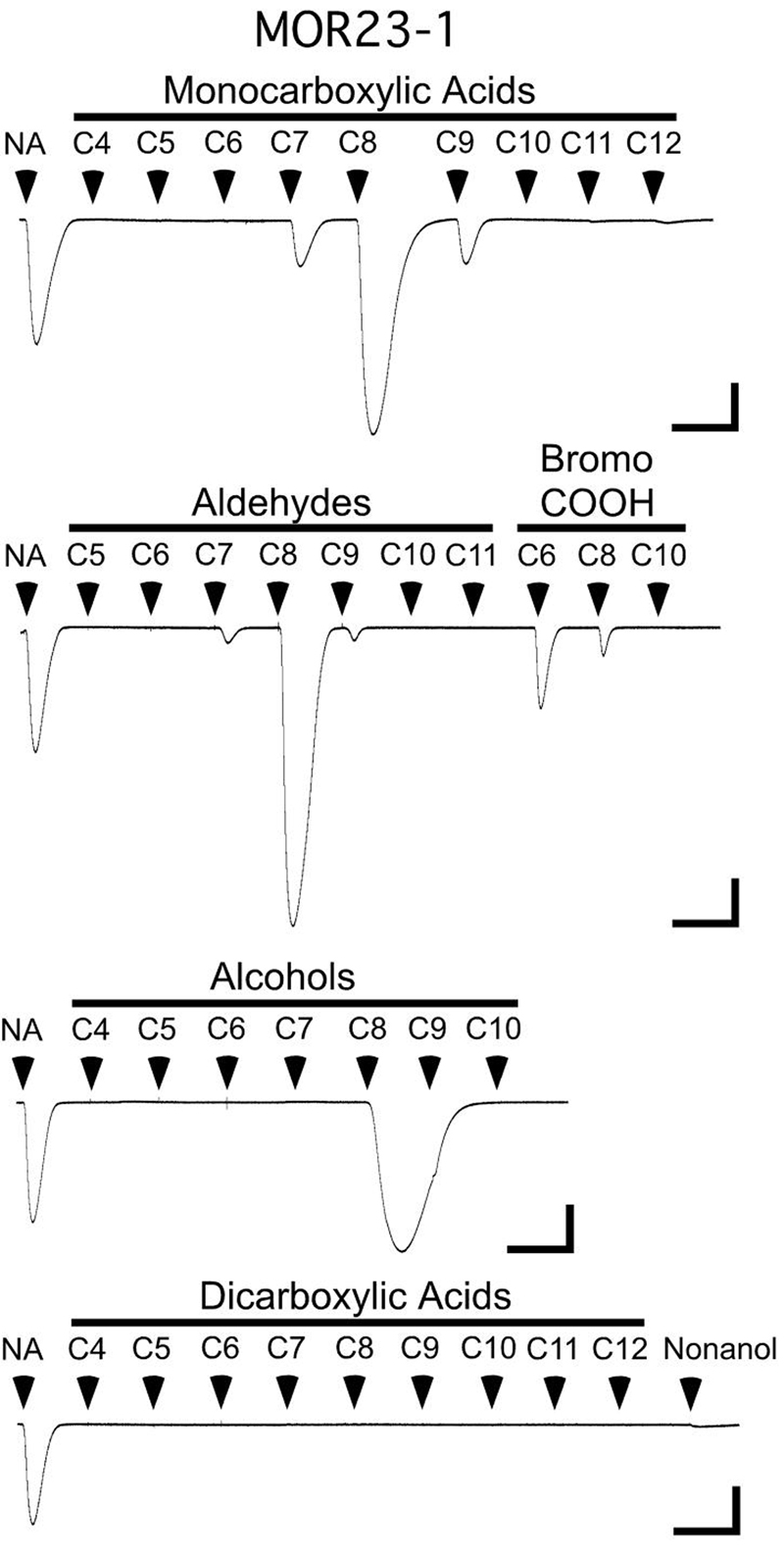
An oocyte expressing MOR23-1, RTP1, Gαolf and CFTR is challenged with 15 sec applications of 35 ligands from our odorant panel (each at 100 μM). Each trace starts with an application of 100μM nonanoic acid, which serves as a normalization standard. All traces are from the same oocyte. Because the nonanol application occurred before the octanol response had ended, nonanol was retested at the end of the dicarboxylic acid screen (bottom trace). Scale bars: 0.5 μA, 10 min.
To explore the molecular receptive range, each MOR was screened with a panel of odorants, each at 100μM. The odorant panel consisted of 41 saturated, aliphatic primary alcohols, aldehydes, monocarboxylic acids, bromocarboxylic acids and dicarboxylic acids, ranging in length from 4 to 12 carbons. Some bromocarboxylic acids in the length series could not be obtained. Odorants were applied for 15 sec (Abaffy et al., 2006), followed by a 10 min wash with ND-96. Each odorant that elicited a statistically significant MOR response at 100 μM was also screened at 30 μM, 10 μM and 3 μM. The statistical significance of receptor responses was determined by comparison to the response of R- oocytes to the same odorant. For each odorant yielding a response, the mean R- response for that odorant was subtracted from the MOR response and the resulting value was then normalized to the response of the same oocyte to 100 μM of the normalizing ligand (nonanoic acid for MOR23-1, MOR32-11 and MOR40-4; undecanoic acid for MOR31-4). These normalized responses are presented as mean ± SEM. We previously observed that small to moderate current responses (≤ 1.5 μA) could be elicited repeatedly, with no loss of amplitude (Abaffy et al., 2006). However, when current amplitudes are large (> 1.5 μA), subsequent current responses can be partially suppressed for many minutes (data not shown). This suppression does not correlate with odorant concentration, but instead appears to be associated with large current amplitudes, suggesting that this temporary suppression occurs within the signal transduction pathway. Thus, during ligand screening, applications that occurred less than 30 minutes following a large response (> 1.5 μA) were redone with a separate set of oocytes. For dose-response analysis, each odorant response was normalized to an immediately preceding normalizing application (3 μM octanoic acid for MOR23-1; 3 μM dodecanoic acid for MOR31-4; 30 μM octanoic acid for MOR32-11; 30 μM undecanal for MOR40-4). Normalized data were fit using Prism 4 (Graphpad, San Diego, CA) according to the equation: I = Imax/(1+(EC50/X)n) where I represents the current response at a given concentration of odorant, X; Imax is the maximal response; EC50 is the concentration of odorant yielding a half maximal response; n is the apparent Hill coefficient. Statistical significance was assessed with Prism 4 (Graphpad, San Diego, CA) using a two-tailed unpaired t-test, or a one-way ANOVA followed by the Dunnett's post-test, as appropriate.
Results
MORs expressed in Xenopus oocytes display appropriate odorant specificity
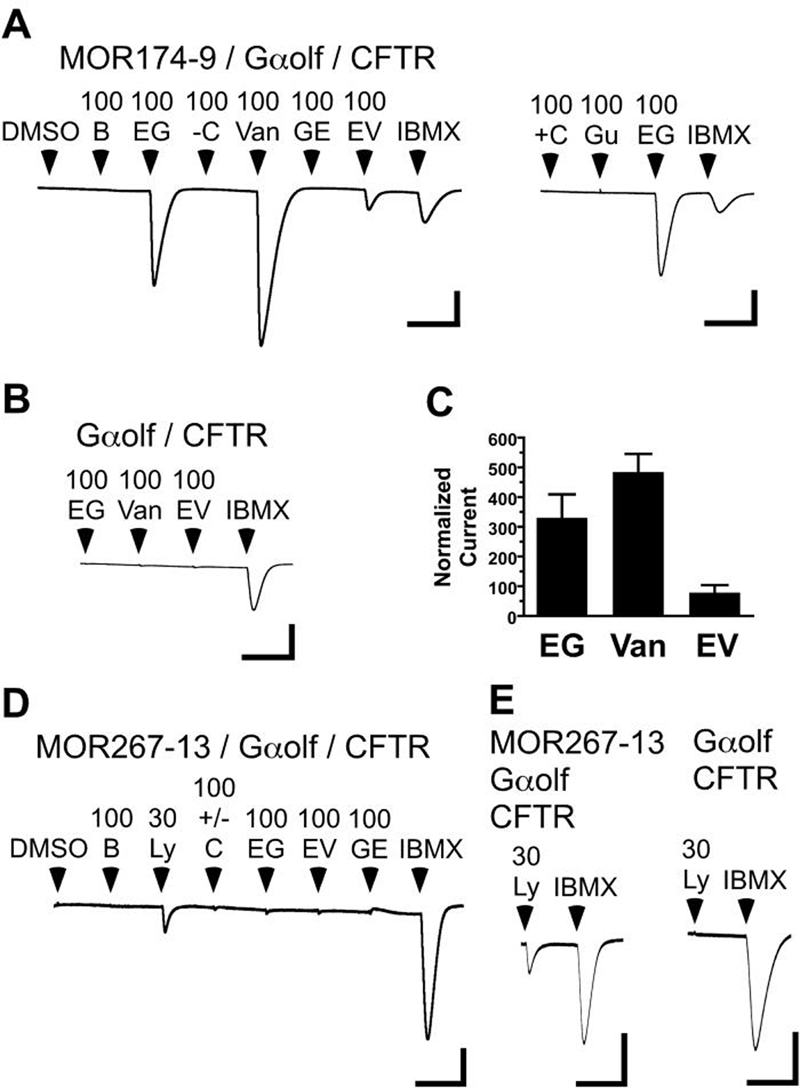
Functional assay of G-protein coupled receptors, such as mammalian ORs, can be conducted using the Xenopus oocyte expression system with the installation of a signal transduction pathway that can provide a measurable output in response to OR activation. Expression of Gαolf can provide linkage to a co-expressed Cl− channel, the cystic fibrosis transmembrane regulator (CFTR) (Wetzel et al., 1999; Katada et al., 2003; Abaffy et al., 2006). However, the use of Xenopus oocytes to express and functionally characterize mammalian ORs (Abaffy et al., 2006, 2007) raises the question of whether the functional properties of MORs expressed in Xenopus oocytes are an accurate reflection of the properties that the receptors would display in a native mouse olfactory sensory neuron (OSN) context. In addition, we are employing the “rhodopsin-tag” expression strategy (Krautwurst et al. 1998) and each MOR construct in this study contains an N-terminal extension consisting of the N-terminal 20 amino acid residues of human rhodopsin (Saito et al., 2004; Abaffy et al., 2006). To examine the functional accuracy of our expression system, we expressed and characterized “rhodopsin-tagged” versions of two MORs that have been characterized in a native neuronal context, MOR174-9 (mOR-EG) and MOR267-13 (MOR23) (Touhara et al., 1999; Kajiya et al., 2001, Oka et al., 2006).
MOR174-9 (mOR-EG) has been characterized in a wide variety of cellular contexts, making it an ideal OR for validation of the oocyte assay. When expressed in the oocyte system, we find that MOR174-9 responds to eugenol, vanillin and ethyl vanillin (each at 100 μM), but does not respond to butanol, (−) carvone, (+) carvone, geraniol, or guaiacol (each at 100 μM) (Fig. 1A). Oocytes expressing Gαolf and CFTR, but no receptor, do not respond to eugenol, vanillin or ethyl vanillin (Fig. 1B). We have also previously observed that methylisoeugenol can antagonize the response to eugenol of MOR174-9 expressed in oocytes (Abaffy et al., 2006). Thus, the agonist specificity, rank order of agonist responsiveness (vanillin>eugenol>ethyl vanillin) and sensitivity to antagonism by methylisoeugenol that we observe for MOR174-9 expressed in Xenopus oocytes are identical to the properties of this receptor when expressed in HEK cells (Kajiya et al., 2001; Katada et al., 2005; Oka et al., 2004; Oka et al., 2006) and more importantly when natively expressed in isolated olfactory sensory neurons (Kajiya et al., 2001; Oka et al., 2004; Oka et al., 2006).
Figure 1. Functionally accurate expression of MOR174-9 (mOR-EG) and MOR267-13 (MOR23) in Xenopus oocytes.
A) Left trace, an oocyte expressing MOR174-9, Gαolf and CFTR is challenged with 15 sec applications of 0.02% DMSO, 100μM butanol (B), eugenol (EG), −carvone (−C), vanillin (Van), Geraniol (GE), ethyl vanillin (EV) and 1mM IBMX. Right trace, a different oocyte expressing MOR174-9, Gαolf and CFTR is challenged with 100μM +carvone (+C), guaiacol (Gu), eugenol (EG) and 1mM IBMX. Scale: 1μA, 10min. B) An oocyte expressing Gαolf and CFTR, but no receptor, is challenged with 15 sec applications of 100μM eugenol (EG), vanillin (Van), ethyl vanillin (EV) and 1mM IBMX. Scale: 1μA, 10min. C) Responses of MOR174-9 expressing oocytes to eugenol, vanillin and ethyl vanillin (each at 100μM). Current responses to each ligand are normalized as a percent of the response of the same oocyte to 1 mM IBMX (mean ± SEM, n = 6). D) An oocyte expressing MOR267-13, Gαolf and CFTR is challenged with 15 sec applications of 0.02% DMSO, 100μM butanol (B), 30μM lyral (Ly), a mix of 100μM −carvone and 100μM +carvone (100 +/− C), 100μM eugenol (EG), ethyl vanillin (EV), geraniol (GE) and 1mM IBMX. Scale: 0.5μA, 10min. E) Left trace, an oocyte expressing MOR267-13, Gαolf and CFTR is challenged with a 15 sec application of 30μM lyral, followed by 1mM IBMX. Scale: 0.5μA, 10min. Right trace, an oocyte expressing Gαolf and CFTR, but no receptor, is challenged with a 15 sec application of 30μM lyral, followed by 1mM IBMX. Scale: 0.5μA, 10min.
MOR267-13 (MOR23) has also been characterized in an OSN context (Touhara et al., 1999). When expressed in the oocyte system, we find that MOR267-13 responds to lyral (30μM), but not to butanol, (−) carvone, (+) carvone, eugenol, ethyl vanillin, or geraniol (each at 100μM) (Fig 1D,E). Current responses to 30 μM lyral are 40±8% of the response to 1 mM IBMX (n=10). Oocytes expressing Gαolf and CFTR, but no receptor, do not respond to lyral (Fig 1E). Thus, the agonist specificity that we observe for MOR267-13 in Xenopus oocytes is identical to the properties of this receptor when natively expressed in OSNs (Touhara et al., 1999).
Molecular receptive range analysis of four mouse odorant receptors
We chose to examine in detail the MRRs of 4 MORs that have been previously shown to have somewhat similar response profiles to carboxylic acids. MOR23-1, MOR31-4 and MOR32-11 have been shown to respond to carboxylic acids in the 7-10 carbon range (Saito et al., 2004). MOR40-4 (S83) has been shown to respond to octanoic and nonanoic acid, as well as 8-bromooctanoic acid and nonanol (Malnic et al., 1999). A microarray approach has been used to demonstrate preferential expression of RNA encoding each of these receptors in the mouse olfactory epithelium (OE), supporting the idea that these receptors function as olfactory receptors in vivo (Zhang et al., 2004). MOR32-11 gene expression in the mouse OE has also been demonstrated by RT-PCR (Young et al., 2003) and the functional expression of MOR40-4 (S83) in a mouse OSN has been demonstrated by calcium imaging and single cell RT-PCR (Malnic et al., 1999). Each MOR was screened with a panel of 41 saturated, aliphatic primary alcohols, aldehydes, monocarboxylic acids, bromocarboxylic acids and dicarboxylic acids, ranging in length from 4 to 12 carbons.
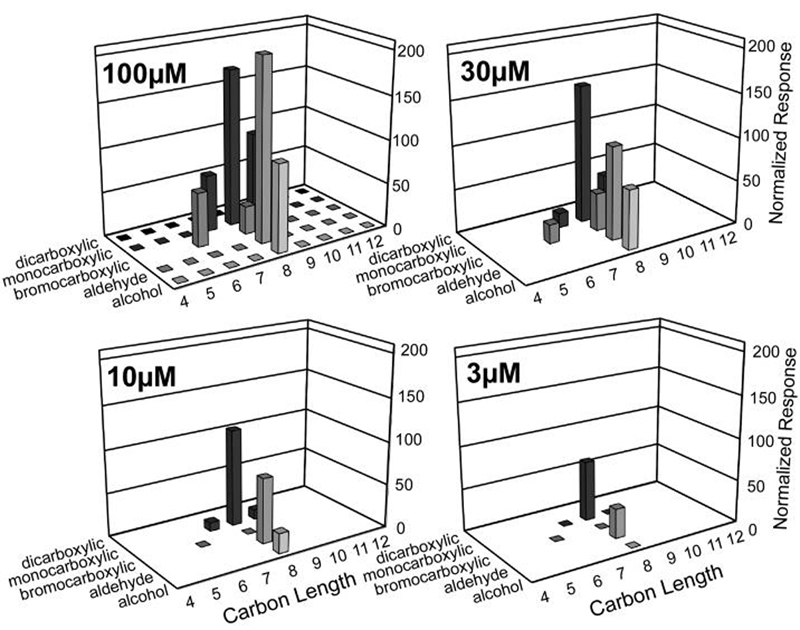
As an example, a screen of MOR23-1 with 35 compounds from our panel is displayed in Figure 2. Each trace represents a continuous 110 min recording. Each recording begins with the application of a normalizing ligand application, in the case of MOR23-1 this is 100μM nonanoic acid. We have found that large responses (> 1.5 μA) can cause temporary suppression of subsequent responses (see Experimental Procedures). Thus, during ligand screening, odorant applications that occurred less than 30 minutes following a large response (> 1.5 μA) were redone with a separate set of oocytes (not shown). Current responses were compared to the responses obtained from R- oocytes (generally small or non-existent), in order to identify receptor specific responses (see Experimental Procedures). The results of this screen are displayed in Figure 3 (upper left). When screened with 100μM concentrations, MOR23-1 responds to several different classes of aliphatic ligands, including monocarboxylic acids, bromocarboxylic acids, an aldehyde and an alcohol, showing a preference for odorants of 8 carbons in length. The bromocarboxylic acids appear to deviate somewhat from this length preference, with the receptor responding better to 6-bromohexanoic acid than to 8-bromooctanoic acid. However, when considering ligand length, the presence of the bromine means that 6-bromohexanoic acid is more comparable in length to heptanoic acid, than to hexanoic acid (and 8-bromooctanoic acid is comparable in length to nonanoic acid). Thus, the ligand length preference of MOR23-1 is well defined. To identify the most potent ligands, each odorant yielding a response at 100 μM was also tested at 30 μM, 10 μM and 3 μM (Figure 3, Table 1). This analysis identified octanoic acid and octanal as the most potent odorant agonists for MOR23-1. Dose-response analysis (see Experimental Procedures) yielded an EC50 of 10 ± 6 μM for octanoic acid and 22 ± 9 μM for octanal at MOR23-1 (Fig. 5A).
Figure 3. Molecular receptive range for MOR23-1 at several ligand concentrations.
Oocytes expressing MOR23-1, RTP1, Gαolf and CFTR were screened with a panel of 41 saturated, aliphatic primary alcohols, aldehydes, monocarboxylic acids, bromocarboxylic acids and dicarboxylic acids, ranging in length from 4 to 12 carbons (15 sec applications of 100 μM, as in Fig. 2). Compounds yielding responses at 100 μM were also screened at 30 μM, 10 μM and 3 μM. Values are the mean of results from 4-8 oocytes (SEM values may be found in Table 1). Flat squares at the base of the graph indicate tested compounds or concentrations that did not yield a response. Blank areas at the base of the graph indicate compounds or concentrations that were not tested.
Table 1. Responses of MORs to a panel of odorants.
Responses are normalized to the response of the same oocyte to 100 μM nonanoic acid (MORs 23-1, 32-11, 40-4) or undecanoic acid (MOR31-4). Values are the mean ± SEM (n = 4-16). 0, no response; nt, not tested. Compounds that did not yield responses from any MOR at 100 μM are not shown (butanol, pentanol, hexanol, heptanol, nonanol, decanol, dodecanol, butanal, pentanal, hexanal, heptanal, butanoic, pentanoic and hexanoic acids, butanedioic, pentanedioic, hexanedioic, heptanedioic, octanedioic, nonanedioic, decanedioic and dodecanedioic acids).
| Ligand | μM | MOR23-1 | MOR31-4 | MOR32-11 | MOR40-4 |
|---|---|---|---|---|---|
| Octanol | 100 | 100 ± 6 | 0 | 0 | 0 |
| 30 | 68 ± 13 | nt | nt | nt | |
| 10 | 23 ± 2 | nt | nt | nt | |
| 3 | nt | nt | nt | nt | |
| Undecanol | 100 | 0 | 0 | 6 ± 2 | 0 |
| 30 | nt | nt | nt | nt | |
| 10 | nt | nt | nt | nt | |
| 3 | nt | nt | nt | nt | |
| Octanal | 100 | 198 ± 27 | 0 | 22 ± 6 | 0 |
| 30 | 107 ± 7 | nt | 0 | nt | |
| 10 | 76 ± 13 | nt | 0 | nt | |
| 3 | 35 ± 5 | nt | 0 | nt | |
| Nonanal | 100 | 0 | 0 | 32 ± 6 | 0 |
| 30 | nt | nt | 0 | nt | |
| 10 | nt | nt | 0 | nt | |
| 3 | nt | nt | 0 | nt | |
| Decanal | 100 | 0 | 0 | 23 ± 5 | 0 |
| 30 | nt | nt | 0 | nt | |
| 10 | nt | nt | 0 | nt | |
| 3 | nt | nt | 0 | nt | |
| Undecanal | 100 | 0 | 0 | 16 ± 3 | 109 ± 18 |
| 30 | nt | nt | 0 | 35 ± 12 | |
| 10 | nt | nt | 0 | 3 ± 1 | |
| 3 | nt | nt | 0 | 0 | |
| Dodecanal | 100 | 0 | 44 ± 11 | 25 ± 3 | 0 |
| 30 | nt | 49 ± 25 | 0 | nt | |
| 10 | nt | 13 ± 4 | 0 | nt | |
| 3 | nt | 0 | 0 | nt | |
| Undecanedioic acid |
100 | 0 | 108 ± 6 | 0 | 0 |
| 30 | nt | 29 ± 7 | nt | nt | |
| 10 | nt | 32 ± 10 | nt | nt | |
| 3 | nt | 0 | nt | nt | |
| 6-Bromo- hexanoic acid |
100 | 61 ± 7 | 0 | 0 | 0 |
| 30 | 22 ± 6 | nt | nt | nt | |
| 10 | 0 | nt | nt | nt | |
| 3 | 0 | nt | nt | nt | |
| 8-Bromo- octanoic acid |
100 | 32 ± 6 | 0 | 0 | 0 |
| 30 | 43 ± 8 | nt | nt | nt | |
| 10 | 0 | nt | nt | nt | |
| 3 | 0 | nt | nt | nt | |
| 10-Bromo- decanoic acid |
100 | 0 | 130 ± 16 | 60 ± 4 | 33 ± 18 |
| 30 | nt | 58 ± 26 | 0 | 0 | |
| 10 | nt | 0 | 0 | 0 | |
| 3 | nt | 0 | 0 | 0 | |
| 11-Bromo- undecanoic acid |
100 | 0 | 37 ± 5 | 27 ± 10 | 0 |
| 30 | nt | 0 | 0 | nt | |
| 10 | nt | 0 | 0 | nt | |
| 3 | nt | 0 | 0 | nt | |
| 12-Bromo- dodecanoic acid |
100 | 0 | 18 ± 9 | 6 ± 2 | 0 |
| 30 | nt | 0 | nt | nt | |
| 10 | nt | 0 | nt | nt | |
| 3 | nt | 0 | nt | nt | |
| Heptanoic acid | 100 | 64 ± 10 | 0 | 35 ± 6 | 0 |
| 30 | 17 ± 4 | nt | 40 ± 12 | nt | |
| 10 | 10 ± 3 | nt | 13 ± 5 | nt | |
| 3 | 0 | nt | 0 | nt | |
| Octanoic acid | 100 | 178 ± 29 | 0 | 76 ± 6 | 0 |
| 30 | 158 ± 12 | nt | 83 ± 14 | nt | |
| 10 | 112 ± 17 | nt | 27 ± 7 | nt | |
| 3 | 70 ± 11 | nt | 0 | nt | |
| Nonanoic acid | 100 | 100 | 0 | 100 | 100 |
| 30 | 48 ± 8 | nt | 61 ± 8 | 61 ± 12 | |
| 10 | 11 ± 4 | nt | 25 ± 4 | 0 | |
| 3 | 0 | nt | 0 | 0 | |
| Decanoic acid | 100 | 0 | 0 | 122 ± 8 | 108 ± 12 |
| 30 | nt | nt | 0 | 72 ± 10 | |
| 10 | nt | nt | 13 ± 3 | 55 ± 14 | |
| 3 | nt | nt | 0 | 0 | |
| Undecanoic acid | 100 | 0 | 100 | 98 ± 22 | 34 ± 8 |
| 30 | nt | 17 ± 6 (7) | 26 ± 8 | 35 ± 3 | |
| 10 | nt | 0 | 0 | 18 ± 4 | |
| 3 | nt | 0 | 0 | 0 | |
| Dodecanoic acid | 100 | 0 | 169 ± 22 | 28 ± 6 | 0 |
| 30 | nt | 94 ± 26 | 0 | nt | |
| 10 | nt | 66 ± 16 | 0 | nt | |
| 3 | nt | 0 | 0 | nt |
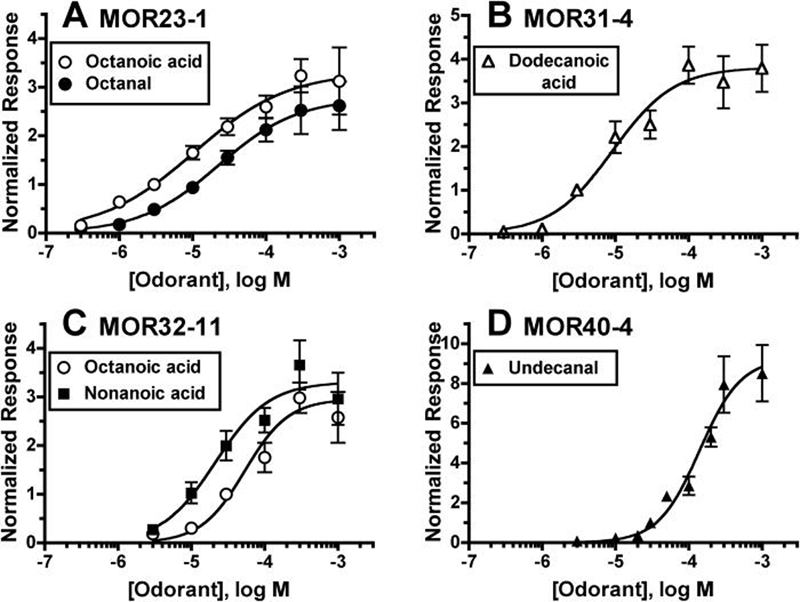
Figure 5. Dose-response analysis for MOR23-1, MOR31-4, MOR32-11 and MOR40-4.
A) Responses of MOR23-1 to a range of octanoic acid and octanal concentrations. Responses were normalized to 3 μM octanoic acid (see Experimental Procedures). Data are the mean ± SEM (n = 4-13). B) Responses of MOR31-4 to a range of dodecanoic acid concentrations. Responses were normalized to 3 μM dodecanoic acid (see Experimental Procedures). Data are the mean ± SEM (n = 5-13). C) Responses of MOR32-11 to a range of octanoic and nonanoic acid concentrations. Responses were normalized to 30 μM octanoic acid (see Experimental Procedures). Data are the mean ± SEM (n = 7-21). D) Responses of MOR40-4 to a range of undecanal concentrations. Responses were normalized to 30 μM undecanal (see Experimental Procedures). Data are the mean ± SEM (n = 5-14).
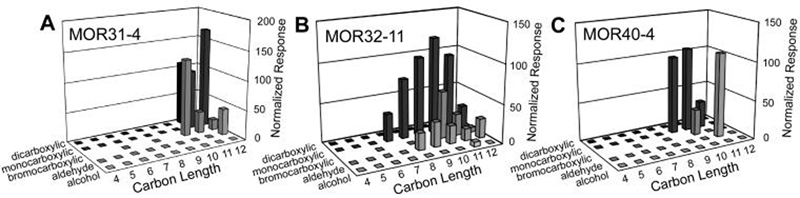
Similar to MOR23-1, MOR31-4 is activated by monocarboxylic acids, bromocarboxylic acids, and an aldehyde. In contrast, MOR31-4 is activated by much longer ligands, preferring 11 and 12 carbon compounds (Fig. 4, Table 1). Again, the bromocarboxylic acids seem to deviate from this ligand length preference, but (as described above) 10-bromodecanoic acid should be considered similar in length to undecanoic acid. Another difference from MOR23-1 is that MOR31-4 is activated by a dicarboxylic acid (undecanedioic acid), but not by any alcohols. Screening with lower odorant concentrations identified dodecanoic acid and undecanedioic acid as most potent ligands for MOR31-4. Dose-response analysis for dodecanoic acid yielded an EC50 of 9 ± 3 μM (Fig. 5B).
Figure 4. Molecular receptive ranges for MOR31-4, MOR32-11 and MOR40-4.
Oocytes expressing an MOR (MOR31-4 in panel A, MOR32-11 in panel B or MOR40-4 in panel C), Gαolf and CFTR were screened with a panel of 41 saturated, aliphatic primary alcohols, aldehydes, monocarboxylic acids, bromocarboxylic acids and dicarboxylic acids, ranging in length from 4 to 12 carbons (15 sec applications of 100 μM as in Fig. 2). Values are the mean of results from 4-16 oocytes (SEM values may be found in Table 1). Flat squares at the base of the graph indicate tested compounds or concentrations that did not yield a response. Blank areas at the base of the graph indicate compounds or concentrations that were not tested.
MOR32-11 is also activated by a variety of ligands classes, including monocarboxylic acids, bromocarboxylic acids, aldehydes and an alcohol. However, MOR32-11 differs from both MOR23-1 and MOR31-4 in that it responds to a wide range of ligand lengths (Fig. 4, Table 1). This is particularly evident for the monocarboxylic acids, with MOR32-11 responding to heptanoic, octanoic, nonanoic, decanoic, undecanoic and dodecanoic acids. Screening with lower odorant concentrations identified heptanoic, octanoic, nonanoic acids as the most potent ligands for MOR32-11. Dose-response analysis yielded EC50's of 53 ± 25 μM for octanoic acid and 21 ± 9 μM for nonanoic acid (Fig. 5C). Responses to heptanoic acid failed to saturate, even at a concentration of 1 mM (data not shown), suggesting that heptanoic acid is a low potency agonist at MOR32-11.
MOR40-4 displayed the most limited MRR, responding to only 5 odorants: nonanoic acid, decanoic acid, undecanoic acid, 10-bromodecanoic acid and undecanal (Fig. 4, Table 1). Screening with lower odorant concentrations identified decanoic acid and undecanal as the most potent ligands for MOR40-4. Dose-response analysis for undecanal yielded an EC50 of 140 ± 36 μM (Fig. 5D).
Discussion
We have examined, in some detail, the MRRs of several MORs that have been previously shown to be responsive to aliphatic carboxylic acids. By providing detailed receptive range information for odorant receptors, we hope to improve our understanding of the way in which MORs contribute to the sampling and perception of odor space. It is the sum of the MRRs of the entire OR family that provides a coverage of odor space that is relevant to a particular species (Zhang and Firestein, 2002; Godfrey et al., 2004). However, coverage of odor space may be uneven, with only a few ORs devoted to unimportant regions and large numbers of ORs devoted to important regions of odor space. From the limited information available it appears that closely related ORs (those within the same subfamily) have related MRRs (Kajiya et al., 2001; Bozza et al., 2002; Feinstein et al., 2004; Abaffy et al., 2006; Schmiedeberg et al., 2007). For example, the three members of the MOR42 subfamily have unique, but overlapping MRRs for aliphatic carboxylic acids (Abaffy et al., 2006). There may also be considerable MRR overlap among ORs from distinct subfamilies.
Each of the MORs we have examined in this study is in a different subfamily and the degree of sequence identity among them is low, ranging from 34% (MOR31-4 and MOR40-4) to 51% (MOR31-4 and MOR32-11). An alternate method of OR classification, based on sequence analysis of predicted binding pocket residues, also indicates that these MORs are not closely related (Man et al., 2007). Despite these receptors being relatively unrelated, there is extensive overlap among their MRRs. Yet within this very small region of odor space, each receptor has a distinctive MRR. For example, while MOR32-11 is a general detector of medium to long (7-12 carbon) carboxylic acids, MORs 23-1, 40-4 and 31-4 can parse this group of odorants into three subgroups: 7-9 carbons, 9-11 carbons, and 11-12 carbons, respectively. These MORs can also perform a similar function in distinguishing among aldehydes. Thus, these receptors contribute to the depth of coverage of this region of odor space and can participate in making fine distinctions among complex mixtures of closely related odorant compounds.
Our use of Xenopus oocytes as an expression system for mammalian ORs, as well as the “rhodopsin-tag” expression strategy, can raise concern about the accuracy of MRR data obtained in our assay system. Thus, it is important that we compare results obtained in our assay system with those obtained in a native neuronal context. We show that when expressed in Xenopus oocytes, the ligand specificities of “rhodopsin-tagged” MOR174-9 (mOR-EG) and MOR267-13 (MOR23) are identical to what has been reported for these receptors when natively expressed in dissociated OSNs (Touhara et al., 1999; Kajiya et al., 2001). This result indicates that MRR information obtained from ORs expressed in Xenopus oocytes can be considered an accurate reflection of the in vivo properties of the receptor. An interesting “discrepancy” can be seen when examining the MOR174-9 response at the glomerular level (Oka et al., 2006). In contrast to results in heterologous system and dissociated OSNs, the MOR174-9 glomerulus fails to respond to vanillin. This is due to enzymatic alteration of the vanillin by the mucus layer of the olfactory epithelium (Oka et al., 2006), adding another layer of complexity to odor coding. A further complication is the observation that individual odorants can be agonists at some ORs and antagonists at other ORs (Oka et al., 2004; Oka et al., 2006). Thus, it is important to remember that while obtaining information about the MRRs of ORs is vitally important for developing an understanding of odor coding, additional factors are at play. The apparent MRR seen at the glomerular level is derivative of, but not necessarily identical to, the MRR at the receptor level.
We have identified the most potent odorant ligands, from among a panel of 41 compounds, for each of four MORs. The low to mid micromolar EC50 values for ligands activating MOR23-1 (octanoic acid and octanal), MOR31-4 (dodecanoic acid) and MOR32-11 (nonanoic and octanoic acid) suggest that these odorants may be important ligands for these MORs. The odorant ligand for MOR40-4 (undecanal) is less potent, suggesting lesser importance. However, a necessary caveat when screening ORs with a finite panel of odorants is that it is difficult to gauge the relative importance of the active odorants that are identified; higher potency ligands may reside outside the screening panel, awaiting identification. But what level of potency should we expect for the important odorant ligands activating a particular OR? Ligand potencies in the low to mid micromolar range appear typical for mammalian ORs expressed in heterologous systems (Kajiya et al., 2001; Saito et al., 2004; Abaffy et al., 2006). However, mammalian olfaction is at least several orders of magnitude more sensitive (Mombaerts, 2004). This difference does not appear to be due to the particular cell type in which an OR is assayed, because ORs assayed in isolated olfactory sensory neurons also display this relatively low sensitivity to odorants (Touhara et al., 1999; Bozza et al., 2002; Oka et al., 2006). More extensive screening may reveal additional ligands with higher affinities. However, ligand affinities higher than mid nanomolar should probably not be expected, because such high affinities would involve ligand off rates that are too slow to be physiologically relevant.
Thus, the mammalian olfactory system appears to use an array of relatively low sensitivity odorant receptors to achieve high sensitivity odor detection. A variety of potential mechanisms have been proposed to explain how the intact mammalian olfactory system can be so much more sensitive than the individual ORs. These include a role for the olfactory mucus and the airflow properties within the nasal cavity (Pelosi, 1998; Oka et al., 2006). Also, isolated olfactory neurons may be less sensitive than olfactory neurons residing in intact olfactory epithelium due to an impaired ability to accumulate intracellular Cl−, which would diminish the major signal amplification mechanism in these neurons (Lowe and Gold, 1993; Kaneko et al., 2004; Reisert et al., 2005). Furthermore, the convergence, amplification and noise reduction resulting from the circuit properties of the olfactory system may allow the sensitivity of mammalian olfaction to be higher than the sensitivity of any individual OR (Bozza et al., 2002; Bhandawat et al., 2005).
Detailed surveillance of important regions of odor space may be accomplished by employing groups of ORs with extensively overlapping, but individually distinctive, molecular receptive ranges (Araneda et al., 2004). This would allow fine distinctions to be made among complex mixtures containing varying ratios of closely related odorant compounds. A group of closely related ORs (an OR subfamily) might be expected to provide such overlapping coverage and several studies support this idea (Kajiya et al., 2001; Bozza et al., 2002; Feinstein et al., 2004; Abaffy et al., 2006; Schmiedeberg et al., 2007). We find that a group of unrelated ORs can also play a role in providing overlapping coverage of a small region of odor space. In addition, the other members of the various subfamilies to which these ORs belong may also be involved, resulting in a large group of ORs being devoted to providing a very dense coverage of this region of odor space.
Acknowledgements
This work was supported by National Institutes of Health grants DC008119 and MH66038 (CWL). SER was supported in part by T32 HL07188. We would like to thank Ana Castro and Floyd Maddox for excellent technical assistance and Drs. Hiroaki Matsunami (Duke University) and Ian Dickerson (University of Rochester) for cDNA constructs.
Abbreviations
- CFTR
cystic fibrosis transmembrane regulator
- GPCR
G-protein coupled receptor
- MOR
mouse odorant receptor
- RR
molecular receptive range
- OR
odorant receptor
- RTP
receptor transporting protein
References
- Abaffy T, Matsunami H, Luetje CW. Functional analysis of a mammalian odorant receptor subfamily. J. Neurochem. 2006;97:1506–18. doi: 10.1111/j.1471-4159.2006.03859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abaffy T, Malhotra A, Luetje CW. The molecular basis for ligand specificity in a mouse olfactory receptor: A network of functionally important residues. J. Biol. Chem. 2007;282:1216–1224. doi: 10.1074/jbc.M609355200. [DOI] [PubMed] [Google Scholar]
- Araneda RC, Kini AD, Firestein S. The molecular receptive range of an odorant receptor. Nat. Neurosci. 2000;3:1248–1255. doi: 10.1038/81774. [DOI] [PubMed] [Google Scholar]
- Araneda RC, Peterlin Z, Zhang X, Chesler A, Firestein S. A pharmacological profile of the aldehyde receptor repertoire in rat olfactory epithelium. J. Physiol. 2004;555.3:743–756. doi: 10.1113/jphysiol.2003.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandawat V, Reisert J, Yau K. Elementary response of olfactory receptor neurons to odorants. Science. 2005;308:1931–34. doi: 10.1126/science.1109886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza T, Feinstein P, Zheng C, Mombaerts P. Odorant receptor expression defines functional units in the mouse olfactory system. J. Neurosci. 2002;22:3033–43. doi: 10.1523/JNEUROSCI.22-08-03033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the β2 adrenergic receptor. Cell. 2004;117:833–846. doi: 10.1016/j.cell.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Godfrey PA, Malnic B, Buck LB. The mouse olfactory receptor gene family. Proc. Natl. Acad. Sci. USA. 2004;101:2156–2161. doi: 10.1073/pnas.0308051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiya K, Inaki K, Tanaka M, Haga T, Kataoka H, Touhara K. Molecular bases of odor discrimination: Reconstitution of olfactory receptors that recognize overlapping sets of odorants. J. Neurosci. 2001;21:6018–6025. doi: 10.1523/JNEUROSCI.21-16-06018.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Nakagawa T, Kataoka H, Touhara K. Odorant response assays for a heterologously expressed olfactory receptor. Biochem. Biophys. Res. Commun. 2003;305:964–969. doi: 10.1016/s0006-291x(03)00863-5. [DOI] [PubMed] [Google Scholar]
- Katada S, Hirokawa T, Oka Y, Suwa M, Touhara K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. J. Neurosci. 2005;25:1806–1815. doi: 10.1523/JNEUROSCI.4723-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautwurst D, Yau KW, Reed RR. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell. 1998;95:917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]
- Lowe G, Gold GH. Nonlinear amplification by calcium-dependent chloride channels in olfactory receptor cells. Nature. 1993;366:283–286. doi: 10.1038/366283a0. [DOI] [PubMed] [Google Scholar]
- Lu M, Staszewski L, Echeverri F, Xu H, Moyer BD. Endoplasmic reticulum degradation impedes olfactory G-protein coupled receptor functional expression. BMC Cell Biol. 2004;5:34. doi: 10.1186/1471-2121-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Vetrivel L, Yang H, Pedemonte N, Zegarra-Moran O, Galietta LJ, Verkman AS. High-affinity activators of cystic fibrosis transmembrane conductance regulator (CFTR) chloride conductance identified by high-throughput screening. J. Biol. Chem. 2002;277:37235–37241. doi: 10.1074/jbc.M205932200. [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- Man O, Willhite DC, Crasto CJ, Shepherd GM, Gilad Y. A framework for exploring functional variability in olfactory receptor genes. PLoS One. 2007;2:e682. doi: 10.1371/journal.pone.0000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock TS, Sammeta N. Trafficking prerogatives of olfactory receptors. Neuroreport. 2003;14:1547–1552. doi: 10.1097/00001756-200308260-00001. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat. Rev. Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- Oka Y, Omura M, Kataoka H, Touhara K. Olfactory receptor antagonism between odorants. EMBO J. 2004;23:120–126. doi: 10.1038/sj.emboj.7600032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, Katada S, Omura M, Suwa M, Yoshihara Y, Touhara K. Odorant receptor map in the mouse olfactory bulb: In vivo sensitivity and specificity of receptor-defined glomeruli. Neuron. 2006;52:857–869. doi: 10.1016/j.neuron.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Pelosi P. Odorant-binding proteins: structural aspects. Ann. N.Y. Acad. Sci. 1998;855:281–293. doi: 10.1111/j.1749-6632.1998.tb10584.x. [DOI] [PubMed] [Google Scholar]
- Reed RR. After the holy grail: establishing a molecular basis for mammalian olfaction. Cell. 2004;116:329–336. doi: 10.1016/s0092-8674(04)00047-9. [DOI] [PubMed] [Google Scholar]
- Reisert J, Lai J, Yau K-Y, Bradley J. Mechanism of the excitatory Cl− response in mouse olfactory receptor neurons. Neuron. 2005;45:553–561. doi: 10.1016/j.neuron.2005.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Schmiedeberg K, Shirokova E, Weber H-P, Schilling B, Meyerhof W, Krautwurst D. Structural determinants of odorant recognition by the human olfactory receptors OR1A1 and OR1A2. J. Struct. Biol. 2007;159:400–412. doi: 10.1016/j.jsb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Schultz BD, Singh AK, Devor DC, Bridges RJ. Pharmacology of CFTR chloride channel activity. Physiol. Rev. 1999;79:S109–144. doi: 10.1152/physrev.1999.79.1.S109. [DOI] [PubMed] [Google Scholar]
- Speca DJ, Lin DM, Sorensen PW, Isacoff EY, Ngai J, Dittman AH. Functional identification of a goldfish odorant receptor. Neuron. 1999;23:487–498. doi: 10.1016/s0896-6273(00)80802-8. [DOI] [PubMed] [Google Scholar]
- Touhara K, Sengoku S, Inaki K, Tsuboi A, Hirono J, Sato T, Sakano H, Haga T. Functional identification and reconstitution of an odorant receptor in single olfactory neurons. Proc. Natl. Acad. Sci. USA. 1999;96:4040–45. doi: 10.1073/pnas.96.7.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezono Y, Bradley J, Min C, McCarty NA, Quick M, Riordan JR, Chavkin C, Zinn K, Lester HA, Davidson N. Receptors that couple to 2 classes of G proteins increase cAMP and activate CFTR expressed in Xenopus oocytes. Receptors Channels. 1993;1:233–241. [PubMed] [Google Scholar]
- Wetzel CH, Oles M, Wellerdieck C, Kuczkowiak M, Gisselmann G, Hatt H. Specificity and sensitivity of a human olfactory receptor functionally expressed in human embryonic kidney 293 cells and Xenopus Laevis oocytes. J. Neurosci. 1999;19:7426–7433. doi: 10.1523/JNEUROSCI.19-17-07426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel CH, Behrendt HJ, Gisselmann G, Stortkuhl KF, Hovemann B, Hatt H. Functional expression and characterization of a Drosophila odorant receptor in a heterologous cell system. Proc. Natl. Acad. Sci. USA. 2001;98:9377–9380. doi: 10.1073/pnas.151103998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Friedman C, Williams EM, Ross JA, Tonnes-Priddy L, Trask BJ. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum. Mol. Genet. 2002;11:535–546. doi: 10.1093/hmg/11.5.535. [DOI] [PubMed] [Google Scholar]
- Young JM, Shykind BM, Lane RP, Tonnes-Priddy L, Ross JA, Walker M, Williams EM, Trask BJ. Odorant receptor expressed sequence tags demonstrate olfactory expression of over 400 genes, extensive alternate splicing and unequal expression levels. Genome Biology. 2003;4:R71. doi: 10.1186/gb-2003-4-11-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rogers M, Tian H, Zhang X, Zou D-J, Liu J, Ma M, Shepherd GM, Firestein S. High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc. Natl. Acad. Sci. USA. 2004;101:14168–14173. doi: 10.1073/pnas.0405350101. [DOI] [PMC free article] [PubMed] [Google Scholar]