Abstract
Complement is an essential component of the innate and acquired immune system1, and consists of a series of proteolytic cascades that are initiated by the presence of micro-organisms. In health, activation of complement is precisely controlled through membrane-bound and soluble plasma-regulatory proteins including factor H (fH)2, a 155 kDa protein composed of twenty domains (termed complement control protein repeats, or CCPs). Many pathogens have evolved the ability to avoid immune- killing by recruiting host complement regulators3 and several pathogens have adapted to avoid complement-mediated killing by sequestering fH to their surface4. Here we present the first structure of a complement regulator in complex with its pathogen surface-protein ligand. This reveals how the important human pathogen Neisseria meningitidis subverts immune responses by mimicking the host, using protein instead of charged-carbohydrate chemistry to recruit the host complement regulator, factor H. The structure also indicates the molecular basis of the host-specificity of the interaction between factor H and the meningococcus, and informs attempts to develop novel therapeutics and vaccines.
Neisseria meningitidis is a human adapted pathogen of global importance as a leading cause of bacterial meningitis and septic shock5. Due to Neisserial strain variation, the vaccines currently available for meningococcal disease are only effective against subsets of strains and do not provide universal protection6,7. However, promising novel antigens have been identified8, including factor H binding protein (known as fHbp, GNA1870, or R2086), a 27 kDa surface lipoprotein which is present on the surface of all strains of N. meningitidis and elicits protective bactericidal antibodies9,10. The structure of a portion of fHbp has been determined by NMR11, and its function studied by subdividing it into a series of regions, termed “A”, “B” and “C” 12. fHbp is the sole receptor for fH on the meningococcus, and recruitment of fH contributes to the ability of the meningococcus to avoid innate immune responses by inhibiting complement-mediated lysis in human plasma13,14. Individuals with polymorphisms in the promoter of the gene encoding fH that are associated with increased plasma fH levels are at increased risk from meningococcal disease15.
We have used a range of approaches to dissect the fH binding sites on fHbp (Fig. 1). High affinity interactions between fH and fHbp were only observed with fHbp constructs containing all three of the previously defined (see above) regions of fHbp (Fig. 1a), implying that fHbp has an extended recognition site for fH across its entire surface. We then sought to identify which of the 20 CCP domains of fH mediate the interaction with fHbp. Far Western, FACS (Fig. 1b) and Surface Plasmon Resonance (Fig. 1c) analyses demonstrated that the key regions of fH recognised by fHbp are the sixth and seventh domains, CCPs 6 & 7 (known hereafter as fH67) and that a construct containing both these domains was capable of inhibiting the fHbp-dependent interaction between fH and N. meningitidis (Fig. 1d). Quantification of the interaction demonstrates that the dissociation constant is ~5nM (Fig. 1c & 1e, Tab. S1, Fig. S1) for any construct containing CCPs 6 & 7. This interaction was not dissociated by high salt (1M NaCl, not shown) or pH ranging from 4 to 8 (not shown), providing additional support for the high affinity nature of the binding event.
Figure 1. The fHbp binding site is localised to CCP6 of fH and requires the complete extracellular portion of fHbp.
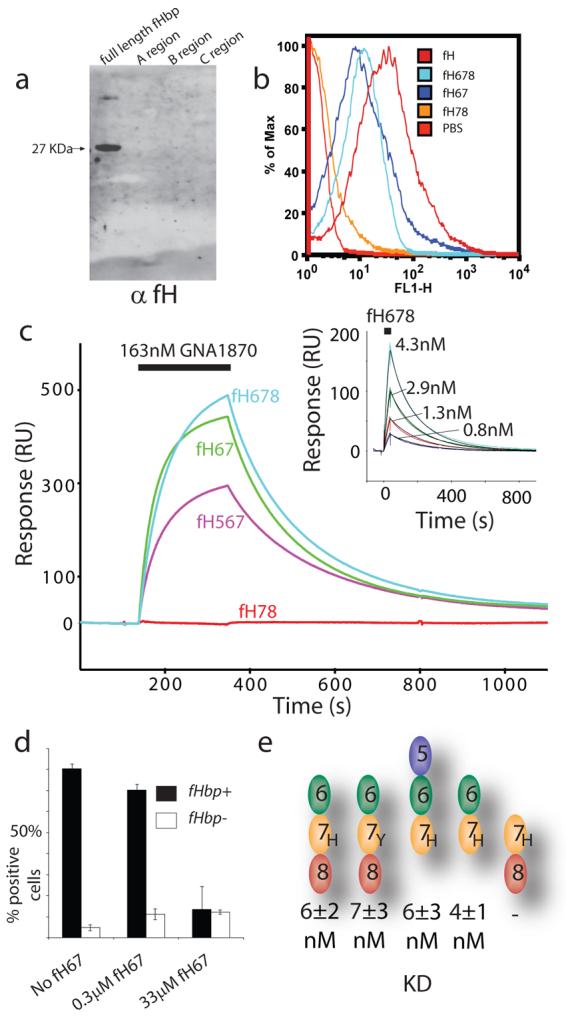
(a) Far Western analysis of fH binding to intact fHbp and truncated versions (as indicated). Membranes were incubated with purified fH which was detected with α-fH pAbs. Binding was only observed to the intact 27 kDa fHbp (shown with an arrow). (b) FACS analysis with α-fH pAbs detected binding between N. meningitidis and fH constructs (all of the His form) containing CCPs 6 & 7 (note that where binding is seen the relative magnitude of the response is not significant since the amount of pAb bound is approximately proportional to the size of the fH construct rather than the tightness of the binding event) and (c) SPR demonstrates that fHbp is only able to bind fH constructs containing CCPs 6 and 7 (the number of molecules of the fH fragments coupled on the sensor channels is in the ratio 1:1:1:3 for fh78:fh567:fh678:fh67, respectively). Signals shown are after subtraction of a control trace from a mock-coupled channel. With the experiment reversed the inset shows a 1:1 Langmuir fit (black lines) to a series of fH678 dilutions (coloured lines) injected over a fHbp surface to determine kinetic parameters (Fig. 1e & Tab. S1). (d) FACS competition studies (using an α-fH mAb directed against CCP5 (MRC OX2428) and therefore unable to recognize the fH67 construct) show that the short fH67 construct (between 0.3 and 33 μM) can compete away binding of full length fH demonstrating that this construct contains the entire fHbp binding site. Values shown are percentage of fluorescence positive cells in a population from three experiments ± s.d. The gate value was set using a control with no fH added. (e) Quantitation with SPR confirms that the presence of CCPs 6 and 7 is necessary and sufficient for high affinity binding to fHbp and that the common fH polymorphism in CCP7 (402His/Tyr) does not significantly alter the affinity of fHbp binding.
To better understand fH:fHbp recognition, we crystallised and determined crystal structures of the complex between fH67 and an fHbp construct comprising regions A, B and C but lacking the lipid anchor. Models for fHbp and fH67 were built and refined to 2.35 Å in two crystal forms with a total of seven independent copies of the complex (Fig. 2). The structure reveals that fHbp folds to form two ß–barrels (Fig. 2a) with the N-terminal barrel consisting of the “A” and part of the “B” regions, whilst the C-terminal barrel (as previously seen in the NMR structure of the “BC” region11, overlaid in Fig. S2) is composed of the rest of the “B” and the “C” regions. Searching structural databases with the N-terminal barrel reveals no close structural homologues (Supplemental Analyses), and the distinct topologies of the ß-barrels (Fig. 2c) suggest that they have not arisen by a gene duplication event.
Figure 2. Structure of fHbp and its complex with fH67.
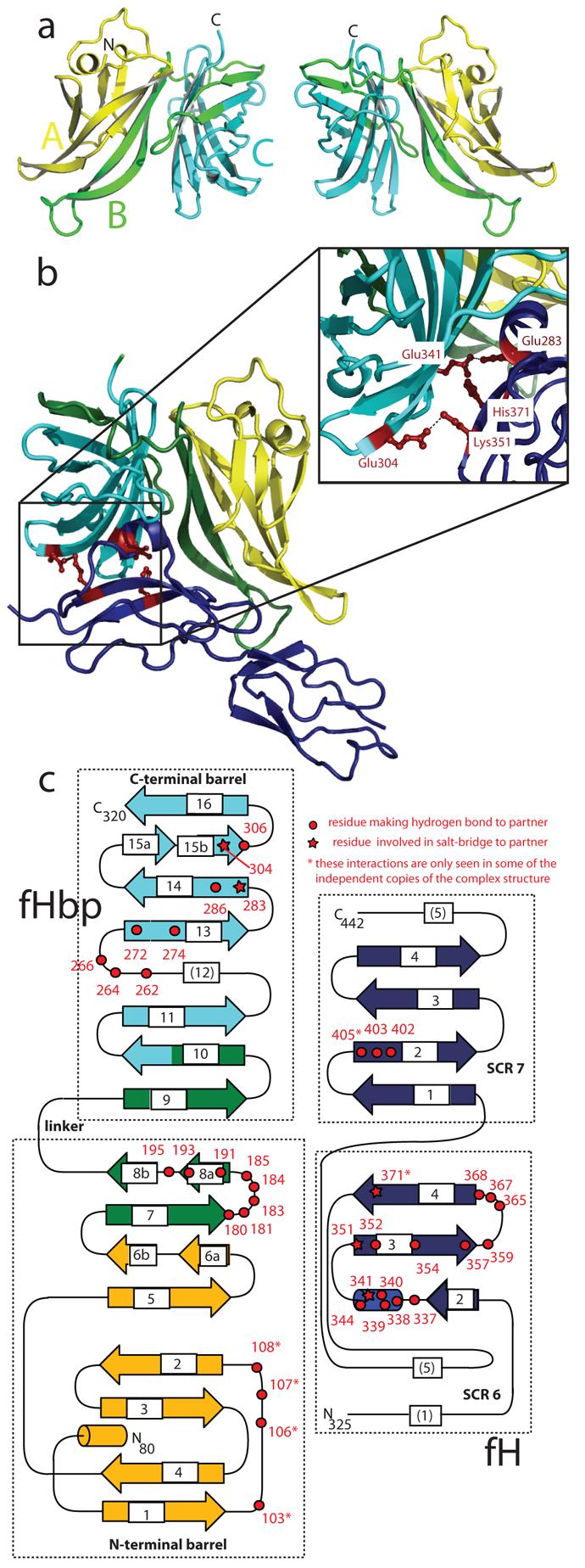
(a) Two views of a cartoon representation of fHbp (residues 80 to 320). The cartoon is coloured according to the regions previously used to study fHbp (“A” region yellow, “B” region green, “C” region cyan) illustrating the way in which these constructs do not reflect the overall architecture. (b) Cartoon of the fHbp:fH67 complex with fHbp coloured as in (a) and fh67 in dark blue. Side chains from both proteins involved in forming salt bridges across the interaction surface are shown in red as ball-and-stick representations (zoomed and reoriented in inset box). (c) Topology of fHbp and fh67 coloured as in (b) with the number of the residues involved in either H-bond or salt-bridge interactions between the proteins highlighted in red. The numbering scheme used throughout is as per Uniprot sequence Q9JXV4. This scheme is offset by +164 from the numbering used for the earlier NMR structure11 (which was numbered from the start of their fragment) and by −65 from the scheme used in Masignani et al.10 where numbering started from residue 1 of the mature protein without the signal sequence.
The fH67:fHbp complex is held together by extensive interactions between both ß-barrels of fHbp and fH CCP6 (Fig. 2b), with more minor contacts to CCP7, consistent with our binding studies. In particular, the helical insertion into the second ß-strand of CCP6 (an unusual feature of this CCP domain) is centrally located in the complex. Analysis by the PISA server gives all seven independent complexes a significance score of 1.0 (i.e. extremely likely to be biologically significant) with the average surface area of fHbp buried in the complex (2860±177Å2) greater than that buried in most antibody:antigen complexes16. Furthermore, the ΔG predicted on the basis of the structure for the formation of the complex (−6 kcal/mol) is in good agreement with the affinity derived from binding studies (−11 kcal/mol, calculated from KD presented in Fig. 1e), providing additional support for the physiological relevance of the crystallised complex. The interaction surface shows good shape complementarity with numerous electrostatic interactions (Fig. 2c, Fig. S3) including multiple hydrogen bonds and salt bridges. To further probe the interaction surface we generated doubly mutated forms of both proteins with charged side chains being replaced by the small hydrophobic residue Ala (fH67R341A,H337A and fHbpE283A,E304A). Using SPR to study the interactions between the mutated proteins and their wild-type partners revealed that the affinity of both mutant forms was reduced by more than two orders of magnitude so that almost no interaction could be seen at analyte concentrations (~50 nM) around ten times the wild-type KD (Fig. 3a). At a thousand-fold higher analyte concentration (μM range) the mutant forms of both proteins did interact with the wild-type partners: fHbpE283A,E304A retained a qualitatively similar on-rate but an increased off-rate with respect to wild-type, whilst fH67R341A,H337A was more similar to the wild-type interaction in its off-rate but had a vastly reduced on-rate (Tab. S1). It is interesting to note that E304 is not conserved in all Neisserial strains with a Thr also found at this position10. Since these strains also sequester fH it is likely that either the loss of a single interaction (rather than the two lost in the mutant) does not decrease the affinity sufficiently to preclude functional fH sequestration or that subtle rearrangements of the local side chains allow the Thr side chain to interact with residues in fH. The ability of the interaction to tolerate some variation in the residues at the interface would clearly be immunologically advantageous.
Figure 3. Interference with fHbp binding of fH.
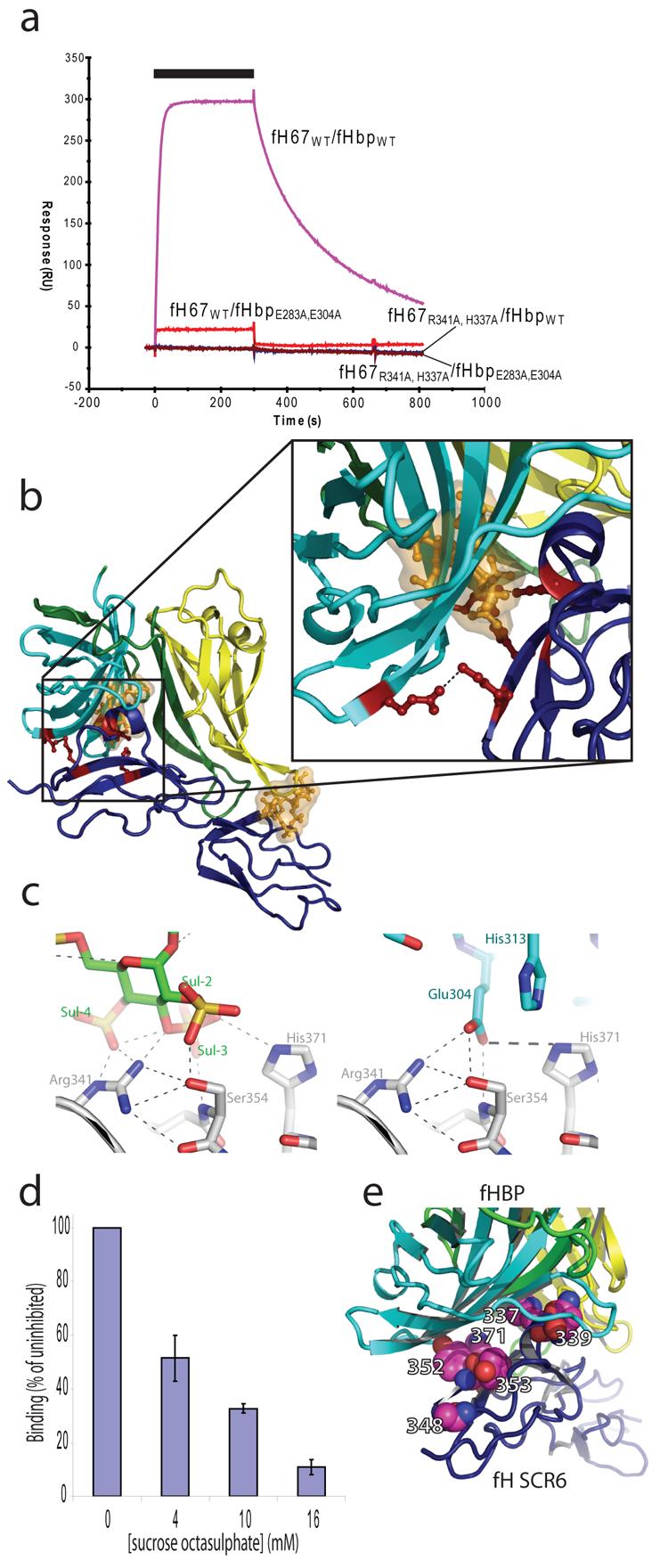
(a) Site-directed mutagenesis to use Ala to substitute charged side-chains, shown by the structure to be involved in complex-formation, abolishes binding to the wild-type form of their partner at concentrations around the wild-type KD. The black bar indicates the time period for which the fH analytes were injected (at 50 nM, 40 μl.min−1) over the fHbp surfaces.(b) Overlay of the fH-bound SOS (in gold as ball-and-stick and semi-transparent surface) from the structure of fH678 in complex with SOS21 onto our fHbp:fH67 structure (coloured as in 2b). This demonstrates that the sites of SOS and fHbp binding overlap, suggesting that SOS could inhibit the interaction. (c) Mimicry of GAG binding to fH via sulphate interactions (LH panel, SOS – green carbons, fH – grey carbons) by charged side chain (Glu 304) of fHbp (RH panel, fHbp – cyan carbons, fH – grey carbons). (d) SPR demonstrates that SOS inhibits binding of fHbp to fH in the mM concentration range (fH678 injected at 4nM, 40μl/min in presence of varying amounts of SOS) Values +/− one SD, n=3. (e) Mapping of sites of amino acid differences in CCP6 of fH between human (UniProt P08603) and rhesus macaque (UniProt O19279). Residues which are altered shown in pink, space filling representation, with fHbp and fh67 shown as in panel (2b).
A common fH polymorphism (fH 402His/Tyr), recently shown to be a major susceptibility factor for age related macular degeneration17-19, lies at the periphery of the interaction site and the side chain of residue 402 (His in this structure) is within hydrogen bonding distance of residues in fHbp (Fig. S4 & S5). However, our interaction studies do not detect any differences in binding (Fig. 1e, Tab. S1) between the fH 402His/Tyr isoforms. This lack of functional consequence suggests, as for variation at the 304 position (see above) that either the loss of a single hydrogen bond from this extensive interaction surface is not sufficient to produce a functional effect or that the small amount of flexibility at the CCP 6/7 junction seen between the crystallographically independent copies of the complex allows subtle rearrangement of the fH CCP domains to enable both polymorphic variants to hydrogen bond to fHbp (Fig. S4).
Mapping published epitopes of fHbp that elicit bactericidal antibodies11,12 onto the structure demonstrates that none of the sites characterised to date lie directly in the fH recognition site. Instead, epitopes recognised by antibodies that affect fH binding20 are located around the edge of the recognition site and so are likely to inhibit fH binding by steric hindrance (Fig. S5).
Our earlier structure21 of CCPs 6, 7 & 8 of fH in complex with sucrose octasulphate (SOS), a highly sulphated analogue of glycosaminoglycans (GAGs), revealed a charged groove on fH that forms an extended interaction site for binding of GAGs on the surface of endothelial cells. This, combined with an additional GAG-binding site in CCPs 19-2022, provides the mechanism by which fH localises to mammalian cells23 to prevent inappropriate complement activation. Examination of the fHbp:fH67 complex demonstrates that the Neisserial protein binding site lies within this extended GAG interaction site, overlapping the CCP6 SOS-binding site (Fig. 3b) with no fH rearrangement upon binding fHbp (Fig S6). There are several examples of pathogens imitating their host by expressing directly related molecules (e.g. sialic acid or glycolipids24). Here, however, N. meningitidis mimics the mechanism by which host cells regulate complement activation on their surface using amino acid side-chains instead of charged sugars. Since the binding sites of fHbp and SOS on fH are overlapping, we tested whether SOS can act as a competitive inhibitor of the interaction between fH and fHbp. Strikingly, SOS prevented the fH/fHbp interaction (assessed by SPR Fig. 3d, and by FACS Fig. S7) at millimolar concentrations, supporting the relevance of the crystal structure to complex formation in solution and providing a lead compound for development of small molecule inhibitors of fH:fHbp interactions.
N. meningitidis has evolved to avoid innate responses at sites of colonisation and during disease. It is also host-restricted, with the human nasopharynx its only natural reservoir5. This host specificity can limit the significance of animal models for studying certain aspects of pathogenesis and immunity. It has previously been shown that N. meningitidis only binds human fH and not fH from other primates or other animals25. The amino acid sequences of fH from human and other primates are highly conserved (~90% identity) whilst other animal fHs are more sequence divergent. It is therefore of note that even the limited number of differences between human and primate sequence changes within CCP6 cluster to the precise site of interaction with fHbp. For instance, mapping the differences between human and rhesus macaque fH26 shows that all six differences within CCP6 lie in or around the interface (Fig. 3e). This indicates the likely structural basis for the species specificity of complement evasion25 by N. meningitidis through fH recruitment, and suggests that specific modification of fH could provide a novel approach to develop more biologically relevant models of meningococcal disease.
Interestingly, the high affinity interaction between fH and fHbp suggests that N. meningitidis could rapidly sequester fH in the plasma, depleting the circulating levels and de-regulating complement, rendering host cells in the vascular compartment more susceptible to complement-mediated damage. This might contribute to the dramatic haemorrhagic rash seen in meningococcal sepsis, and suggests that inhibitors specifically designed to block the interaction between CCP6 and fHbp might both promote complement-mediated clearance of bacteria and prevent sequestration of fH and consequent vascular damage27.
The success of meningococcal vaccines containing fHbp which are currently being evaluated may be hampered by binding of fH which, due to high affinity and the extensive sites of interaction, could cloak critical, conserved epitopes on the antigen. Structural analysis of this complex paves the way for the development of engineered fHbp vaccines in which fH binding is abolished to unveil the full array of protective epitopes including those that could generate responses to inhibit recruitment of fH by this important human pathogen.
Summary Methods
Full methods are described in the supplementary material. Recombinant proteins were expressed and purified using affinity columns or size exclusion chromatography and protein concentration estimated using absorbance at λ=280nm. Binding assays (FACS, far Western and SPR) were performed according to published protocols. The complex was crystallised by vapour diffusion and X-ray diffraction data collected at the European Synchrotron Radiation Facility (ESRF, France) and DIAMOND (UK). The structure was solved by a combination of molecular replacement for the fH67 portion21 (1/3 of the structure) and isomorphous replacement/anomalous dispersion methods, with seven independent copies refined to 2.35Å in two different crystal forms.
Supplementary Material
Supplementary Information accompanies the paper on www.nature.com/nature.
Acknowledgments
Thanks are due to Gérard Bricogne and the Global Phasing Consortium for access to a β version of autoSHARP and autoBUSTER, to the beamline staff of the European Synchrotron Radiation Source (particularly Dr. G. Leonard) and Diamond and to many members of the Laboratory of Molecular Biophysics, Oxford and the Lea Group for help with X-ray data collection. B.E.P. was funded by a Wellcome Trust studentship P.R. & S.J. by Medical Research Council grants to S.M.L., J.E.D. by a Wellcome Trust grant to SM.L. and J.E.L. by an Engineering and Physical Sciences Research Grant to S.M.L. Work in C.M.T.'s laboratory is funded by the Wellcome Trust and the Medical Research Council. E.K. is an EMBO fellow. B.E.P, J.E.D. & J.J.E.C. performed SPR experiments, B.E.P., J.J.E.C., J.E.L., S.Q expressed proteins and crystallised the complex, M.C.S., E.K & Q.Z. performed all assays with Neisseria and expressed fHbp, S.J., P.R. and S.M.L. collected and analysed X ray data, S.M.L. phased the X-ray data and built/refined the models, S.M.L. and C.M.T. designed the research and wrote the paper.
Footnotes
Coordinates and X-ray data have been deposited in the protein databank with accession identifiers 2w80 & 2w81.
References
- 1.Lachmann PJ. Biological functions of the complement system. Biochem Soc Trans. 1990;18:1143–5. doi: 10.1042/bst0181143. [DOI] [PubMed] [Google Scholar]
- 2.Zipfel PF, Jokiranta TS, Hellwage J, Koistinen V, Meri S. The factor H protein family. Immunopharmacology. 1999;42:53–60. doi: 10.1016/s0162-3109(99)00015-6. [DOI] [PubMed] [Google Scholar]
- 3.Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nat Rev Microbiol. 2008;6:132–42. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jozsi M, Zipfel PF. Factor H family proteins and human diseases. Trends Immunol. 2008;29:380–7. doi: 10.1016/j.it.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Stephens DS, Greenwood B, Brandtzaeg P. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet. 2007;369:2196–210. doi: 10.1016/S0140-6736(07)61016-2. [DOI] [PubMed] [Google Scholar]
- 6.Feavers IM. ABC of meningococcal diversity. Nature. 2000;404:451–2. doi: 10.1038/35006546. [DOI] [PubMed] [Google Scholar]
- 7.Gray SJ, et al. Epidemiology of meningococcal disease in England and Wales 1993/94 to 2003/04: contribution and experiences of the Meningococcal Reference Unit. J Med Microbiol. 2006;55:887–96. doi: 10.1099/jmm.0.46288-0. [DOI] [PubMed] [Google Scholar]
- 8.Harrison LH. Prospects for vaccine prevention of meningococcal infection. Clin Microbiol Rev. 2006;19:142–64. doi: 10.1128/CMR.19.1.142-164.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher LD, et al. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect Immun. 2004;72:2088–100. doi: 10.1128/IAI.72.4.2088-2100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masignani V, et al. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J Exp Med. 2003;197:789–99. doi: 10.1084/jem.20021911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantini F, et al. Solution structure of the immunodominant domain of protective antigen GNA1870 of Neisseria meningitidis. J Biol Chem. 2006;281:7220–7. doi: 10.1074/jbc.M508595200. [DOI] [PubMed] [Google Scholar]
- 12.Giuliani MM, et al. The region comprising amino acids 100 to 255 of Neisseria meningitidis lipoprotein GNA 1870 elicits bactericidal antibodies. Infect Immun. 2005;73:1151–60. doi: 10.1128/IAI.73.2.1151-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madico G, et al. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J Immunol. 2006;177:501–10. doi: 10.4049/jimmunol.177.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider MC, et al. Functional significance of factor H binding to Neisseria meningitidis. J Immunol. 2006;176:7566–75. doi: 10.4049/jimmunol.176.12.7566. [DOI] [PubMed] [Google Scholar]
- 15.Haralambous E, et al. Factor H, a regulator of complement activity, is a major determinant of meningococcal disease susceptibility in UK Caucasian patients. Scand J Infect Dis. 2006;38:764–71. doi: 10.1080/00365540600643203. [DOI] [PubMed] [Google Scholar]
- 16.Davies DR, Padlan EA, Sheriff S. Antibody-antigen complexes. Annu Rev Biochem. 1990;59:439–73. doi: 10.1146/annurev.bi.59.070190.002255. [DOI] [PubMed] [Google Scholar]
- 17.Edwards AO, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–4. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 18.Hageman GS, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102:7227–32. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haines JL, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–21. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 20.Beernink PT, Granoff DM. Bactericidal antibody responses induced by meningococcal recombinant chimeric factor H-binding protein vaccines. Infect Immun. 2008;76:2568–75. doi: 10.1128/IAI.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prosser BE, et al. Structural basis for complement factor H linked age-related macular degeneration. J Exp Med. 2007;204:2277–83. doi: 10.1084/jem.20071069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt CQ, et al. A new map of glycosaminoglycan and C3b binding sites on factor H. J Immunol. 2008;181:2610–9. doi: 10.4049/jimmunol.181.4.2610. [DOI] [PubMed] [Google Scholar]
- 23.Meri S, Pangburn MK. Discrimination between activators and nonactivators of the alternative pathway of complement: regulation via a sialic acid/polyanion binding site on factor H. Proc Natl Acad Sci U S A. 1990;87:3982–6. doi: 10.1073/pnas.87.10.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fedtke I, Gotz F, Peschel A. Bacterial evasion of innate host defenses--the Staphylococcus aureus lesson. Int J Med Microbiol. 2004;294:189–94. doi: 10.1016/j.ijmm.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 25.Granoff DM, Welsch JA, Ram S. Binding of Complement factor H to Neisseria meningitidis is Specific for Human fH and Inhibits Complement Activation by Rat and Rabbit Sera. Infect Immun. 2008 doi: 10.1128/IAI.01191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibbs RA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–34. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 27.Heckenberg SG, et al. Clinical features, outcome, and meningococcal genotype in 258 adults with meningococcal meningitis: a prospective cohort study. Medicine (Baltimore) 2008;87:185–92. doi: 10.1097/MD.0b013e318180a6b4. [DOI] [PubMed] [Google Scholar]
- 28.Jokiranta TS, et al. Analysis of the recognition mechanism of the alternative pathway of complement by monoclonal anti-factor H antibodies: evidence for multiple interactions between H and surface bound C3b. FEBS Lett. 1996;393:297–302. doi: 10.1016/0014-5793(96)00905-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


