Abstract
In order to accurately replicate its viral genome, the Herpes Simplex Virus 1 (HSV-1) DNA polymerase usually polymerizes the correct dNTP opposite the template base being replicated. We employed a series of purine-dNTP analogues to determine the chemical features of the base necessary for the herpes polymerase to avoid polymerizing incorrect dNTPs. The enzyme uses N-3 to prevent misincorporation of purine dNTPs, but does not require N-3 for correct polymerization. A free pair of electrons on N-1 also helps prevent misincorporation opposite A, C, and G, and strongly drives polymerization opposite T. N6 contributes a small amount both for preventing misincorporation and for correct polymerization. Within the context of guanine in either the incoming dNTP or the template base being replicated, N2 prevents misincorporation opposite adenine but plays at most a minor role for incorporation opposite C. In contrast, adding N2 to dNTPs of either adenine, purine, 6-chloropurine or 1-deazapurine greatly enhances incorporation opposite C, likely via the formation of a hydrogen bond between N2 of the purine and O2 of the pyrimidine. Herpes polymerase is very sensitive to the structure of the base-pair at the primer 3′-terminus since eliminating N-1, N-3, or N6 from a purine nucleotide at the primer 3′-terminus interfered with polymerization of the next two dNTPs. The biological and evolutionary implications of these data are discussed.
Keywords: Fidelity, Misincorporation, Selectivity, Kinetics, Nucleotide
The Herpes Simplex Virus 1 (HSV-1)1 DNA polymerase is essential for viral replication and shows significant homology to other polymerases in the Herpesviridae family. The polymerase complex consists of two subunits: UL30, the subunit possessing the polymerase activity as well as a 3′-5′ proofreading exonuclease activity, and UL42, an accessory factor that increases the processivity of the polymerase by tethering the UL30 subunit directly to the DNA (1, 2). Both origin-dependent DNA synthesis and productive viral replication require this two subunit enzyme (3-5). The herpes polymerase misincorporates dNTPs once every 103-104 nucleotides polymerized (6-8). This frequency is at the low end typically observed for replicative polymerases such that the polymerase appears to rely heavily on its associated exonuclease activity to accurately replicate DNA (9, 10).
A key issue regarding DNA polymerases is how they avoid misincorporating dNTPs. Several theories have been proposed to explain the mechanism of polymerase fidelity including hydrogen bonding to form the appropriate base pair, shape and size of the nucleotides to fit the active site, and the used of specific chemical features to prevent misincorporation and enhance correct incorporation (11, 12). Insights into the various models have come from studies using a variety of approaches, including structural studies, protein mutagenesis, and substrate mutagenesis (13-15). Even with these different data, however, the precise mechanism(s) used by any one polymerase to minimize polymerization of incorrect dNTPs have yet to be completely clarified. Moreover, polymerases from different evolutionary families clearly use different mechanisms for choosing a correct (d)NTP (8, 15, 16).
Herpes polymerase is a B family polymerase, as are the replicative eukaryotic polymerases α, δ, and ε, DNA polymerases from T4 and RB69 viruses, among others (17, 18). All of these enzymes contain a remarkably highly conserved P helix – related to the O helix in A family polymerases – whose amino acids surround the incipient base-pair between the incoming dNTP and the template being replicated (19).
Previously, we elaborated on the rules employed by human polymerase α (pol α) to ensure accurate incorporation of dATP followed by polymerization of additional dNTPs. To minimize the frequency with which pol α incorporates an incorrect dNTP followed by additional nucleotides, this enzyme uses two orthogonal screens. During selection for either correct or incorrect polymerization of dATP, pol α uses N-1 and N-3 to prevent misincorporation opposite A, C, and G (i.e. as negative selectors) and N-1 and N6 to enhance correct incorporation opposite T (i.e. as positive selectors) (20). However, to incorporate additional dNTPs onto a primer terminus, pol α monitors the shape of the base pair at the 3′-terminus (20). If the 3′-terminus of the primer:template is incorrectly shaped, pol α polymerizes further dNTPs extremely slowly compared to the rate when the 3′-terminus is correctly shaped (20).
While polymerases from different families clearly use different mechanisms, it is unclear how similar (or different) the mechanisms between different enzymes within the same evolutionary family will be. Here we investigate the screens for dNTP incorporation and extension that herpes DNA polymerase (exo−) utilizes. While the general mechanism of herpes polymerase and pol α appears similar, consistent with their evolutionary association, the enzymes do exhibit some significant differences. We consider the biological implications of these results.
Experimental Procedures
Materials
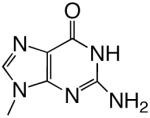
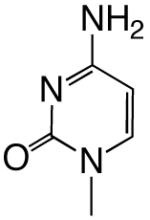
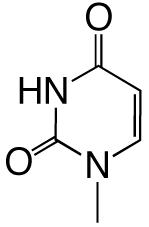
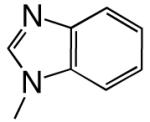
All reagents were of the highest quality commercially available. Unlabeled dNTPs were purchased from Invitrogen and radiolabeled dNTPs were purchased from PerkinElmer. Purined-dNTP analogues (Figure 1) were synthesized as previously described (20, 21). Synthetic DNA oligonucleotides of defined sequence were acquired from either Oligos, etc. or Biosearch Technologies and their concentrations determined spectrally.
Figure 1.
Structures of the bases and designated abbreviations. The numbering systems for a benzimidazole and a purine (6-methyl-1-deazapurine) are shown..
Herpes polymerase (UL30/UL42 complex, exo−) was expressed and purified as described previously (22).
5′-end labeling of primers and annealing of primer/template pairs
DNA primers were 5′-[32P]-labeled using polynucleotide kinase and [ -32P] ATP, gel purified, and annealed to the appropriate template as previously described (23, 24).
Polymerization assays
All kinetic data were obtained under steady-state conditions. Assays typically contained 1 uM 5′-[32P]-primer/template, 50 mM TRIS-HCl (pH 8.0), 50 mM MgCl2, 1 mM dithiothreitol, 0.1 mg/mL bovine serum albumin, 5% glycerol, and varying concentrations of dNTPs and/or nucleotide analogues, in a total volume of 10 uL (20, 22). Polymerization reactions were initiated by the addition of enzyme, incubated at 37°C, and quenched by adding an equal volume of gel loading buffer (90% formamide in 1X Tris/Borate/EDTA buffer, 0.05% xylene cyanol and bromophenol blue). Products were separated by denaturing gel electrophoresis (20% acrylamide, 7.5 M urea) and analyzed by phosphorimagery (Molecular Dynamics). Kinetic parameters were obtained by fitting the data to the Michaelis-Menten equation using Prism software. All rates were normalized to the same final enzyme concentration of 1 nM. The reported discrimination factor values (DF) were determined by comparing the efficiency of incorporation for the analog (Vmax/KM) to the efficiency of incorporation for the correct dNTP (Vmax/KM normalized to 1).
Polymerization of additional dNTPs after incorporation of a normal or analogue nucleotide.
Elongation reactions contained the analogue triphosphate, at a fixed concentration >> KM, and increasing concentrations of the next correct dNTP to be polymerized (i.e. TTP for DNA C,G,T and dATP for DNAA), and were quantitated using the running start methodology developed by Goodman and coworkers (25). The steady-state kinetic parameters were calculated using Molecular Dynamics and Prism software as described above. The discrimination factor values (DF) were determined by comparing the efficiency of polymerization after the analogue (Vmax/KM) to the efficiency of polymerization after the correct dNTP (Vmax/KM, normalized to 1). Polymerization of the second correct dNTP was measured similarly, except assays now contained the analogue triphosphate at a fixed concentration >> KM, the next correct dNTP at a fixed concentration >> KM, and increasing concentrations of the second correct dNTP.
Results
We examined polymerization of the dNTP analogues shown in Figure 1 by the herpes DNA polymerase (UL30/UL42 complex) using primer-templates of defined sequence (Table 1). The enzyme lacked exonuclease activity, hence different rates reflect changes in the polymerization activity. Similar to previous studies showing removal of N-1, N-3, and N6 from adenine resulted in dNTPs that pol rapidly incorporated opposite all 4 natural template bases (20), herpes polymerase incorporated benzimidazole- and 4-methylbenzimidazole-dNTP opposite all 4 natural template bases much more rapidly than the enzyme misincorporated a natural dNTP (Tables 2 and 3).
Table 1.
DNA Primer-Templates
| DNAA | 5′ – TCCATATCACAT – 3′ |
| 3′ – AGGTATAGTGTAATTCTTATCATCT – 5′ | |
| DNAT | 5′ – TCCATATCACAT – 3′ |
| 3′ – AGGTATAGTGTATATCTTATCATCT – 5′ | |
| DNAT2 | 5′ – TCCATATCACAT – 3′ |
| 3′ – AGGTATAGTGTATAGCTTATCATCT – 5′ | |
| DNAC | 5′ – TCCATATCACAT – 3′ |
| 3′ – AGGTATAGTGTACATCTTATCATCT – 5′ | |
| DNAG | 5′ – TCCATATCACAT – 3′ |
| 3′ – AGGTATAGTGTAGATCTTATCATCT – 5′ | |
| DNAI | 5′ – TCCATATCACAT – 3′ |
| 3′ – AGGTATAGTGTAITTCTTATCATCT – 5′ |
Table 2.
Polymerization of natural dNTPs.
| dNTP | Base Structure | DNAN | Vmax (SD) | Km (SD) | Vmax/Km | Discrimination a |
|---|---|---|---|---|---|---|
 |
DNAT | 2.30 (0.05) | 0.49 (0.05) | 4.7 | 1 | |
| dATP | DNAA | 0.73 (0.04) | 1500 (300) | 0.00049 | 2700 | |
| DNAC | 0.98 (0.02) | 1100 (80) | 0.00089 | 3300 | ||
| DNAG | 0.27 (0.01) | 3600 (300) | 0.000075 | 11000 | ||
 |
DNAT | 2.7 (0.1) | 1400 (300) | 0.0019 | 2500 | |
| dGTP | DNAA | 0.27 (0.04) | 3100 (700) | 0.000087 | 15000 | |
| DNAC | 4.4 (0.7) | 1.5 (0.4) | 2.9 | 1 | ||
| DNAG | 0.55 (0.04) | 1500 (200) | 0.00037 | 2300 | ||
 |
DNAT | 1.1 (0.2) | 4000 (900) | 0.00028 | 3000 | |
| dCTP | DNAA | 0.59 (0.07) | 2200 (400) | 0.00027 | 3100 | |
| DNAC | b | b | 0.0000049 | >100,000 | ||
| DNAG | 0.94 (0.01) | 1.1 (0.1) | 0.85 | 1 | ||
 |
DNAT | 0.43 (0.03) | 1800 (300) | 0.00024 | 5400 | |
| dTTP | DNAA | 1.71 (0.06) | 1.3 (0.2) | 1.3 | 1 | |
| DNAC | 1.4 (0.1) | 1800 (400) | 0.00078 | 1700 | ||
| DNAG | 0.57 (0.02) | 1300 (90) | 0.00044 | 3000 | ||
Discrimination values are defined as Vmax/KM for the correct dNTP opposite that template base (i.e. dATP:T, dCTP:G, etc.) divided by Vmax/KM for the noted dNTP (or dNTP analogue) opposite that template base. Thus, for example, the discrimination value for dATP opposite A would be:
Some Vmax and KM values could not be obtained from a Michaelis-Menten curve under our reaction conditions. Instead, the Vmax/KM ratio was obtained from a linear plot of the kinetic data.
Table 3.
Polymerization of hydrophobic dNTP analogues
| dNTP | Base Structure | DNAN | Vmax (SD) | Km (SD) | Vmax/Km | Discriminationa |
|---|---|---|---|---|---|---|
 |
DNAT | 1.33 (0.07) | 55 (16) | 0.024 | 200 | |
| dBTP | DNAA | 0.99 (0.04) | 97 (17) | 0.010 | 130 | |
| DNAC | 0.94 (0.04) | 120 (20) | 0.0078 | 370 | ||
| DNAG | 0.23 (0.01) | 130 (17) | 0.0018 | 470 | ||
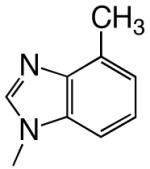 |
DNAT | 2.3 (0.1) | 120 (20) | 0.019 | 250 | |
| dZTP | DNAA | 0.53 (0.05) | 490 (80) | 0.0011 | 1200 | |
| DNAC | 0.60 (0.07) | 280 (70) | 0.0021 | 1400 | ||
| DNAG | 0.078 (0.005) | 290 (60) | 0.00027 | 3100 | ||
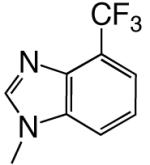 |
DNAT | 1.09 (0.07) | 370 (60) | 0.0029 | 1600 | |
| d4CF3BTP | DNAA | 0.42 (0.05) | 200 (60) | 0.0021 | 620 | |
| DNAC | 0.93 (0.11) | 180 (50) | 0.0052 | 560 | ||
| DNAG | b | b | 0.000087 | 9800 | ||
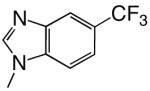 |
DNAT | 1.30 (0.07) | 24 (4) | 0.054 | 87 | |
| d5CF3BTP | DNAA | 0.17 (0.01) | 26 (7) | 0.0065 | 200 | |
| DNAC | 0.43 (0.04) | 79 (22) | 0.0054 | 540 | ||
| DNAG | 0.150 (0.006) | 290 (30) | 0.00052 | 1600 | ||
| d6CF3BTP | 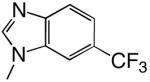 |
DNAT | 1.7 (0.1) | 14 (4) | 0.12 | 39 |
| d6CF3BTP | DNAA | 0.29 (0.03) | 220 (50) | 0.0013 | 1000 | |
| DNAC | 0.51 (0.04) | 48 (13) | 0.011 | 260 | ||
| DNAG | 0.39 (0.04) | 120 (30) | 0.0031 | 270 | ||
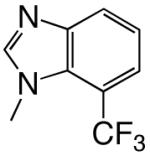 |
DNAT | b | b | 0.00031 | 15000 | |
| d7CF3BTP | DNAA | 0.0045 (0.0008) | 40 (21) | 0.00011 | 12000 | |
| DNAC | 0.0060 (0.0007) | 31 (12) | 0.00019 | 15000 | ||
| DNAG | 0.0031 (0.0009) | 120 (90) | 0.000026 | 33000 | ||
Discrimination values are defined as Vmax/KM for the correct dNTP opposite that template base (i.e. dATP:T, dCTP:G, etc.) divided by Vmax/KM for the noted dNTP (or dNTP analogue) opposite that template base.
Some Vmax and KM values could not be obtained from a Michaelis-Menten curve under our reaction conditions. Instead, the Vmax/KM ratio was obtained from a linear plot of the kinetic data.
Shape can influence the rate of unnatural dNTP incorporation
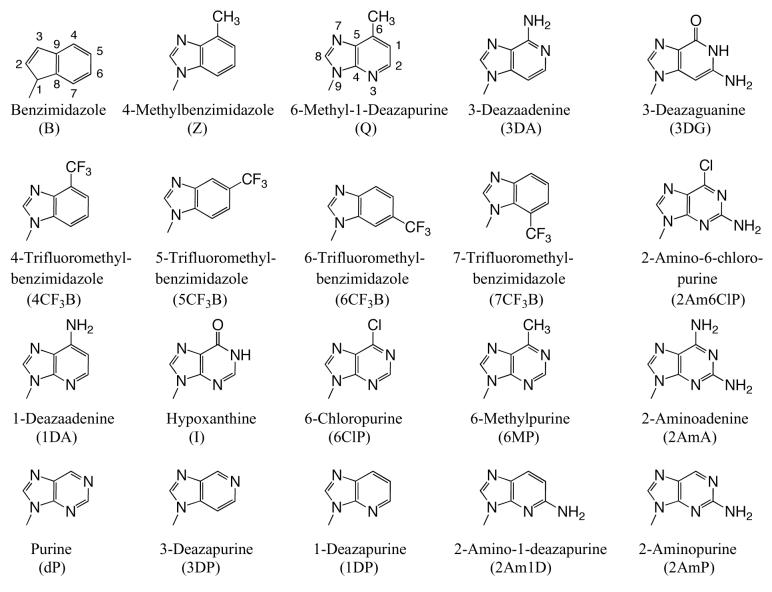
To explore the role, if any, that base shape plays during the polymerization reaction, we compared polymerization of 4-, 5-, 6-, and 7-trifluoromethylbenzimidazole dNTPs. These four different bases have similar hydrophobicities and, therefore, should have similar stacking abilities (12, 20), thus minimizing this potentially confounding variable. Table 3 shows that herpes polymerase incorporates these 4 dNTPs with very different efficiencies depending on the location of the trifluoromethyl group in the 6-membered ring. While herpes polymerase well-tolerated (<2-fold average decrease in incorporation efficiency compared to benzimidazole dNTP) the trifluoromethyl group at the 5- and 6-positions of benzimidazole (i.e. on the Watson-Crick hydrogen bonding face of the base), a trifluoromethyl group at the 4-position was less favored (efficiency decreased an average of 11-fold opposite all 4 DNA templates compared to benzimidazole, Table 3 and Figure 2). Notably, the enzyme very strongly discriminated against 7-trifluoromethylbenzimidazole dNTP (efficiency decreased an average of 64-fold), indicating that herpes polymerase does not effectively tolerate a trifluoromethyl group in the minor groove of the incipient base pair. Furthermore, since this set of bases all have similar hydrophobicity, these results indicate that hydrophobicity/stacking ability alone cannot account for the rapid polymerization of the 5- and 6-trifluoromethylbenzimidazole dNTPs. To elucidate why the polymerase misincorporates dNTPs containing base analogues such as benzimidazole so rapidly but not the canonical dNTPs such as dATP, we systematically examined the roles of N-1, N-3, and N6, the 3 nitrogens that distinguish benzimidazole and adenine (Figure 1).
Figure 2.
Misincorporation frequency of benzimidazole and trifluoromethylbenzimidazole compounds. The values of the average discrimination factor (opposite all 4 DNA templates) are provided.
Herpes polymerase uses N-3 of a purine to identify incoming dNTPs as incorrect
Firstly, we examined the effect of removing N-3 from two high-fidelity bases, adenine and guanine. Herpes polymerase incorporated 3-deazaadenine dNTP and 3-deazaguanine dNTP opposite the correct template bases (T and C, respectively), almost identically to dATP and dGTP (2.2- and 1.1-fold less efficiently, Table 4). Thus, herpes polymerase clearly does not require N-3 of a purine for efficient, correct polymerization. Similarly, 3-deazapurine dNTP was incorporated only 2.6-fold slower opposite a template T as compared to purine dNTP.
Table 4.
Polymerization of dNTP analogues containing modifications at N-1, N2, N-3, and N6
| dNTP | Base Structure | DNAN | Vmax (SD) | Km (SD) | Vmax/Km | Discriminationa |
|---|---|---|---|---|---|---|
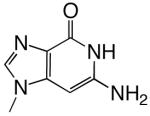 |
DNAT | 1.4 (0.2) | 700 (200) | 0.0020 | 2400 | |
| d3DGTP | DNAA | 0.17 (0.02) | 270 (80) | 0.00063 | 2100 | |
| DNAC | 4.0 (0.1) | 1.5 (0.2) | 2.7 | 1.1 | ||
| DNAG | 2.3 (0.1) | 190 (20) | 0.012 | 77 | ||
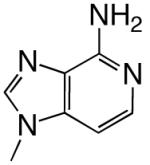 |
DNAT | 2.62 (0.05) | 1.2 (0.1) | 2.2 | 2.2 | |
| d3DATP | DNAA | 0.30 (0.03) | 860 (180) | 0.00035 | 3800 | |
| DNAC | 0.68 (0.05) | 1600 (200) | 0.00043 | 6800 | ||
| DNAG | 0.24 (0.03) | 1200 (300) | 0.00020 | 4600 | ||
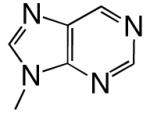 |
DNAT | 2.98 (0.05) | 2.1 (0.3) | 1.4 | 3.4 | |
| dPTP | DNAA | 0.11 (0.01) | 87 (26) | 0.0013 | 1000 | |
| DNAC | 0.74 (0.02) | 320 (20) | 0.0023 | 1300 | ||
| DNAG | 0.030 (0.003) | 200 (50) | 0.00015 | 6100 | ||
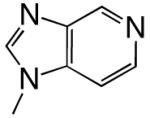 |
DNAT | 4.2 (0.1) | 8.0 (0.8) | 0.53 | 8.9 | |
| d3DPTP | DNAA | 0.0224 (0.0003) | 7.9 (0.4) | 0.0028 | 480 | |
| DNAC | 0.078 (0.003) | 62 (6) | 0.0013 | 2300 | ||
| DNAG | 0.0078 (0.0002) | 9.2 (0.9) | 0.00085 | 1100 | ||
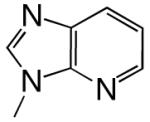 |
DNAT | 0.32 (0.02) | 430 (70) | 0.00074 | 6400 | |
| d1DPTP | DNAA | 0.10 (0.01) | 220 (50) | 0.00047 | 2900 | |
| DNAC | 0.15 (0.01) | 220 (60) | 0.00068 | 4300 | ||
| DNAG | 0.0088 (0.0005) | 230 (40) | 0.000038 | 24000 | ||
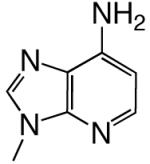 |
DNAT | 1.1 (0.1) | 260 (60) | 0.0042 | 1100 | |
| d1DATP | DNAA | 0.66 (0.04) | 640 (70) | 0.0010 | 1300 | |
| DNAC | 0.41 (0.02) | 520 (50) | 0.00080 | 3800 | ||
| DNAG | 0.084 (0.004) | 440 (50) | 0.00020 | 4600 | ||
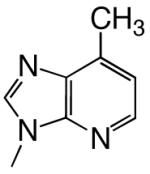 |
DNAT | b | b | 0.00026 | 18000 | |
| dQTP | DNAA | 1.11 (0.09) | 1600 (200) | 0.00069 | 1900 | |
| DNAC | 0.82 (0.08) | 590 (120) | 0.0014 | 2100 | ||
| DNAG | 0.069 (0.006) | 720 (130) | 0.000096 | 8900 | ||
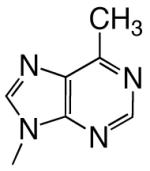 |
DNAT | 4.8 (0.1) | 6.4 (0.5) | 0.75 | 6.3 | |
| d6MPTP | DNAA | 0.11 (0.01) | 96 (27) | 0.0011 | 1100 | |
| DNAC | 0.50 (0.08) | 1100 (250) | 0.00045 | 6400 | ||
| DNAG | 0.079 (0.008) | 310 (70) | 0.00025 | 3700 | ||
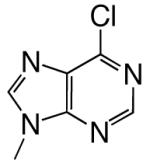 |
DNAT | 2.09 (0.06) | 24 (2) | 0.087 | 54 | |
| d6ClPTP | DNAA | 0.11 (0.01) | 310 (70) | 0.00035 | 3700 | |
| DNAC | 0.89 (0.08) | 67 (15) | 0.013 | 220 | ||
| DNAG | 0.026 (0.002) | 230 (40) | 0.00011 | 7700 | ||
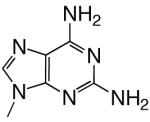 |
DNAT | 2.28 (0.05) | 0.8 (0.1) | 2.9 | 1.6 | |
| d2AmATP | DNAA | b | b | 0.000055 | 24000 | |
| DNAC | 1.93 (0.05) | 63 (8) | 0.031 | 94 | ||
| DNAG | 0.11 (0.02) | 1400 (400) | 0.000079 | 11000 | ||
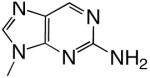
| d2Am1DP | 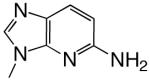 |
DNAT | 2.16 (0.23) | 673 (165) | 0.0032 | 1500 |
| DNAA | 0.49 (0.06) | 2250 (420) | 0.00022 | 6200 | ||
| DNAC | 4.44 (0.07) | 119 (7.7) | 0.037 | 78 | ||
| DNAG | 0.42 (0.05) | 2630 (470) | 0.00016 | 5300 | ||
 |
DNAT | 2.72 (0.10) | 1.95 (0.37) | 1.4 | 3.4 | |
| d2AmPTP | DNAA | b | b | 0.000061 | 21000 | |
| DNAC | 2.24 (0.10) | 141 (20) | 0.016 | 180 | ||
| DNAG | b | b | 0.000075 | 11000 | ||
| d2Am6ClP | 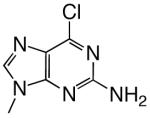 |
DNAT | 2.50 (0.05) | 11.4 (0.9) | 0.22 | 21 |
| DNAA | 0.00013 | 10000 | ||||
| DNAC | 1.95 (0.08) | 45 (5) | 0.043 | 67 | ||
| DNAG | 0.11 (0.02) | 1300 (300) | 0.000085 | 10000 | ||
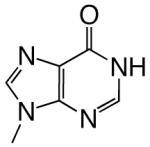 |
DNAT | 0.52 (0.06) | 270 (60) | 0.0019 | 2500 | |
| dITP | DNAA | 0.60 (0.09) | 41 (19) | 0.015 | 87 | |
| DNAC | 3.5 (0.1) | 0.6 (0.1) | 5.8 | 0.5 | ||
| DNAG | 0.022 (0.006) | 320 (150) | 0.000069 | 12000 | ||
Discrimination values are defined as Vmax/KM for the correct dNTP opposite that template base (i.e. dATP:T, dCTP:G, etc.) divided by Vmax/KM for the noted dNTP (or dNTP analogue) opposite that template base.
Some Vmax and KM values could not be obtained from a Michaelis-Menten curve under our reaction conditions. Instead, the Vmax/KM ratio was obtained from a linear plot of the kinetic data.
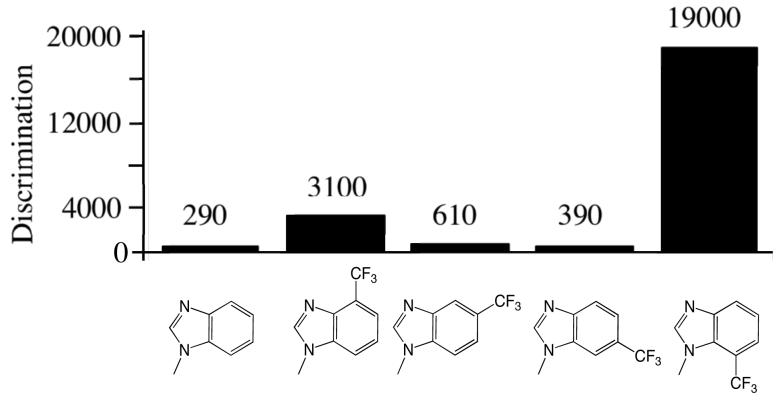
Whereas N-3 does not play an important role during correct dNTP incorporation, it performs a critical function to help prevent misincorporation. Removing N-3 from dGTP significantly increased misincorporation opposite both A and G (Table 4, 7-fold and 30-fold, respectively), with the average misincorporation frequency opposite A, C, and G increasing 4.4-fold (Figure 3). Removing N-3 from purine dNTP and dATP had much smaller effects on misincorporation than the effects on dGTP, in most cases <2-fold increases or decreases in misincorporation efficiency. The only notable change was the 5.5-fold increase in 3-deazapurine dNTP misincorporation opposite G as compared to purine dNTP.
Figure 3.
Effects of adding N-3 to low-fidelity bases and removing N-3 from high-fidelity bases. Panel A shows the effect of adding N-3 to benzimidazole and 4-methylbenzimidazole on the average discrimination factor for polymerization opposite A, T, C, and G. Panel B shows the effect of removing N-3 from guanine, adenine, and purine for misincorporation opposite either A, G, and T (dGTP/d3DGTP) or A, G, and C (dPTP/d3DPTP and dATP/d3DATP).
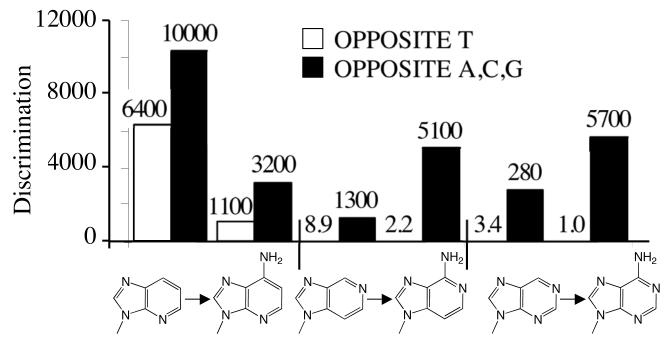
Adding the equivalent of purine N-3 to “low-fidelity bases” such as benzimidazole and 4-methylbenzimidazole dramatically reduces misincorporation by the herpes polymerase. Adding N-3 to benzimidazole- and 4-methylbenzimidazole-dNTP, thereby generating 1-deazapurineand 1-deaza-6-methylpurine-dNTPs, respectively, decreased their misincorporation opposite all 4 natural bases by an average of 30-fold (Tables 3 and 4 and Figure 3). Thus, N-3 helps prevent inappropriate dNTP polymerization in multiple contexts – removing it from the natural, high fidelity bases increases misincorporation, and adding N-3 to a low fidelity, unnatural base decreases misincorporation.
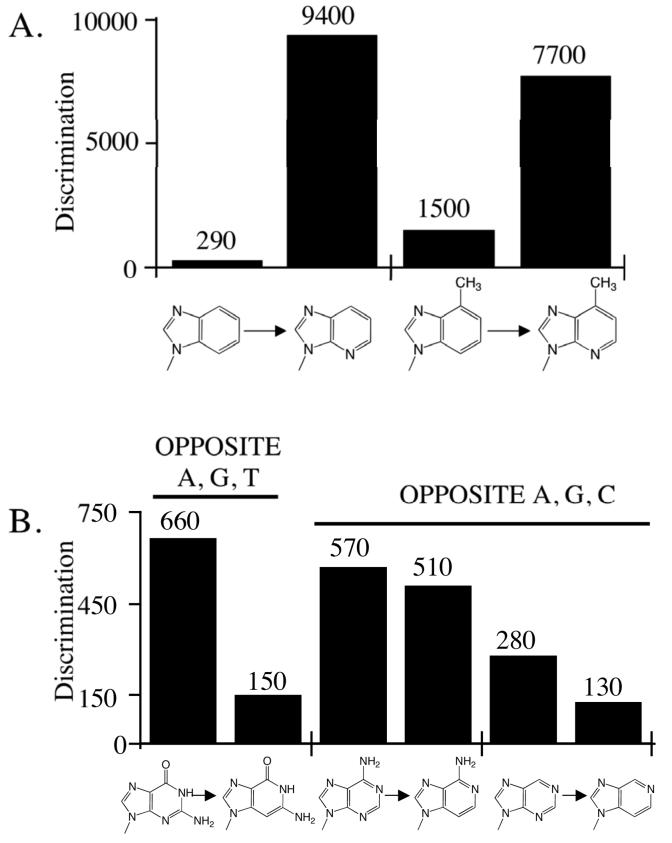
N-1 Primarily Enhances Polymerization Opposite T
We determined the effects of adding N-1 to a series of bases (i.e. converting benzimidazole to 3-deazapurine, 1-deaza-6-methylpurine to 6-methylpurine, 1-deazapurine to purine, 2-amino-1-deazapurine to 2-aminopurine, and 1-deazaadenine to adenine) and measured polymerization of each pair of dNTPs. In all 5 cases, N-1 will act as a hydrogen bond acceptor with the potential to form a Watson-Crick hydrogen bond with N(H)-3 of thymine. Consistent with this possibility, adding N-1 greatly enhanced polymerization opposite T (by 22- to 2,900-fold, Table 4 and Figure 4).
Figure 4.
Effect of adding N-1 to 1-deaza-6-methylpurine and benzimidazole (Panel A), and 1-deazapurine and 1-deazaadenine (Panel B). The discrimination factor for polymerization opposite T and the average discrimination factor for polymerization opposite A, C, and G are shown.
N-1 appears to perform a smaller role than N-3 in preventing misincorporation opposite A, C, and G. In three cases examined – benzimidazole dNTP vs. 3-deazapurine dNTP, 1-deazaadenine dNTP vs. dATP, and 2-amino-1-deazapurine dNTP vs. 2-aminopurine dNTP – the presence of N-1 slightly enhanced the ability of herpes polymerase to prevent incorporation opposite all three template bases (by 2- to 4-fold, Table 4 and Figure 4). However, adding N-1 to 1-deazapurine dNTP and forming purine dNTP slightly increased the likelihood of misincorporation opposite A, C, and G (by 4-fold), while converting 1-deaza-6-methylpurine dNTP to 6-methylpurine dNTP had little effect on misincorporation.
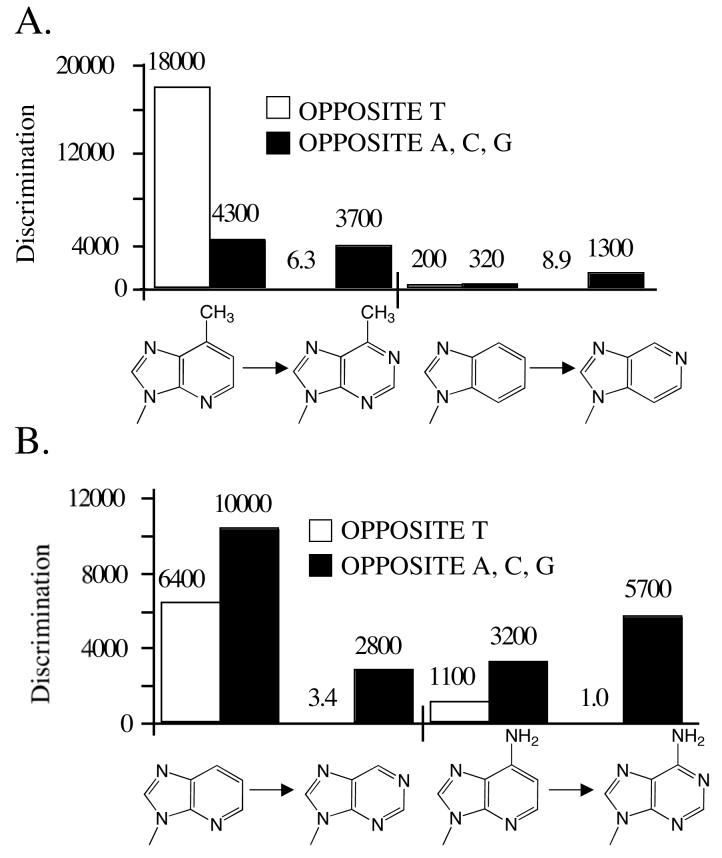
N6 enhances polymerization opposite T and helps prevent misincorporation opposite A, C, and G
The exocyclic amine of a purine, N6, can form a hydrogen bond with O4 of thymine. In the four cases examined (i.e. converting 1-deazapurine to 1-deazaadenine, purine to adenine, 3-deazapurine to 3-deazaadenine, and 2-aminopurine to 2-aminoadenine), herpes polymerase incorporated the aminated dNTPs 2- to 6-fold more efficiently opposite T than the analogous dNTPs lacking N6, consistent with this hydrogen bond acting as a positive selector for polymerization opposite T (Table 4 and Figure 5).
Figure 5.
Effect of adding N6 to 1-deazapurine, 3-deazapurine, and purine. The average discrimination factors (opposite A, C, and G) are T are shown.
Comparing rates of misincorporation of each of the pairs of compounds suggests that N6can also help prevent misincorporation. Addition of N6 in two cases (i.e. converting 3-deazapurine dNTP into 3-deazaadenine dNTP, and purine dNTP into dATP) slightly increased the ability of herpes polymerase to identify the resulting dNTPs as wrong for polymerization opposite A, C, and G (by 2- to 4-fold, Table 4 and Figure 5). Interestingly, adding N6 in the absence of N-1 (i.e. converting 1-deazapurine dNTP into 1-deazaadenine dNTP) increased the frequency of misincorporation 3-fold opposite A, C, and G, and in the case of converting 2-aminopurine to 2-aminoadenine, N6 had little effect on incorporation opposite A, C, and G, suggesting that the effects of N6 are context dependent.
To better understand how varying the exocyclic group at C-6 affects the polymerase, we compared the effects of the slightly electron donating CH3 (i.e. converting purine dNTP into 6-methylpurine dNTP, benzimidazole into 4-methylbenzimidazole, and 1-deazapurine dNTP into 1-deaza-6-methylpurine dNTP) and the electron withdrawing Cl (converting purine dNTP into 6-chloropurine dNTP and 2-aminopurine dNTP into 2-amino-6-chloropurine dNTP) groups. The methyl group had minimal impact on both correct polymerization opposite T and misincorporation opposite A, C and G. In contrast, the chlorine significantly decreased correct polymerization opposite T (6- to 16-fold less efficiently, Table 4), but again had minimal effects on misincorporation.
Role of N2
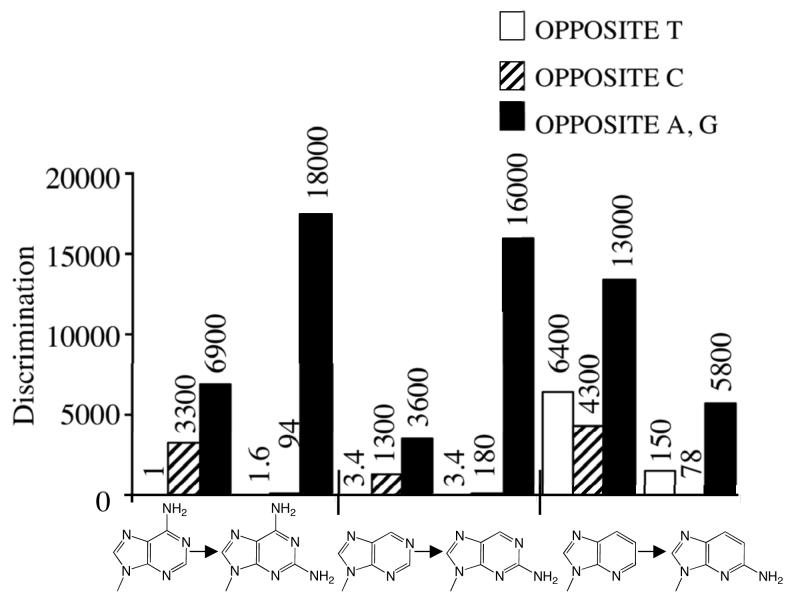
We explored the role of N2 by both removing it from dGTP and adding it to a series of dATP derivatives – adenine, purine, 6-chloropurine, and 1-deazapurine dNTPs. Opposite a template T, adding N2 to the adenine derivatives had relatively modest effects, ranging from a slight inhibition of polymerization for 2-aminoadenine-dNTP compared to dATP, to a 4-fold increase in polymerization for 2-amino-1-deazapurine-dNTP compared to 1-deazapurine-dNTP (Figure 6). Adding N2 to these bases generally decreased polymerization of the resulting aminated dNTPs opposite A and G, in some cases by rather substantial amounts – up to a 21-fold decrease in polymerization opposite a template A upon converting purine-dNTP into 2-aminopurine-dNTP. The most dramatic and consistent effects, however, occurred during polymerization opposite C. The addition of N2 to dATP, purine-dNTP, 6-chloropurine-dNTP, and 1-deazapurine-dNTP increased the efficiency of incorporation by 35-, 7-, 3-, and 55-fold, respectively (Table 4 and Figure 6). Importantly, the largest increase occurred with the one base lacking N-1, 1-deazapurine-dNTP.
Figure 6.
Effect of adding N2 to adenine, purine, and 1-deazapurine. The values of the average discrimination factors (opposite A and G), along with those opposite T and C, are shown.
While removing N2 from dGTP, thereby forming dITP, had little effect during polymerization opposite T and a small effect on incorporation opposite G, it dramatically increased polymerization opposite A (>170-fold increase, Table 4). Adding N2 had minimal impacts during incorporation of dGTP and dITP opposite C, in contrast to the large effects of N2 during polymerization of adenine-dNTP analogues opposite C. Similarly, when inosine replaced guanine as the template base (Table 1), herpes polymerase incorporated dCTP only 1.3-fold less efficiently than with guanine as the template base (Vmax/KM, data not shown). However, this replacement increased the efficiency of dATP incorporation by a factor of 80, similar to the effects on dITP versus dGTP incorporation. Thus, removing N2 from guanine profoundly affects formation of the incorrect hypoxanthine:adenine base-pair.
Modifying the structure of the base-pair at the primer 3′-terminus impairs further dNTP polymerization
We examined the role of base-pair structure at the primer 3′-terminus by measuring the addition of the next correct dNTP onto primer 3′-termini that consisted of either a natural, correct base-pair or a modified base-pair.3 When the template base was T, the absence of a single Watson-Crick hydrogen bond, either at N-1 or N6, impaired polymerization of the next correct dNTP (39- to 130-fold decrease, Table 5), while the absence of both hydrogen bonds very severely hindered polymerization (a 34,000-fold decrease upon converting adenine into benzimidazole, Table 5). The loss of N-3 and the associated electron density in the minor groove also moderately impaired polymerization of the next correct dNTP (12- to 25-fold decrease, Table 5). Similarly, when the template base was C, the polymerase incorporated the next dNTP 6-fold less efficiently than after incorporating 3-deaza-dGTP as compared to dGTP (Table 7).
Table 5.
Insertion of the Next Correct dNTP on DNAT by Herpes Polymerase (dTTP insertion following analogue X)
| X in 5′-Primer-X-3′ | Vmax (SD) | Km (SD) | Vmax/Km | Discrimination |
|---|---|---|---|---|
| Adenine | 20 (1) | 8 (1) | 2.5 | 1 |
| Benzimidazole | 0.019 (0.003) | 260 (130) | 0.000073 | 34000 |
| Purine | 1.21 (0.05) | 19 (4) | 0.064 | 39 |
| 3-Deazapurine | 1.01 (0.07) | 190 (50) | 0.0053 | 470 |
| 3-Deazaadenine | 7.2 (0.2) | 72 (9) | 0.10 | 25 |
| 1-Deazaadenine | 0.25 (0.01) | 13 (3) | 0.019 | 130 |
| 6-Methylpurine | 2.5 (0.1) | 51 (12) | 0.049 | 51 |
| 6-Chloropurine | 4.6 (0.3) | 460 (70) | 0.010 | 250 |
| 2-Amino-6-Chloropurine | 4.1 (0.4) | 270 (70) | 0.015 | 170 |
| 2-Aminopurine | 4.5 (0.3) | 33 (8) | 0.14 | 18 |
| 2-Aminoadenine | 3.0 (0.1) | 9 (2) | 0.33 | 7.6 |
Table 7.
Insertion of the First and Second Correct dNTP on DNAC by Herpes Polymerase
| X in 5′-Primer-X-3′ | Vmax (SD) | Km (SD) | Vmax/Km | Discrimination |
|---|---|---|---|---|
| Guanine | 4.9 (0.1) | 14 (2) | 0.35 | 1 |
| 3-Deazaguanine | 11.0 (0.2) | 190 (10) | 0.058 | 6 |
| X in 5′-Primer-X-T-3′ | Vmax (SD) | Km (SD) | Vmax/Km | Discrimination |
| Guanine | 1.91 (0.06) | 2.7 (0.3) | 0.71 | 1 |
| 3-Deazaguanine | 0.27 (0.03) | 440 (140) | 0.00061 | 1200 |
The identity of the group at C-6 of purines, not just whether or not it can form a hydrogen bond with T, also affected polymerization of the next correct dNTP. Whereas the polymerase tolerated a hydrogen (i.e. purine) or methyl (i.e. 6-methylpurine) equally well at C-6, the presence of an electron-withdrawing chlorine further decreased addition of the next correct dNTP by 6- to 9-fold (Table 5).
We examined the effects of increasing the number of hydrogen bonds involving the base-pair at the primer 3′ terminus by comparing the rates of addition of the next correct dNTP onto X:T base-pairs where X either contained or lacked N2. In the 3 cases examined – adenine vs. 2-aminoadenine, purine vs. 2-aminopurine, and 2-amino-6-chloropurine – adding an additional hydrogen bonding group had relatively modest effects. In the case of the two unnatural bases, the presence of N2 increased the efficiency of adding the next dNTP by around 2-fold, while adding N2 to an A:T base-pair impaired polymerization of the next correct dNTP by 8-fold (Table 5).
The identity of the base at the primer 3′-terminus affected not only polymerization of the next correct dNTP (n+1 addition), but also addition of the second dNTP (n+2 addition). The absence of a single Watson-Crick hydrogen bond in the base-pair (loss of N-1 or N6) at the primer terminus still inhibited addition of the second dNTP (by 10- to 64-fold, Table 6). Just as it typically hindered polymerization of the next correct dNTP, adding N2 to either purine or adenine to form an extra hydrogen bond hindered n+2 incorporation (2- to 9-fold, Table 6). In contrast, the absence of N-3 greatly decreased the efficiency of polymerization of the second dNTP, much more so than addition of the first dNTP (by 450- to 930-fold, Table 6). Similarly, n+2 incorporation after incorporation of 3-deaza-dGTP on DNAC was 1200-fold less efficient than after incorporation of dGTP (Table 7).
Table 6.
Insertion of the Second Correct dNTP on DNAT2 by Herpes Polymerase (dCTP insertion following polymerization of analogue X and T)
| X in 5′-Primer-X-T-3′ | Vmax (SD) | Km (SD) | Vmax/Km | Discrimination |
|---|---|---|---|---|
| Adenine | 1.43 (0.04) | 1.0 (0.1) | 1.4 | 1 |
| Purine | 1.32 (0.05) | 9.5 (2.2) | 0.14 | 10 |
| 3-Deazapurine | 0.46 (0.05) | 1500 (300) | 0.00031 | 4500 |
| 3-Deazaadenine | 0.76 (0.09) | 500 (100) | 0.0015 | 930 |
| 1-Deazaadenine | 0.44 (0.03) | 20 (5) | 0.022 | 64 |
| 6-Methylpurine | 1.9 (0.1) | 16 (4) | 0.12 | 12 |
| 2-Aminopurine | 0.36 (0.01) | 8.1 (1.3) | 0.044 | 32 |
| 2-Aminoadenine | 0.39 (0.01) | 2.5 (0.5) | 0.16 | 8.8 |
Discussion
We investigated the chemical features of the base that herpes DNA polymerase uses to distinguish between right and wrong purine dNTPs. N-1, N-3, and N6 play distinct yet complementary roles to ensure accurate polymerization of purine dNTPs by both enhancing polymerization of correct dNTPs (i.e. as positive selectors) and reducing polymerization of wrong dNTPs (i.e. as negative selectors). These functional groups, and their ability to interact with the template base, also affect polymerization of at least the next two dNTPs.
Herpes polymerase, as well as other B family polymerases such as pol α and T4 DNA polymerase, do not use the shape of the incipient base-pair formed between the incoming dNTP and template base as a major determinant to prevent misincorporation. Both herpes polymerase and pol α can incorporate dNTPs bearing various misshapen bases (Ex., benzimidazole, and 5-and 6-trifluoromethylbenzimidazole) orders of magnitude faster than they misincorporate a natural dNTP (12, 20). Likewise, T4 DNA polymerase and pol α both incorporate misshapen dNTPs such as 5-nitroindole dNTP opposite natural template bases at rates that approach those for a natural, correct dNTP (11, 26). Accommodation of these modified bases opposite template purines also indicates that the active site is likely very flexible, and does not form a tight fit around the incipient base-pair. Furthermore, while pol α and T4 DNA polymerase both generate A:difluorotoluene base-pairs with at least modest selectivity, the reactions are much less efficient than formation of a canonical base-pair (27, 28).
N6 affects both correct and incorrect polymerization, although in each case the effects are relatively small (Figure 5). These effects likely reflect the chemical properties of N6 (Eg. hydrogen bonding capacity), since neither a methyl nor a Cl had a similar effect. Furthermore, within the context of purine and 2-aminopurine, adding Cl to C-6 both decreased incorporation opposite T and increased incorporation opposite C. These effects of Cl are consistent with the hydrogen bonding capacity of N-1 playing a role both for enhancing correct polymerization and preventing incorrect incorporation. The electron withdrawing Cl likely reduces the electron density at N-1, thus interfering with the ability of N-1 to form a hydrogen bond with N(H)-3 of T and reducing the electronic repulsion between purine N-1 and the electron rich N-3 of C. Analogously, the addition of a halogen (chlorine or fluorine) to C-2 of adenine reduces hydrogen bonding to thymine, likely due to a reduced pKa of N-1 (26).
N-1 of a purine dNTP plays a critical role to ensure accurate replication. In every case examined, adding N-1 dramatically increased polymerization opposite T – up to factors of 1100 and 1900 in the cases of 1-deazaadenine dNTP and 1-deazapurine dNTP, respectively (Figure 4). Since the hybridization state of N-1 is appropriate for forming a Watson-Crick hydrogen bond with N(H)-3 of T, these data suggest that formation of this hydrogen bond enhances correct polymerization. Energetically, this hydrogen bond appears to contribute >4 kcal mol−1. However, the actual strength of the bond is probably weaker than this since replacing the electron rich N-1 with a relatively electron deficient C-H may now result in a repulsive interaction with N(H)-3 of T. Although not quite as large as the effects of adding N-3, adding N-1 also decreases misincorporation of the resulting dNTPs opposite A, C, and G.
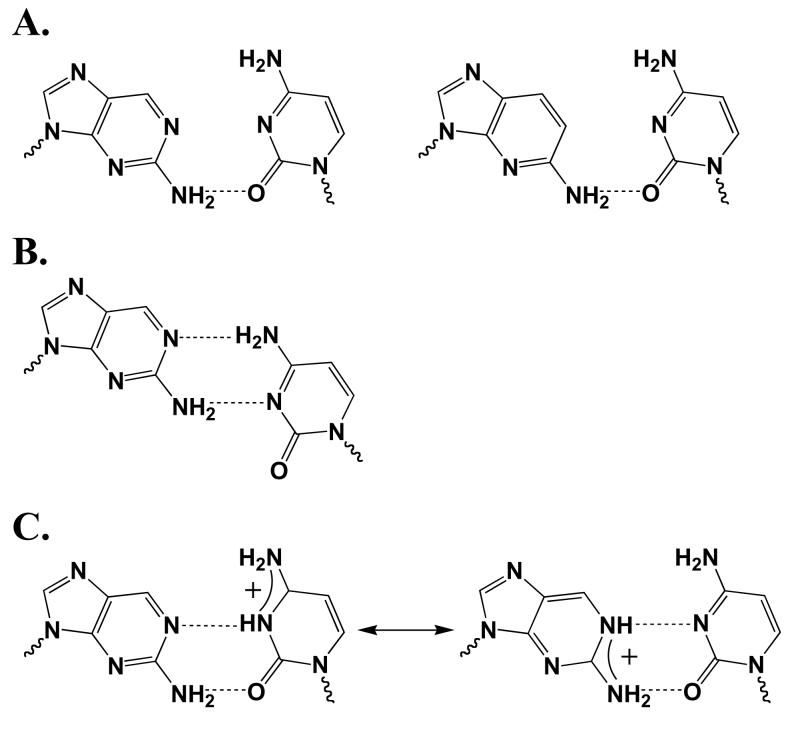
While adding N2 to purine dNTPs had relatively small effects on polymerization opposite T, this same modification significantly and specifically increased incorporation opposite C (3.3-to 55-fold, Table 4 and Figure 6). Enhanced incorporation opposite C likely results from formation of a new hydrogen bond between N2 of the aminated purine and O2 of cytosine. While structural studies have posited that protonation/tautomerization of the aminated purine enhances misincorporation opposite C (Figure 7 and (27-29)), this possibility is not feasible in the case of 2-amino-1-deazapurine due to the absence of N-1. Rather, the simplest mechanism consistent with all of the purine dNTP analogues is formation of a new hydrogen bond involving N2 of the purine and O2 of cytosine. Importantly, this mechanism would be identical to that observed for pol α, another B family polymerase (21).
Figure 7.
Potential structures for the 2-aminopurine:cytosine and 1-deaza-2-aminopurin base-pair via; (A) just a H-bond between N2 of the purine and O4 of C; (B) formation of a wobble base-pair, and; (C) protonation of the base-pair.
While N2 of guanine plays a minimal role during formation of G:C base-pairs, it has a key role in fidelity. Removing N2 from guanine nucleotides, either in the context of the dNTP or the template base, specifically promoted formation of hypoxanthine:adenine mismatches. Herpes polymerase incorporated dITP 170-fold more efficiently than dGTP opposite A (Tables 2 and 4), and polymerized dATP 80-fold more efficiently opposite I than opposite G (data not shown). Thus, herpes polymerase clearly uses N2 of guanine to specifically prevent formation of a hypoxanthine:adenine base-pair.
The highly conserved Tyr818 may help mediate the effects of both N2 and N-3. Both N2 and N-3 lay in the major groove of the base-pair formed by the incoming dNTP and template base. Based on the structure of the closed E-DNA-dNTP ternary complex determined for RB69 polymerase, the electron-deficient edge of the herpes Tyr818 phenol (the equivalent of Tyr567 in RB69 polymerase) will be near these nitrogens (33). As such, replacing the electron rich N-3 with an electron deficient CH will alter the electronic environment of Tyr818. Likewise, the presence or absence of N2 will also alter this environment. Importantly, mutating the equivalent Tyr567 of RB69 polymerase to Ala compromises fidelity with only small effects on correct incorporation of dNTPs (34). These qualitatively similar effects of mutating either N2 or N-3 of the purine dNTP or Tyr567 suggest that they may interact mechanistically.
Both herpes polymerase and pol α are members of the B superfamily of polymerases, and share tremendous homology between their active sites (30). The P helix surrounds the incipient base pair between the incoming dNTP and the template base being replicated, and the only difference between these enzymes is replacement of the leucine (pol α) that resides in the major groove of the incipient base pair with valine (herpes polymerase) (20). Not surprisingly, therefore, both polymerases use similar general mechanisms involving both positive and negative selectivity to distinguish right from wrong dNTPs. Adding N2 to adenine and related purines or removing N2 from guanine has similar effects with both enzymes – increased polymerization opposite C in the former and increased misincorporation opposite A in the latter (21). Likewise, N-3 performs the identical role in both enzymes. Neither enzyme required N-3 for efficient, correct polymerization of purine dNTPs. Adding N-3 to low fidelity bases prevented misincorporation by both polymerases (7- to 30-fold for the herpes pol and 5- to 40-fold for pol α), whereas removing N-3 from high fidelity bases increased misincorporation by both polymerases (Table 4, Figure 3 and (20). With both enzymes, removing N-3 from adenine had much less effect than removing N-3 from guanine, and the effects were very mismatch dependent.
Notably, the enzymes also differ in several important respects. Herpes polymerase discriminates much more effectively against dNTPs whose bases lack N-1, N-3, and N6 than does pol α. In the case of benzimidazole and 4-trifluorobenzimidazole, herpes polymerase discriminates only 1.7- and 4.4-fold more strongly, respectively, than does pol α. However, herpes polymerase discriminates 10- to 19-fold more strongly against 5-, 6-, and 7-trifluoromethylbenzimidzole, as well as 4-methylbenzimidazole dNTPs, respectively (12, 20). Additionally, N-1 and N6 also have somewhat different roles for the two enzymes. Pol α uses N6 solely to enhance polymerization opposite T, by a factor of 8-34 (20), while herpes polymerase uses N6 both to prevent misincorporation of dATP and to only slightly enhance polymerization opposite T (2- to 6-fold). While both enzymes employ N-1 to prevent misincorporation opposite A, C, and G, the effects on correct polymerization opposite T are significantly different. Adding N-1 to dNTPs containing the bases benzimidazole, 1-deazapurine, 1-deaza-6-methylpurine, and 1-deazaadenine stimulated polymerization opposite T by both enzymes, but the effects on herpes polymerase were 3,1-, 25-, 41-, and 55-fold larger, respectively, than on pol α ((20) and vide infra).
Based on these data for herpes polymerase and pol α in conjunction with the substantial amino acid homology between B superfamily polymerases (30), we anticipate that all of the B family enzymes differentiate right and wrong dNTPs via similar general mechanisms. However, there will likely be substantial variability in how the enzymes use specific atoms of the bases of the dNTP and, we suspect, the template base. In the case of pol α and herpes polymerase, these differences could be driven by leucine-valine replacement, or be the much larger differences in amino acids that surround the P helix (35).
Eliminating even one hydrogen bond from the base-pair at the primer 3′ terminus significantly decreased the rate of addition of the next correct dNTP. Adding an additional, unnatural, hydrogen bond involving N2 either inhibited polymerization of the next correct dNTP (i.e. 2-aminoadenine:T), or slightly stimulated polymerization (i.e., 2-aminopurine:T and 6-chloro-2-aminopurine:T). Since these changes in hydrogen bonding likely affect the shape of the base-pair at the primer 3′-terminus, herpes polymerase probably has an exquisite sensitivity to the shape of the primer 3′-terminus when deciding how efficiently to polymerize the next correct dNTP. Functionally, the ability of herpes polymerase to so readily differentiate between a correct and incorrect base-pair would allow the enzyme's associated 3′-5′ exonuclease to remove just added, incorrect nucleotides.
Effects of an unusual base-pair at a primer 3′-terminus extend to at least the next two polymerization events. Potentially, herpes polymerase could sense even minor structural changes two base-pairs removed from the replication site. The sensor would have to be extremely sensitive since even base-pairs that should have a normal structure (i.e. purine:T, 2-aminoadenine:T, etc.) affect latter polymerization events. Alternatively, the enzyme could sense unusual π-electron distributions in the template and/or primer bases. Since the bases in duplex DNA form an extended π-electron cloud (31), changes in electronic distribution at one position will be transmitted to neighboring positions. Furthermore, the amino acids that interact with the planar faces and associated π-electron clouds of both the template base being replicated and the base of the incoming dNTP are extremely conserved among B family polymerases (20, 30, 32), suggesting that they are doing more than just “filling space” by interacting with these bases.
Herpes polymerase appears to use two orthogonal screens to replicate DNA with high fidelity. During incorporation, shape appears to play a minimal role, as evidenced by the rapid polymerization of dNTPs containing bases such as benzimidazole, 5-trifluorobenzimidazole, and 6-trifluorobenzimidazole. Rather, the enzyme uses specific chemical features of the base – N-1, N-3, and to a lesser extent, N2 (in the case of guanine) and N6 to prevent misincorporation. In the presence of these “negative selectors”, herpes polymerase then uses the formation of Watson-Crick hydrogen bonds to promote correct incorporation. In contrast to the low importance of shape for determining whether or not to incorporate a dNTP, shape of the base-pair at the primer-terminus appears to play a critical role in the addition of subsequent dNTPs.
Footnotes
This work was supported by National Institutes of Health Grant AI059764 (R.D.K.).
Abbreviations used: d1DATP, 9-β-D-2′-deoxyribofuranosyl-(1-deazaadenine)-5′-triphosphate; d1DPTP, 9-β-D-2′-deoxyribofuranosyl-(1-deazapurine)-5′-triphosphate; d3DATP, 9-β-D-2′-deoxyribofuranosyl-(3-deazaadenine)-5′-triphosphate; d3DGTP, 9-β-D-2′-deoxyribofuranosyl-(3-deazaguanine)-5′-triphosphate; d3DPTP, 9-β-D-2′-deoxyribofuranosyl-(3-deazapurine)-5′-triphosphate; dBTP, 1-β-D-2′-deoxyribofuranosyl-(benzimidazole)-5′-triphosphate; dNTP, natural 2′-deoxy-5′-triphosphate; dPTP, 9-β-D-2′-deoxyribofuranosyl-(purine)-5′-triphosphate; dQTP, 9-β-D-2′-deoxyribofuranosyl-(1-deaza-6-methylpurine)-5′-triphosphate; dZTP, 1-β-D-2′-deoxyribofuranosyl-(4-methylbenzimidazole)-5′-triphosphate; d4CF3BTP, 1-β-D-2′-deoxyribofuranosyl-(4-trifluoromethylbenzimidazole)-5′-triphosphate; d5CF3BTP, 1-β-D-2′-deoxyribofuranosyl-(5-trifluoromethylbenzimidazole)-5′-triphosphate; d6CF3BTP, 1-β-D-2′-deoxyribofuranosyl-(6-trifluoromethylbenzimidazole)-5′-triphosphate; d7CF3BTP, 1-β-D-2′-deoxyribofuranosyl-(7-trifluoromethylbenzimidazole)-5′-triphosphate; d6MPTP, 9-β-D-2′-deoxyribofuranosyl-(6-methylpurine)-5′-triphosphate; d6ClPTP, 9-β-D-2′-deoxyribofuranosyl-(6-chloropurine)-5′-triphosphate; d2AmATP, 9-β-D-2′-deoxyribofuranosyl-(2-aminoadenine)-5′-triphosphate; d2AmPTP, 9-β-D-2′-deoxyribofuranosyl-(2-aminopurine)-5′-triphosphate; d2Am6ClPTP, 9-β-D-2′-deoxyribofuranosyl-(2-amino-6-chloropurine)-5′-triphosphate; d2Am1DPTP, 9-β-D-2′-deoxyribofuranosyl-(2-amino-1-deazapurine)-5′-triphosphate; dITP, 9-β-D-2′-deoxyribofuranosyl-(hypoxanthine)-5′-triphosphate; HSV-1, Herpes Simplex Virus 1; pol α, DNA polymerase α; exo−, exonuclease deficient; Tris-HCl, Tris(hydroxymethyl)aminomethane; DTT, dithiothreitol; BSA, bovine serum albumin; and EDTA, ethylenediaminetetraacetic acid.
For all of these studies, we have compared the specificity parameter, VMAX/KM, to understand how changing the structure of a base affects polymerization. Because herpes polymerase is a processive polymerase, it is not possible to directly compare the VMAX and KM values. The rate-limiting step during steady-state turnover is dissociation of the product DNA from the E-DNA complex. Consequently, VMAX values for correct dNTPs will reflect the DNA dissociation rate, and not phosphodiester bond formation. Furthermore, the existence of a slow step after chemistry will also result in the KD and KM for dNTPs likely being very different. In contrast, this may or may not be the case for the analogues, thereby obviating the possibility of comparing these rates in a mechanistic sense.
References
- 1.Lehman IR, Boehmer PE. Replication of herpes simplex virus DNA. J Biol Chem. 1999;274:28059–28062. doi: 10.1074/jbc.274.40.28059. [DOI] [PubMed] [Google Scholar]
- 2.Boehmer PE, Lehman IR. Herpes simplex virus DNA replication. Annu Rev Biochem. 1997;66:347–384. doi: 10.1146/annurev.biochem.66.1.347. [DOI] [PubMed] [Google Scholar]
- 3.Johnson PA, Best MG, Friedmann T, Parris DS. Isolation of a herpes simplex virus type 1 mutant deleted for the essential UL42 gene and characterization of its null phenotype. J Virol. 1991;65:700–710. doi: 10.1128/jvi.65.2.700-710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stow ND. Herpes simplex virus type 1 origin-dependent DNA replication in insect cells using recombinant baculoviruses. J Gen Virol. 1992;73(Pt 2):313–321. doi: 10.1099/0022-1317-73-2-313. [DOI] [PubMed] [Google Scholar]
- 5.Wu CA, Nelson NJ, McGeoch DJ, Challberg MD. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J Virol. 1988;62:435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts JD, Kunkel TA. DNA Replication in Eukaryotic Cells: Concepts, Enzymes and Systems. 1996:217–247. [Google Scholar]
- 7.Kunkel TA, Bebenek K. Recent studies of the fidelity of DNA synthesis. Biochim Biophys Acta. 1988;951:1–15. doi: 10.1016/0167-4781(88)90020-6. [DOI] [PubMed] [Google Scholar]
- 8.Bebenek K, Abbotts J, Wilson SH, Kunkel TA. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J Biol Chem. 1993;268:10324–10334. [PubMed] [Google Scholar]
- 9.Chaudhuri M, Song L, Parris DS. The herpes simplex virus type 1 DNA polymerase processivity factor increases fidelity without altering pre-steady-state rate constants for polymerization or excision. J Biol Chem. 2003;278:8996–9004. doi: 10.1074/jbc.M210023200. [DOI] [PubMed] [Google Scholar]
- 10.Song L, Chaudhuri M, Knopf CW, Parris DS. Contribution of the 3′- to 5′-exonuclease activity of herpes simplex virus type 1 DNA polymerase to the fidelity of DNA synthesis. J Biol Chem. 2004;279:18535–18543. doi: 10.1074/jbc.M309848200. [DOI] [PubMed] [Google Scholar]
- 11.Chiaramonte M, Moore CL, Kincaid K, Kuchta RD. Facile polymerization of dNTPs bearing unnatural base analogues by DNA polymerase alpha and Klenow fragment (DNA polymerase I) Biochemistry. 2003;42:10472–10481. doi: 10.1021/bi034763l. [DOI] [PubMed] [Google Scholar]
- 12.Kincaid K, Beckman J, Zivkovic A, Halcomb RL, Engels JW, Kuchta RD. Exploration of factors driving incorporation of unnatural dNTPS into DNA by Klenow fragment (DNA polymerase I) and DNA polymerase alpha. Nucleic Acids Res. 2005;33:2620–2628. doi: 10.1093/nar/gki563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beard WA, Wilson SH. Structural design of a eukaryotic DNA repair polymerase: DNA polymerase beta. Mutat Res. 2000;460:231–244. doi: 10.1016/s0921-8777(00)00029-x. [DOI] [PubMed] [Google Scholar]
- 14.Minnick DT, Liu L, Grindley ND, Kunkel TA, Joyce CM. Discrimination against purine-pyrimidine mispairs in the polymerase active site of DNA polymerase I: a structural explanation. Proc Natl Acad Sci U S A. 2002;99:1194–1199. doi: 10.1073/pnas.032457899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 16.Echols H, Goodman MF. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 17.Knopf CW. Evolution of viral DNA-dependent DNA polymerases. Virus Genes. 1998;16:47–58. doi: 10.1023/a:1007997609122. [DOI] [PubMed] [Google Scholar]
- 18.Wong SW, Wahl AF, Yuan PM, Arai N, Pearson BE, Arai K, Korn D, Hunkapiller MW, Wang TS. Human DNA polymerase alpha gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988;7:37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hopfner KP, Eichinger A, Engh RA, Laue F, Ankenbauer W, Huber R, Angerer B. Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc Natl Acad Sci U S A. 1999;96:3600–3605. doi: 10.1073/pnas.96.7.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beckman J, Kincaid K, Hocek M, Spratt T, Engels J, Cosstick R, Kuchta RD. Human DNA polymerase alpha uses a combination of positive and negative selectivity to polymerize purine dNTPs with high fidelity. Biochemistry. 2007;46:448–460. doi: 10.1021/bi061243s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patro JN, Milan U, Kuchta RD. Role of the 2-amino group of purines during dNTP polymerization by human DNA polymerase α. Biochemistry. 2009;48:180–918. doi: 10.1021/bi801823z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavanaugh NA, Kuchta RD. Initiation of new DNA strands by the herpes simplex virus-1 primase-helicase complex and either herpes DNA polymerase or human DNA polymerase alpha. J Biol Chem. 2009;284:1523–1532. doi: 10.1074/jbc.M805476200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. 1982 [Google Scholar]
- 24.Kuchta RD, Mizrahi V, Benkovic PA, Johnson KA, Benkovic SJ. Kinetic mechanism of DNA polymerase I (Klenow) Biochemistry. 1987;26:8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- 25.Goodman MF, Creighton S, Bloom LB, Petruska J. Biochemical basis of DNA replication fidelity. Crit Rev Biochem Mol Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 26.Reineks EZ, Berdis AJ. Evaluating the contribution of base stacking during translesion DNA replication. Biochemistry. 2004;43:393–404. doi: 10.1021/bi034948s. [DOI] [PubMed] [Google Scholar]
- 27.Hariharan C, Bloom LB, Helquist SA, Kool ET, Reha-Krantz LJ. Dynamics of nucleotide incorporation: snapshots revealed by 2-aminopurine fluorescence studies. Biochemistry. 2006;45:2836–2844. doi: 10.1021/bi051644s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morales JC, Kool ET. Varied molecular interactions at the active sites of several DNA polymerases: nonpolar nucleoside isosteres as probes. J Am Chem Soc. 2000;122:1001–1007. doi: 10.1021/ja993464+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore CL, Zivkovic A, Engels JW, Kuchta RD. Human DNA primase uses Watson-Crick hydrogen bonds to distinguish between correct and incorrect nucleoside triphosphates. Biochemistry. 2004;43:12367–12374. doi: 10.1021/bi0490791. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Cao W, Zakharova E, Konigsberg W, De La Cruz EM. Fluorescence of 2-aminopurine reveals rapid conformational changes in the RB69 DNA polymerase-primer/template complexes upon binding and incorporation of matched deoxynucleoside triphosphates. Nucleic Acids Res. 2007;35:6052–6062. doi: 10.1093/nar/gkm587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sowers LC, Boulard Y, Fazakerley GV. Multiple structures for the 2-aminopurine-cytosine mispair. Biochemistry. 2000;39:7613–7620. doi: 10.1021/bi992388k. [DOI] [PubMed] [Google Scholar]
- 32.Sowers LC, Fazakerley GV, Eritja R, Kaplan BE, Goodman MF. Base pairing and mutagenesis: observation of a protonated base pair between 2-aminopurine and cytosine in an oligonucleotide by proton NMR. Proc Natl Acad Sci U S A. 1986;83:5434–5438. doi: 10.1073/pnas.83.15.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 34.Yang G, Wang J, Konigsberg W. Base selectivity is impaired by mutants that perturb hydrogen bonding networks in the RB69 DNA polymerase active site. Biochemistry. 2005;44:3338–3346. doi: 10.1021/bi047921x. [DOI] [PubMed] [Google Scholar]
- 35.Wang TS, Wong SW, Korn D. Human DNA polymerase alpha: predicted functional domains and relationships with viral DNA polymerases. Faseb J. 1989;3:14–21. doi: 10.1096/fasebj.3.1.2642867. [DOI] [PubMed] [Google Scholar]
- 36.Kelley SO, Barton JK. Electron transfer between bases in double helical DNA. Science. 1999;283:375–381. doi: 10.1126/science.283.5400.375. [DOI] [PubMed] [Google Scholar]
- 37.Liu S, Knafels JD, Chang JS, Waszak GA, Baldwin ET, Deibel MR, Jr., Thomsen DR, Homa FL, Wells PA, Tory MC, Poorman RA, Gao H, Qiu X, Seddon AP. Crystal structure of the herpes simplex virus 1 DNA polymerase. J Biol Chem. 2006;281:18193–18200. doi: 10.1074/jbc.M602414200. [DOI] [PubMed] [Google Scholar]