Abstract
Tryptophan hydroxylase-2 (TPH2) catalyzes the synthesis of neuronal serotonin, a major neurotransmitter involved in many brain functions and psychiatric disorders. We have previously revealed a critical role of the human TPH2 (hTPH2) 5′-UTR in gene expression regulation. This study aimed to further characterize mechanism(s) by which the hTPH2 5′-UTR regulates gene expression. An internal ribosome entry site (IRES) activity in hTPH2 5′-UTR was suggested by the conventional bicistronic reporter assay; however, further stringent experiments, including in vitro translation, quantitative real-time PCR, Northern blot, ribonuclease protection assay, and monocistronic reporter assay, demonstrated that the hTPH2 5′-UTR harbors a bidirectional promoter, but not IRES, within its downstream segment (61~141). The antisense promoter is much stronger than the sense promoter, but the strength of both promoters are cell-line dependent, with the highest and lowest activities being observed in HEK-293T and SK-N-MC cells, respectively. In accordance with our previous findings, the upstream segment (1~60) of hTPH2 5′-UTR suppresses the neighboring promoter of both direction, independent of the cell line and its location in the 5′- or 3′-flanking regions of the gene. In summary, this study demonstrates that no IRES but an asymmetric bidirectional promoter is present in the downstream segment of hTPH2 5′-UTR, and this promoter is susceptible to a gene silencing effect caused by the upstream segment (1~60) of hTPH2 5′-UTR. Our findings point to the potential involvement of antisense transcription and non-coding RNA in the regulation of TPH2 gene expression.
Keywords: Tryptophan hydroxylase-2, bidirectional promoter, internal ribosome entry site, serotonin, antisense transcript, gene expression
1. Introduction
Serotonin (5-HT) is a major neurotransmitter involved in many brain functions, and a number of pharmaceuticals that modulate 5-HT neurotransmission are widely used for the treatment of various psychiatric disorders. The synthesis of 5-HT is initiated by the hydroxylation of the amino acid tryptophan, which is the rate-limiting step catalyzed by tryptophan hydroxylase (TPH). Two isoforms of TPH (TPH1 and TPH2) have been identified, among which TPH2 is exclusively expressed in the brain while TPH1 is predominantly expressed in peripheral tissues (Walther and Bader 2003; Walther et al., 2003; Côté et al., 2003; Zhang et al., 2004). Thus, genetic or epigenetic factors affecting TPH2 gene expression or catalytic properties may alter 5-HT neurotransmission and thereby modify behavioral traits, drug responses, and disease susceptibility. In fact, genetic variance in human TPH2 (hTPH2) has been reportedly associated with a growing number of psychiatric diseases and behavioral traits, including major depression (Zill et al., 2004a; Zhou et al., 2005), affective disorders (Harvey et al., 2004; Lopez et al., 2007; Cichon et al., 2008), suicidality (Zill et al., 2004b; Jollant et al., 2007; Lopez de Lara et al., 2007), autism (Coon et al., 2005), early onset obsessive-compulsive disorder (Mössner et al., 2006), attention deficit hyperactivity disorder (ADHD) (Sheehan et al., 2005; Walitza et al., 2005), panic disorder (Maron et al., 2007), chronic fatigue syndrome (Goertzel et al., 2006; Smith et al., 2006), drug addiction (Reuter et al., 2007a; Nielsen et al., 2008), as well as the amygdala responsiveness (Brown et al., 2005; Canli et al., 2005), emotional processing (Herrmann et al., 2007), and personality traits (Gutknecht et al., 2007; Reuter et al. 2007b). In addition, the antidepressant response to the selective serotonin reuptake inhibitors (SSRIs) in human and rodent is also associated with TPH2 genetic variance (Peters et al., 2004; Crowley et al., 2005; Cervo et al., 2005; Tzvetkov et al., 2008).
TPH2 gene expression exhibits a circadian rhythm and is regulated by various hormones and stressors (Liang et al., 2004; Clark et al., 2005; Sanchez et al. 2005; Malek et al., 2005 and 2007; Hiroi et al., 2006; Brown et al., 2006). Particularly, the rhythmic expression of TPH2 is induced by the daily surge of glucocorticoids, which are terminal products of the hypothalamic-pituitary-adrenal (HPA) axis, a critical neuroendocrine system that responds to stress and also shows a circadian rhythm of activity (Malek et al., 2007). These characteristics of TPH2 expression supports the notion of reciprocal interactions between the 5-HT system and HPA axis (Dinan, 1996; Lowry, 2002), and implies that modulation of TPH2 gene expression by specific hormones and stressors may underlie the activation and feedback control of HPA axis. In other words, the regulation of HPA axis function may depend on a normal flexibility of TPH2 gene expression. In support of this presumption, our previous study has demonstrated that functional polymorphisms in the 3′-untranslated region (UTR) of rhesus monkey TPH2 is predictive of HPA axis activity, including the morning cortisol level and dexamethasone suppression of cortisol release, which reflect the HPA axis activation and feedback control, respectively (Chen et al., 2006). Since HPA axis dysfunction is involved in a variety of stress-related psychiatric diseases, such as depression and anxiety, HPA axis reactivity differentiated by TPH2 variants might thus underlie the associations between TPH2 and such disorders, just like the case for the serotonin transporter (5-HTT), another serotoninergic gene (Gotlib et al., 2007). In addition, it has been reported in a rodent model that TPH2 gene expression is modulated by the treatment of SSRI fluoxetine, and the fluoxetine regulation of TPH2 expression correlates with its antidepressant effect (Shishkina et al., 2007). Hence, the knowledge of TPH2 gene expression regulation will greatly advance our understanding of the pathophysiology of psychiatric disorders, but also help to elucidate mechanisms underlying individual variations in behavioral traits, drug responses and disease susceptibility related to 5-HT neurotransmission.
During our previous study of the functional significance of the hTPH2 polymorphisms, we revealed that the upstream segment (8~53) of TPH2 5′-UTR exerts a strong inhibitory effect on gene expression (Chen et al., 2008). This finding is in accordance with the identification of a repressive bipartite REST/NRSF binding motif in this region (Patel et al., 2007); however, both transcriptional and post-transcriptional mechanisms are involved in gene expression regulation by the TPH2 5′-UTR (Chen et al., 2008). In the present study, we performed a series of experiments to further explore the mechanisms by which the 5′-UTR regulates TPH2 gene expression. We found that an asymmetric bidirectional promoter is present in the downstream 5′-UTR of TPH2. As an abundance of bidirectional promoters exist in the human genome and may induce RNA-directed DNA methylation and gene silencing (Mette et al., 2000; Trinklein et al., 2004; Chen et al., 2004; Tufarelli et al., 2003), our findings provide new insights into the regulation of TPH2 gene expression.
2. Materials and Methods
2.1. Construction of bicistronic and monocistonic reporter systems
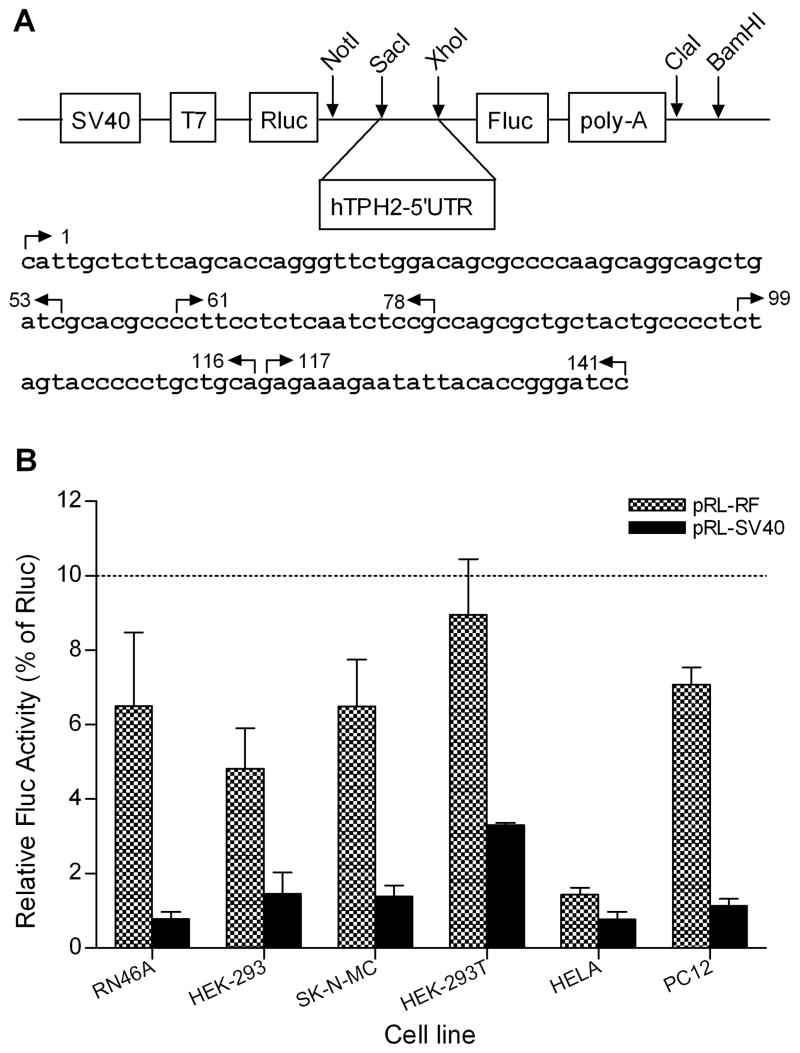
To make the bicistronic reporter system, we first amplified the whole firefly luciferase (Fluc) coding region plus the poly-A signal from pGL4.14 (Promega, Madison, WI) by using the NotI-pGL(7)F and BamHI-pGL(2031)R primers (Table 1). The PCR product, as well as another renilla luciferase (Rluc) vector pRL-SV40 (Promega), was subjected to sequential digestion by NotI and BamHI, followed by the ligation of the PCR product and pRL-SV40 large fragment. The resultant pRL-RF construct is supposed to yield a single mRNA transcript comprising both reporter cistrons (Rluc as the first and Fluc as the second), between which there is a 101-bp space containing the SacI and XhoI restriction sites (shown in Fig. 1A). To clone the whole and partial 5′-UTR of hTPH2, a series of primers (Table 1) were designed based on the hTPH2 mRNA sequence (Genbank accession number: NM_173353). The whole 5′-UTR (1~141) was first amplified from human genomic DNA (Promega) using the SacI-hTPH2(1)F and XhoI-hTPH2(141)R primers. Following double digestion by SacI and XhoI, the PCR product was cloned into the pRL-RF construct between Rluc and Fluc, and the resultant construct was then used as the template to amplify the partial 5′-UTR fragments except the 117~141, which was cloned into the pRL-RF construct by using the 5′-phosphorylated linkers hTPH2(117)F and hTPH2(141)R (Table 1). The whole and partial fragments of the 5′-UTR were also cloned into the pGL4.14 vector (monocistronic reporter). In addition, the whole (141~1) and partial (141~61) fragments of the 5′-UTR were conversely cloned into the pRL-RF and pGL4.14 vectors. All of the constructs were sequence-verified and correct orientation was confirmed.
Table 1.
Primers used for the cloning and quantitative RT-PCR.
| Name | Sequence |
|---|---|
| Cloning | |
| NotI-pGL(7)F | 5′-ttctGCGGCCGCactggccggtacctgagctc-3′ |
| BamHI-pGL(2031)R | 5′-cgcaaacGGATCCttatcgat-3′ |
| SacI-hTPH2(1)F | 5′-tgagGAGCTCcattgctcttcagcaccagg-3′ |
| SacI-hTPH2(61)F | 5′-atcgGAGCTCccttcctctcaatctccg-3′ |
| SacI-hTPH2(99)F | 5′-tactGAGCTCctagtaccccctgctgca-3′ |
| XhoI-hTPH2(53)R | 5′-agagCTCGAGgatcagctgcctgcttgg-3′ |
| XhoI-hTPH2(78)R | 5′-gcagCTCGAGcggagattgagaggaagg-3′ |
| XhoI-hTPH2(116)R | 5′-attcCTCGAGtgcagcagggggtactag-3′ |
| XhoI-hTPH2(141)R | 5′-ctggCTCGAGggatcccggtgtaatattctttc-3′ |
| XhoI-hTPH2(1)F | 5′-tgagCTCGAGcattgctcttcagcaccagg-3′ |
| SacI-hTPH2(141)R | 5′-ctggGAGCTCggatcccggtgtaatattctttc-3′ |
| hTPH2(117)F* | 5′-CgagaaagaatattacaccgggatccC-3′ |
| hTPH2(141)R* | 5′-TCGAGggatcccggtgtaatattctttctcGAGCT-3′ |
| qRT-PCR | |
| Rluc(735)F | 5′-gataactggtccgcagtggt-3′ |
| Rluc(957)R | 5′-accagatttgcctgatttgc-3′ |
| Fluc(594)F | 5′-caccttcgtgacttcccatt-3′ |
| Fluc(761)R | 5′-tgactgaatcggacacaagc-3′ |
5′-phosphorylated linker. Upper case - designed to introduce the restriction site. Numbers in the parentheses represent the starting positions in target sequences (hTPH2, pGL4.14, and pRL-SV40). Positions in hTPH2 are relative to the transcription start site (TSS), while those in the reporter vectors are based on the sequences of pGL4.14 (AY864928, Fluc) and pRL-SV40 (AF025845, Rluc), respectively.
Fig. 1.
Schematic structure of the pRL-RF (Rluc/Fluc) bicistronic reporter system (A) and the relative Fluc activity (% of Rluc) of the pRL-RF control without hTPH2–5′UTR insert (B). The sequence between the NotI and BamHI sites was amplified from the pGL4.14 vector. The whole and partial sequences of the hTPH2 5′-UTR were cloned into the pRL-RF between the Rluc and Fluc by using the SacI and XhoI digestion. The start locations of the cloning primers were indicated by arrows, with the right and left directions representing the forward and reverse primers, respectively. The predicted terminal oligopyrimidine tracts (TOP) are underlined. The relative Fluc activities (Means±SE) of the pRL-RF and pRL-SV40 are expressed as percentage (%) of the Rluc activity. pRL-SV40 encodes only Rluc but not Fluc.
2.2. Cell Culture
RN46A, HEK-293, HEK-293T, PC12, SK-N-MC, and HELA cells were employed in the present study. RN46A cells were cultured in Neurobasal™Medium (Invitrogen, Carlsbad, CA) complemented with 8% heat-inactivated FBS and 0.05% L-glutamine. HEK-293, HEK-293T and HELA cells were maintained in Dulbecco’s Modified Eagle’s Medium containing 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg streptomycin, and 0.1 mM non-essential amino acids. SK-N-MC cells were grown in Ham’s F12 Medium with 10% FBS, while PC12 cells were cultured in Ham’s F12 Medium with 15% FBS and 2.5% horse serum. RN46A cells were maintained at 33°C in a 5% CO2 incubator, while other cells were cultured in an atmosphere of 5% CO2 at 37°C.
2.3. Transient transfection and luciferase assays
Transfections for RN46A, PC12 and HELA cells were performed using Lipofectamine™ 2000 (Invitrogen), while the HEK-293, HEK-293T and SK-N-MC cells were transfected with ProFection® Mammalian Transfection System (Promega). The day before transfection, cells were seeded in 24-well plates at approximately 2×105 cells/well. For each well, 0.8 μg of each bicistronic (pRL-RF/hTPH2–5′UTR) or monocistronic (pGL/hTPH2–5′UTR) construct was transfected according to the manufacturer’s protocol. For the monocistronic constructs, 0.2 μg of a second pRL-TK reporter vector was co-transfected as the control to determine transfection efficiency. The transfected cells were maintained in culture and the growth medium was changed 4–6 h after transfection. Cells were harvested 24h after transfection, and the Fluc and Rluc activities were measured using the Dual Luciferase Reporter Assay System (Promega) on a Victor3™V Multilabel Counter (Wallac-PerkinElmer, Turku, Finland). Transfections and assays were performed in triplicate at least three times, with different DNA preparations of the same clone to ensure validity. For the bicistronic constructs, both Fluc and Rluc activities are expressed relative to those of the control vector (pRL-RF without insert) and are used to conduct the statistics. For the monocistronic constructs, the normalized luciferase data (firefly/Renilla) were used to perform the statistics and are expressed relative to the control pGL4.14 vector without insert.
2.4. RNA isolation and quantative real-time PCR (qRT-PCR)
HEK-293T cells were seeded in P100 dishes at approximately 2×106 cells/dish and transfection of the bicistronic constructs was performed the next day using 16 μg of construct per dish. Cells were lysed at 24 h after transfection by the addition of 5 ml of TRIzol Reagent (Invitrogen) per dish and total RNA was isolated according to the manufacturer’s protocol. To avoid DNA contamination, RNA samples were treated with RQ1 RNase-free DNaseI (Promega) for 1 h at 37°C. For the qRT-PCR, an aliquot of total RNA was reverse transcribed into cDNA using Superscript™III reverse transcriptase and oligo-dTs (Invitrogen), and the synthesized cDNA was diluted to 50ng/μl for use. Real-time PCR was performed on a Roche LightCycler 2.0 system (Roche Diagnostics, Indianapolis, IN) using 50 ng of cDNA in each reaction. Quantification of the luciferase mRNA was based on an external standard curve using serial 10-fold dilutions (107~100 copies) of the pRF construct. The firefly and Renilla luciferase mRNA levels were determined by SYBR Green RT-PCR using primer sets Fluc(594)F/(761)R and Rluc(735)F/(957)R (Table 1), respectively, with serial dilutions of the pRL-RF construct as the external standard. The PCR reactions were run in duplicate and the entire experiment was triplicated.
2.5. In vitro transcription
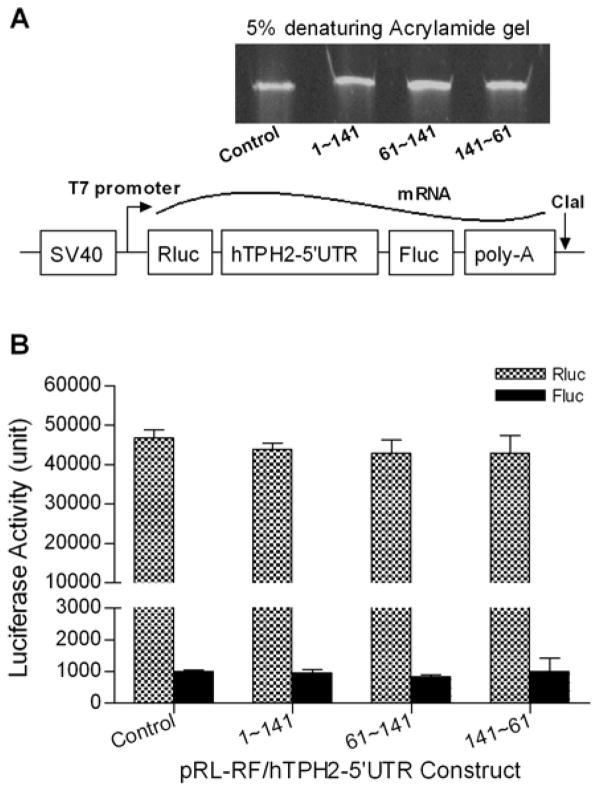
In vitro transcription was performed to generate RNA probes for Northern blot and Ribonuclease protection assay, as well as to make bicistronic mRNA for in vitro translation. The sense and antisense RNA probes for the hTPH2 5′-UTR was transcribed as previously described (Chen et al., 2008). For the generation of luciferase RNA probes, partial coding regions of the Fluc and Rluc were amplified using the primer sets Fluc(594)F/(761)R and Rluc(735)F/(957)R (Table 1), respectively, and the purified PCR products were then cloned into pGEM-T vector (Promega). The constructs were sequence-verified, followed by linearization using either NcoI or NotI, depending on the insert direction and the purpose of generating the sense or antisense probe. To make the bicistronic mRNA, specific pRL-RF/hTPH2–5′UTR constructs were linearized by ClaI digestion. In vitro transcription reactions with the linearized templates were carried out with a MAXIscript®T7/SP6 Kit (Ambion, Austin, TX) in the presence (for probes) or absence (for bicistronic mRNA) of biotinylated UTP (Ambion) according to the manufacturer’s instructions. The mRNA products were purified with Ambion’s MEGAclear™kit and an aliquot of each was run on 5% denaturing (8M urea) acrylamide gels in 0.5× TBE, followed by detection with ethidium bromide.
2.6. In vitro translation
The Ambion’s Retic Lysate IVT™ kit was employed to perform the in vitro translation by using the in vitro transcribed bicistronic mRNA as the template. Reactions were performed in a total volume of 25 μl comprising 1× Translation Mix (methionine-free), 17 μl of RBC Lysate (Ambion), 2.5 mM of methionine (Sigma-Aldrich, St. Louis, MO), and 0.5 pmol of RNA template. Following the incubation of the mixture for 90 min in a water bath at 30°C, the reactions were placed on ice for 5 min. Two aliquots (10 μl for each) of the reaction were taken out for the measurement of the Fluc and Rluc activities by using the Dual Luciferase Reporter Assay System (Promega) on a Victor3™ V Multilabel Counter (Wallac-PerkinElmer).
2.7. Northern blot analysis
Total RNA (20 μg) from HEK-293T cells transfected with specific pRL-RF/hTPH2–5′UTR constructs were fractionated in a 1.2% agarose-formaldehyde gel and then transferred onto a ZetaProbe® membrane (Bio-Rad Laboratories, Hercules, CA), followed by a cross-link to the membrane. The membrane was then hybridized to the biotinylated antisense Fluc or sense Rluc probe by using Ambion’s NorthernMax® Kit according to the manufacturer’s protocol with minor modification. Briefly, following a 1-hour pre-hybridization of the membrane in ULTRAhyb Hybridization Buffer, 1 nM of RNA probe was added and hybridized overnight at 60°C. Signals of hybridization bands were detected by chemiluminescence using horseradish peroxidase-conjugated streptavidin (Pierce, Rockford, IL) and a LAS-3000 imaging system (Fujifilm Life Science, New Haven, CT). The antisense Fluc and sense Rluc probe is complementary to the sense and antisense mRNA transcript, respectively.
2.8. Ribonuclease protection assay (RPA)
Total RNA isolated from HEK-293T cells transfected with specific pRL-RF/hTPH2–5′UTR constructs, as well as the biotinylated strand-specific RNA probe for the partial 5′-UTR (61~141), were used to perform RPA by using Ambion’s RPA III™ kit according to the manufacturer’s protocol (streamlined). Briefly, 6 μg of denatured total RNA and 20 pmol of RNA probe were mixed, denatured and hybridized in 10 μl of Hybridization Buffer III overnight at 42°C. 150 μl of RNase Digestion Buffer III with or without RNase A/T1 was added to each tube and incubated at 37°C for 30 min. Following inactivation of the RNase by adding 225 μl RNase Inactivation Solution III, the reactions were centrifuged and the supernatant was discarded. The pellet was then dissolved in 12 μl of Gel Loading Buffer II (Ambion) and loaded on a 6% denaturing (8M urea) acrylamide gel and electrophoresed at 120 V at room temperature (RT). The RNAs were then transferred, cross-linked, and detected by chemiluminescence.
2.9. Bioinformatics and data analysis
Transcription factor (TF) binding sites within the hTPH2 5′-UTR were predicted by the MatInspector (http://www.genomatix.de/products/MatInspector/index.html) and Match™ (http://www.gene-regulation.com/pub/programs.html) programs, as well as a novel FMI tool (http://promoterplot.fmi.ch/cgi-bin/dep.html). Statistics were carried out using StatView 5.0 software (SAS Institute Inc., USA). Comparisons of the luciferase activities and mRNA levels between the constructs were performed by an analysis of variance (ANOVA), followed by Fisher’s protected least-squares difference (PLSD) test
3. Results
3.1. An IRES activity in hTPH2 5′-UTR was suggested by the bicistronic reporter assay but not supported by in vitro translation
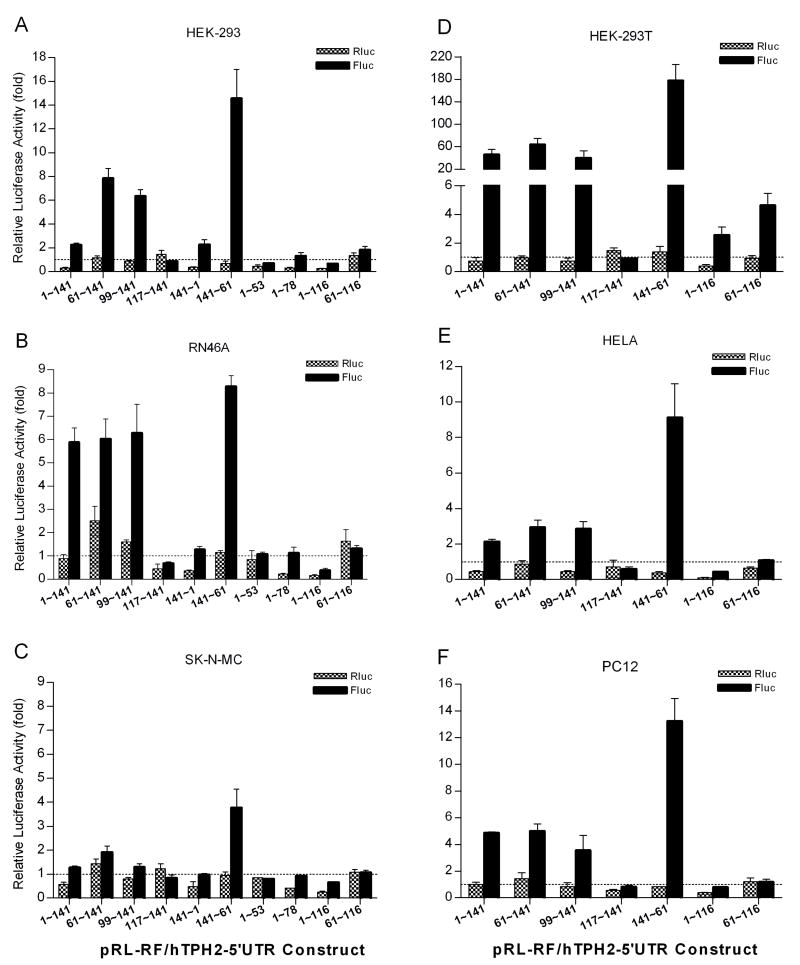
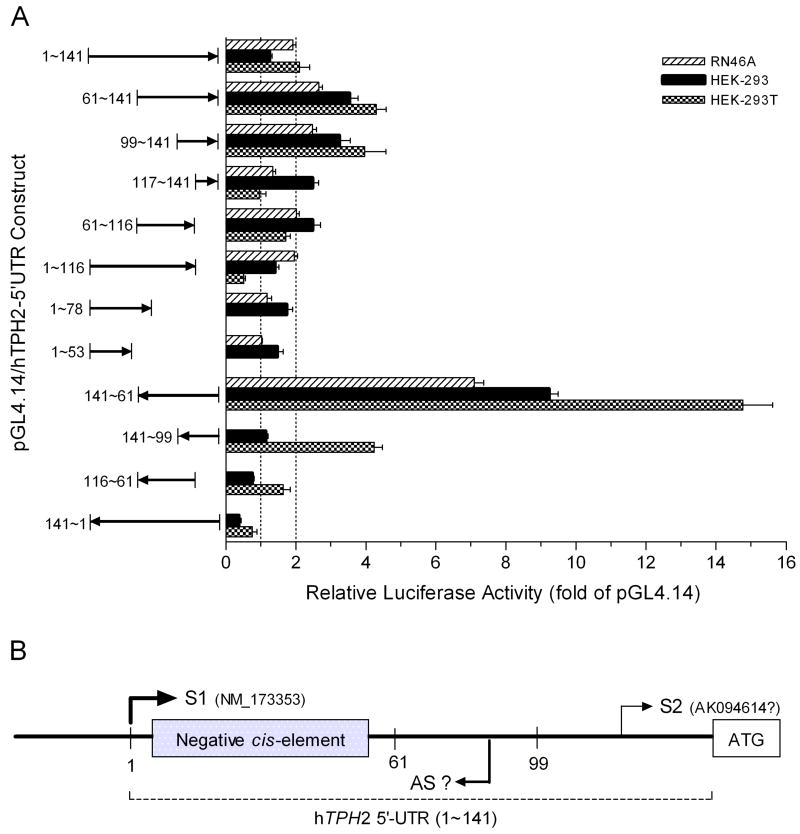
A conventional bicistronic reporter assay was first employed to test whether the hTPH2 5′-UTR harbors an IRES activity. A series of hTPH2 5′-UTR segments were cloned into a pRL-RF construct between Rluc and Fluc coding regions (Fig. 1A), and the expressed luciferase activities were measured following transient transfection in a diversity of cell lines, including RN46A, HEK-293, HEK-293T, SK-N-MC, PC12, and HELA cells. For the pRL-RF construct without the insertion of hTPH2 5′-UTR segments, the Fluc activity which represents a background expression (or noise) was significantly higher than that of the pRL-SV40 which does not encodes Fluc, but was only about 2% to 9% of the Rluc (first cistron) activity in all the cell lines (Fig. 1B), conforming to the expression feature of a bicistronic reporter without IRES activity. So, the pRL-RF construct can serve as the negative control (IRES-free), and the Fluc and Rluc activities of the pRL-RF/hTPH2–5′UTR constructs were then expressed relative to the negative control and are shown in Fig. 2. Unexpectedly, the partial antisense (141~61) construct showed increased Fluc activity ranging from about 4 to 180 fold (3.8 fold in SK-N-MC, 177.3 fold in HEK-293T, and >9 fold in other cells) above the control. Three sense constructs (1~141, 61~141, and 99~141) which contain the minimum 99~141 segment showed about 6 or more fold Fluc activity above the control in RN46A, HEK-293 and HEK-293T cells, except for the 1~141 construct in HEK-293 (only ~2 fold), while about a 2 to 5 fold increase above control was observed in HELA and PC12 cells, and even less change (<2 fold) was found in SK-N-MC cells. Similar to the 141~61 antisense construct, the three sense constructs showed the highest and lowest Fluc activity in HEK-293T (~50 fold) and SK-N-MC (<2 fold) cells, respectively. Thus, it appeared that both strands of the downstream segment (61~141) of hTPH2 5′-UTR, especially the antisense strand, could harbor a cell-dependent IRES activity.
Fig. 2.
Expression of the first (Rluc) and second (Fluc) reporters following transient transfection of pRL-RF/hTPH2–5′-UTR constructs into the HEK-293 (A), RN46A (B), SK-N-MC (C), HEK-293T (D), HELA (E) and PC12 (F) cells. The Fluc and Rluc activities (Means±SE) of the constructs are expressed as folds relative to the Fluc and Rluc activity of the control (pRL-RF without insert), respectively. The Fluc activity (%) relative to Rluc for each construct can be estimated as Fluc(fold)×Rc(%)/Rluc(fold), where Rc is the relative Fluc activity (% of Rluc) shown in Fig. 1B.
However, it has been recently documented that splicing or a cryptic promoter may affect the bicistronic reporter assay (Kozak, 2005; Holcik et al., 2005). To exclude the potential effect of such transcriptional processes on the interpretation of IRES activity, we generated the single bicistronic mRNA from four specific constructs (1~141, 61~141, 141~61, and the control) by using in vitro transcription (Fig. 3A), and performed in vitro translation by using them as the template. As shown in Fig. 3B, the control transcript expressed considerable activity of the first Rluc reporter but very low activity of the second Fluc reporter, with the Fluc/Rluc activity ratio being about 0.02 (2%). The insertion of hTPH2 5′-UTR segments in both direction between the two reporters did not enhance the activity of the downstream Fluc reporter. Hence, in conflict with the bicistronic reporter assay, the in vitro translation of single bicistronic mRNA did not support the presence of IRES activity within the hTPH2 5′-UTR.
Fig. 3.
In vitro transcription of the bicistronic mRNA (A) and the expression of luciferase activities by in vitro translation (B). The in vitro transcribed mRNAs were verified by running on a 5% denaturing (8M urea) acrylamide gel.
3.2. Multiple reporter mRNAs were transcribed in cells transfected with the bicistronic constructs
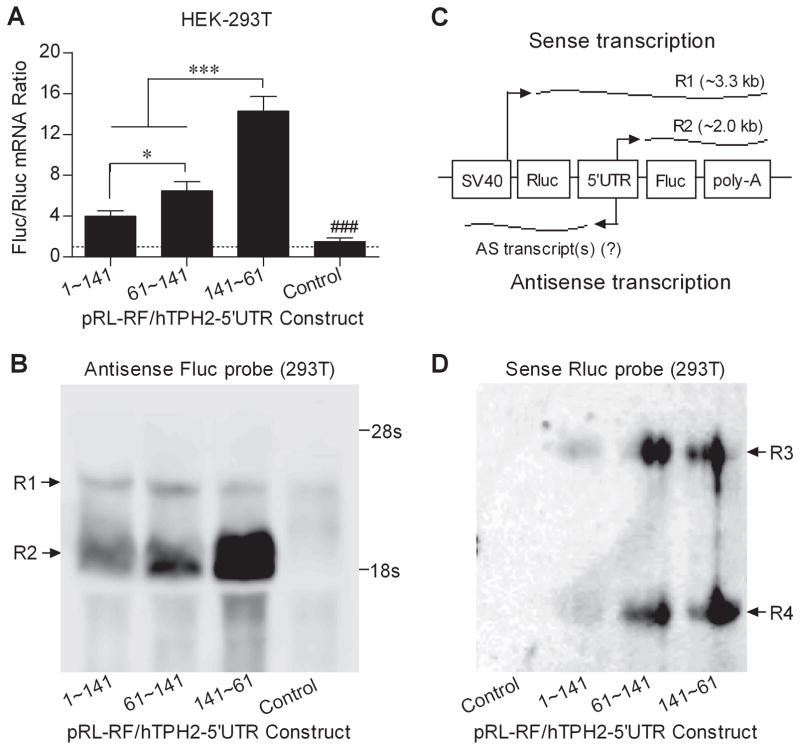
Because it can be inferred that Fluc and Rluc should share the same mRNA level if only a single mRNA comprising both reporters was transcribed, we performed quantitative real-time PCR to measure the mRNA levels of both Fluc and Rluc in HEK-293T cells transfected with specific bicistronic constructs (1~141, 61~141, 141~61, and the pRL-RF control). As shown in Fig. 4A, only the pRL-RF control showed an Flu/Rluc ratio of about 1, while the three 5′-UTR constructs expressed a much higher Fluc mRNA level than Rluc, with the Fluc/Rluc ratio ranging from about 4 to 14, and the Fluc mRNA level generally paralleled the Fluc activity. Accordingly, it is very likely that additional mRNA transcript(s) were transcribed for the second Fluc reporter.
Fig. 4.
Detection of the reporter mRNA levels and transcripts in HEK-293T cells transiently transfected with specific pRL-RF/hTPH2–5′-UTR constructs. (A) Fluc/Rluc mRNA ratio in HEK-293T cells; (B) Northern blot analysis using the antisense Fluc probe (complementary to sense Fluc); (C) Illustration of the mRNA transcripts derived from the pRL-RF/hTPH2–5′-UTR constructs; (D) Northern blot analysis using the sense Rluc probe. Levels of the reporter mRNA ratio are shown in Means±SE. The overall F=35.79 (P<0.0001) for ANOVA post-hoc test. *P<0.05, ***P<0.001 compared between the indicated constructs, ###P<0.001 compared with the constructs containing hTPH2 5′-UTR insert. Two sense transcripts (R1 and R2) reactive to antisense Fluc probe, as well as two antisense transcripts (R3 and R4) reactive to sense Rluc probe were observed. The sense transcript R2 is introduced by the insertion of hTPH2 5′-UTR with an expected size, and its abundance parallels the mRNA and activity (protein) levels of Fluc.
Northern blot was then carried out to detect the Fluc mRNA transcripts using an antisense Fluc probe. As shown in Fig. 4B, one mRNA band (R1) was observed for each of the four tested constructs, while another band (R2) of smaller size was found for the 5′-UTR constructs but not the control, and the abundance of R2 correlated well with the detected levels of Fluc mRNA and activity. While the R1 band presumably transcribed by the SV40 promoter showed an expected size of about 3.3 kb, the R2 band had an estimated size of about 2 kb, suggesting it is likely transcribed by the hTPH2 5′-UTR segments inserted between the two reporters. Hence, as illustrated in Fig. 4C, in addition to the normal bicistronic mRNA spanning both Rluc and Fluc reporters, a second mRNA comprising the Fluc reporter only was introduced by both sense and antisense hTPH2 5′-UTR segments (i.e., the hTPH2 5′-UTR exhibited a bidirectional promoter activity). Northern blot using a sense Rluc probe was also performed to detect the potential antisense transcript(s) comprising the Rluc coding region. As shown in Fig. 4D, two reactive bands (R3 and R4) with parallel abundance were observed for the 5′-UTR constructs but not for the control, and the abundance of both bands was much lower for the 1–141 construct than for the 61~141 and 141~61 constructs, generally paralleling the observed Fluc activity.
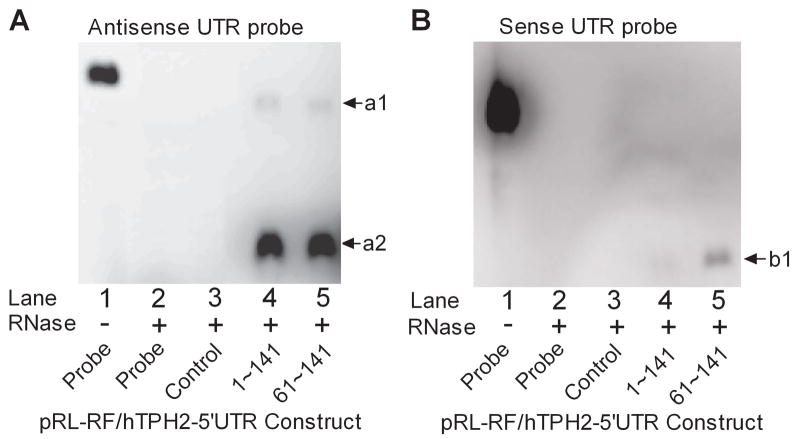
By using strand-specific RNA probes that target the hTPH2 5′-UTR (61~141), ribonuclease protection assay (RPA) was performed to validate the sense and antisense transcripts. When the antisense probe was employed, as shown in Fig. 5A, two bands (a1 and a2) protected from RNase digestion were observed for the 1–141 and 61–141 sense constructs but not for the pRL-RF control. According to the relative size and abundance, it is very likely that the larger band a1 is derived from the bicistronic mRNA (R1 in Fig. 4B) which hybridizes with the full-length probe, while the smaller band a2 represents the additional mRNA comprising only Fluc (R2 in Fig. 4B) which hybridizes with partial antisense probe. The a2 band showed much higher abundance than a1, but no remarkable abundance difference was observed between the two constructs for both a1 and a2 bands. For the RPA with the sense probe, as shown in Fig. 5B, one band (b1) protected from RNase digestion was observed for the 61–141 construct but showed weak signal for the 1–141 construct, in consistence with the Northern blot data, suggesting there may exist either one single antisense transcript or multiple antisense transcripts that yield a protected band of the same size owing to the same region of hybridization with the probe.
Fig. 5.
Ribonuclease protection assay (RPA) using the antisense (A) and sense (B) RNA probe spanning the 61~141 segment of hTPH2 5′-UTR. Total mRNA used here were isolated from HEK-293T cells transiently transfected with pRL-RF/hTPH2–5′UTR constructs. Note that both the sense and antisense probes contain partial sequence of the pGEM-T vector. An exposure time of 25 min was used so that the band a1 can be observed.
3.3. An asymmetric bidirectional promoter in hTPH2 5′-UTR was verified using a monocistronic reporter system
To confirm the promoter activity within the hTPH2 5′-UTR, we cloned specific segments of hTPH2 5′-UTR into the pGL4.14 reporter vector and measured the Fluc expression of these constructs in transiently transfected RN46A, HEK-293, and HEK-293T cells. Similar to the bicistronic reporter assay, the 141~61 antisense construct showed the highest Fluc expression, with about 7, 10, and 15 fold Fluc activity of pGL4.14 being expressed in RN46A, HEK-293, and HEK-293 cells, respectively (Fig. 6A). In addition, the sense constructs containing the minimum 99~141 segment showed Fluc expression of about 2~4 fold of pGL4.14 in all three cell lines, except for the transfection of the 1~141 construct in HEK-293. Thus, as illustrated in Fig. 6B, the downstream segment (61~141) of hTPH2 5′-UTR harbors a cell-line dependent, asymmetric bidirectional promoter, with the antisense promoter being much stronger than the sense promoter. Accordingly, the Fluc activity of each bicistronic construct shown in Fig. 2 simply reflects the corresponding promoter activity of the inserted 5′-UTR segment.
Fig. 6.
The firefly luciferase expression of the pGL4.14/hTPH2–5′UTR constructs in RN46A, HEK-293, and HEK-293T cells. Luciferase activities driven by the hTPH2 5′-UTR are expressed as folds (Means±SE) relative to the control pGL4.14 vector.
The reporter activities of the bicistronic and monocistronic constructs (Fig. 2 and 6A, respectively) were further compared between specific hTPH2 5′-UTR segments, so as to clarify segments critical for gene expression regulation. For both kinds of constructs, the 1~116 segment showed significantly lower Fluc expression than the 61~116 segment in all cell lines, except for the monocistronic construct in RN46A. Similarly, the 1~141 segment showed significantly lower Fluc activity than the 61~141 segment in all the three tested cell lines for the monocistronic constructs, and in most of cell lines (HEK-293, HEK-293T, SK-N-MC and HELA) for the bicistronic constructs. Thus, the 1~60 segment exerts a negative effect on the activity of the downstream promoter. Moreover, the 141~61 antisense construct displayed a strong promoter activity in all tested cell lines while the 141~1 antisense did not, suggesting that the 1~60 segment can silence the upstream promoter, in accordance with our previous findings (Chen et al., 2008). Interestingly, the 1~141, 1~116 and 141~1 bicistronic constructs showed significantly lower Rluc activity than the 61~141, 61~116 and 141~61 constructs, respectively, indicating that the Rluc expression was also significantly reduced by the 1~60 segment of either direction. Thus, the segment 1~60 of hTPH2 5′-UTR exerts a bidirectional negative effect on gene expression, regardless of whether it is located in the 5′ or 3′ region of the gene. We then predicted the potential transcription factor binding sites within the 61~141 segment that harbors the bidirectional promoter. Unfortunately, the prediction varied among the program algorithms (data not shown) and the exact transcription factor(s) involved in the promoter activity could not be reliably determined by this means.
4. Discussion
5′-UTRs play an important role in the regulation of gene expression by mechanisms involving both transcriptional and post-transcriptional processes. In particular, the 5′-UTRs of a number of cellular mRNAs have been reported to harbor IRES activity, which allows translation initiation under cellular stress in a cap-independent manner (Baird et al., 2006). The hTPH2 5′-UTR is only 141 bp in length and contains no upstream AUGs, but it has a GC content of ~60% and predicted free energy of −36.12 kcal/mol, and its upstream segment (8~53) strongly inhibits gene expression by both transcriptional and post-transcriptional mechanisms (Chen et al., 2008). In addition, it is known that TPH2 gene expression is modulated by specific hormones and stressors (Liang et al., 2004; Clark et al., 2005; Sanchez et al. 2005; Malek et al., 2005 and 2007; Hiroi et al., 2006; Brown et al., 2006). With these considerations, we assessed whether the hTPH2 5′-UTR harbors an IRES which can alternatively initiate mRNA translation. The existence of an IRES activity in hTPH2 5′-UTR was suggested by a conventional bicistronic reporter assay, but was not supported by in vitro translation of single bicistronic mRNA. Further investigation by qRT-PCR, Northern blot, RPA, and monocistronic reporter assay revealed that the false IRES activity suggested by the bicistronic reporter assay was primarily due to the transcription of an additional mRNA for the second reporter only, driven by a promoter within the downstream segment (61~141) of hTPH2 5′-UTR. Similarly, IRES activity was previously claimed in the 5′-UTR of some other cellular mRNAs by the bicistronic reporter assay, but it was later denied by further stringent experiments (Han and Zhang, 2002; Wang et al., 2005; Liu et al., 2005). Actually, both splicing and the presence of cryptic promoters can confound the bicistronic reporter assay and thereby result in the appearance of a false IRES activity (Kozak, 2005). In this regard, our present study strengthens the proposal that previously claimed IRES activity based on the bicistronic reporter assay should be re-examined. Further in silico analysis of the hTPH2 5′-UTR indicates that it contains terminal oligopyrimidine tracts (TOP) (shown in Fig. 1A), suggesting that hTPH2 mRNA might belong to TOP mRNAs whose translation is regulated in a growth-dependent manner (Davuluri et al., 2000).
It has been established that an abundance of bidirectional promoters exist in eukaryotic genes, with an estimate of over 20% human mRNA transcripts forming sense-antisense pairs (Trinklein et al., 2004; Chen et al., 2004). An increasing body of evidence suggests that natural antisense transcripts may play a pivotal role in eukaryotic gene expression. The sense and antisense transcript pairs potentially lead to overlapping, perfectly matching RNA-RNA hybrids, and this double-stranded RNA (dsRNA) may induce DNA methylation and gene silencing (Tufarelli et al., 2003; Mette et al., 2000). In our present study, all the reporter assay, qRT-PCR, Northern blot, and RPA consistently indicated that the downstream segment (61~141) of hTPH2 5′-UTR displays an asymmetric bidirectional (weak sense but strong antisense) promoter activity. Both the sense and antisense promoters are cell-line dependent, with the highest and lowest activity being observed in HEK-293T and SK-N-MC cells, respectively. Since a search of the Genbank database showed no coding mRNA overlapping the promoter of hTPH2, the antisense transcript of hTPH2, if it exists in vivo, is likely to be a non-coding RNA. Regardless of the splicing, this potential antisense transcript and the normal hTPH2 mRNA are expected to overlap at least for the upstream 1~60 segment of the 5′-UTR, and may thus form a dsRNA and thereby lead to DNA methylation and gene silencing. Although the existence of any natural antisense transcript is yet to be verified, it deserves our attention that the antisense promoter exhibits considerable strength in all the cell lines tested, including HEK-293, HEK-293T and HELA cells that are derived from human tissues. It is noteworthy that some genes such as c-fos, c-myb, L-myc and c-myc have antisense transcription but no stable transcript was detected (Bender et al., 1987; Krystal, 1988; Spicer and Sonenshein, 1992), and recent studies suggest the existence of numerous yet undiscovered small RNAs that are derived from natural antisense transcript (Henz et al., 2007; Kawaji and Hayashizaki, 2008; Kawaji et al., 2008). As for the hTPH2 5′-UTR, it is possible that the upstream negative segment (1~60) silences the antisense promoter, as well as the transcription of the normal hTPH2 mRNA under basal conditions. Thus, further studies are required to validate the antisense transcription of TPH2 in vivo and its role in gene expression regulation.
It is now recognized that a large fraction of human genes possess multiple promoters driving gene expression from distinct transcription start sites, and aberrant use of one promoter over another has been found to be associated with various diseases (Sandelin et al., 2007; Davuluri et al., 2008). Accordingly, the existence of a sense promoter in hTPH2 5′-UTR suggests that alternative promoter usage might be involved in the regulation of TPH2 gene expression. In fact, a second isoform of hTPH2 mRNA (Genbank accession number: AK094614) whose 5′ terminal starts from nucleotide 132 in the downstream 5′-UTR of hTPH2, has been cloned from brain tissues including the amygdala, brainstem, prefrontal cortex and hippocampus (Ota et al., 2004; Haghighi et al., 2008). In this regard, our finding of the promoter in hTPH2 5′-UTR is supported by in vivo evidence. Similarly, it has been reported that some other genes also use promoters of different strength to direct tissue type and/or developmental stage specific expression (Davuluri et al., 2008). For example, the human brain-derived neurotrophic factor (BDNF) gene has multiple functional promoters that are used tissue- and brain-region specifically, and like TPH2, it also has bidirectional transcription, resulting in the formation of dsRNA duplexes in the brain in vivo (Pruunsild et al., 2007). Both TPH2 and BDNF are neuronal genes whose expression is highly inducible. Increasing evidence suggests that the development and function of the nervous system is heavily dependent on RNA editing and the intricate spatiotemporal expression of a wide repertoire of non-coding RNAs, including micro RNAs, small nucleolar RNAs, and longer non-coding RNAs. Non-coding RNAs may provide the key to understanding the multi-tiered links between neural development, nervous system function, and neurological diseases (Mehler and Mattick, 2006). Thus, the bidirectional promoter identified in human TPH2 and BDNF genes might represent an important mechanism for the regulation of gene expression by non-coding RNA in neurons.
In agreement with our previous findings (Chen et al., 2008), this study provides further evidence for the suppression of gene expression by the upstream segment (1~60) of hTPH2 5′-UTR. Interestingly, this negative segment can inhibit the activity of both sense (downstream) and antisense (upstream) promoters within the hTPH2 5′-UTR. In other words, the inhibition of gene expression by this segment is bidirectional, albeit in vitro. The 141~61 construct (both bicistronic and monocistronic) displayed a strong promoter activity in most cell lines while the 141~1 construct did not, suggesting the 1~60 segment can completely suppress the upstream promoter, just like the case in our previous study (Chen et al., 2008). For the sense constructs, significant but <2 fold difference were observed for the Fluc activity between the 61~141 and 1~141 segments in most cases (except for the bicistronic constructs in HEK-293 and RN46A cells). The situation is very similar for the comparison between the 61~116 and 1~116 segments, indicating that the 1~60 segment can just partly, but not completely, inhibit the downstream promoter activity. Thus, the 1~60 segment of hTPH2 5′-UTR can silence the upstream promoter but has less inhibitory effect on the downstream promoter. It deserves our attention that in bicistronic constructs the expression of the first reporter Rluc was also substantially but not completely repressed by the 1~60 segment of either direction, irrespective of the cell line, suggesting that this segment can also hamper the expression of the upstream cistron. However, the 1–141 and 61–141 constructs displayed a similar level of the bicistronic mRNA (band R1 in Fig.4B), but apparently more abundant than the empty control, suggesting that the repression of Rluc expression by the 1–60 segment may involve posttranscriptional processes including mRNA stability and translation efficiency. Because it has been well-documented that 3′-UTR plays a critical role in the regulation of mRNA stability and translation efficiency, we speculate that the repressed Rluc activity in the 1–141 construct might be conferred by the alteration in the length and context of the 3′-UTR of Rluc following the insertion of TPH2 5′-UTR downstream the Rluc. It is possible that this insertion makes the mRNA more stable but hinders the translation of mRNA. Thus, the inhibition of gene expression by the 1~60 segment is independent of the cell line and its location in 5′ and 3′ region of the gene, indicating that this segment may function as a critical cis-element for the silencing of the upstream (especially the neighboring) promoter, just like an on-off switch for gene expression regulation. The mechanism(s) by which this segment inhibits gene expression is yet to be determined; however, epigenetic mechanisms such as RNA-directed DNA methylation and chromatin modification might be involved (Tufarelli et al., 2003; Mette et al., 2000), because a dsRNA might be formed due to the bidirectional promoter within the downstream 5′-UTR. Also, the 16~45 segment is predicted to be a target site of a miRNA named hsa-mir-370 (miRBase accession number: MI0000778) by the RegRNA program (http://regrna.mbc.nctu.edu.tw/). Consistently, a search of CpG island using the updated rules from Takai and Jones (http://cpgislands.usc.edu/cpg.aspx) suggested a number of potential CpG DNA-methylation sites within the TPH2 5′-UTR, implying that DNA methylation may be involved in the regulation of TPH2 gene expression.
In summary, our present study has provided evidence for the presence of an asymmetric bidirectional promoter, but not IRES, in the downstream segment (61~141) of the hTPH2 5′-UTR. It also provides further evidence for a potent suppression of gene expression by the upstream segment (1~60) of hTPH2 5′-UTR. These findings shed light on the regulation of TPH2 gene expression, and further advance our understanding of brain functions involved in psychiatric disorders and behavioral traits related to the 5-HT system.
Acknowledgments
Funding
This study was supported by AA016194 (GMM), DA016606 (GMM), MH077995 (GMM), and RR00168.
The authors would like to thank Mrs. Hong Yang for the excellent technician support. We also thank Dr. Bin Jia, Dr. Shuji Sato, Dr. Swee Kee Wong, and Dr. I-Chueh Huang at HMS/NEPRC for their helpful suggestion and kind help for this study. We are grateful to Mrs. Jennifer Carter for the administrative support.
Footnotes
Conflict of Interest Statement
Dr. Guo-Lin Chen and Dr. Gregory M. Miller do not have any conflict of interest to report.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA. 2006;12:1755–85. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender TP, Thompson CB, Kuehl WM. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987;237:1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- Brown HJ, Henderson LA, Keay KA. Hypotensive but not normotensive haemorrhage increases tryptophan hydroxylase-2 mRNA in caudal midline medulla. Neurosci Lett. 2006;398:314–318. doi: 10.1016/j.neulet.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Brown SM, et al. A regulatory variant of the human tryptophan hydroxylase-2 gene biases amygdala reactivity. Mol Psychiatry. 2005;10:884–888. doi: 10.1038/sj.mp.4001716. [DOI] [PubMed] [Google Scholar]
- Calcagno E, Canetta A, Guzzetti S, Cervo L, Invernizzi RW. Strain differences in basal and post-citalopram extracellular 5-HT in the mouse medial prefrontal cortex and dorsal hippocampus, relation with tryptophan hydroxylase-2 activity. J Neurochem. 2007;103:1111–20. doi: 10.1111/j.1471-4159.2007.04806.x. [DOI] [PubMed] [Google Scholar]
- Canli T, Congdon E, Gutknecht L, Constable RT, Lesch KP. Amygdala responsiveness is modulated by tryptophan hydroxylase-2 gene variation. J Neural Transm. 2005;112:1479–1485. doi: 10.1007/s00702-005-0391-4. [DOI] [PubMed] [Google Scholar]
- Cervo L, et al. Genotype-dependent activity of tryptophan hydroxylase-2 determines the response to citalopram in a mouse model of depression. J Neurosci. 2005;25:8165–72. doi: 10.1523/JNEUROSCI.1816-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GL, Novak MA, Hakim S, Xie Z, Miller GM. Tryptophan hydroxylase-2 gene polymorphisms in rhesus monkeys, association with hypothalamic-pituitary-adrenal axis function and in vitro gene expression. Mol Psychiatry. 2006;11:914–928. doi: 10.1038/sj.mp.4001870. [DOI] [PubMed] [Google Scholar]
- Chen GL, Vallender EJ, Miller GM. Functional characterization of the human TPH2 5′ regulatory region, untranslated region and polymorphisms modulate gene expression in vitro. Hum Genet. 2008;122:645–57. doi: 10.1007/s00439-007-0443-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, et al. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004;32:4812–20. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichon S, et al. Brain-specific tryptophan hydroxylase 2 (TPH2), a functional Pro206Ser substitution and variation in the 5′-region are associated with bipolar affective disorder. Hum Mol Genet. 2008;17:87–97. doi: 10.1093/hmg/ddm286. [DOI] [PubMed] [Google Scholar]
- Clark JA, Pai LY, Flick RB, Rohrer SP. Differential hormonal regulation of tryptophan hydroxylase-2 mRNA in the murine dorsal raphe nucleus. Biol Psychiatry. 2005;57:943–946. doi: 10.1016/j.biopsych.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Coon H, et al. Possible association between autism and variants in the brain-expressed tryptophan hydroxylase gene (TPH2) Am J Med Genet B Neuropsychiatr Genet. 2005;135:42–46. doi: 10.1002/ajmg.b.30168. [DOI] [PubMed] [Google Scholar]
- Côté F, et al. Disruption of the nonneuronal tph1 gene demonstrates the importance of peripheral serotonin in cardiac function. Proc Natl Acad Sci USA. 2003;100:13525–30. doi: 10.1073/pnas.2233056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Suzuki Y, Sugano S, Plass C, Huang TH. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008;24:167–77. doi: 10.1016/j.tig.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Davuluri RV, Suzuki Y, Sugano S, Zhang MQ. CART classification of human 5′ UTR sequences. Genome Res. 2000;10:1807–1816. doi: 10.1101/gr.gr-1460r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinan TG. Serotonin and the regulation of hypothalamic-pituitary-adrenal axis function. Life Sci. 1996;58:1683–1694. doi: 10.1016/0024-3205(96)00066-5. [DOI] [PubMed] [Google Scholar]
- Goertzel BN, Pennachin C, de Souza Coelho L, Gurbaxani B, Maloney EM, Jones JF. Combinations of single nucleotide polymorphisms in neuroendocrine effector and receptor genes predict chronic fatigue syndrome. Pharmacogenomics. 2006;7:475–483. doi: 10.2217/14622416.7.3.475. [DOI] [PubMed] [Google Scholar]
- Gotlib IH, Joormann J, Minor KL, et al. HPA axis reactivity: a mechanism underlying the associations among 5-HTTLPR, stress, and depression. Biol Psychiatry. 2008;63:847–851. doi: 10.1016/j.biopsych.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutknecht L, et al. Tryptophan hydroxylase-2 gene variation influences personality traits and disorders related to emotional dysregulation. Int J Neuropsychopharmacol. 2007;10:309–20. doi: 10.1017/S1461145706007437. [DOI] [PubMed] [Google Scholar]
- Haghighi F, et al. Genetic architecture of the human tryptophan hydroxylase 2 Gene: existence of neural isoforms and relevance for major depression. Mol Psychiatry. 2008;13:813–20. doi: 10.1038/sj.mp.4002127. [DOI] [PubMed] [Google Scholar]
- Han B, Zhang JT. Regulation of gene expression by internal ribosome entry sites or cryptic promoters, the eIF4G story. Mol Cell Biol. 2002;22:7372–84. doi: 10.1128/MCB.22.21.7372-7384.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey M, et al. Support for the involvement of TPH2 gene in affective disorders. Mol Psychiatry. 2004;9:980–981. doi: 10.1038/sj.mp.4001557. [DOI] [PubMed] [Google Scholar]
- Henz SR, et al. Distinct expression patterns of natural antisense transcripts in Arabidopsis. Plant Physiol. 2007;144:1247–55. doi: 10.1104/pp.107.100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann MJ, et al. Additive effects of serotonin transporter and tryptophan hydroxylase-2 gene variation on emotional processing. Cereb Cortex. 2007;17:1160–3. doi: 10.1093/cercor/bhl026. [DOI] [PubMed] [Google Scholar]
- Hiroi R, McDevitt RA, Neumaier JF. Estrogen selectively increases tryptophan hydroxylase-2 mRNA expression in distinct subregions of rat midbrain raphe nucleus, association between gene expression and anxiety behavior in the open field. Biol Psychiatry. 2006;60:288–295. doi: 10.1016/j.biopsych.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Holcik M, Graber T, Lewis SM, Lefebvre CA, Lacasse E, Baird S. Spurious splicing within the XIAP 5′ UTR occurs in the Rluc/Fluc but not the betagal/CAT bicistronic reporter system. RNA. 2005;11:1605–9. doi: 10.1261/rna.2158605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jollant F, et al. The influence of four serotonin-related genes on decision-making in suicide attempters. Am J Med Genet B Neuropsychiatr Genet. 2007;144:615–24. doi: 10.1002/ajmg.b.30467. [DOI] [PubMed] [Google Scholar]
- Kawaji H, Hayashizaki Y. Exploration of small RNAs. PLoS Genet. 2008;4:e22. doi: 10.1371/journal.pgen.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33:6593–602. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal G, et al. Multiple mechanisms for transcriptional regulation of the myc gene family in small-cell lung cancer. Mol Cell Biol. 1988;8:3373–3381. doi: 10.1128/mcb.8.8.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Wessel JH, 3rd, Iuvone PM, Tosini G, Fukuhara C. Diurnal rhythms of tryptophan hydroxylase 1 and 2 mRNA expression in the rat retina. Neuroreport. 2004;15:1497–1500. doi: 10.1097/01.wnr.0000131007.59315.66. [DOI] [PubMed] [Google Scholar]
- Liu Z, Dong Z, Han B, Yang Y, Liu Y, Zhang JT. Regulation of expression by promoters versus internal ribosome entry site in the 5′-untranslated sequence of the human cyclin-dependent kinase inhibitor p27kip1. Nucleic Acids Res. 2005;33:3763–71. doi: 10.1093/nar/gki680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Lara C, et al. Effect of tryptophan hydroxylase-2 gene variants on suicide risk in major depression. Biol Psychiatry. 2007;62:72–80. doi: 10.1016/j.biopsych.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Lopez VA, Detera-Wadleigh S, Cardona I, Kassem L, McMahon FJ The National Institute of Mental Health Genetics Initiative Bipolar Disorder Consortium. Nested association between genetic variation in tryptophan hydroxylase II, bipolar affective disorder, and suicide attempts. Biol Psychiatry. 2007;61:181–6. doi: 10.1016/j.biopsych.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Lowry CA. Functional subsets of serotonergic neurones: implications for control of the hypothalamic-pituitary-adrenal axis. J Neuroendocrinol. 2002;14:911–23. doi: 10.1046/j.1365-2826.2002.00861.x. [DOI] [PubMed] [Google Scholar]
- Malek ZS, Dardente H, Pevet P, Raison S. Tissue-specific expression of tryptophan hydroxylase mRNAs in the rat midbrain, anatomical evidence and daily profiles. Eur J Neurosci. 2005;22:895–901. doi: 10.1111/j.1460-9568.2005.04264.x. [DOI] [PubMed] [Google Scholar]
- Malek ZS, Sage D, Pevet P, Raison S. Daily rhythm of tryptophan hydroxylase-2 mRNA within Raphe neurons is induced by corticoid daily surge and modulated by enhanced locomotor activity. Endocrinology. 2007;148:5165–72. doi: 10.1210/en.2007-0526. [DOI] [PubMed] [Google Scholar]
- Maron E, et al. Association study of tryptophan hydroxylase 2 gene polymorphisms in panic disorder. Neurosci Lett. 2007;411:180–184. doi: 10.1016/j.neulet.2006.09.060. [DOI] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mössner R, et al. Transmission disequilibrium of polymorphic variants in the tryptophan hydroxylase-2 gene in children and adolescents with obsessive-compulsive disorder. Int J Neuropsychopharmacol. 2006;9:437–442. doi: 10.1017/S1461145705005997. [DOI] [PubMed] [Google Scholar]
- Nielsen DA, et al. TPH2 and TPH1: Association of Variants and Interactions with Heroin Addiction. Behav Genet. 2008;38:133–50. doi: 10.1007/s10519-007-9187-7. [DOI] [PubMed] [Google Scholar]
- Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–5. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- Patel PD, Bochar DA, Turner DL, Meng F, Mueller HM, Pontrello CG. Regulation of tryptophan hydroxylase-2 gene expression by a bipartite RE-1 silencer of transcription/neuron restrictive silencing factor (REST/NRSF) binding motif. J Biol Chem. 2007;282:26717–24. doi: 10.1074/jbc.M705120200. [DOI] [PubMed] [Google Scholar]
- Peters EJ, Slager SL, McGrath PJ, Knowles JA, Hamilton SP. Investigation of serotonin-related genes in antidepressant response. Mol Psychiatry. 2004;9:879–89. doi: 10.1038/sj.mp.4001502. [DOI] [PubMed] [Google Scholar]
- Reuter M, Hennig J, Amelang M, Montag C, Korkut T, Hueweler A, Stürmer T. The role of the TPH1 and TPH2 genes for nicotine dependence: a genetic association study in two different age cohorts. Neuropsychobiology. 2007a;56:47–54. doi: 10.1159/000110728. [DOI] [PubMed] [Google Scholar]
- Reuter M, Kuepper Y, Hennig J. Association between a polymorphism in the promoter region of the TPH2 gene and the personality trait of harm avoidance. Int J Neuropsychopharmacol. 2007b;10:401–4. doi: 10.1017/S1461145706007073. [DOI] [PubMed] [Google Scholar]
- Sanchez RL, Reddy AP, Centeno ML, Henderson JA, Bethea CL. A second tryptophan hydroxylase isoform, TPH-2 mRNA, is increased by ovarian steroids in the raphe region of macaques. Brain Res Mol Brain Res. 2005;135:194–203. doi: 10.1016/j.molbrainres.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet. 2007;8:424–36. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- Sheehan K, et al. Tryptophan hydroxylase 2 (TPH2) gene variants associated with ADHD. Mol Psychiatry. 2005;10:944–949. doi: 10.1038/sj.mp.4001698. [DOI] [PubMed] [Google Scholar]
- Shishkina GT, Kalinina TS, Dygalo NN. Up-regulation of tryptophan hydroxylase-2 mRNA in the rat brain by chronic fluoxetine treatment correlates with its antidepressant effect. Neuroscience. 2007;150:404–12. doi: 10.1016/j.neuroscience.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Smith AK, White PD, Aslakson E, Vollmer-Conna U, Rajeevan MS. Polymorphisms in genes regulating the HPA axis associated with empirically delineated classes of unexplained chronic fatigue. Pharmacogenomics. 2006;7:387–394. doi: 10.2217/14622416.7.3.387. [DOI] [PubMed] [Google Scholar]
- Spicer DB, Sonenshein GE. An antisense promoter of the murine c-myc gene is localized within intron 2. Mol Cell Biol. 1992;12:1324–9. doi: 10.1128/mcb.12.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–6. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufarelli C, et al. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet. 2003;34:157–65. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- Tzvetkov MV, Brockmöller J, Roots I, Kirchheiner J. Common genetic variations in human brain-specific tryptophan hydroxylase-2 and response to antidepressant treatment. Pharmacogenet Genomics. 2008;18:495–506. doi: 10.1097/FPC.0b013e3282fb02cb. [DOI] [PubMed] [Google Scholar]
- Walitza S, et al. Transmission disequilibrium of polymorphic variants in the tryptophan hydroxylase-2 gene in attention-deficit/hyperactivity disorder. Mol Psychiatry. 2005;10:1126–1132. doi: 10.1038/sj.mp.4001734. [DOI] [PubMed] [Google Scholar]
- Walther DJ, Bader M. A unique central tryptophan hydroxylase isoform. Biochem Pharmacol. 2003;66:1673–1680. doi: 10.1016/s0006-2952(03)00556-2. [DOI] [PubMed] [Google Scholar]
- Walther DJ, et al. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science. 2003;299:76. doi: 10.1126/science.1078197. [DOI] [PubMed] [Google Scholar]
- Wang Z, Weaver M, Magnuson NS. Cryptic promoter activity in the DNA sequence corresponding to the pim-1 5′-UTR. Nucleic Acids Res. 2005;33:2248–58. doi: 10.1093/nar/gki523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Beaulieu JM, Sotnikova TD, Gainetdinov RR, Caron MG. Tryptophan hydroxylase-2 controls brain serotonin synthesis. Science. 2004;305:217. doi: 10.1126/science.1097540. [DOI] [PubMed] [Google Scholar]
- Zhou Z, et al. Haplotype-based linkage of tryptophan hydroxylase 2 to suicide attempt, major depression, and cerebrospinal fluid 5-hydroxyindoleacetic acid in 4 populations. Arch Gen Psychiatry. 2005;62:1109–1118. doi: 10.1001/archpsyc.62.10.1109. [DOI] [PubMed] [Google Scholar]
- Zill P, et al. SNP and haplotype analysis of a novel tryptophan hydroxylase isoform (TPH2) gene provide evidence for association with major depression. Mol Psychiatry. 2004a;9:1030–1036. doi: 10.1038/sj.mp.4001525. [DOI] [PubMed] [Google Scholar]
- Zill P, Buttner A, Eisenmenger W, Moller HJ, Bondy B, Ackenheil M. Single nucleotide polymorphism and haplotype analysis of a novel tryptophan hydroxylase isoform (TPH2) gene in suicide victims. Biol Psychiatry. 2004b;56:581–586. doi: 10.1016/j.biopsych.2004.07.015. [DOI] [PubMed] [Google Scholar]