Abstract
Vesicle transport is essential for the movement of proteins, lipids and other molecules between membrane compartments within the cell. The role of the class VI myosins in vesicular transport is especially intriguing because they are the only class that has been shown to move “backwards” towards the minus end of actin filaments1. Myosin VI is found in distinct intracellular locations and implicated in processes such as endocytosis2,3, exocytosis, maintenance of Golgi morphology4,5 and cell movement6. We have shown that the C-terminal tail is the key targeting region and have identified three binding sites: a WWY motif for Dab2 binding, a RRL motif for GIPC/Optineurin binding and a site that binds specifically and with high affinity (Kd 0.3 μM) to PIP2-containing liposomes. This is the first demonstration that myosin VI binds lipid membranes. Lipid binding induces a large structural change in the tail (31% increase in helicity) and when associated with lipid vesicles it can dimerise. In vivo targeting and recruitment of myosin VI to clathrin-coated structures (CCSs) at the plasma membrane is mediated by Dab2 and PIP2 binding.
Dab2 is a myosin VI binding partner present on endocytic CCSs at the plasma membrane7,8. To establish whether binding to Dab2 is involved in targeting myosin VI to CCSs we tested an extensive series of myosin VI tail deletion fragments and point mutants using the mammalian 2-hybrid assay7. A relatively conservative single amino acid change from a tryptophan to a leucine (W1184L, WWY→WLY) was found to abolish myosin VI binding to Dab2 (Fig. 1a). ‘Pull down’ experiments using GST-tagged wild type myosin VI tail or tail containing the WWY→WLY mutation together with in vitro translated Dab2 confirmed the identity of the Dab2 binding site (Fig. 1c). To check whether the Dab2 binding site was essential for targeting myosin VI to CCSs in vivo we over-expressed GFP-tagged mutant tail constructs in HeLa cells. We observed that myosin VI containing a mutated Dab2 binding site (WWY→WLY) was not targeted to CCSs (Fig. 1d,e). It was previously shown2,8 that the presence of a large insert just before the globular C-terminal domain (Fig. S1) in conjunction with Dab2 binding was also required for targeting myosin VI to CCSs at the plasma membrane.
Figure 1.
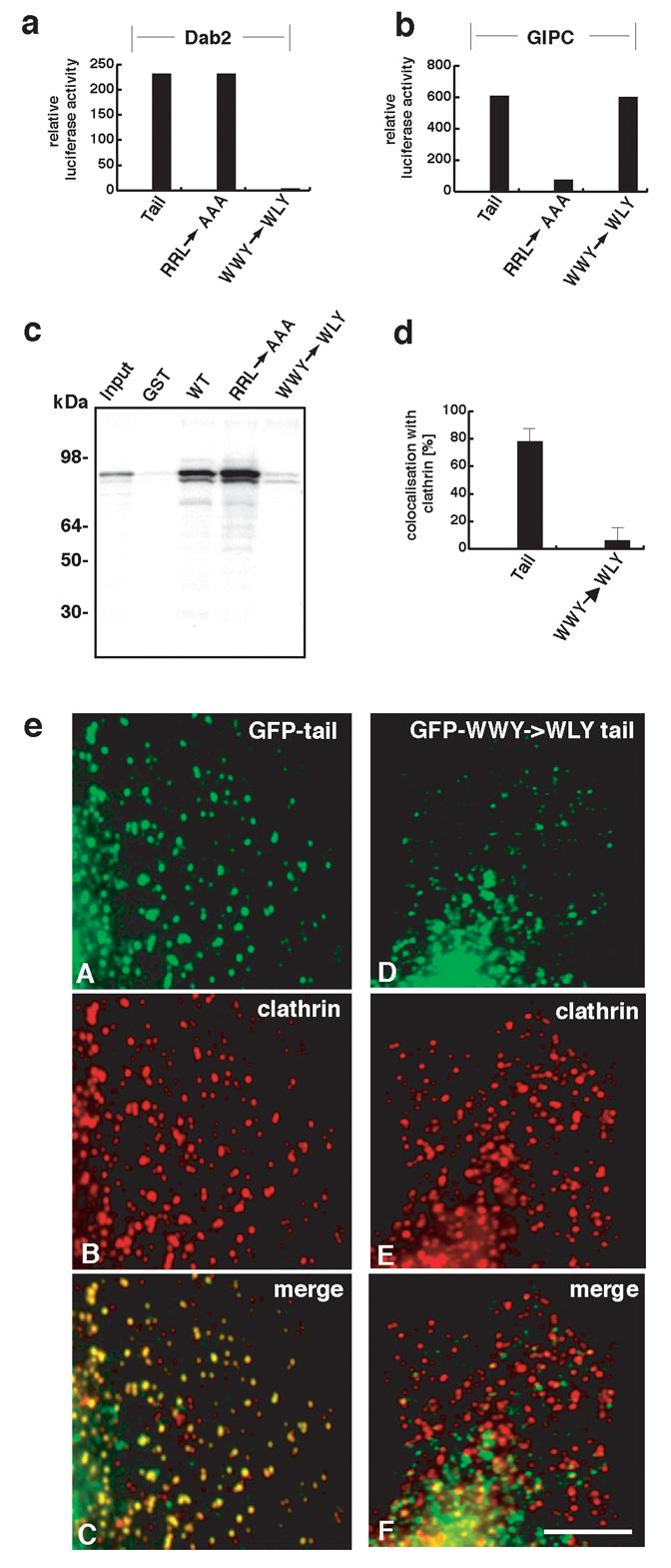
Two distinct “hot spots” for binding Dab2 and GIPC were identified on the myosin VI tail and it is shown that an intact Dab2 binding site is required for targeting myosin VI to CCSs. (a) and (b) Deletion and mutated constructs of the chicken myosin VI tail were screened for binding to Dab2 and GIPC using the mammalian 2-hybrid system. The mutation W1184L (WWY→WLY) in the tail abolishes binding to Dab2 whilst preserving binding to GIPC. Mutation of the GIPC binding site (amino acids 1107-1109, RRL→AAA) abolishes GIPC binding in addition to optineurin binding but has no effect on Dab2 binding. These results indicate that there are two distinct sites for binding partners on the myosin VI tail. (c) Autoradiograph of [35S]Dab2 “pull-down” fractions. Recombinant, purified GST-tagged myosin VI tail containing the WWY→WLY mutation does not bind [35S]Dab2 whereas GST-tagged myosin VI tail (both wild-type and containing the mutations for GIPC/optineurin binding (RRL→AAA) show binding to [35S]Dab2. GST alone was included as a control. (d) Quantitation of the colocalisation of endogenous clathrin with GFP-tagged myosin VI tail (+ LI) (both wild type and Dab2 binding mutant) by measuring the immunofluorescence staining shown in (e). Myosin VI containing the mutated Dab2 binding site (WWY→WLY) does not colocalise with clathrin. (e) Immunofluorescence staining of over-expressed GFP-labelled myosin VI whole tail (+LI) constructs and endogenous clathrin in HeLa cells. Panels A and D show the GFP-labelled wild-type tail and GFP-tail containing WWY→WLY mutation respectively. Panels B and E show the endogenous clathrin staining and C and F show the merged images. Dab2 binding is required for targeting the myosin VI isoform with the large insert (+LI) to clathrin-coated pits and vesicles. Note the tail with the WWY→WLY mutation localises to punctate structures but since they are not clathrin-coated vesicles they are probably secretory or uncoated vesicles. Scale bar = 5 μm.
GIPC is another myosin VI binding partner9 that is found in both clathrin-coated10 and uncoated endocytic vesicles3. To test the involvement of GIPC in targeting myosin VI to CCSs we mapped the GIPC binding site on the myosin VI tail by deletion and alanine scanning mutagenesis. We observed that the mutation RRL to AAA in the C-terminal tail domain (aa 1107-1109) (Fig. 1b) abolished GIPC binding, but had no effect on Dab2 binding (Fig. 1a,b). Optineurin, a myosin VI binding protein associated with the Golgi complex and secretion, also specifically binds to the RRL binding site in the myosin VI tail4. Although phosphorylation of the threonines in the TINT sequence 15 residues upstream of RRL (Fig. S1b) regulated Optineurin binding to the RRL site4 we observed that TINT phosphorylation had no effect on GIPC binding to this site (data not shown).
Since a number of endocytic and actin cytoskeleton proteins such as AP-2, Dab2, gelsolin and cofilin bind to phospholipids we tested whether the myosin VI tail would bind lipids. “Mixed brain’ liposomes were incubated with whole myosin VI tail (tail), C-terminal globular tail fragment (CT), and full-length myosin VI (with Ca2+) and after centrifugation these proteins were present in the lipid pellet fraction, indicating strong lipid binding (Fig. 2a, b). Without liposomes, all the myosin VI constructs remained in the supernatant after centrifugation. To map the lipid-binding region, myosin VI tail constructs were tested for liposome binding using the sedimentation assay (Fig. 2c). Although N-terminal helical tail fragments with and without the large insert (NT+LI, NT) scored a low but significant level of homology with the lipid binding BAR domain of D-amphiphysin (Brian Peter & Harvey McMahon, personal communication) they did not bind to liposomes. Similarly a construct comprising the last 142 aa of the C-terminal tail (CT 1134-1276) (Fig. S1) did not bind liposomes (Fig. 2c). Thus we searched for potential liposome binding sequences between aa 1059 and 1134 in the myosin VI C-terminal tail. Deletion of this 75aa region (ΔPIP2 tail) leads to a 70% reduction in lipid binding. Within this region there is a stretch of basic/hydrophobic residues (RRLKVYHAWKSKNKKR, aa 1107-1122) and a series of single point mutations in the basic residues were made but they had no effect on liposome binding, nor did double mutations in this region (data not shown). However, both myosin VI C-terminal tail and whole tail constructs containing four point mutations in this region (WKSKNKKR→WASANNNR, aa 1115-1122) were found to have severely reduced liposome binding (by at least 70%) (Fig. 2c).
Figure 2.
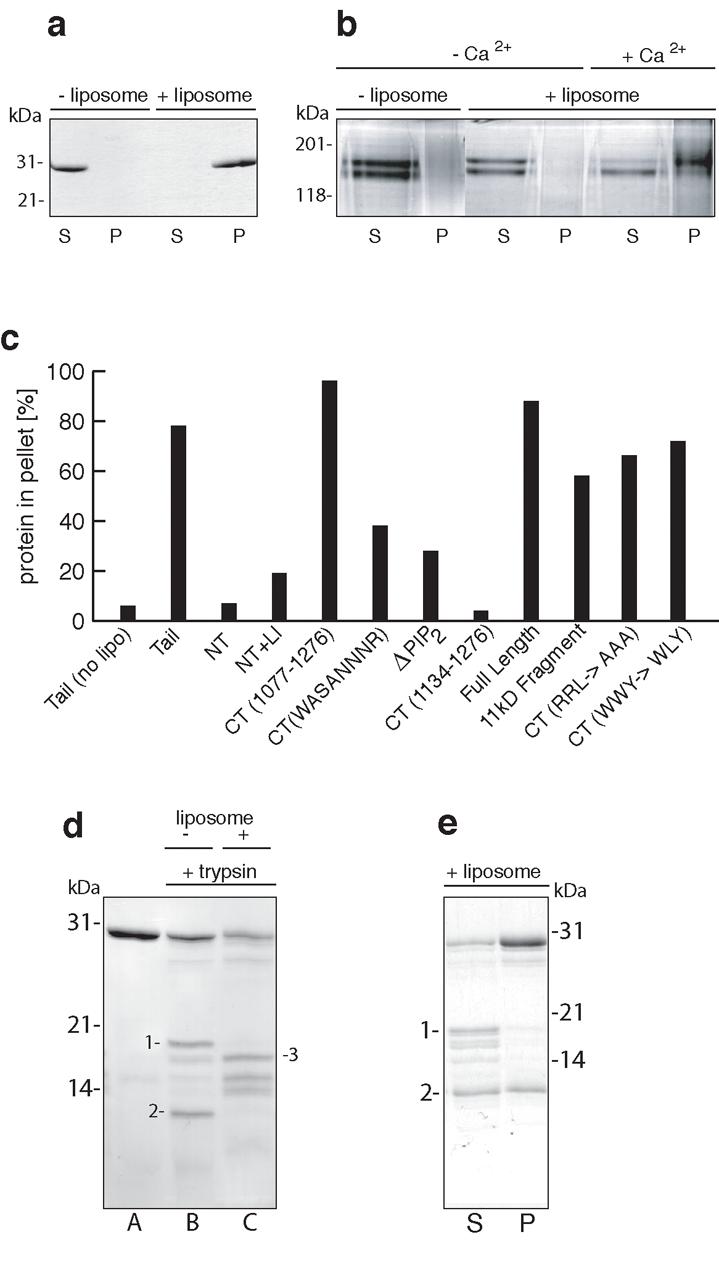
The C-terminal half of the myosin VI tail binds to liposomes and is composed of two independent domains. (a), (b), and (e) myosin VI tail fragments binding to mixed brain liposomes were tested in sedimentation assays and analysed by 10 or 15% acrylamide SDS-PAGE. “P” denotes pellet and “S” supernatant. (a) The C-terminal half of the tail without the large insert (CT) binds liposomes. The CT tail (aa1077-1276) is in the pellet plus liposomes, and in the supernatant without liposomes. (b) Full-length myosin VI binds to liposomes only in the presence of calcium (0.3 mM free Ca2+). Note in the presence of calcium the upper band of the doublet (intact myosin VI) preferentially binds to liposomes. (c) The lipid binding site is within aa 1077-1133 of the myosin VI C-terminal tail. The % total protein in pellet fractions after sedimentation with mixed brain liposomes were measured by quantitation of SDS-PAGE gels. Each binding experiment was repeated at least three times and a typical experiment is shown. Tail fragments tested were: N-terminal tail without or with the large insert (NT: aa 845-1032, NT+LI: aa 845-1060), the C-terminal tail without the large insert (CT aa1077-1276), the ΔPIP2 tail (aa 845-1276 with residues 1059-1134 deleted) and the following mutations in CT: RRL→AAA, WWY→WLY, WKSKNKKR→WASANNNR (aa 1115-1122), the last 142 amino acids of the C-terminal tail (CT 1134-1276), and the 11 kD tryptic digest fragment (domain 2). (d) SDS-PAGE (15% acrylamide) gel showing C-terminal myosin VI tail (CT) (lane A) and tryptic digests of CT in the absence (lane B) and presence (lane C) of liposomes. Digestion without liposomes results in cleavage at site 1 (see Fig. S1b) and yields two fragments (18kDa and 11kDa: labelled 1 and 2 in lane B) whereas digestion in the presence of liposomes leads to cleavage at site 2 (Fig. S1b) and new fragments (fragment 3 is labelled in lane C). (e) Domain 2 in the C-terminal tail of myosin VI contains the lipid binding site. The tryptic digest of CT (in the absence of liposomes as in figure 2d, lane B) was incubated with liposomes and centrifuged. Both undigested CT and domain 2 are in the pellet fraction indicating they bind to liposomes.
We probed the structural organization of the C-terminal cargo-binding domain of myosin VI using limited proteolysis. We observed that, like the myosin V C-terminal tail16, the myosin VI C-terminal domain is extremely susceptible to tryptic digestion, rapidly yielding 2 fragments (bands 1 and 2 in Fig. 2d). Sequence analysis revealed that cleavage occurred at R1122 (site 1 in Fig. S1b) leading to 18 kDa and 11 kDa fragments (bands 1 and 2 in Fig. 2d) that can be separated under non-dissociating conditions (see methods). In the presence of liposomes, the tryptic cleavage pattern is different; cleavage does not occur at R1122, but at K1132 (lane C, band 3 in Fig. 2d and site 2 in Fig. S1b) presumably because lipid binding to the potential binding site (KSKNKKR) protects site 1 from tryptic cleavage. In the lipid binding assay we observed that the 11 kDa fragment (domain 2) binds to liposomes whereas the 18 kDa fragment (domain 1) does not (Fig. 2e) confirming the identity of the lipid binding region previously established in the mutational analysis (Fig. 2c).
When lipid binding to full-length myosin VI was tested it was observed that myosin VI only bound to liposomes in the presence of calcium (Fig. 2b). Since calcium has no apparent affect on lipid binding to the myosin VI tail or tail fragments it is likely that calcium must have some effect on the conformation of the whole molecule, probably binding to the calmodulin molecules in the neck/linker region in the motor domain. Frequently, full-length myosin VI preparations expressed and purified from baculovirus/insect cells migrate as a closely spaced doublet on SDS-PAGE (Fig. 2b) and over time the top band decreases and the bottom band increases, most likely reflecting proteolysis. Since the N-terminus of the myosin VI molecule with its His-tag is still intact, proteolysis must occur at the extremely sensitive tryptic cleavage site (R1122) identified near the lipid-binding region in the C-terminal tail domain (Fig. S1b). This proposal is supported by the observation that more of the top (intact) myosin VI band (90%) is bound to liposomes compared with the lower band (40%) in the sedimentation assays (Fig.2b) and this is consistent with the lipid binding site being localised to this region of the C-terminal tail.
To determine whether myosin VI exhibits specificity for different phosphoinositides, lipid binding to the C-terminal tail was tested using liposomes containing 10% of the series of phosphoinositides shown in Fig. 3a. The C-terminal tail binds with high specificity to PIP2-containing liposomes, and exhibits little or no binding to the other phosphoinositides (Fig. 3a). To establish the concentration of PIP2 necessary for liposome binding to myosin VI, synthetic liposomes were prepared with a range of PIP2 concentrations. The minimal concentration of PIP2 in the synthetic liposomes required for myosin VI binding was about 2-5% (Fig. 3b). Liposomes prepared from mixed brain lipids (“folch” fraction I) contain a minimum of 10% PIP2 and exhibit the strongest binding to the myosin VI tail. The strength of lipid binding was determined by FRET (fluorescence resonance energy transfer) between trytophan groups on the myosin VI tail and dansyl-labelled PIP2-containing liposomes (Fig. 3c). Addition of increasing amounts of tail protein to a fixed amount of liposomes results in a hyberbolic curve, indicating that the binding is non-cooperative (Fig. 3c). A Kd of 0.3μM was measured indicating that the C-terminal tail region binds strongly to PIP2-liposomes. To investigate whether lipid binding had any effect on the secondary structure of the C-terminal myosin VI tail we measured its circular dichroic (CD) spectrum with and without liposomes. The CD spectrum of the C-terminal tail alone indicates a protein with a mixed secondary structure composition (Fig. 4a dashed line). However with liposomes, the CD spectrum altered to that of an α-helical protein (with double minima at 208 and 222 nm) within 5 minutes (Fig. 4a solid line). The α-helical signal (ellipticity at wavelength 222nm) increased by 31% on addition of liposomes, indicating that lipid binding induces significant a-helix formation in the C-terminal region of the myosin VI tail. The liposomes themselves showed no circular dichroic signal in the range of wavelength used (200-250 nm) and as expected there was no significant change in the CD spectra of the N-terminal tail region with liposomes (Fig. 4a).
Figure 3.
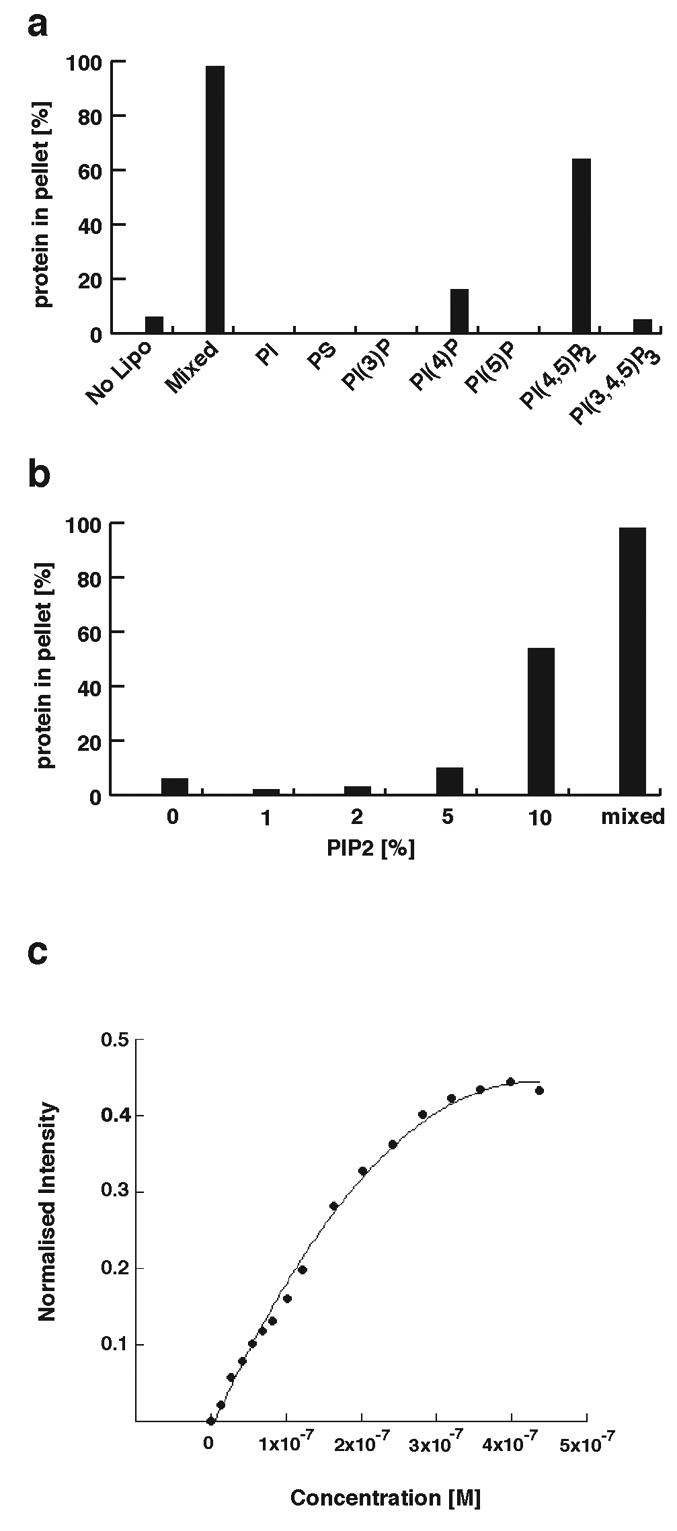
Myosin VI binds specifically to PIP2 (PI(4,5)P2)-containing liposomes. (a) The binding of the myosin VI C-terminal (CT) tail was tested with liposomes of varying compositions (liposomes contained 40% phosphatidylcholine (PC), 40% phosphatidylethanalomine (PE), 10% cholesterol and 10% of a variable phosphoinositide), and the percentage of protein in the pellet after sedimentation was measured as in figure 2c. The binding experiments were repeated 3 times and a typical experiment where the % binding relative to the binding to mixed brain liposomes (100%) is shown. The tail binds strongly to PI(4,5)P2 (PIP2) (phosphatidylinositol-4,5-bisphosphate)-containing liposomes and to mixed brain liposomes (Folch fraction I) whereas no or little binding was observed to liposomes containing PI (phosphatidylinositol), PS (phosphatidylserine), PI(3)P (phosphatidylinositol-3-phosphate), PI(4)P (phosphatidylinositol-4-phosphate), PI(5)P (phosphatidylinositol-5-phosphate), or PI(3,4,5)P3 (phosphatidylinositol-3,4,5-trisphosphate). (b) The CT tail was tested against liposomes containing 10% cholesterol, 40% PC and 40% PE and varying amounts of PIP2 using the sedimentation assay in figure 2c. A minimum of 2-5% PIP2 was required for binding, while maximal binding was seen using mixed liposomes that contain a minimum of 10% PIP2. (c) Fluorescence resonance energy transfer between tryptophan groups on the CT tail and dansylated liposomes (composed of 10% cholesterol, 10% PIP2, 40% phosphatidylcholine and 40% dansylated phosphatidylethanolamine) was measured and the resulting curve was fit using Kaleidograph to obtain the Kd (0.3 μM).
Figure 4.
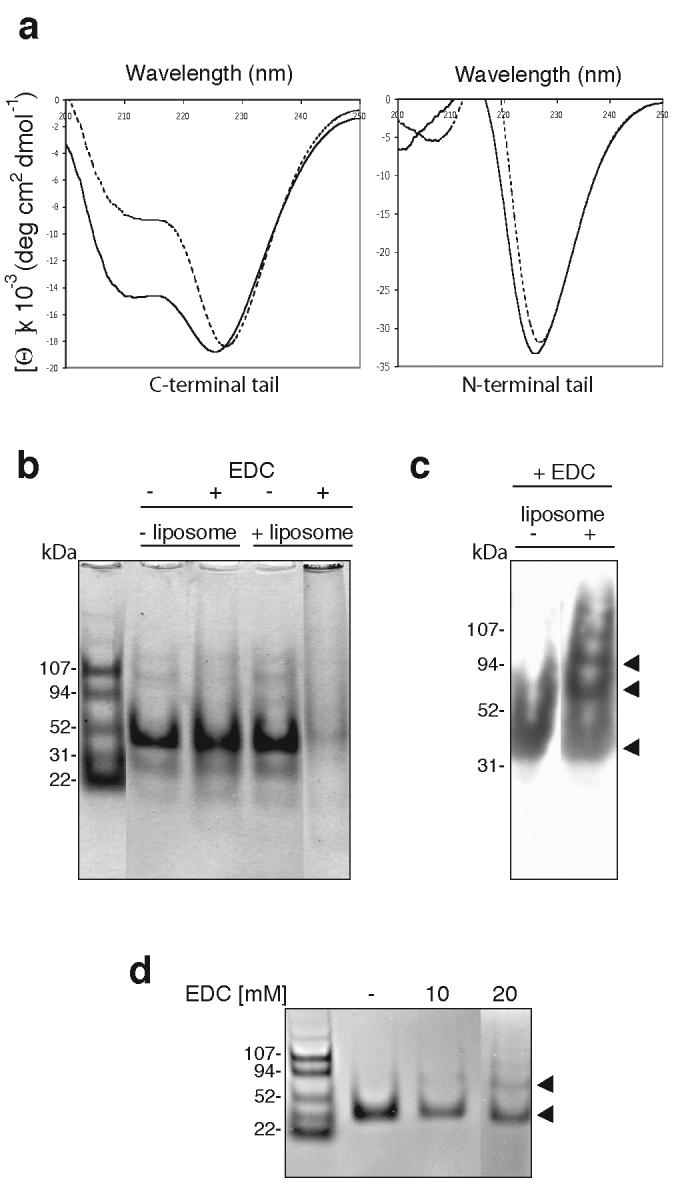
Liposome binding increases the helical structure of the C-terminal tail of myosin VI and induces dimerisation. (a) Circular dichroic spectra of the myosin VI C-terminal tail (CT) and the N-terminal tail with (solid lines: + 0.2 mg ml-1 liposomes) or without mixed brain liposomes (dashed lines). An average of five spectra are shown, normalised against spectra of buffer alone or buffer + liposomes. With liposomes CT adopts a highly helical spectrum and the (helical) signal at 222nm increases by 31% whereas the spectrum of the N-terminal tail is not altered (this construct does not bind liposomes in sedimentation assays: see figure 2c). (b) Liposome binding induces multimerisation of the myosin VI C-terminal tail. 12 μM CT was incubated with or without 0.4 mg ml-1 mixed brain liposomes and/or 90 mM EDC, a zero-length crosslinker, and analysed by SDS-PAGE. Without liposomes the monomer CT tail (30kDa) is not crosslinked by EDC but with both liposomes and EDC, the monomer band multimerises and remains at the top of the gel. BSA was used as a negative control and was not crosslinked upon addition of liposomes and EDC (data not shown). (c) Western blot of the C-terminal tail (+/− liposomes, + 90 mM EDC) using a polyclonal myosin VI C-terminal antibody. With liposomes, only a monomer band is visible whereas without liposomes, dimer, tetramer (indicated by the arrows) and trace higher order bands can be detected. (d) Liposome binding induces dimerisation of the myosin VI C-terminal tail (CT). The SDS-PAGE gel shows CT (12 μM) after a one-hour incubation with 0.4 mg ml-1 mixed brain liposomes and: lane 1, no EDC; lane 2, 10 mM EDC; lane 3, 20 mM EDC. The positions of monomer and dimer bands are indicated with black arrowheads.
Since the microtubule based kinesin motor KIF1A/Unc104 appears to dimerise upon lipid binding12 we tested whether liposome binding induces dimerisation of the myosin VI C-terminal tail (CT) by using the zero-length crosslinker EDC (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide). Without liposomes the CT is still a monomer on SDS/PAGE after incubation with EDC for one hour (Fig. 5b, c) in agreement with our previous data using full-length expressed or native myosin VI13. However, after incubation with EDC in the presence of increasing amounts of liposomes, an obvious dimer band and then tetramer and higher order multimers were detected (Fig. 4b,c). To determine the minimal concentration of EDC crosslinker required for dimerisation, CT with liposomes was incubated with increasing amounts of EDC. Using 0.1mM-2mM EDC only monomer bands are seen, whereas at 10mM EDC the monomer band decreases and a faint dimer band begins to emerge. Upon addition of 20 mM EDC, a stronger dimer band is visible (Fig. 4d). How precisely the C-terminal tail is cross-linked on the surface of the liposomes is unclear since it does not contain any predicted coiled-coil regions or other recognizable dimerisation motifs (Fig. S1b). Whether myosin VI attached to lipid vesicles functions in the cell as monomers, dimers or even as multimers is still an unresolved question.
Figure 5.
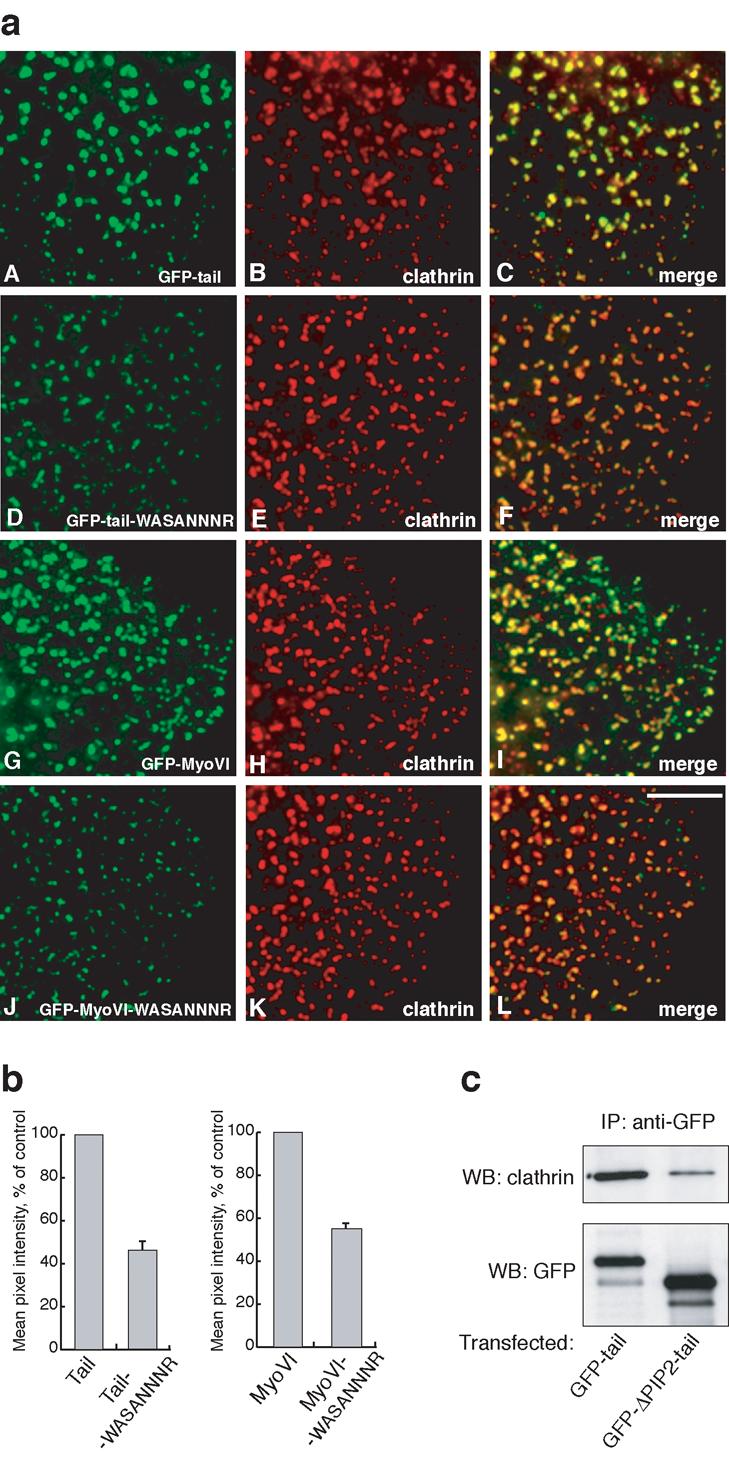
Mutations in the PIP2 binding site reduce the targeting of myosin VI to clathrin-coated structures in vivo. In a) HeLa cells were transfected with full-length wild-type GFP-myosin VI (G-I) or GFP-tail (A-C) or mutants (J-L and D-F respectively) containing the –WASANNNR- point mutation in the PIP2 binding site and processed for immunofluorescence with anti-GFP and anti-clathrin antibodies. Both tail domain and full-length myosin VI with mutations in the PIP2 binding site show significantly reduced recruitment to clathrin-coated structures at the plasma membrane (F and L). The fluorescence intensity of the different GFP-labelled constructs on at least 500 individual CCSs was measured using IP-Lab software. Scale bar = 5 μm. In b) the bar graphs show the mean pixel intensity of GFP fluorescence on CCSs of the mutant constructs relative to the wild-type controls. The results shown are the mean values of two independent experiments (± range). In a separate experiment c) HeLa cells were transfected with intact GFP-tagged myosin VI tail or GFP-tails with a 75aa deletion of the PIP2 binding region (ΔPIP2-tail) and the expressed GFP-proteins were immunoprecipitated with anti-GFP antibodies. The immunoprecipitated (IP) complexes were run on SDS-PAGE and blotted with antibody against clathrin (top panel). One fifth of each of the IP samples were run on the same gel and blotted with GFP antibody to ensure that equal over-expression and immunoprecipitation of both tail constructs had occurred (bottom panel).
To investigate the role of PIP2-lipid binding on the in vivo functions of myosin VI, we expressed GFP-labelled full-length myosin VI and tail and the corresponding mutants containing PIP2 binding site mutations (-WASANNNR-). We observed that both the full-length myosin VI and tail with the PIP2 binding site mutations showed considerably less recruitment to CCSs compared to their wild-type constructs (Fig. 5a). Quantitation of the immunofluorescence images shows that mutation of the PIP2 binding site reduces the recruitment of both full-length myosin VI and its tail domain by about 50% (Fig. 5b). As an alternative approach to quantify the localisation of myosin VI to CCSs we expressed GFP-labelled wild type myosin VI tail and the GFP-labelled mutant tail missing the PIP2 binding region (ΔPIP2 tail) and immunoprecipitated the GFP-tail constructs using GFP antibodies to “pull down” the tail-clathrin complexes. Although slightly higher levels of the mutant tail are expressed in the cells far less clathrin is associated with the ΔPIP2 tail compared with the wild type tail (Fig. 5c). Thus the targeting and functional role of full-length myosin VI in CCSs in the early endocytic pathway is mediated by the C-terminal domain binding to both Dab2 and PIP2 and requires the presence of the large insert in the tail domain.
We have demonstrated that the C-terminal tail of myosin VI is composed of two key domains that contain ‘hot spots’ for binding adaptor proteins [the WWY (1183-1185) and RRL (1107-1110) sites] and a PIP2-lipid binding region that are crucial for targeting/recruiting myosin VI to distinct compartments within the cell. Previous co-immunoprecipitation studies indicated that the binding between the myosin VI tail and its adaptor proteins was relatively weak4. Low affinity protein-protein interactions as observed in many clathrin and AP-2 binding proteins14,15 are essential for the rapid and accurate coordination between the vast range of components involved in membrane trafficking pathways such as endocytosis. The hydrophobic Dab2-binding site (WWY) is in an extensive region in the myosin VI tail that is predicted to be unstructured by multiple secondary structure prediction algorithms (performed using the NPS@.Network Protein Sequence Analysis web server16) (Fig. S1b). Such unfolded regions are believed to play important roles in many endocytic proteins14,17,18; for example it has been suggested that these unfolded regions containing binding sites that act as a flexible “string” to “fish for and hook” binding partners and upon binding these unstructured regions may become structured14. The large increase in helicity (31%) in the myosin VI tail when it binds PIP2 would fit in with this type of model. The observation that the C-terminal tail region of myosin VI contains a flexible proteolytically susceptible linker region connecting two functional domains (Fig. S2) provides further support for such a model. PIP2 is a crucial second messenger19,20 that is known to bind and recruit many endocytic proteins to the plasma membrane (see reviews21,22) and regulate the formation, scission and uncoating of clathrin-coated vesicles. Thus myosin VI by binding not only to Dab2 but also to PIP2 concentrated in the plasma membrane at active sites of endocytosis may be recruited to the membrane at a very early stage of clathrin-coated pit assembly (Fig. S2). The region in the myosin VI tail that binds PIP2 contains a motif of basic residues (R/K) alternating with hydrophobic residues very similar to the region identified in a number of cytoskeletal proteins that have been shown to bind to PIP223. The monomeric kinesin motor KIF1A/Unc104 dimerises upon liposome binding and the resulting dimer is able to transport the liposome processively along microtubules24. Could such a scenario occur with monomeric myosin VI 13 i.e. could it dimerise upon liposome binding and then work as a processive motor? The most recent in vitro results on the functional state of myosin VI are intriguing but contradictory. Full-length myosin VI monomers can be induced to form a few processive dimers (17%) when clustered on actin filaments25 on the other hand, a single myosin VI monomer when coupled to a polystyrene bead (of a similar size to an intracellular vesicle) can move processively with a large step (40 nm)26. Thus whether myosin VI exists and functions in the cell as a monomer and/or dimer remains to be established. However if myosin VI plays a major role in vesicular trafficking then its dimerisation on the surface of a lipid vesicle to generate a processive motor would be an efficient mechanism for regulating this transport function. Thus one of our next major challenges is to establish how the protein binding partners, inserts and PIP2 affect the structure, properties and functional (monomer/dimer/multimer) state of myosin VI and determine its role(s) in vivo in membrane trafficking pathways such as clathrin- mediated endocytosis (see speculative model in Fig. S2).
METHODS
Human myosin VI isoforms and antibodies
The human myosin VI (+ large insert) was generated from KIAA 0389 clone from Human cDNA Bank Section, Kazusa DNA Research Institute, Japan, and cloned into the pEGFP-C3 vector for mammalian transient expression. Intact chicken myosin VI (+large insert) was expressed in Sf9 insect cells using the Baculovirus expression system and purified and checked for activity as described13. Mouse monoclonal anti-clathrin X-22 and rabbit polyclonal anti-GFP antibodies were from AbCam and the rabbit polyclonal myosin VI tail antibody as previously described2. Rabbit anti-clathrin heavy chain antibody used for the Western blot was from M.S. Robinson (CIMR, Cambridge).
Cell culture, transfection, immunofluorescence and immunoblotting
HeLa cells grown on cover-slips to 50% confluency were transfected using FuGENE (Roche Diagnostics) over night with 2 μg of intact human myosin VI (+large insert) or only the tail domain in pEGFP vector. To visualize myosin VI associated with clathrin-coated or uncoated vesicles, cells were pre-permeabilized with 0.05% saponin in PBS for 30s before fixing with paraformaldehyde. Cells were visualised in a Zeiss Axioplan microscope and data analysed with IP-Lab software (Zeiss). Colocalisation of GFP-myosin VI with clathrin was quantitated in the cell periphery of GFP-myosin VI transfected cells double-labelled for clathrin. 50 CCSs on average were counted in at least 5-10 cells for each construct. In the quantitative immuno-staining experiments identical acquisition settings were used and circles were manually drawn around single CCSs and the mean pixel intensity was determined for the region of interest. At least 500 CCSs were measured for each construct (20-30 CCSs per cell and at least 10 cells per experiment) and the experiment was repeated twice. The measurements were normalized for the intensity of clathrin staining in each sample. Immunoprecipitation assays using GFP antibodies to pull down GFP-myosin VI tail-clathrin complexes were carried out as previously described27.
Identification of Dab2 and GIPC Binding Sites
Chicken brush border myosin VI intact tail (+LI) (aa 840-1277) was used as a template to generate a series of myosin VI tail deletion mutants by PCR. To narrow down the binding sites in the tail, alanine scanning mutations (mutating 3 consecutive residues at a time to A) and binding site mutations were generated by PCR using the QuikChange Site-Directed Mutagenesis kit (Stratagene). All these tail mutants were cloned into the pM “bait” vector while binding partners (Dab2, GIPC, and optineurin) were cloned into the pV16 “prey” vector (Clontech) and the mammalian-2-hybrid assay was performed in CHO cells as described7. For the pull-down assays GST-tagged chicken myosin VI intact tail (+LI) (wild-type or W1184L or RRL->AAA mutants) were expressed in E.coli and purified as described27. Full length Dab2 was cloned into pcDNA3 (Invitrogen) and in vitro translated and labelled with [35S]methionine using the TNT-coupled Reticulocyte Lysate system (Promega). The pull-down assays were performed as previously described4.
Expression, purification and digestion of myosin VI tail constructs
Chicken myosin VI intact tail (+LI) constructs were generated by PCR and mutants prepared using the QuikChange Site-Directed Mutagenesis kit (Stratagene). The tail fragments were cloned into the pRSET vector, expressed in E.coli C41 cells and purified essentially as described previously27 with the following modifications. Soluble C-terminal tail fragments after cell lysis were first passed over an anion exchange matrix (Whatman DE53) and then applied to a Ni-NTA column (Qiagen), washed and His-tagged tail fragments eluted with TBS (Tris-buffered saline) + 300 mM Imidazole pH 7.4. Insoluble full-length chicken myosin VI tail constructs were dissolved in 8M urea, 10 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM DTT before applying to the DE53 resin. The flow-through was applied to the Ni-NTA column, washed with 8M Urea in TBS, and the protein was refolded on the column by stepwise washes (4M Urea, 2M Urea, no Urea in TBS) before elution with TBS+300 mM Imidazole pH 7.4. The proteins were dialysed and diluted with HEPES/salt solution (20 mM Hepes pH 7.4, 150 mM NaCl, 1 mM DTT). The C-terminal myosin VI tail domain (14 μM) in HEPES/salt solution without and with (0.4mg ml-1) liposomes was digested with trypsin (Sigma, type T1426) for 30 min at 0° C (protein to trypsin molar ratio of 500 to 1). Digestion was stopped by the addition of a 2 fold molar excess of trypsin inhibitor (Sigma type T0256). The resulting fragments were electro-blotted onto Immobilon™-PVDF membrane (Millipore) and their N-terminal sequences determined using an Applied Biosystems Procise 494 Sequencer and further analyzed using MALDI-TOF mass spectrometry. Since 11kDa domain-2 retains the His tag it can be separated from 18kDa domain-1 by chromatography on a Ni-NTA column in HEPES/salt solution (non-dissociating conditions) as described above.
Lipid binding assay
“Mixed brain liposomes” (Folch fraction 1, Sigma B1502) and liposomes made from 40% phosphatidylcholine, 40% phosphatidylethanolamine, 10% cholesterol and 10% of a variable phosphoinositol (ex PtdIns(4,5)P2) obtained from Avanti Polar Lipids were treated with chloroform, aspirated under Argon and then dried under vacuum for 10 minutes before resuspension by sonication in 20 mM HEPES, pH 7.4, 150 mM NaCl and 1 mM DTT to a final concentration of 1 mg ml-128. 4 μM protein and 0.4 mg ml-1 liposomes were mixed, incubated and centrifuged for 15 min at 160,000×g at 4°C. Supernatant and pellet fractions were analysed on SDS-PAGE (10 or 15% acrylamide) gels using prestained High or Broad range standards (Biorad) for calibration. Gels were quantitated using a Molecular Dynamics densitometer and ImageQuant software (the intensity of the tail band in the pellet was divided by the total intensity (pellet + supernatant) for each sample to determine the percent of protein in the pellet).
Fluorescence resonance energy transfer
A Perkin Elmer LS55 Luminescence Spectrometer was used to measure fluorescence resonance energy transfer (FRET) between tryptophan groups on the myosin VI C-terminal tail and dansylated liposomes consisting of 10% cholesterol, 10% PIP2, 40% PC and 40% Dansyl - PE (Avanti Polar Lipids). 11 μM protein was titrated into 16 μg ml-1 liposomes in buffer (20 mM Hepes, 150 mM NaCl, 1 mM DTT) under constant temperature (20°C) using a Hamilton Microlab 500 syringe pump. An excitation of 340 nm was used, and the emission intensity at 520 nm (I) normalised against the intensity without addition of protein (I0) (resulting in (I−I0)/I0) which was plotted against protein concentration. Kaleidograph was used to obtain the Kd from the following curve-fit:
where Imax is maximum intensity, Kd is the binding constant, and c is a constant. The measured Kd was 0.3 μM, or 1.4 μM upon incorporation of a linear decay in the curve fit. The non-binding tail fragment (1134-1276) was used as a control.
Circular dichroism and crosslinking experiments
Five spectra for each protein (8-14 μM in 20 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM DTT) at 20°C were measured and averaged on a Jobin Yvon CD6 spectrometer with the spectrum of salt/buffer alone subtracted. 50 μl of 1mg ml-1 mixed brain liposomes (in above HEPES/salt solution) were added to 200 μl of the protein sample and the spectra were measured immediately.
The zero length cross-linker EDC (1-Ethyl-3-(3dimethylaminopropyl) carbodiimide) (Sigma, E-1769) (final concentrations 20-90 μM) was added to 50 μl aliquots of myosin VI C-terminal tail (12 μM) in HEPES/salt solution alone or after addition of 0.4 mg ml-1 mixed brain liposomes. After incubation at RT for 1h, a phosphate loading buffer (pH 6.2) was added (25 mM NaH2PO4, 37 mM Na2HPO4, 3.5 mM SDS, 10% b-mercaptoethanol, 10% glycerol, 4M Urea, 0.01% bromophenol blue) and samples analysed by SDS-PAGE using phosphate buffered gels (25 mM NaH2PO4, 72 mM Na2HPO4, pH7.4, 3.5 mM SDS, 4.5% acrylamide) and phosphate running buffer (25 mM NaH2PO4, 72 mM Na2HPO4, 3.5 mM SDS, pH7.4). Cross-linked phosphorylase-b molecular weight marker (Sigma) was used to calibrate the gels.
Supplementary Material
ACKNOWLEDGEMENTS
We thank B. Peter and H. McMahon for help with lipid binding assays and discussion; R. Williams, D. Veprintsev and O. Perisic for programs and help with the FRET assay, S. Peak-Chew and F. Begum for N-terminal sequencing and Mass Spectrometry and D. Owen for critical reading of the manuscript. The work was funded by a USA Royal Society Postdoctoral Fellowship (GS), a Croucher Foundation (Hong Kong) Student Scholarship (JSA), a Wellcome Trust Senior Fellowship (FB) and was supported by the Medical Research Council. The Cambridge Institute for Medical Research is in receipt of a strategic award from the Wellcome Trust.
References
- 1.Wells AL, et al. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–8. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 2.Buss F, Arden SD, Lindsay M, Luzio JP, Kendrick-Jones J. Myosin VI isoform localized to clathrin-coated vesicles with a role in clathrin-mediated endocytosis. Embo J. 2001;20:3676–84. doi: 10.1093/emboj/20.14.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aschenbrenner L, Lee T, Hasson T. Myo6 facilitates the translocation of endocytic vesicles from cell peripheries. Mol Biol Cell. 2003;14:2728–43. doi: 10.1091/mbc.E02-11-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahlender DA, et al. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J Cell Biol. 2005;169:285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warner CL, et al. Loss of myosin VI reduces secretion and the size of the Golgi in fibroblasts from Snell's waltzer mice. Embo J. 2003;22:569–79. doi: 10.1093/emboj/cdg055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geisbrecht ER, Montell DJ. Myosin VI is required for E-cadherin-mediated border cell migration. Nat Cell Biol. 2002;4:616–20. doi: 10.1038/ncb830. [DOI] [PubMed] [Google Scholar]
- 7.Morris SM, et al. Myosin VI binds to and localises with Dab2, potentially linking receptor-mediated endocytosis and the actin cytoskeleton. Traffic. 2002;3:331–41. doi: 10.1034/j.1600-0854.2002.30503.x. [DOI] [PubMed] [Google Scholar]
- 8.Dance AL, et al. Regulation of myosin-VI targeting to endocytic compartments. Traffic. 2004;5:798–813. doi: 10.1111/j.1600-0854.2004.00224.x. [DOI] [PubMed] [Google Scholar]
- 9.Bunn RC, Jensen MA, Reed BC. Protein interactions with the glucose transporter binding protein GLUT1CBP that provide a link between GLUT1 and the cytoskeleton. Mol Biol Cell. 1999;10:819–32. doi: 10.1091/mbc.10.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lou X, McQuistan T, Orlando RA, Farquhar MG. GAIP, GIPC and Galphai3 are concentrated in endocytic compartments of proximal tubule cells: putative role in regulating megalin's function. J Am Soc Nephrol. 2002;13:918–27. doi: 10.1681/ASN.V134918. [DOI] [PubMed] [Google Scholar]
- 11.Pashkova N, Jin Y, Ramaswamy S, Weisman LS. Structural basis for myosin V discrimination between distinct cargoes. Embo J. 2006;25:693–700. doi: 10.1038/sj.emboj.7600965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klopfenstein DR, Tomishige M, Stuurman N, Vale RD. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 2002;109:347–58. doi: 10.1016/s0092-8674(02)00708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lister I, et al. A monomeric myosin VI with a large working stroke. Embo J. 2004;23:1729–38. doi: 10.1038/sj.emboj.7600180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans PR, Owen DJ. Endocytosis and vesicle trafficking. Curr Opin Struct Biol. 2002;12:814–21. doi: 10.1016/s0959-440x(02)00395-0. [DOI] [PubMed] [Google Scholar]
- 15.Dell'Angelica EC. Clathrin-binding proteins: got a motif? Join the network! Trends Cell Biol. 2001;11:315–8. doi: 10.1016/s0962-8924(01)02043-8. [DOI] [PubMed] [Google Scholar]
- 16.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–50. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 17.Brett TJ, Traub LM, Fremont DH. Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure (Camb) 2002;10:797–809. doi: 10.1016/s0969-2126(02)00784-0. [DOI] [PubMed] [Google Scholar]
- 18.Kalthoff C, Alves J, Urbanke C, Knorr R, Ungewickell EJ. Unusual structural organization of the endocytic proteins AP180 and epsin 1. J Biol Chem. 2002;277:8209–16. doi: 10.1074/jbc.M111587200. [DOI] [PubMed] [Google Scholar]
- 19.Sakisaka T, Itoh T, Miura K, Takenawa T. Phosphatidylinositol 4,5-bisphosphate phosphatase regulates the rearrangement of actin filaments. Mol Cell Biol. 1997;17:3841–9. doi: 10.1128/mcb.17.7.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Czech MP. PIP2 and PIP3: complex roles at the cell surface. Cell. 2000;100:603–6. doi: 10.1016/s0092-8674(00)80696-0. [DOI] [PubMed] [Google Scholar]
- 21.Cremona O, De Camilli P. Phosphoinositides in membrane traffic at the synapse. J Cell Sci. 2001;114:1041–52. doi: 10.1242/jcs.114.6.1041. [DOI] [PubMed] [Google Scholar]
- 22.Itoh T, Takenawa T. Regulation of endocytosis by phosphatidylinositol 4,5-bisphosphate and ENTH proteins. Curr Top Microbiol Immunol. 2004;282:31–47. doi: 10.1007/978-3-642-18805-3_2. [DOI] [PubMed] [Google Scholar]
- 23.Janmey PA, Xian W, Flanagan LA. Controlling cytoskeleton structure by phosphoinositide-protein interactions: phosphoinositide binding protein domains and effects of lipid packing. Chem Phys Lipids. 1999;101:93–107. doi: 10.1016/s0009-3084(99)00058-4. [DOI] [PubMed] [Google Scholar]
- 24.Klopfenstein DR, Vale RD. The lipid binding pleckstrin homology domain in UNC-104 kinesin is necessary for synaptic vesicle transport in Caenorhabditis elegans. Mol Biol Cell. 2004;15:3729–39. doi: 10.1091/mbc.E04-04-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park H, et al. Full-length myosin VI dimerizes and moves processively along actin filaments upon monomer clustering. Mol Cell. 2006;21:331–6. doi: 10.1016/j.molcel.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 26.Iwaki M, et al. Cargo binding makes a wild-type single-headed myosin-VI move processively. Biophys J. 2006;90:3643–3652. doi: 10.1529/biophysj.105.075721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buss F, et al. The localization of myosin VI at the Golgi complex and leading edge of fibroblasts and its phosphorylation and recruitment into membrane ruffles of A431 cells after growth factor stimulation. J Cell Biol. 1998;143:1535–45. doi: 10.1083/jcb.143.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peter BJ, et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 303:495–9. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


