Abstract
Gap junctions play critical roles in tissue function and homeostasis. Connexin43 (Cx43) is a major gap junction protein expressed in the mammalian heart and other tissues and may be regulated by its interaction with other cellular proteins. Using the yeast two-hybrid screen, we identified a novel Cx43-interacting protein of 85-kDa, CIP85, which contains a single TBC, SH3, and RUN domain, in addition to a short coiled coil region. Homologues containing this unique combination of domains were found in human, D. melanogaster, and C. elegans. CIP85 mRNA is expressed ubiquitously in mouse and human tissues. In vitro interaction assays and in vivo co-immunoprecipitation experiments confirmed the interaction of endogenous CIP85 with Cx43. In vitro interaction experiments using CIP85 mutants with in-frame deletions of the TBC, SH3, and RUN domains indicated that the SH3 domain of CIP85 is involved in its interaction with Cx43. Conversely, analysis of Cx43 mutants with proline to alanine substitutions in the two proline-rich regions of Cx43 revealed that the P253LSP256 motif is an important determinant of the ability of Cx43 to interact with CIP85. Laser-scanning confocal microscopy showed that CIP85 colocalized with Cx43 at the cell periphery, particularly in areas reminiscent of gap junction plaques. The functional importance of the interaction between CIP85 and Cx43 was suggested by the observation that CIP85 appears to induce the turnover of Cx43 through the lysosomal pathway.
Gap junctions are formed by specialized proteins termed connexins that are arranged in the cell membrane and allow for the passive intercellular exchange of small molecules, ions, and second messengers of 1000 daltons or less (1). Connexins are a multigene family encoding membrane-spanning proteins that are highly homologous in their extracellular and transmembrane domains but are more structurally diverse in the cytoplasmic loops and C-terminal regions (2). Connexin43 (Cx43)1 is a 43-kDa protein and is one of the most widely expressed members of the conserved family of gap junction proteins (2).
Cell-to-cell communication mediated through gap junctions is known to be essential for the synchronization of rhythmic contractions in the myocardium and uterus (3, 4). Cx43 is the major gap junction protein expressed in the heart and mutations of the Cx43 gene are associated with cardiac abnormalities (5), and have been identified in patients with hypoplastic left heart syndrome (6). In addition, decreased levels of Cx43 transcript and protein have been reported in human congestive heart failure (7). A number of other human diseases appear to result from mutations in other connexin genes, such as peripheral neuropathies, deafness, cataracts, and skin diseases (8). In addition, gap junctions are thought to play a critical role in regulating cell growth and differentiation. The down-regulation of gap junctional communication (GJC) has been noted in many primary tumors and established tumor cell lines, and several oncogenes are known to disrupt GJC (9-13). A greater understanding of the regulation of Cx43 may help to suggest novel ways to intervene in these connexin-associated disease processes.
In the past decade, a number of distinct modular domains that govern protein–protein interactions have been discovered. These include the SH3, PTB, PH, PDZ, WW, and other domains, each of which recognizes and interacts with specific binding domains within a protein partner (14). Characterization of protein–protein binding mediated by these domains has demonstrated the critical role they play in assembling proteins whose interaction is required for the coupling of intracellular signaling pathways, recruitment of substrates to enzymes, and specific subcellular localization of proteins (14). The SH3 domain is a small noncatalytic domain of 50 to 60 amino acids present in many intracellular proteins, and it mediates protein–protein interactions that are important in intracellular signaling and cytoskeletal architecture (15). Characterization of SH3-binding proteins has demonstrated that they bind to specific proline-rich sequences with a preference for arginine and leucine residues (16). The C-terminus of Cx43 has two proline-rich regions, one of which interacts with the SH3 domain of v-Src involved in the closure of the Cx43 channels (13, 17).
The trafficking and assembly of Cx43 channels and its removal from the plasma membrane and subsequent degradation represent important regulatory mechanisms. After translation in the rough endoplasmic reticulum (ER), Cx43 is trafficked through the Golgi apparatus where Cx43 forms connexon hexamers, followed by transport to the plasma membrane and assembly into gap junction channels (18, 19). It has been revealed that newly synthesized Cx43 is incorporated at the periphery of existing gap junctions and older Cx43 is removed from the center of the plaques into pleiomorphic vesicles (20). As a transmembrane protein, Cx43 has a surprisingly short half-life of approximately 2−5 h (21), and both lysosomes and proteasomes have been reported to be involved in Cx43 degradation (22, 23). However, the key proteins involved in the endocytosis of Cx43 leading to lysosomal degradation or the degradation of Cx43 by the proteasome are still elusive.
Ypt/Rabs constitute the largest group within the Ras GTPase superfamily and are key regulators of protein transport in the exocytic and endocytic pathways of eukaryotic cells (24). These proteins function as molecular switches that cycle between inactive GDP-bound and active GTP-bound forms, a process tightly regulated by several classes of proteins including guanine nucleotide exchange factors (GEFs), GTPase-activating proteins (GAPs), and guanine nucleotide dissociation inhibitors (GDIs) (25). Yeast Gyp1p and Gyp7p have been demonstrated to function as Ypt GAPs (26), and they have a TBC domain named from the Tre-2, Bub2p, and Cdc16p proteins, which contain the domain (27, 28). The TBC domains of Gyp1p and Gyp7p contain a catalytic arginine finger that is critical in the stimulation of GTP hydrolysis by Ypt/Rabs (26). TBC domains have also been found in some Rab GAP proteins such as RN-tre and GAPCenA (29, 30).
In the activated state, Ras superfamily GTPases interact with various effector molecules to activate downstream pathways. The recently identified RUN domain, named from the RPIP8, UNC-14, and NESCA proteins, is present in several proteins deduced from the genomes of human, D. melanogaster and C. elegans, and could function as a specific effector for some proteins of the Ras superfamily (31). The frequent occurrence of RUN domains together with TBC and FYVE domains in several hypothetical proteins suggests that they could have specific functions related to Rab signaling pathways (31).
We employed the yeast two-hybrid screen to search for putative Cx43-interacting proteins and identified a novel protein, termed CIP85 (Cx43-interacting protein of 85-kDa), which contained a SH3 domain (32, 33). We discovered that CIP85 also contained a single TBC and RUN domain, in addition to a short coiled coil region. Our data suggested that the CIP85–Cx43 interaction is dependent on the SH3 domain of CIP85 and a proline-rich region of Cx43. CIP85 appears to stimulate the turnover of Cx43 through the lysosomal pathway.
EXPERIMENTAL PROCEDURES
Materials, Growth Conditions, and Media
All the chemicals were purchased from either Sigma (St. Louis, MO) or Fisher Scientific (Tustin, CA) except when indicated. AmpliTaq DNA polymerase was purchased from Applied Biosystems (Foster City, CA). All the restriction enzymes were purchased from New England Biolabs (Beverly, MA), and the T4 DNA ligase was purchased from Promega (Madison, WI). Escherichia coli was grown at 37 °C in Luria-Bertani medium (Qbiogene, Carlsbad, CA) containing 100 μg/mL ampicillin. The genotype of the Saccharomyces cerevisiae yeast reporter strain L40 is MATa trp1 leu2 his3 LYS::lexA-His3 URA3::lexA-lacZ (gift from Stanley Hollenberg). Yeast cells were grown at 30 °C in YPDA medium containing 1% yeast extract, 2% bactopeptone (both from Difco, Kansas City, MO), 2% glucose, and 0.1 mg/mL adenine or in synthetic minimal medium supplemented with appropriate amino acids. HEK293 cells and Hela cells were grown in high glucose Dulbecco's modified Eagle's medium (Gibco, Carlsbad, CA) supplemented with 10% fetal calf serum (HyClone, Walkersville, MD) and 2 mM l-glutamine (Gibco) at 37 °C in 5% CO2.
Identification of CIP85 by the Yeast Two-Hybrid Screen
The yeast two-hybrid screen was performed as previously described (33). Briefly, the cDNA fragment encoding the C-terminal cytoplasmic tail of Cx43 (Cx43CT), was prepared by PCR and cloned into the LexA fusion vector pBTM116. The yeast reporter strain L40 was transformed with this construct to express the LexA-Cx43CT fusion protein. The yeast transformants were further transformed with a 9−10 day mouse embryo cDNA library in the pVP16 vector (gift from Stanley Hollenberg) (34). Several clones encoding polypeptides that interacted with Cx43CT were identified and further tested by directed two-hybrid assays as previously described (33). Briefly, the yeast strain L40 was co-transformed with the individual library plasmid along with pLexA-Cx43CT or a negative control plasmid, pLexA-lamin. The yeast co-transformants were then analyzed for interaction by using the β-galactosidase filter assay. A positive clone, CID62, was chosen for further investigation. To obtain its full-length cDNA, the CID62 cDNA fragment was labeled with [α-32P]-dCTP as a probe to screen an oligo-dT primed cDNA library derived from a 16-day mouse embryo (gift from Megan Brown). A positive clone encoding CIP85 was isolated and sequenced at the University of Hawaii Biotechnology Core Facility. The cDNA sequence of CIP85 was deposited in the GenBank database under accession number AY382616.
Subcloning of CIP85 cDNA and Construction of CIP85 In-Frame Deletion Mutants
A pET-32LIC (Novagen, Madison, WI) construct (gift from Bonnie Warn-Cramer) was digested with KpnI and EcoRV to remove the insert and the resulting blunt ends were filled in with Klenow enzyme for self-ligation. The resulting plasmid termed pET-32.1 was used for the subcloning of CIP85 cDNA. The CIP85 cDNA clone was used as the template for a PCR reaction. Primer Bam-Flag-CIP85 (5′-TATATTGGATCCATGGACTACAAGGACGACGATGACAAGATGATGTCAGGAAACCACACGCC-3′) containing a BamHI site (italic) and encoding a Flag epitope (N-MDYKDDDDKM-C) at the NH2-terminus was paired with primer VPCIP853 (5′-CAATAGAATTCTCTGCCTTCTGTTGAAGGGA-3′) containing an EcoRI site (italic). The PCR product was digested with BamHI and EcoRI and cloned into pET-32.1 and pcDNA3.1 (Invitrogen, Carlsbad, CA). To prepare an in-frame deletion of the TBC domain, the Flag-CIP85 cDNA was used as the template in PCR reactions with primer pairs: Bam-Flag-CIP85 with TBCD1(5′-CTGAGAAACTCCGTTCCCTAGAGAACTCAGCTTCCATCTTC-3′), and TBCD2 (5′-GAAGATGGAAGCTGAGTTCTCTAGGGAACGGAGTTTCTCAG-3′)withVPCIP853, respectively. The two PCR products were purified with Qiaquick purification kit (Qiagen, Valencia, CA) and combined for the second round of PCR supplemented with primers Bam-Flag-CIP85 and VPCIP853. The expected PCR product containing the deletion was purified with Qiaquick gel extraction kit (Qiagen) and cloned into pcDNA3.1 (Invitrogen). The insert was sequenced completely to confirm its authenticity before subcloning into pET-32.1 and pcDNA3.1. To prepare the in-frame deletion of the SH3 domain, primers TBCD1 and TBCD2 were replaced by primers SH3D1 (5′-CATGCTTACGCAGCCACCGGGATGAGCGAAGCAAAGAGTAC-3′)andSH3D2(5′-GTACTCTTTGCTTCGCTCATCCCGGTGGCTGCGTAAGCATG-3′), respectively. For the in-frame deletion of the RUN domain, primers TBCD1 and TBCD2 were replaced by primers RUND1 (5′-GGACGACTCTGTGACAGAACTCCCTGCTAGGAGAGAG-3′) and RUND2 (5′-CTCTCTCCTAGCAGGGAGTTCTGTCACAGAGTCGTCC-3′), respectively. The RUN domain deletion mutant prepared by PCR with the Taq DNA polymerase showed a single base substitution of A by C at nucleotide 2167 in the coding sequence, which did not change the predicted protein sequence (data not shown).
Northern Blotting
The CID62 cDNA fragment was labeled with [α-32P]-dCTP (NEN, Boston, MA) using Prime-it random primer labeling kit (Stratagene, La Jolla, CA), and purified with a STE G-50 column (Eppendorf, Boulder, CO). The probe was hybridized to a commercially prepared membrane containing 2 μg/lane of mRNA derived from various mouse tissues following the vendor's recommendations (product 77621, Clontech, Palo Alto, CA). In addition, a cDNA clone, FLJ00006 (gift from Takahiro Nagase) (35), encoding a human CIP85 homologue, was used as a template for PCR. Primer CIP855 (5′-GAAGGTGATGAGCCTGGCTC-3′) was paired with primer MCIP853 (5′-CCGAATTCGTCCTTCTCGATCTGCTTG-3′) to amplify a 0.3 kb fragment (Figure 1A). The PCR product was labeled with [α-32P]-dCTP and purified as described above. The probe was hybridized to a commercially prepared membrane containing 2 μg/lane of mRNA derived from various human tissues following the vendor's suggestions (product D180550, Invitrogen).
Figure 1.
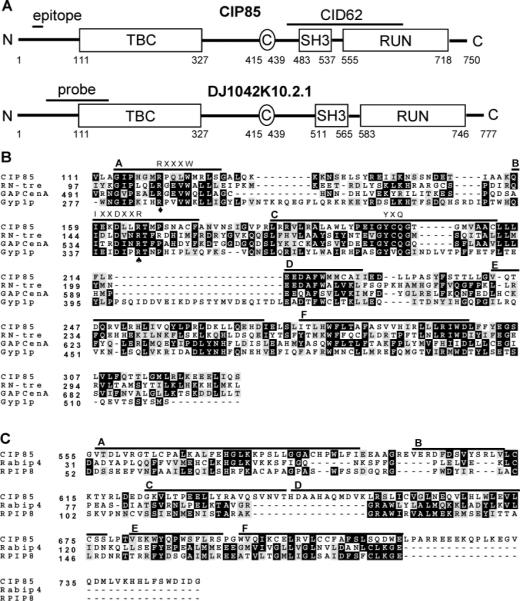
Homologues of CIP85 and their functional domains. Panel A, schematic diagram of CIP85 and a human CIP85 homologue (DJ1042K10.2.1). Both proteins contain TBC, SH3, and RUN domains and coiled coil (C) regions. The region corresponding to CID62 is shown, which encompasses the entire SH3 domain and the NH2-terminal half of the RUN domain of CIP85. The epitope region (Q31-KEESSEQPELCYDE45) used for preparation of the CIP85 antibody is shown. The cDNA probe used to detect the expression of CIP85 mRNA in human tissues by Northern blot analysis is also indicated. Panel B, multiple protein sequence alignment (Clustal W) of TBC domains from CIP85 of mouse (GenBank accession number AY382616), RN-tre of human (GenBank protein accession number NP_055503), GAPCenA of human (GenBank protein accession number NP_036329), and Gyp1p of S. cerevisiae (GenBank protein accession number Q08484). The black shaded areas represent residues matching the consensus sequence, and the gray shaded areas represent residues of conservative substitution. Three conserved “fingerprint” motifs (RXXXW, IXXDXXR, and YXQ) among the members of the Gyp protein family (42), in six shared regions named A–F, are shown (28). A spade denotes R165 in CIP85 that is conserved, and the corresponding site is critical for Gyp1p and Gyp7p GAP activity (26). A diamond denotes R120 in CIP85 that is conserved, and the corresponding site may play a role in the stabilization of the Gyp1p GAP domain (42). Panel C, multiple protein sequence alignment (Clustal W) of RUN domains from CIP85, Rabip4 (GenBank protein accession number NP_058039), and RPIP8 (GenBank protein accession number NP_058039) of mouse. Six shared regions named A–F are shown (31).
Preparation of a CIP85 Antibody and Antibody Preblocking
A region in the NH2-terminus of CIP85 (K32EESSE37) was identified by PCGENE software to exhibit high antigenicity and, thus, may serve as an effective antibody epitope. Rabbits were immunized with a synthetic 15-mer peptide (Q31KEESSEQPELCYDE45) containing this possible epitope to prepare a CIP85 antibody (Washington Biotechnology, Simpsonville, MD). This polyclonal antibody was specific for CIP85 in immunoblotting (Figure 4A), immunoprecipitation (Figure 4B), and immunofluorescence microscopy experiments (Figures 7A). For control experiments, the CIP85 antibody was preblocked for 30 min with the 15-mer CIP85 epitope peptide at 0.3 μg/μL prior to use. In addition, a monoclonal Cx43 antibody, P2D12 (Fred Hutchinson Cancer Research Center, Seattle, WA), was preblocked for 30 min with the 15-mer Cx43 epitope peptide (S368-SRASSRPRPDDLEI382) at 40 μg/mL prior to use.
Figure 4.
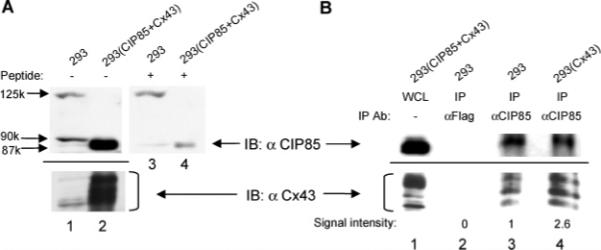
Interaction between endogenous CIP85 and Cx43 in vivo. Panel A, detection of endogenous CIP85 and Cx43 in 5 mg of lysates from HEK293 cells using the CIP85 or Cx43 antibodies. The CIP85 antibody was incubated without (lanes 1 and 2) or with (lanes 3 and 4) 0.3 μg/μL CIP85 epitope peptide before immunoblotting. Lysates (0.1 mg) of HEK293 transfectants coexpressing exogenous CIP85 and Cx43 were used as positive controls (lanes 2 and 4). Panel B, nontransfected HEK293 cells or HEK293 transfectants expressing exogenous Cx43 were lysed in 300 μL of 0.2% NP-40 lysis buffer. A Flag antibody or the CIP85 antibody was used to immunoprecipitate the protein complexes, and the associated Cx43 was detected with a Cx43 antibody (lanes 2−4). Lysates of HEK293 transfectants coexpressing CIP85 and Cx43 were used as positive controls (lane 1). The increased amount of Cx43 co-immunoprecipitated with CIP85 in HEK293 cells transfected with Cx43 was quantitated by densitometry, and the signal intensity was normalized to that obtained from nontransfected HEK 293 cells for comparison.
Figure 7.
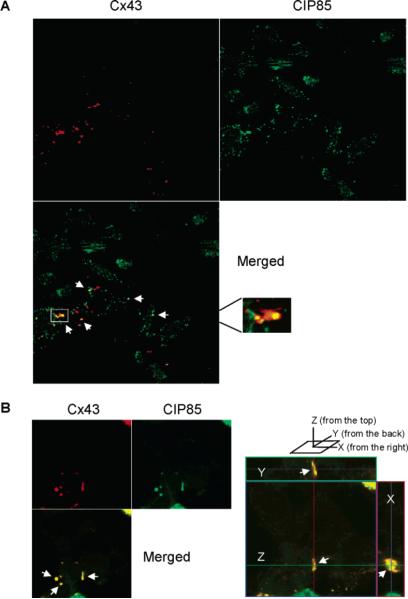
Subcellular colocalization of Cx43 and CIP85. Panel A, laser-scanning confocal microscopy was performed to determine the subcellular localization of Cx43 and endogenous CIP85 in Hela cells (clone C4) utilizing TRITC-conjugated or FITC-conjugated secondary antibodies. The white arrows indicate regions of colocalization of CIP85 and Cx43. The boxed area in the merged image was magnified to show the regional colocalization of Cx43 and CIP85 in a gap junction plaque. Panel B, HEK293 cells were transiently transfected with Cx43 and Flag-CIP85. Laser-scanning confocal microscopy was performed to determine the subcellular localization of Cx43 and CIP85 with TRITC-conjugated or FITC-conjugated secondary antibodies. The white arrows indicate regions of colocalization of CIP85 and Cx43. Right panel section: an orthoimage showing the colocalization of CIP85 and Cx43 in a gap junction plaque in the X-, Y-, and Z-axes.
Interaction of CIP85 with Cx43 in Vitro
E. coli BL21 cells (Novagen) transformed with pET-32.1-Flag-CIP85 (wild type or the deletion mutants) were grown at 30 °C in LB medium containing 100 μg/mL ampicillin with shaking to reach OD600 = 0.5. IPTG (1 mM) was added, and the bacteria were grown for an additional 6 h. Cell cultures were centrifuged at 3000g, and the pellets were resuspended in PBS (137 mM NaCl, 2.7 mM KCl, 8.2 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.4) before brief sonication (Heat Systems) and recentrifugation. The resulting supernatant contained whole cell lysates (WCL) of Flag-CIP85. Alternatively, Flag-CIP85 was further purified with anti-Flag M2 affinity gel and eluted with Flag peptide following the vendor's suggestions (product A2220, Sigma). As a negative control, BL21 cells hosting pET-32.1 alone were treated under the same conditions. A BL21 strain hosting the plasmid hemagglutinin (HA)-pTAT-Cx43 cDNA (gift from Rui Lin) was used for the production of WCL containing HA-Cx43. Ten ml of WCL from BL21 cells containing either Flag-CIP85 or pET-32.1 alone was incubated with or without 4 mL of WCL containing HA-Cx43 in PBS at 4 °C. Glutathione-agarose beads (20 μL, Sigma) were used to collect Flag-CIP85 because CIP85 was discovered to adhere to the beads, which were then washed in PBS and pelleted by centrifugation. The proteins were resolved by electrophoresis on SDS-containing 10% polyacrylamide gels and electrotransferred onto Immobilon-P membranes (Millipore). Flag-CIP85 or HA-Cx43 was detected using a polyclonal Flag antibody (Santa Cruz, CA) or a monoclonal Cx43 antibody (P2D12) by immunoblotting as previously described (17). To study the interaction of the CIP85 deletion mutants with Cx43, 10 mL of WCL (wild type or various mutants) was used in the interaction assays as described above with 1.5 mL of HACx43 WCL, respectively. Alternatively, 50 μg purified Flag-CIP85 (wild type or the SH3 domain deletion mutant purified from the affinity gel) with 1 mL HA-Cx43 WCL was used in the interaction assays as described above. Quantification of the proteins was determined by densitometry of bands developed on X-Omat XAR-5 films (Kodak, Rochester, NY) using the Quantity One software provided with a Fluor-S Multilmager (Bio-Rad, Hercules, CA).
Co-immunoprecipitation Studies of the CIP85–Cx43 Interaction
HEK293 cells were transiently transfected with pcDNA3.1-Cx43 (wt or mutants) (17), pcDNA3.1-Flag-CIP85, or both, using Lipofectamine 2000 (Invitrogen). At 24 h post-transfection, cells were washed in PBS, harvested by scraping, and lysed in 0.2% NP-40 lysis buffer supplemented with protease inhibitors (0.2% Nonidet P-40, 150 mM NaCl, 20 mM Tris, pH 8.0, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 10 mM NaF, 160 μM Na3-VO3, 1 mM benzamidine, 1 μg/mL leupeptin, 1 μg/mL pepstatin). The lysates were clarified and immunoprecipitated with a monoclonal Flag antibody (Sigma) at 4 °C. As a negative control, an equal amount of the lysate was immunoprecipitated with a monoclonal GST antibody (Santa Cruz). The immune complexes were collected with protein A/G plus-agarose beads (Santa Cruz) and washed 10 times in PBS. Flag-CIP85 and Cx43 were detected by immunoblotting as described above. To study the interaction between endogenous CIP85 and Cx43, 20 mg of HEK293 cells were lysed in 0.2% NP-40 lysis buffer, precipitated with the CIP85 antibody, or a polyclonal Flag antibody (Santa Cruz) as a negative control, followed by the procedures as described above.
Laser-Scanning Confocal Microscopy
HEK293 cells were transiently co-transfected with pcDNA3.1-Cx43 and pcDNA3.1-Flag-CIP85 using Lipofectamine 2000 (Invitrogen). At 24 h post-transfection, cells were fixed in cold methanol at −20 °C for 30 min. The subcellular localization of Cx43 and CIP85 was detected with a monoclonal Cx43 antibody (P4G9) and the polyclonal CIP85 antibody by confocal microscopy using a Zeiss LSM 510 laser-scanning confocal microscope with a 63x objective as previously described (36). Alternatively, Hela cells stably expressing Cx43 (clone C4) (37), were fixed in cold methanol and used for confocal microscopy. Goat anti-mouse tetramethyl rhodamine isothiocyanate (TRITC)-conjugated secondary antibody (Sigma) and goat anti-rabbit fluorescein isothiocyanate (FITC)-conjugated secondary antibody (Molecular Probes, Eugene, OR) were used in the experiements. TRITC and FITC were excited with a 543-nm or 488-nm laser lines, respectively. The emission filters for TRITC and FITC were HQ 560 and HQ 505−530, respectively. Optical sections were taken at 0.2-μm intervals, and images were captured and digitally processed to produce merged images and images in each of the three dimensions.
Cx43 Turnover Measurements
Hela cells stably expressing Cx43 (clone C4) (37), were transiently transfected with pcDNA3.1-Flag-CIP85 using Lipofectamine 2000 (Invitrogen) or Effectene (Qiagen). The turnover of Cx43 was studied as previously described (38). Briefly, at 24 h post-transfection, cells were pulse-labeled with [35S]methionine/cysteine (ProMix, Amersham, Piscataway, NJ) at 100 μCi/ml in methionine-free medium (Gibco) for 45 min at 37 °C without or with 10 mM NH4Cl, 40 μM MG132 or 100 μg/mL leupeptin. The medium was aspirated and replaced with nonradioactive, serum-free medium supplemented with 10 mM l-methionine and 5 mM l-cysteine. Pulse-labeled cells were harvested immediately as the 0 h controls or chased for 3 h at 37 °C without or with 10 mM NH4Cl, 40 μM MG132, or 100 μg/mL leupeptin. Cells were washed in PBS, lysed in RIPA buffer (150 mM NaCl, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, 10 mM Tris, pH 7.2) supplemented with protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 10 mM NaF, 160 μM Na3VO3), and immunoprecipitated with Cx43-CT368 peptide antiserum. Immunoprecipitated proteins were resolved by SDS–PAGE, exposed to a Cyclone Storage screen and processed on a Cyclone Phosphoimager (Packard, Meridon, CT) using the OptiQuant software. The data were analyzed statistically using the Student's t test.
RESULTS
A Yeast Two-Hybrid Screen Identified CIP85 as a Novel Cx43-Interacting Protein
The cytoplasmically localized Cx43CT (residues 222−382) was chosen for construction of the “bait” in the yeast two-hybrid screen as previously described (32, 33). The yeast reporter strain L40 expressing LexA-Cx43CT was transformed with a 9−10 day mouse embryo cDNA library (34). Out of 12 million yeast transformants, 42 colonies grew on histidine-minus plates and expressed detectable levels of β-galactosidase activity (32, 33). These data indicated that a specific interaction occurred between these cDNA library polypeptides and Cx43CT, thus they were designated as Cx43-interacting domains or CIDs. Selected individual library plasmids were further tested by directed yeast two-hybrid assays. The CID62 clone was found to produce β-galactosidase activity weakly after a 4 h-incubation when co-transformed with pLexA-Cx43CT, but not with the negative control fusion plasmid pLexA-lamin (Table 1). In comparison, CID91 containing the PDZ2 domain of ZO-1 (39, 40), gave a strongly blue colony color after the 4 h-incubation (Table 1). L40 cells co-transformed with the unmodified pVP16 plasmid and pLexA-Cx43CT did not give a blue colony color supporting the conclusion from earlier trial experiments that Cx43CT by itself is not self-activating in the yeast two-hybrid system (Table 1).
Table 1.
Interaction of CID62 with Cx43CT: β-Galactosidase Filter Assaya
| pVP16 derivative | pLexA-Cx43CT (4 h) | pLexA-lamin (ON) |
|---|---|---|
| pVP16 | − | − |
| CID62 | + | − |
| CID91 | ++ | − |
β-galactosidase activity was determined by a filter assay using the L40 yeast strain cotransformed with the indicated plasmids. A minimum of four independent transformants were analyzed. ++ and +, colonies that turned strongly blue and weakly blue, respectively, within a 4 h-incubation; −, colonies that did not turn blue after overnight (ON) incubation.
DNA sequencing of the CID62 clone (569 bp) and analysis of its deduced amino acid sequence showed that it contained a SH3 domain that mediates protein–protein interactions in numerous proteins (15, 16). The presence of a SH3 domain in CID62 and our earlier demonstration that the v-Src-Cx43 interaction is SH3-dependent (17), stimulated further interest in this clone. To obtain the full-length cDNA containing the CID62 sequence, the CID62 cDNA fragment was used as a probe to screen an oligo-dT primed cDNA library derived from a 16-day mouse embryo. A 2.7-kb full-length cDNA was isolated and sequenced. The complete nucleotide sequence contained an open reading frame encoding a protein of 750 amino acids with a predicted molecular mass of 85-kDa, which was designed as Cx43-interacting protein 85, or CIP85.
Identification and Analysis of the Domains of CIP85
A BLAST database search (http://www.ncbi.nlm.nih.gov/BLAST) using the CIP85 sequence revealed several mouse cDNA clones in the NIH Mammalian Gene Collection that encode the same hypothetical proteins (41). Several hypothetical CIP85 homologues were also uncovered: DJ1042K-10.2.1 of human (90% identity), CG12241-PA of D. melanogaster (64% identity) and F41D9.1 of C. elegans (43% identity). In addition to the SH3 domain (residues 483−537), CIP85 contained a TBC domain (residues 111−327) and a RUN domain (residues 555−718) revealed by a Pfam analysis (http://pfamwustl.edu) (Figure 1A). A short coiled coil region (residues 415−439) was also identified in CIP85 using a COILS analysis (http://www.ch.embnet.org/software/COILS_form.html). The presence of similar domains and their relative organization in CIP85, DJ1042K10.2.1 (Figure 1A), CG1224-PA, and F41D9.1 suggested that they belong to a novel gene family.
The TBC domain of CIP85 was aligned with those of RN-tre (30), GAPCenA (29), and yeast Gyp1p (26), which function as GAPs for the mammalian Rab5, Rab6, and yeast Ypt51p GTPases, respectively (Figure 1B). Remarkably, three motifs, R120PQLW124, I159EKDLLR165, and Y200CQ202 in the TBC domain of CIP85 corresponded to three “fingerprint” sequences that are conserved among members of the yeast Gyp protein family, which serve as GAPs for the Ypt GTPases (28, 42). The R165 site in CIP85 may be of particular importance because the corresponding R343 site in Gyp1p and the R458 site in Gyp7p are essential for their GAP activities against the Ypt51p and Ypt7p GTPase, respectively (26). The R343 site and a second site at R286 in Gyp1p may play a role in the stabilization of its GAP domain (42). The presence of these conserved sites and motifs in CIP85 suggested that CIP85 may function as a Rab GAP to inactivate Rab GTPases.
A BLAST database search of the RUN domain of CIP85 showed that it was 95% identical to the RUN domain in the human DJ1042K10.2.1 protein and 77% identical to that in CG12241-PA of D. melanogaster. The RUN domains of Rabip4, a Rab4 effector (43), and RPIP8, a Rap2 effector (44), were aligned with that of CIP85 (Figure 1C). RUN domains have been suggested to be downstream effectors of proteins of the Ras superfamily (31). The frequent occurrence of RUN domains together with TBC domains in proteins such as CIP85 suggests that they could have specific functions related to the Rab signaling pathways involved in exocytic or endocytic pathways (31).
The presence of a coiled coil in CIP85 is intriguing because the coiled coil located downstream of the TBC domain of GAPCenA mediates the interaction between GAPCenA and its substrate, Rab6 (29). A coiled coil is a bundle of α-helices that are wound into a superhelix that may facilitate protein–protein interactions (45).
CIP85 mRNA Is Ubiquitously Expressed in Mouse and Human Tissues
Northern blot analyses were performed to determine the pattern of CIP85 expression in mouse and human tissues. A single CIP85 mRNA of approximately 2.7-kb was highly expressed in mouse testis and brain, moderately expressed in heart, lung, liver, skeletal muscle and kidney, and expressed at low levels in spleen (Figure 2A). Northern blot analysis of human tissues indicated that the human homologues of CIP85 mRNA were most highly expressed in brain and spleen, with lower levels in uterus and placenta, and very low, but still detectable levels in kidney and lung (Figure 2B). It is intriguing that CIP85 mRNA was expressed at high levels in human spleen but at low levels in mouse spleen, but the reason for this apparent species-specific tissue expression is currently unknown. In contrast to the mouse 2.7-kb mRNA, the human CIP85 mRNA was evident as two distinct bands of approximately 2.5-kb and 2.8-kb (Figure 2B), which may arise from alternate splicing events. These data clearly demonstrated that mouse CIP85 and its human homologues are ubiquitously expressed in tissues.
Figure 2.
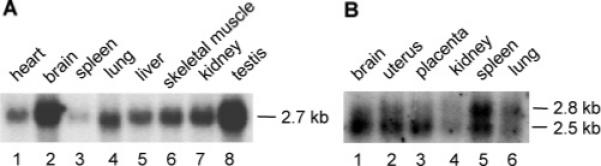
Expression of CIP85 mRNA in mouse and human tissues. Panel A, a [α-32P]-dCTP-labeled CID62 fragment of CIP85 cDNA (Figure 1A) was used to probe a membrane containing 2 μg/lane of mRNA derived from various mouse tissues. Panel B, a [α-32P]-dCTP-labeled probe fragment of human CIP85 cDNA (Figure 1A) was hybridized to a membrane containing 2 μg/lane of mRNA derived from various human tissues.
Full-Length CIP85 Interacts with Cx43 in Vitro and in Vivo
The ability of the Cx43CT “bait” to interact with CID62 in the yeast two-hybrid system, suggested that the full-length wild-type proteins would also interact. To confirm this possibility, the interaction between CIP85 and Cx43 was examined by in vitro interaction assays and co-immunoprecipitation of endogenously expressed CIP85 and Cx43. Full-length Flag epitope-tagged CIP85 and HA epitope-tagged Cx43 were expressed in E. coli BL21 cells. Although the yield of soluble HA-Cx43 in bacteria was low possibly due to the multiple transmembrane regions present in Cx43 (Figure 6A), sufficient amounts of HA-Cx43 were expressed to permit the in vitro interaction assays. HA-Cx43 cell lysates were incubated with either Flag-CIP85 or the vector alone lysates. Because CIP85 could bind to glutathione-agarose beads, these were used to collect the Flag-CIP85 protein complexes. The results showed that Cx43 was only detected in association with CIP85 (Figure 3, lane 3), but not with the vector alone lysates (Figure 3, lane 2). In addition, in vitro interaction assays revealed that Cx43 was associated with the affinity gel-purified Flag-CIP85 (Figure 5C, lane 2). Moreover, the amount of associated Cx43 was increased when additional amount of purified Flag-CIP85 was added to the in vitro interaction assays (data not shown). These data using full-length CIP85 and Cx43 confirmed the CID62-Cx43CT interaction observed in the yeast two-hybrid assays.
Figure 6.
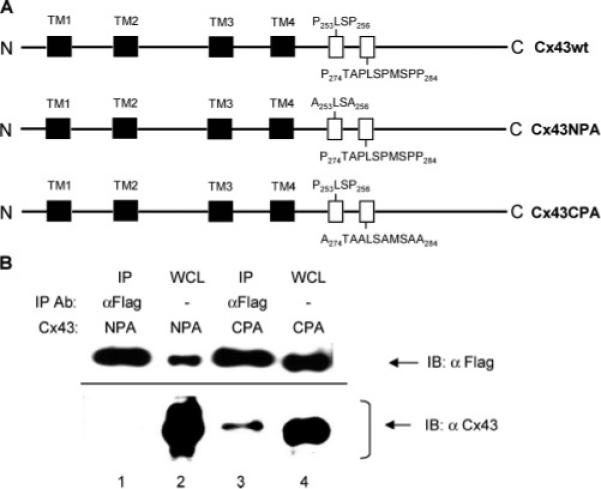
Identification of the CIP85-interacting region in Cx43 by analyses of proline site substitution mutants. Panel A, schematic diagrams of the Cx43 proline to alanine substitution mutants (Cx43NPA and Cx43CPA). The primary structure of Cx43 includes four transmembrane regions (TM1−4) indicated by black shaded boxes and two proline-rich regions (P253LSP256 and P274TAPLSPMSPP284) indicated by the open boxes. Panel B, HEK293 cells transiently coexpressing Flag-CIP85 along with either the Cx43NPA mutant or the Cx43CPA mutant were lysed in 300 μL of 0.2% NP-40 lysis buffer. A Flag antibody was used to immunoprecipitate subpopulations of the protein complexes, and the associated Cx43 was detected by immunoblotting with a Cx43 antibody (lanes 1 and 3). Five microliters of lysates of HEK293 transfectants were used as positive controls (lanes 2 and 4).
Figure 3.
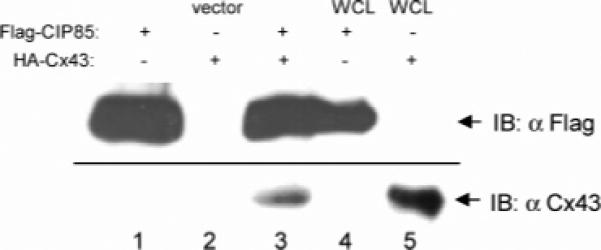
Interaction of CIP85 with Cx43 in vitro. Lysates from bacteria expressing Flag-CIP85 or the vector alone were incubated with or without lysates containing HA-Cx43 (lanes 1−3). Flag-CIP85 complexes were collected on glutathione-agarose beads and immunoblotted with a Cx43 antibody. Lysates containing either Flag-CIP85 (lane 4) or HA-Cx43 (lane 5) were used as positive controls.
Figure 5.
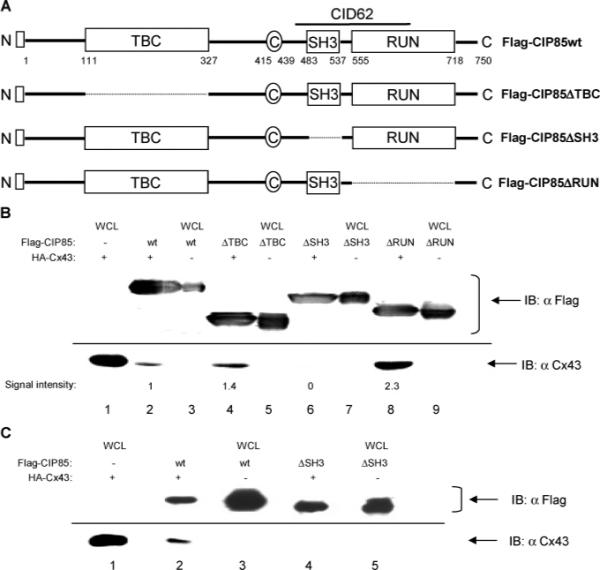
Identification of the Cx43-interacting domain in CIP85 by analyses of in-frame deletion mutants. Panel A, schematic diagrams of the CIP85 mutants with in-frame deletions of the TBC domain (ΔTBC), SH3 domain (ΔSH3), or RUN domain (ΔRUN). □: Flag epitope located at the NH2-terminus. Panel B, bacterial lysates containing Flag-CIP85wt (lane 2), the Flag-CIP85ΔTBC (lane 4), Flag-CIP85ΔSH3 (lane 6), or Flag-CIP85ΔRUN (lane 8) mutants were used in the in vitro interaction assays against HA-Cx43. The Flag-CIP85 present in the assays and the associated Cx43 were detected by immunoblotting with Flag or Cx43 antibodies (lanes 2, 4, 6, and 8). Bacterial lysates containing HA-Cx43 (lane 1), Flag-CIP85wt (lane 3), and the various mutants (lanes 5, 7, and 9) were used as positive controls to show the migration positions of these proteins. The signal intensities quantitated by densitometry were normalized for comparison. Panel C, affinity gel-purified Flag-CIP85wt (lane 2) and the Flag-CIP85ΔSH3 mutant (lane 4) were used in the in vitro interaction assays against HA-Cx43. The Flag-CIP85 present in the assays and the associated Cx43 were detected by immunoblotting with Flag or Cx43 antibodies (lanes 2 and 4). Bacterial lysates containing HA-Cx43 (lane 1), Flag-CIP85wt (lane 3), and Flag-CIP85ΔSH3 (lane 5) were used as positive controls.
We prepared an antibody against CIP85 to detect the endogenously expressed protein. Analysis of CIP85 revealed a region of high antigenicity (K32EESSE37) near the NH2-terminus (Figure 1A). This epitope was outside of the TBC domain, thus an antibody against this epitope was less likely to interfere with any binding or catalytic activities of this domain. In addition, this antigenic site is conserved in DJ1042K10.2.1 (K31EESAE36), the human CIP85 homologue. The resulting CIP85 antibody recognized a ∼90-kDa band from HEK293 cells (Figure 4A, lane 1), which was specifically diminished by the competing immunizing peptide (Figure 4A, lane 3). Human DJ1042K10.2.1 is predicted to encode a 88-kDa product, therefore this antibody-reactive band from HEK293 cells is likely to be encoded by the human DJ1042K10.2.1 gene. Low levels of endogenous Cx43 were also detected in HEK293 cells with a Cx43 antibody (Figure 4A, lane 1). To study the endogenous CIP85–Cx43 interaction, CIP85 antibody immunoprecipitates were prepared from HEK293 cells and the presence of Cx43 was determined by immunoblotting. The CIP85 immunoprecipitates clearly demonstrated the presence of endogenous Cx43 (Figure 4B, lane 3). Transfection of exogenous Cx43 into HEK293 cells resulted in an apparent increase (2.6-fold) in the amount of associated Cx43 detected in the CIP85 immunoprecipitates (Figure 4B, lane 4). The Cx43 bands were confirmed by the ability of the competing peptide to reduce the Cx43 band intensity (data not shown). In addition, co-immunoprecipitation of CIP85 and Cx43 transiently expressed in HEK293 cells also confirmed the interaction of these full-length proteins in mammalian cells (data not shown). Taken together, these results clearly demonstrated the ability of CIP85 and Cx43 to interact under native conditions.
The CIP85–Cx43 Interaction Is Dependent on the SH3 Domain and a Proline-Rich Region
Because the CID62 peptide contained a complete SH3 domain, we postulated that this domain might be involved in the CIP85–Cx43 interaction. However, the presence of the NH2-terminal half of the RUN domain in the CID62 peptide suggested that the SH3 domain might not act alone in the interaction (Figure 5A). To determine which domain(s) in full-length CIP85 was responsible for the interaction with Cx43, in vitro interaction assays were performed using bacterium-expressed Flag-CIP85wt or Flag-CIP85 mutants containing in-frame deletions of the TBC, SH3, or RUN domains (Figure 5A). The interaction assays using CIP85 WCL showed that the deletion of the SH3 domain in CIP85 completely abolished the interaction with Cx43 (Figure 5B, lane 6). Furthermore, the interaction assays using affinity gel-purified CIP85 showed the loss of Cx43 association with the CIP85ΔSH3 mutant (Figure 5C, lane 4), in contrast to the wild type control (Figure 5C, lane 2). These data confirmed that the SH3 domain of CIP85 was the primary determinant of its interaction with Cx43. It is intriguing that the deletion of either the TBC or the RUN domains of CIP85 did not abolish, but apparently increased, the binding of CIP85 to Cx43 as shown in Figure 5B (lanes 4 and 8). Binding of Cx43 to the CIP85ΔTBC and CIP85ΔRUN mutant was about 1.4-fold and 2.3-fold that of the CIP85wt control, respectively. The absence of these domains might relieve constraints on the interaction of the SH3 domain of CIP85 with Cx43.
These data suggested the requirement for a proline-rich region in Cx43 to interact with the SH3 domain of CIP85. The C-terminal tail of Cx43 contains two proline-rich regions: a more N-terminally-located one at P253LSP256 and a more C-terminally-located one at P274TAPLSPMSPP284 (Figure 6A). To determine which of these proline-rich regions was involved in the interaction with CIP85, we used two Cx43 proline to alanine substitution mutants: Cx43NPA (P253LSP256 → A253LSA256) and Cx43CPA (P274TAPLSPMSPP284 → A274TAALSAMSAA284) (Figure 6A) (17). The Cx43NPA mutant or the Cx43CPA mutant were coexpressed with Flag-CIP85 in HEK293 cells. Co-immunoprecipitation was performed with a Flag antibody and the co-immunoprecipitates were blotted with a Cx43 antibody. Although Flag-CIP85 was expressed at lower levels in the HEK293 cells coexpressing the Cx43NPA mutant than in those coexpressing the Cx43CPA mutant (Figure 6B, lanes 2 and 4), only the Cx43NPA mutant failed to associate with an equivalent amount of immunoprecipitated Flag-CIP85 (Figure 6B, lanes 1 and 3), indicating that the P253LSP256 site of Cx43 was the primary proline-rich region involved in binding to the SH3 domain of CIP85. In addition, lysates of HEK293 transfectants containing Cx43wt or the Cx43CPA mutant were used in the in vitro interaction assays with Flag-CIP85. The results showed that both Cx43wt and the Cx43CPA mutant interacted with CIP85 (data not shown). Taken together, these results strongly suggested that the CIP85–Cx43 interaction is dependent upon the SH3 domain of CIP85 and the P253LSP256 proline-rich region of Cx43.
CIP85 Colocalizes with Cx43 at the Plasma Membrane
The physical association of CIP85 with Cx43 suggested that the proteins colocalized within the cell. The subcellular localization of Cx43 and endogenous CIP85 in Hela cells (clone C4) was examined with the use of Cx43 and CIP85 antibodies and laser-scanning confocal microscopy. The results showed the expected localization of Cx43 to the cell periphery in punctate reactions reminiscent of gap junction plaques (Figure 7A). Endogenous CIP85 was localized mainly to the plasma membrane with some intracellular punctuate reactions (Figure 7A). Merged images revealed that CIP85 colocalized with Cx43 in regions of gap junction plaques (Figure 7A). An area in the merged image was magnified to show the regional colocalization of Cx43 and CIP85 in a gap junction plaque (Figure 7A, box). To determine if Cx43 and CIP85 colocalized in three dimensions, overexpressed Flag-CIP85 and Cx43 in HEK293 cells was studied by laser-scanning confocal microscopy. Merged images revealed that CIP85 colocalized with Cx43 in gap junction plaques analyzed in three dimensions (Figure 7B, right panel section). Taken together, these results confirmed the colocalization of CIP85 and Cx43 in mammalian cells.
The Turnover of Cx43 Is Stimulated by CIP85
Cx43 is trafficked to the plasma membrane and removed for subsequent degradation with a short half-life. The physical interaction and colocalization of CIP85 and Cx43 at the plasma membrane and the presence of the Rab GAP-like domain in CIP85 suggested that CIP85 might regulate Cx43 levels. Hela cells stably expressing Cx43 (clone C4), were transiently transfected with the vector alone, Flag-CIP85wt or the Flag-CIP85ΔSH3 mutant. The turnover of Cx43 was measured by pulse-chase labeling and characterized by the use of proteasomal and lysosomal inhibitors. Pulse-chase labeling results indicated that the amount of Cx43 remaining after a 3 h-chase in cells coexpressing CIP85wt was about 31% less than that of the vector alone control (Table 2). However, Cx43 turnover was not significantly altered in Hela cells expressing the CIP85ΔSH3 mutant (Table 2), which no longer interacted with Cx43. These results suggested that the presence of CIP85 and its interaction with Cx43 were associated with the increased turnover of Cx43. In addition, as shown in Table 2, the increased turnover of Cx43 induced by CIP85 was completely blocked by the lysosomal inhibitor, NH4Cl (46), whereas the partial block by the proteasomal inhibitor, MG132 (47), was not statistically significant, suggesting that CIP85 is involved primarily in the lysosomal pathway of Cx43 degradation. Leupeptin, a lysosomal inhibitor (46), also had a similar inhibitory effect as NH4Cl (data not shown).
Table 2.
Measurement of Cx43 Turnovera
| pcDNA3.1 derivative | percentage of Cx43 remaining |
|---|---|
| pcDNA3.1 | 85 ± 2 (5) |
| Flag-CIP85wt | 54 ± 5 (4) |
| Flag-CIP85ΔSH3 | 79 ± 6 (4) |
| Flag-CIP85wt + NH4Cl | 89 ± 2 (5) |
| Flag-CIP85wt + MG132 | 66 ± 2 (3) |
Hela cells stably expressing Cx43 were transiently transfected with pcDNA3.1 alone or Flag-CIP85 constructs (wild type or the SH3 domain deletion mutant). The cells were pulse-labeled with [35S]Met/Cys at 100 μCi/mL for 45 min and chased for 3 h without or with 10 mM NH4Cl or 40 μM MG132. [35S]-labeled Cx43 proteins were immunoprecipitated and quantitatively measured by densitometry of the autoradiographs. The mean values ± SEM (t test) represent the percentages of Cx43 remaining after 3 h-chases compared to 0 h-chase controls. The numbers of independent experiments are indicated in parentheses. Compared to the value of the vector alone in a two-tailed Student's t test, the value of CIP85wt is significantly different (P < 0.013), whereas the value of CIP85ΔSH3 is not significantly different (P > 0.1). Compared to the value of CIP85wt without inhibitor in a two-tailed Student's t test, the value of CIP85wt pulsed and chased with NH4Cl is significantly different (P < 0.004), whereas the value of CIP85wt pulsed and chased with MG132 is not significantly different (P > 0.1).
DISCUSSION
Our initial yeast two-hybrid screen uncovered several clones that were confirmed as in Vivo binding partners of Cx43CT when they were retested in directed yeast two-hybrid assays (32, 33). Many of these proteins were homologous to known proteins, but some represented novel proteins containing interesting domains (data not shown). The CID62 clone reported in this paper was chosen for further study because it contained a SH3 domain and the NH2-terminal portion of the novel RUN domain. The presence of the SH3 domain suggested a possible basis for the observed interaction with the C-terminal tail of Cx43, which is known to contain proline-rich regions. To study this interaction further, the full-length cDNA of CIP85 was isolated from a cDNA library derived from a 16-day mouse embryo.
A BLAST database search uncovered several mouse cDNA clones in the NIH Mammalian Gene Collection that encoded the same hypothetical protein (41). In addition, hypothetical homologues were identified in human (DJ1−042K10.2.1), D. melanogaster (CG12241-PA), and C. elegans (F41D9.1). Besides the SH3 domain, a TBC domain, a RUN domain, and a short coiled coil region were revealed in CIP85 by Pfam and COILS analyses. Although individual TBC, SH3 and RUN domains are found in a large number and variety of proteins, the combination of these three domains are found only in CIP85 and a few other hypothetical proteins (31). CIP85 is a novel protein that is expressed in all mouse and human tissues examined (Figure 2). The expression of CIP85 was particularly high in mouse testis and brain and in human spleen and brain. In addition, CIP85 was expressed endogenously at low levels in HEK293 cells (Figure 4A), Hela and COS7 cells (data not shown). The reasons for the different expression levels of CIP85 in mouse spleen (low) and human spleen (high) are not evident, but it may reflect a species difference in the function of CIP85 in this particular tissue. The ubiquitous expression of CIP85 in mammalian tissues suggests that it may play an important, but currently undefined, role in common cellular and tissue functions. It is also possible that CIP85 plays a role in other cellular activities or signaling pathways that are independent of Cx43 and its function.
CIP85 was predicted to have cytoplasmic/nuclear discrimination as 89 to 11 by a Psort analysis (http://psort.nibb.ac.jp). A NetPhos analysis (http://www.cbs.dtu.dk/services/Netphos) predicted putative phosphorylation at twenty serine sites, five threonine sites, and six tyrosine sites in CIP85, which may be targeted by cAMP- and cGMP-dependent protein kinase, protein kinase C, casein kinase II and other kinases predicted using a ScanProsite analysis (http://ca.expasy.org/cgi-bin/scanprosite). ScanProsite also suggested the possibility of NH2-glycosylation and N-myristoylation sites in CIP85, however, judging from the usually sharp migration of the CIP85 bands on SDS-gels (Figure 4A), this post-translational modification may not actually occur in cells.
The conclusion that CIP85 interacted with Cx43 was supported by results from four distinct experimental approaches. First, the yeast two-hybrid assays demonstrated the interaction between Cx43CT, representing the cytoplasmically-located C-terminal tail of Cx43, and the CIP85-derived CID62 fragment, which contains the SH3 domain and N-terminal half of the RUN domain (Table 1). Second, in vitro interaction assays utilizing crude or purified CIP85 and Cx43 expressed in E. coli showed the ability of these full-length proteins to interact with one another (Figures 3 and 5C). Third, co-immunoprecipitation assays detected the interaction between endogenously expressed CIP85 and Cx43 (Figure 4B), which clearly demonstrated the ability of these proteins to interact under native conditions. And fourth, as predicted by their physical interaction, laser-scanning confocal microscopy images showed an apparent colocalization of subpopulations of endogenous or overexpressed CIP85 and Cx43 at the cell periphery, particularly in areas reminiscent of gap junction plaques (Figure 7). These results were confirmed by the three-dimensional analysis of an area of colocalization (Figure 7B, right panel section). Thus, these combined data indicate that the novel CIP85 protein interacts with the gap junction protein, Cx43. Although these data are consistent with the direct interaction between CIP85 and Cx43, it is possible that an “adapter” protein may mediate the interaction.
Analysis of domain deletion mutants of CIP85 and proline substitution mutants of Cx43 confirmed the importance of the SH3 domain of CIP85 and the proline-rich region located at P253LSP256 of Cx43 in the interaction of these proteins (Figures 5 and 6). To our knowledge this is the first report of the involvement of this proline-rich site in the interaction of Cx43 with another protein partner. This site is distinct from the proline-rich region at P274TAPLSPMSPP284 of Cx43, which was demonstrated previously to partner with the SH3 domain of v-Src (17). We have proposed that this initial interaction is pivotal in the subsequent ability of v-Src to phosphorylate Cx43 and promote the closure of the Cx43 channels (13). It is noteworthy, that the Cx43 P253LSP256 site contains S255, which is one of three serine sites targeted by MAP kinase activated by the EGF receptor (48). It is possible that phosphorylation of S255 by MAP kinase, or other protein kinases, may modulate the binding of CIP85 to Cx43. Stimulation of cells with EGF led to an apparent increased amount of Cx43 associated with CIP85 (Lan and Lau, unpublished observation).
The demonstrated ability of connexins to interact with other proteins raises the important question concerning the functional significance of the interactions. In the case of CIP85, our current data indicated that its transient coexpression with Cx43 in Hela cells was associated with the stimulation of Cx43 turnover, which resulted in a modest, but reproducible, decrease in the amount of Cx43 measured by pulse-chase labeling (Table 2). This effect was dependent upon the interaction of the proteins because the CIP85ΔSH3 mutant lacking the SH3 domain, and thus unable to interact with Cx43, failed to induce the change (Table 2). In addition, the increased turnover of Cx43 induced by CIP85 was completely blocked by the lysosomal inhibitor, NH4Cl (Table 2) or leupeptin (data not shown), indicating that CIP85 may be involved in the degradation of Cx43 through the lysosomal pathway. The moderate decrease of Cx43 should be considered in the context of the transfection efficiencies of CIP85, which varied between 30% and 50% of the total Hela cells. The low transfection efficiencies may be due to the nature of CIP85 because transfection of a GFP control under identical experimental conditions approached 90% (data not shown). Efforts are currently underway to establish cell lines stably expressing CIP85 to be used to further explore the effects of CIP85 on Cx43 levels.
These data suggested the possibility that CIP85 may affect the levels of Cx43 through a currently unknown mechanism. The presence of some conserved sites and motifs in the CIP85 TBC domain suggested that CIP85 may function as a Rab GAP to inactivate Rab GTPases. Recently, it has been demonstrated that internalized Cx43 is accumulated within Rab5 positive endosomes (49), and that lysosomes play a key role in degrading internalized Cx43 (46). Our chemical inhibitor data suggested that CIP85 is involved in the lysosomal pathway of Cx43 degradation (Table 2). Whether CIP85 is involved in the regulation of Rab5 or other Rabs, which might mediate Cx43 disassembly at the plasma membrane or Cx43 degradation through the lysosomal pathway, is an interesting question for future research.
The recently identified RUN domains might function as effectors of proteins of the Ras superfamily (31). The frequent occurrence of RUN domains together with TBC domains in proteins such as CIP85 suggests that they could have specific functions related to the Rab signaling pathways involved in exocytic or endocytic pathways (31). The potential function and roles of the newly discovered RUN domains in proteins such as CIP85 remain to be discovered.
In summary, we describe a novel protein that interacts with Cx43 through a SH3 domain interaction involving the P253-LSP256 site in Cx43. CIP85 is a member of a novel protein family, which contains a unique combination of TBC, SH3, and RUN domains. CIP85 is ubiquitously expressed in mouse and human tissues. It is distributed primarily to the plasma membrane where it colocalizes with Cx43 in apparent gap junction plaques. Interestingly, the interaction of CIP85 with Cx43 appeared to stimulate the degradation of Cx43 through a currently unknown mechanism involving the lysosomal pathway.
ACKNOWLEDGMENT
We thank Takahiro Nagase (Kazusa DNA Research Institute, Chiba, Japan), Bonnie Warn-Cramer (Cancer Research Center of Hawaii), Rui Lin (Scripps Research Institute), and Martha Kanemitsu (Arena Pharmaceuticals, San Diego, CA) for their contribution of molecular reagents. We also wish to thank Tanya Koropatnick (Pacific Biomedical Research Center, Honolulu, HI) for invaluable technical assistance in laser-scanning confocal microscopy and Xinli Li and Darren Park (Cancer Research Center of Hawaii) for helpful suggestions during the course of these studies.
Footnotes
This work was supported in part by Grant CA52098 from the National Institutes of Health (to A.F.L.) and Grant-in-Aid 0051599Z (to A.F.L.) and Predoctoral Fellowship HIFW-29-96 (to C.J.) from the American Heart Association, Hawaii Affiliate.
Abbreviations: Cx, connexin; Cx43, connexin43; Cx43CT, C-terminal cytoplasmic tail of Cx43; CIP85, a Cx43-interacting protein of 85-kDa; GJC, gap junctional communication; SH3, Src-homology 3; Ypt, yeast protein transport; Rab, Ras gene from rat brain; GAP, GTPase-activating protein; GDI, guanine nucleotide dissociation inhibitor; GEF, guanine nucleotide exchange factor; Gyp, GAP for yeast protein transport; GST, glutathione S-transferase; BSA, bovine serum albumin; PBS, phosphate-buffered saline; IB, immunoblotting; IP, immunoprecipitation; WCL, whole cell lysates; wt, wild type; MAP, mitogen-activated protein; EGF, epidermal growth factor.
REFERENCES
- 1.Kumar NM, Gilula NB. The gap junction communication channel. Cell. 1996;84:381–388. doi: 10.1016/s0092-8674(00)81282-9. [DOI] [PubMed] [Google Scholar]
- 2.Goodenough DA, Goliger JA, Paul DL. Connexins, connexons, and intercellular communication. Annu. Rev. Biochem. 1996;65:475–502. doi: 10.1146/annurev.bi.65.070196.002355. [DOI] [PubMed] [Google Scholar]
- 3.Beyer EC, Paul DL, Goodenough DA. Connexin family of gap junction proteins. J. Membr. Biol. 1990;116:187–194. doi: 10.1007/BF01868459. [DOI] [PubMed] [Google Scholar]
- 4.Spray DC, Burt JM. Structure–activity relations of the cardiac gap junction channel. Am. J. Physiol. 1990;258:C195–205. doi: 10.1152/ajpcell.1990.258.2.C195. [DOI] [PubMed] [Google Scholar]
- 5.Britz-Cunningham SH, Shah MM, Zuppan CW, Fletcher WH. Mutations of the Connexin43 gap-junction gene in patients with heart malformations and defects of laterality. N. Engl. J. Med. 1995;332:1323–1329. doi: 10.1056/NEJM199505183322002. [DOI] [PubMed] [Google Scholar]
- 6.Dasgupta C, Martinez AM, Zuppan CW, Shah MM, Bailey LL, Fletcher WH. Identification of connexin43 (alpha1) gap junction gene mutations in patients with hypoplastic left heart syndrome by denaturing gradient gel electrophoresis (DGGE) Mutat. Res. 2001;479:173–186. doi: 10.1016/s0027-5107(01)00160-9. [DOI] [PubMed] [Google Scholar]
- 7.Dupont E, Matsushita T, Kaba RA, Vozzi C, Coppen SR, Khan N, Kaprielian R, Yacoub MH, Severs NJ. Altered connexin expression in human congestive heart failure. J. Mol. Cell. Cardiol. 2001;33:359–371. doi: 10.1006/jmcc.2000.1308. [DOI] [PubMed] [Google Scholar]
- 8.White TW, Paul DL. Genetic diseases and gene knockouts reveal diverse connexin functions. Annu. Rev. Physiol. 1999;61:283–310. doi: 10.1146/annurev.physiol.61.1.283. [DOI] [PubMed] [Google Scholar]
- 9.Yamasaki H, Naus CG. Role of connexin genes in growth control. Carcinogenesis. 1996;17:1199–1213. doi: 10.1093/carcin/17.6.1199. [DOI] [PubMed] [Google Scholar]
- 10.Omori Y, Yamasaki H. Mutated connexin43 proteins inhibit rat glioma cell growth suppression mediated by wild-type connexin43 in a dominant-negative manner. Int. J. Cancer. 1998;78:446–453. doi: 10.1002/(sici)1097-0215(19981109)78:4<446::aid-ijc10>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 11.Loewenstein WR, Rose B. The cell–cell channel in the control of growth. Semin. Cell Biol. 1992;3:59–79. doi: 10.1016/s1043-4682(10)80008-x. [DOI] [PubMed] [Google Scholar]
- 12.Yamasaki H. Gap junctional intercellular communication and carcinogenesis. Carcinogenesis. 1990;11:1051–1058. doi: 10.1093/carcin/11.7.1051. [DOI] [PubMed] [Google Scholar]
- 13.Lin R, Warn-Cramer BJ, Kurata WE, Lau AF. v-Src phosphorylation of connexin 43 on Tyr247 and Tyr265 disrupts gap junctional communication. J. Cell Biol. 2001;154:815–827. doi: 10.1083/jcb.200102027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen GB, Ren R, Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 15.Pawson T. SH2 and SH3 domains in signal transduction. Adv. Cancer Res. 1994;64:87–110. doi: 10.1016/s0065-230x(08)60835-0. [DOI] [PubMed] [Google Scholar]
- 16.Feng S, Chen JK, Yu H, Simon JA, Schreiber SL. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 17.Kanemitsu MY, Loo LW, Simon S, Lau AF, Eckhart W. Tyrosine phosphorylation of connexin 43 by v-Src is mediated by SH2 and SH3 domain interactions. J. Biol. Chem. 1997;272:22824–22831. doi: 10.1074/jbc.272.36.22824. [DOI] [PubMed] [Google Scholar]
- 18.Musil LS, Goodenough DA. Multisubunit assembly of an integral plasma membrane channel protein, gap junction connexin43, occurs after exit from the ER. Cell. 1993;74:1065–1077. doi: 10.1016/0092-8674(93)90728-9. [DOI] [PubMed] [Google Scholar]
- 19.Laird DL, Castillo M, Kasprzak L. Gap junction turnover, intracellular trafficking, and phosphorylation of connexin43 in Brefeldin A-treated rat mammary tumor cells. J. Cell Biol. 1995;131:1193–1203. doi: 10.1083/jcb.131.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- 21.Saffitz JE, Laing JG, Yamada KA. Connexin expression and turnover: implications for cardiac excitability. Circ. Res. 2000;86:723–728. doi: 10.1161/01.res.86.7.723. [DOI] [PubMed] [Google Scholar]
- 22.Laing JG, Beyer EC. The gap junction protein connexin43 is degraded via the ubiquitin proteasome pathway. J. Biol. Chem. 1995;270:26399–26403. doi: 10.1074/jbc.270.44.26399. [DOI] [PubMed] [Google Scholar]
- 23.Laing JG, Tadros PN, Westphale EM, Beyer EC. Degradation of connexin43 gap junctions involves both the proteasome and the lysosome. Exp. Cell Res. 1997;236:482–492. doi: 10.1006/excr.1997.3747. [DOI] [PubMed] [Google Scholar]
- 24.Segev N. Ypt and Rab GTPases: insight into functions through novel interactions. Curr. Opin. Cell Biol. 2001;13:500–511. doi: 10.1016/s0955-0674(00)00242-8. [DOI] [PubMed] [Google Scholar]
- 25.Segev N. Ypt/rab gtpases: regulators of protein trafficking. Science's STKE 2001. 2001:RE11. doi: 10.1126/stke.2001.100.re11. [DOI] [PubMed] [Google Scholar]
- 26.Albert S, Will E, Gallwitz D. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 1999;18:5216–5225. doi: 10.1093/emboj/18.19.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richardson PM, Zon LI. Molecular cloning of a cDNA with a novel domain present in the tre-2 oncogene and the yeast cell cycle regulators BUB2 and cdc16. Oncogene. 1995;11:1139–1148. [PubMed] [Google Scholar]
- 28.Neuwald AF. A shared domain between a spindle assembly checkpoint protein and Ypt/Rab-specific GTPase-activators. Trends Biochem. Sci. 1997;22:243–244. doi: 10.1016/s0968-0004(97)01073-6. [DOI] [PubMed] [Google Scholar]
- 29.Cuif MH, Possmayer F, Zander H, Bordes N, Jollivet F, Couedel-Courteille A, Janoueix-Lerosey I, Langsley G, Bornens M, Goud B. Characterization of GAPCenA, a GTPase activating protein for Rab6, part of which associates with the centrosome. EMBO J. 1999;18:1772–1782. doi: 10.1093/emboj/18.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanzetti L, Rybin V, Malabarba MG, Christoforidis S, Scita G, Zerial M, Di Fiore PP. The Eps8 protein coordinates EGF receptor signaling through Rac and trafficking through Rab5. Nature. 2000;408:374–377. doi: 10.1038/35042605. [DOI] [PubMed] [Google Scholar]
- 31.Callebaut I, de Gunzburg J, Goud B, Mornon JP. RUN domains: a new family of domains involved in Ras-like GTPase signaling. Trends Biochem. Sci. 2001;26:79–83. doi: 10.1016/s0968-0004(00)01730-8. [DOI] [PubMed] [Google Scholar]
- 32.Jin C, Lau AF. In: Gap Junctions. Werner R, editor. IOS Press; Amsterdam: 1998. pp. 230–234. [Google Scholar]
- 33.Jin C, Lau AF, Martyn KD. Identification of connexin-interacting proteins: application of the yeast two-hybrid screen. Methods. 2000;20:219–231. doi: 10.1006/meth.1999.0939. [DOI] [PubMed] [Google Scholar]
- 34.Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix-loop- helix proteins with a two-hybrid system. Mol. Cell. Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hattori A, Okumura K, Nagase T, Kikuno R, Hirosawa M, Ohara O. Characterization of long cDNA clones from human adult spleen. DNA Res. 2000;7:357–366. doi: 10.1093/dnares/7.6.357. [DOI] [PubMed] [Google Scholar]
- 36.Loo LW, Kanemitsu MY, Lau AF. In vivo association of pp60v-src and the gap-junction protein connexin 43 in v-src-transformed fibroblasts. Mol. Carcinog. 1999;25:187–195. [PubMed] [Google Scholar]
- 37.Warn-Cramer BJ, Cottrell GT, Burt JM, Lau AF. Regulation of connexin-43 gap junctional intercellular communication by mitogen-activated protein kinase. J. Biol. Chem. 1998;273:9188–9196. doi: 10.1074/jbc.273.15.9188. [DOI] [PubMed] [Google Scholar]
- 38.Crow DS, Beyer EC, Paul DL, Kobe SS, Lau AF. Phosphorylation of connexin43 gap junction protein in uninfected and Rous sarcoma virus-transformed mammalian fibroblasts. Mol. Cell. Biol. 1990;10:1754–1763. doi: 10.1128/mcb.10.4.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giepmans BN, Moolenaar WH. The gap junction protein connexin43 interacts with the second PDZ domain of the zona occludens-1 protein. Curr. Biol. 1998;8:931–934. doi: 10.1016/s0960-9822(07)00375-2. [DOI] [PubMed] [Google Scholar]
- 40.Toyofuku T, Yabuki M, Otsu K, Kuzuya T, Hori M, Tada M. Direct association of the gap junction protein connexin-43 with ZO-1 in cardiac myocytes. J. Biol. Chem. 1998;273:12725–12731. doi: 10.1074/jbc.273.21.12725. [DOI] [PubMed] [Google Scholar]
- 41.Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA. Generation and initial analysis of more than 15, 000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rak A, Fedorov R, Alexandrov K, Albert S, Goody RS, Gallwitz D, Scheidig AJ. Crystal structure of the GAP domain of Gyp1p: first insights into interaction with Ypt/Rab proteins. EMBO J. 2000;19:5105–5113. doi: 10.1093/emboj/19.19.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mari M, Macia E, Le Marchand-Brustel Y, Cormont M. Role of the FYVE finger and the RUN domain for the subcellular localization of Rabip4. J. Biol. Chem. 2001;276:42501–42508. doi: 10.1074/jbc.M104885200. [DOI] [PubMed] [Google Scholar]
- 44.Janoueix-Lerosey I, Pasheva E, de Tand MF, Tavitian A, de Gunzburg J. Identification of a specific effector of the small GTP-binding protein Rap2. Eur. J. Biochem. 1998;252:290–298. doi: 10.1046/j.1432-1327.1998.2520290.x. [DOI] [PubMed] [Google Scholar]
- 45.Lupas A. Coiled coils: new structures and new functions. Trends Biochem. Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 46.Qin H, Shao Q, Igdoura SA, Alaoui-Jamali MA, Laird DW. Lysosomal and proteasomal degradation play distinct roles in the life cycle of Cx43 in gap junctional intercellular communication-deficient and -competent breast tumor cells. J. Biol. Chem. 2003;278:30005–30014. doi: 10.1074/jbc.M300614200. [DOI] [PubMed] [Google Scholar]
- 47.Leithe E, Cruciani V, Sanner T, Mikalsen SO, Rivedal E. Recovery of gap junctional intercellular communication after phorbol ester treatment requires proteasomal degradation of protein kinase C. Carcinogenesis. 2003;24:1239–1245. doi: 10.1093/carcin/bgg066. [DOI] [PubMed] [Google Scholar]
- 48.Warn-Cramer BJ, Lampe PD, Kurata WE, Kanemitsu MY, Loo LW, Eckhart W, Lau AF. Characterization of the mitogen-activated protein kinase phosphorylation sites on the connexin-43 gap junction protein. J. Biol. Chem. 1996;271:3779–3786. doi: 10.1074/jbc.271.7.3779. [DOI] [PubMed] [Google Scholar]
- 49.Mograbi B, Corcelle E, Defamie N, Samson M, Nebout M, Segretain D, Fenichel P, Pointis G. Aberrant Connexin 43 endocytosis by the carcinogen lindane involves activation of the ERK/mitogen-activated protein kinase pathway. Carcinogenesis. 2003;24:1415–1423. doi: 10.1093/carcin/bgg093. [DOI] [PubMed] [Google Scholar]