Abstract
In the 30 years since its discovery, tyrosine phosphorylation has emerged as a fundamentally important mechanism of signal transduction and regulation in all eukaryotic cells, governing many processes, including cell proliferation, cell cycle progression, metabolic homeostasis, transcriptional activation, neural transmission, differentiation and development, and aging. Perturbations in tyrosine phosphorylation underlie many human diseases, and in particular cancer, and this has prompted the development of inhibitors of tyrosine kinases implicated in disease, a number of which have been approved for clinical use. The following is a brief personal reflection on some of the salient findings over the past 30 years that led to the development of tyrosine kinase inhibitors for disease therapy.
Introduction
A fortuitous observation made in the summer of 1979 during our studies of polyomavirus middle T and v-Src associated kinase activities led to the discovery of tyrosine phosphorylation as a new type of protein modification [1-3]. Over the past 30 years amazing progress has been made in elucidating how tyrosine phosphorylation regulates protein function, and in developing therapeutic drugs that antagonize the activity of tyrosine kinases that are aberrantly activated in cancer.
Historical perspective
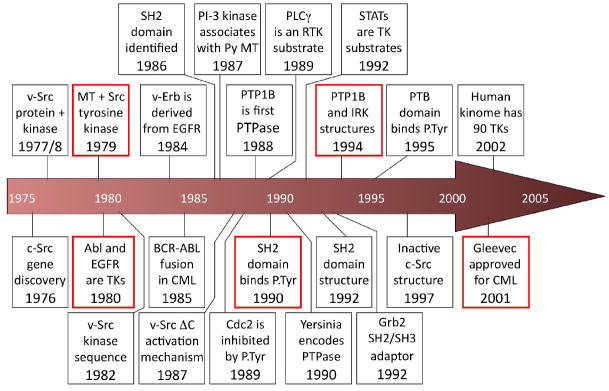
Following the 1979 discovery that Src is a tyrosine kinase, the number of distinct tyrosine kinases grew rapidly, accelerated by the advent of rapid DNA sequencing technology and PCR. All the cloned tyrosine kinases proved to have a related catalytic domain sequence containing a series of conserved motifs, later shown to be involved in catalysis, which are largely shared with those in the catalytic domains of the serine/threonine kinases. Progress in understanding the function of tyrosine phosphorylation was equally rapid (see time line in Figure 1). The early observation that ligand binding induced a rapid increase in EGF and PDGF receptor tyrosine kinase (RTK) autophosphorylation implied an important role for tyrosine phosphorylation in growth factor signaling and proliferation, and by extension in oncogenesis through hijacking of growth factor tyrosine phosphorylation signaling pathways. A key step in understanding how RTKs initiate intracellular signaling was the discovery that phosphotyrosine (P.Tyr) residues on activated RTKs are recognized by a phosphodependent-binding domain, the SH2 domain, in a manner dictated by the primary sequence of the amino acids immediately downstream of the P.Tyr [4]. The recruitment of SH2 domain proteins to autophosphorylated RTKs at the plasma membrane is critical for initiating and propagating downstream signaling. SH2 domain proteins can have a variety of functions, including adaptor proteins to recruit other signaling proteins, enzymes that act on membrane molecules, such as phospholipases, cytoplasmic tyrosine kinases that relay signals, E3 ubiquitin ligases, and transcription factors. Proteins containing a second type of P.Tyr-binding domain, PTB, were also found to be involved in RTK signaling. Subsequently, other P.Tyr-binding domains have been identified, including a subset of catalytically-dead protein phosphatases in the protein-tyrosine phosphatase (PTP) and the dual-specificity phosphatase (DSP) families, the C2 domain of PKCδ, and pyruvate kinase M2.
Figure 1.
Timeline of some important discoveries in tyrosine phosphorylation
There are two classes of tyrosine kinase. Receptor tyrosine kinases are type I transmembrane proteins possessing an N-terminal extracellular domain, which can bind activating ligands, a single transmembrane domain, and a C-terminal cytoplasmic domain that includes the catalytic domain. Nonreceptor tyrosine kinases lack a transmembrane domain; most are soluble intracellular proteins, but a subset associate with membranes via a membrane-targeting posttranslational modification, such as an N-terminal myristoyl group, and can act as the catalytic subunit for receptors that lack their own catalytic domain. RTKs comprise 58 of the 90 tyrosine kinases in the human genome [5]. Most of them are activated through binding of their extracellular domain to specific protein ligands, such as growth factors and cytokines, some of which are themselves membrane anchored proteins. Ligand binding elicits RTK oligomerization and subsequent activation of the cytoplasmic catalytic domain by transphosphorylation of Tyr in the activation loop or juxtamembrane domain, resulting in tyrosine phosphorylation of recruited cytoplasmic proteins, thereby transducing extracellular signals across the plasma membrane. Nonreceptor tyrosine kinases, acting as catalytic subunits of receptors that lack a kinase domain, are activated in a similar fashion through ligand-induced oligomerization or conformational reorganization of the receptors they are bound to.
Signaling specificity downstream of each receptor tyrosine kinase system is dictated by the sequences embedding the tyrosine phosphorylation sites on the receptor proteins themselves or their associated subunits, whose phosphorylation leads to recruitment of a spectrum of SH2/PTB proteins dependent on the repertoire expressed in the cell. Specific cellular responses result from the integration of the constellation of signaling pathways that are activated downstream of the different SH2/PTB proteins, and include both cytoplasmic and nuclear responses. Important signaling pathways activated by RTKs include the Ras/ERK MAP kinase cascade and the PI-3 kinase pathway. Indeed, the discovery of PI-3 kinases emerged from the study of a lipid kinase activity associated with tyrosine kinases, and the first MAP kinase cascade was discovered biochemically as series of activities stimulated by EGF or insulin upon activation of their respective receptor RTKs.
Tyrosine phosphorylation proved to be not only involved in growth factor receptor signaling, but also in many other cellular processes, including cell adhesion via integrin signaling, metabolic regulation through the insulin RTK, in cell cycle control through inhibitory tyrosine phosphorylation of CDKs, transcriptional activation through tyrosine phosphorylation of latent cytoplasmic STAT family transcription factors, in neural transmission, and even in aging via the IGF-1R, to mention but a few processes regulated by tyrosine phosphorylation. Tyrosine kinases play a particularly important role in development and cellular differentiation in metazoans, and in evolutionary terms, tyrosine kinases proper are found in all metazoans, consistent with their playing major role in cell-cell signaling and transmembrane signal transduction, a critical function for establishment of a multicellular organism.
The Tyrosine Kinome
The availability of complete genome sequences afforded the possibility of using bioinformatics tools to predict the total numbers of protein kinase genes in an organism, and in particular the complement of tyrosine kinases. In general, tyrosine kinases represent 10-15% of total protein kinase genes in a typical metazoan organism. For instance, in C. elegans 90 out of 454 total protein kinase genes encode tyrosine kinases, and in humans 90 of the 525 protein kinase genes are tyrosine kinases [5,6]. Of the 90 human tyrosine kinases 58 are RTKs; four of these RTKs are predicted to lack catalytic activity (e.g. ErbB3) [5]. Opposing the action of the tyrosine kinases are 108 protein phosphatases that can remove phosphate from P.Tyr in proteins [7]. Tyrosine kinases do not have a strong preference for the primary sequence embedding the target tyrosine, although a predilection for acidic residues within the four amino acids in either side of the Tyr has been deduced for individual kinases using degenerate peptide library approaches [8]. Instead, the target specificity of tyrosine kinases is dictated in large part by second site interactions between the kinase and a potential substrate, such as that mediated by SH2 domains, and also by a shared subcellular localization. In general, PTPs also lack a tight primary sequence specificity for the target P.Tyr, although in a few cases the catalytic domain has a pocket for a second acidic residue. Moreover, there is not a one to one relationship between tyrosine kinases and phosphatases, and, like the tyrosine kinases, the action of PTPs is limited by their secondary interactions with potential targets and by their subcellular localization.
The Evolution of Tyrosine Phosphorylation
Some intriguing insights into the evolutionary origins of tyrosine phosphorylation have come from the recent analysis of the kinome of the choanoflagellate Monosiga brevicollis, a single-cell ocean-dwelling organism [9-11]. Surprisingly, M. brevicollis has a tyrosine phosphorylation network as complex as that of humans with 128 predicted tyrosine kinases. However, only seven of these tyrosine kinases are orthologous to the tyrosine kinases found in metazoans [9]. There had been speculation that tyrosine phosphorylation must have arisen as a signaling system in a single cell organism that then gave rise to the first metazoans. Indeed, choanoflagellates lie close to the base of the metazoan lineage, and could have given rise to metazoans. However, the enormous divergence in tyrosine phosphorylation networks between choanoflagellates and simple metazoans suggests it is more likely that both choanoflagellates and metazoans arose from a common progenitor with a simpler tyrosine phosphorylation system. Of course, this begs the question as to how tyrosine phosphorylation arose and acquired its signaling function [11]. Tyrosine phosphorylation is known in prokaryotes, and a class of bacterial tyrosine (BY) kinase has been characterized [12]. However, the BY kinases are not related to the eukaryotic tyrosine kinases in either sequence or structure. Conventional PTPs and Cdc25 family PTPs are present in the yeasts, which lack true tyrosine kinases, but their function is to dephosphorylate the regulatory tyrosine phosphates on CDKs and MAPKs. Nonetheless, the existence of PTPs may have been critical in allowing the evolution of a new class of protein kinase with specificity for tyrosine, which in the absence of enzymes that dephosphorylate P.Tyr could be deleterious. The other key component of tyrosine phosphorylation-based signaling is a domain that can bind specifically to P.Tyr residues in other proteins. Like PTPs, proteins with SH2 domains are also found in organisms, such as yeast and slime molds, that lack tyrosine kinases proper, but whether the SH2 domain arose de novo as a P.Tyr-binding domain, or whether it was derived from a domain that binds other acidic moieties is unknown. In this regard, it is interesting to note that the conserved yeast Spt6 protein has a divergent SH2 domain that binds to P.Ser2 in the CTD repeat of the large subunit of RNA polymerase II [13].
In discussions about the evolution of tyrosine phosphorylation, a common question is “Why tyrosine?”. Like Ser/Thr phosphate esters, the P.Tyr phosphate ester linkage has a relatively high bond energy, and the equilibrium constant for most protein kinases is close to 1, meaning that kinases can catalyze the reverse reaction in the presence of ADP and remove phosphate from the phosphorylated residue in a target protein to generate ATP. Perhaps the major reason why P.Tyr was selected is that the SH2 domain binding energy for a P.Tyr residue is higher than that for P.Ser or P.Thr, because of the contribution of contacts that can be made with the phenolic ring in addition to interactions with the phosphate. Despite the relatively large number of tyrosine kinases, P.Tyr accounts for <1% of phosphate esterified to proteins [2]. There are several reasons for this. Unlike P.Ser/Thr, P.Tyr rarely plays a structural role in proteins, and is primarily regulatory. In addition, most tyrosine kinases are tightly negatively regulated, and only activated under specific conditions. Finally, PTPs have a very high turnover rate, and, in consequence, P.Tyr residues have a very short half life, unless protected by binding to SH2/PTP domains.
Structural Basis of Signal Transduction by Tyrosine Phosphorylation
Tremendous insights into the mechanistic underpinnings of tyrosine phosphorylation signaling have come from three-dimensional structures obtained for tyrosine kinases and phosphatases, SH2 and PTP domains, etc. These structures have illuminated the mechanisms of activation of tyrosine kinases, through dimerization, and auto/transphosphorylation-induced conformational changes, the novel catalytic mechanism used by PTPs, and the selectivity of SH2 domains and PTP domains for specific primary sequences containing P.Tyr. Insights into how tyrosine kinase activity is negatively regulated by phosphorylation and by other proteins has also come from structures; for example, the Grb14 BPS domain bound to the insulin receptor catalytic domain [14], and a fragment of MIG6 bound to the EGF receptor catalytic domain [15]. As yet there are few structures of tyrosine kinases interacting with their substrates, although the Csk/c-Src structure [16], the APS SH2 domain bound to the P.Tyr residues in the insulin receptor activation loop [17], and FGFR2 catalytic domain dimer caught in the act of transphosphorylation [18], and the twin SH2 domains of PLCγ1 bound to the FGFR1 catalytic domain are examples [19]. As structures of multidomain fragments of tyrosine kinases bound to their substrates are reported, another emerging theme is the intricacy of the intra- and intermolecular interactions between the domains, which has important implications for regulation and substrate specificity. For instance, SH2 domains not only interact with specific P.Tyr residues, but also with proteins via other surface loops (e.g. the Abl SH3 domain binds to the Crk SH2 domain via a PXXP motif in the DE loop [20], and the SAP SH2 domain binds to the SH3 domain of FynT [21]).
Tyrosine Kinase Substrates
A true understanding of the roles of tyrosine phosphorylation requires a knowledge of the proteins that are phosphorylated by the activated tyrosine kinases in question. Early attempts to identify substrates for tyrosine kinases involved screening for alkali-stable phosphoproteins on two-dimensional gels and analysis of candidate substrates from 32P-labeled cells expressing an oncogenic tyrosine kinase or after stimulation with a growth factor that activates an RTK. The development of anti-P.Tyr antibodies was a major advance, and allowed rapid identification of potential substrates in unlabeled cells and tissues. Affinity purification with P.Tyr-binding domains also yielded substrates. In each case, the site of tyrosine phosphorylation had to be identified, usually by mutagenesis and peptide mapping, and then functional studies carried out to determine the effects of tyrosine phosphorylation. Excluding autophosphorylation sites on tyrosine kinases, which tend to predominate, a catalogue of over a hundred substrates was established. In general, tyrosine phosphorylation has been found to induce protein-protein interaction through the binding of proteins with SH2/PTB domains to the tyrosine phosphorylated protein, thus propagating a signal. However, there are some examples where tyrosine phosphorylation regulates catalytic activity; this may involve intramolecular interactions between the P.Tyr moiety and an SH2 domain (e.g. c-Src and SHP2), but there are cases where tyrosine phosphorylation acts to change activity through an allosteric mechanism, such as PLCγ1.
The current catalogue of characterized tyrosine phosphorylated protein, however, appears to represent a significant underestimate of the prevalence of tyrosine phosphorylation. Recent mass spectrometry-based phosphoproteomic analyses for P.Tyr-containing peptides in different cell types and tissues under different conditions have revealed hundreds of tyrosine phosphorylation sites occupied in a single cell type in response to a stimulus that activates a tyrosine kinase [22-26](see also the PhosphositePlus, Phosida, and Phospho.ELM web resources, among other phosphoproteomic databases). Among these are sites in previously identified signaling proteins, but there are also cytoskeletal proteins, proteins involved in RNA metabolism, and even glycolytic enzymes, such as enolase, which ironically was among the first v-Src substrates to be identified 25 years ago. While the consequences of phosphorylation of these Tyr in the new targets await functional characterization, the roles of tyrosine phosphorylation are obviously more varied than we originally thought.
Tyrosine Kinases and Disease: The Development of Inhibitors
Hand in hand with our understanding of the functions of tyrosine phosphorylation came the realization that aberrant tyrosine phosphorylation plays a critical role in a number of diseases. Indeed, 51 of the 90 tyrosine kinases are implicated in cancer either through mutation or under-or overexpression [27], and a fair number of PTPs and DSPs have also been implicated in disease. The initial discovery that several viral oncoproteins are constitutively active tyrosine kinases that transform cells through tyrosine phosphorylation, led to an immediate interest in the possibility of developing inhibitors that would block the action of such kinases [28]. This interest was heightened with the discovery that the oncoprotein driving chronic myelogenous leukemia is a fusion protein between BCR and c-ABL, BCR-ABL, which has constitutive tyrosine kinase activity. This was followed by the finding that ERBB2 is overexpressed in a subset of breast cancers. Natural product tyrosine kinase inhibitors (TKIs) were identified in the early 1980’s, and programs to develop synthetic small molecule tyrosine kinase inhibitors were initiated in industry at CIBA-Geigy and in academic groups, including Levitzki’s, in the mid-1980’s [29]. Progress was encouraging, but developing inhibitors with true drug-like properties proved more difficult. The first success grew out of an earlier program at CIBA-GEIGY to develop protein kinase C inhibitors, which provided the 2-phenylaminopyrimidine scaffold. An early tyrosine kinase target was the PDGF receptor, where an inhibitor could be useful in treating restenosis. Synthesis of a series of 2-phenylaminopyrimidine derivatives led to the PDGFR-selective CGP53716, and to CGP57148B, which inhibits PDGFR and v-Abl in vitro and in vivo equally potently, as well as c-KIT. CGP57148B became known as STI571 (Signal Transduction Inhibitor), and once it entered clinical trials for CML was given the generic name imatinib (when formulated as the mesylate salt, imatinib is marketed by Novartis as Gleevec™ (USA) or Glivec™ (Europe). Imatinib went into clinical trials in 1998, and because of the stunning effectiveness in treating chronic phase CML, imatinib was approved for the treatment of CML on May 10, 2001 [30].
In the interim, other pharmaceutical and biotech companies had been developing inhibitors for tyrosine kinases known to be activated in cancer or which are critical for the development of tumor vasculature. In relatively, quick succession a series of TKIs have been approved for treatment of cancer: gefitinib (Iressa™) and erlotinib (Tarceva™), both EGFR inhibitors approved for NSCLC therapy, sunitinib (Sutent™); a split RTK inhibitor that was approved for treatment of renal carcinoma and imatinib-resistant CML, dasatinib (Sprycel™), a mixed Src/Abl inhibitor approved as an alternative therapy that works for some imatinib-resistant CML; lapatinib (Tykerb™), an EGFR/ERBB2 inhibitor approved for breast cancer therapy; and nilotinib (Tasigna™), a derivative of imatinib that is significantly more potent and selective. In addition to the TKIs, humanized monoclonal antibody (MAb) drugs that counter the activity of RTKs involved in cancer have also been approved: trastuzumab (Herceptin™), an anti-ERBB2 MAb that was the first approved drug that antagonizes a tyrosine kinase (1998); cetuximab (Erbitux™)and panitumumab (Vectibix™), EGFR neutralizing MAbs; and cevacizumab (Avastin™), an anti-VEGF MAb. Many additional TKIs directed against other target tyrosine kinases are currently in cancer clinical trials. For instance, HGF receptor (MET), FAK, IGF-1R, SYK, FLT3, SRC, VEGFR2, and JAK2 inhibitors, among others, are in cancer trials, and many of these are sure to be approved for clinical use within the next few years.
Tyrosine Kinase Inhibitors; Looking Forward
Many of the first generation TKIs are not very selective, often having many kinase targets in addition to the target tyrosine kinase [31-33]. In large part this stems from the fact that appropriate counterscreens were not available for the majority of protein kinases at the time when these inhibitors were being developed. In addition, the technology for developing selective kinase inhibitors has improved significantly, particularly with the use of structure- and fragment-based design methods. Nevertheless, off-target specificities did not preclude the development of these first generation TKIs as drugs, and indeed a combination of on-target and off-target effects may partly underlie the effectiveness of these TKIs in some circumstances, particularly when there is redundancy in the tyrosine kinases that are activated in a tumor. The multikinase specificity of imatinib has been put to great use in the treatment of tumors that depend on mutationally activated c-KIT or PDGFRα RTKs, such as gastrointestinal stromal tumors (GIST). Moreover, a virtue is now being made of broad inhibitor specificity through the development of multikinase inhibitors.
The effectiveness of TKIs in cancer therapy provides strong support for the idea that individual tumors can be dependent on a single activated tyrosine kinase. However, there is also emerging evidence for redundancy among driver RTKs in cancer, such that inhibition of one tyrosine kinase may result in upregulation or activation of a second RTK that can serve the same function [34,35]. This suggests that the use of combinations of TKIs or multikinase inhibitors may be more effective. The “addiction” of cancers to specific signaling pathways is now being explored by screening the sensitivity of large numbers of tumor cell lines to approved protein kinase inhibitors [36]. In general, the most sensitive lines prove to contain activating mutations in the target kinase. For instance, the NSCLC cell lines most sensitive to erlotinib have activating EGFR mutations, and the melanoma, thyroid, and colorectal carcinoma lines sensitive to a RAF inhibitor have mutant B-RAF [36]. Kinome-wide resequencing studies on large numbers of tumor cell lines and primary tumors have become another avenue for pinpointing mutant kinases driving the tumor phenotype that have not previously been implicated in cancer [37-39]. The use of phosphoproteomic analysis of P.Tyr-containing peptides also has the potential to uncover activated tyrosine kinases in cancer [40]. Finally, kinome-wide siRNA screens of tumor cell lines harboring defined oncogenic mutations for protein kinase dependence are yielding interesting and distinct patterns of kinase dependency that highlight essential signaling pathways and provide kinase targets for inhibition that may not necessarily be mutant in the cancer [41-45].
Despite the success of the first generation of TKIs , resistance to the drugs is a frequent event, and in the great majority of cases this is found to be due to point mutations within the catalytic domain that render it resistant to inhibition. This has been particularly well worked out in the case of imatinib-resistant CML, where a large number of point mutations that render BCR-ABL resistant to imatinib has been identified through sequencing [46]. The most common is T315I, a mutation in the so-called gatekeeper residue, which blocks imatinib binding to the catalytic cleft because the bulkiness of the Ile side chain results in a steric clash. Mutation of the equivalent residue in EGFR, T790, to Met also commonly underlies the resistance of NSCLC with mutant EGFR [47,48] to treatment with the EGFR inhibitor erlotinib [49]. The propensity of tumors to generate resistant variants due to their genetic instability has demanded the generation of TKIs that can inhibit the mutant forms (e.g. sunitinib for imatinib-resistant GIST, and dasatinib and nilotinib for imatinib-resistant CML). In addition, prospective studies are now routinely carried out to determine what sort of mutations will render a target tyrosine kinase resistant to an inhibitor that is in clinical trials, so that a head start on developing second generation inhibitors can be made. In practice, it may prove beneficial to start therapy with a combination of inhibitors targeting the same tyrosine kinase, analogous to triple therapy for HIV, to reduce the chances of the tumor developing a resistant form of the kinase.
Looking forward, we can expect continuing development of second generation TKIs to circumvent the problem of resistance to first generation compounds. Allosteric inhibitors, which interact with sites outside the kinase domain catalytic cleft, are likely to become commonplace. In this regard, small molecule inhibitors that interact with the extracellular domains of RTKs and prevent dimerization are likely to be developed. Molecular diagnosis of individual tumors for activated tyrosine kinases will be used to define which TKIs to use for treatment. For therapeutic purposes, we need to learn how to use TKIs in the right combination with other signal transduction inhibitors, and with chemotherapeutic drugs, and to optimize dosing and scheduling to treat cancers more effectively. It will also be critical to elucidate the rules underlying the crosstalk and feedback mechanisms that lead to activation of redundant tyrosine kinase pathways upon inhibition of a single tyrosine kinase in cancer. Undoubtedly, TKIs will also be approved for use in the treatment of other types of disease, e.g. inflammatory and immune-related conditions, although this raises issues about the safety profiles appropriate for long term use of TKIs.
Coda
The last thirty years have been an exciting ride and we have learned an amazing amount about the functions and evolution of tyrosine phosphorylation. However, there is still much more to do and the next thirty years of research on tyrosine phosphorylation promises to hold more insights and surprises, as well as applications of this knowledge to benefit the human condition.
Acknowledgments
It is obviously impossible to write even a brief review of the past thirty years of research on tyrosine phosphorylation and cite all of the important papers that have made progress in our understanding and in clinical translation so rapid, and I apologize to all those whose work has been omitted from the reference list.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eckhart W, Hutchinson MA, Hunter T. An activity phosphorylating tyrosine in polyoma T antigen immunoprecipitates. Cell. 1979;18:925–933. doi: 10.1016/0092-8674(79)90205-8. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T, Sefton BM. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci U S A. 1980;77:1311–1315. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter T, Eckhart W. The discovery of tyrosine phosphorylation: it’s all in the buffer! Cell. 2004;116:S35–39. doi: 10.1016/s0092-8674(04)00049-2. [DOI] [PubMed] [Google Scholar]
- 4.Pawson T. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- *5.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762.. This paper describes the first complete catalogue of human protein kinases, and defines the set of 90 tyrosine kinases encoded by the human genome.
- 6.Plowman GD, Sudarsanam S, Bingham J, Whyte D, Hunter T. The protein kinases of Caenorhabditis elegans: a model for signal transduction in multicellular organisms. Proc Natl Acad Sci U S A. 1999;96:13603–13610. doi: 10.1073/pnas.96.24.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 8.Turk BE. Understanding and exploiting substrate recognition by protein kinases. Curr Opin Chem Biol. 2008;12:4–10. doi: 10.1016/j.cbpa.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *9.Manning G, Young SL, Miller WT, Zhai Y. The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan. Proc Natl Acad Sci U S A. 2008;105:9674–9679. doi: 10.1073/pnas.0801314105.. This paper and reference 10 identify the proteins involved in tyrosine phosphorylation-based signaling in the choanoflagellate Monosiga brevicollis, an ocean-dwelling protist. The complexity of the tyrosine phosphorylation network in this single-cell organism is unexpected and has implications for the evolution of tyrosine kinases.
- *10.Pincus D, Letunic I, Bork P, Lim WA. Evolution of the phospho-tyrosine signaling machinery in premetazoan lineages. Proc Natl Acad Sci U S A. 2008;105:9680–9684. doi: 10.1073/pnas.0803161105.. This paper and reference 19 identify the proteins involed in tyrosine phosphorylation-based signaling in the choanoflagellate Monosiga brevicollis, an ocean-dwelling protist. The complexity of the tyrosine phosphorylation network in this single-cell organism is unexpected and has implications for the evolution of tyrosine kinases.
- 11.Mayer BJ. Clues to the evolution of complex signaling machinery. Proc Natl Acad Sci U S A. 2008;105:9453–9454. doi: 10.1073/pnas.0804669105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grangeasse C, Cozzone AJ, Deutscher J, Mijakovic I. Tyrosine phosphorylation: an emerging regulatory device of bacterial physiology. Trends Biochem Sci. 2007;32:86–94. doi: 10.1016/j.tibs.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007;21:160–174. doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Depetris RS, Hu J, Gimpelevich I, Holt LJ, Daly RJ, Hubbard SR. Structural basis for inhibition of the insulin receptor by the adaptor protein Grb14. Mol Cell. 2005;20:325–333. doi: 10.1016/j.molcel.2005.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741–744. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levinson NM, Seeliger MA, Cole PA, Kuriyan J. Structural basis for the recognition of c-Src by its inactivator Csk. Cell. 2008;134:124–134. doi: 10.1016/j.cell.2008.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J, Liu J, Ghirlando R, Saltiel AR, Hubbard SR. Structural basis for recruitment of the adaptor protein APS to the activated insulin receptor. Mol Cell. 2003;12:1379–1389. doi: 10.1016/s1097-2765(03)00487-8. [DOI] [PubMed] [Google Scholar]
- 18.Chen H, Xu CF, Ma J, Eliseenkova AV, Li W, Pollock PM, Pitteloud N, Miller WT, Neubert TA, Mohammadi M. A crystallographic snapshot of tyrosine trans-phosphorylation in action. Proc Natl Acad Sci U S A. 2008;105:19660–19665. doi: 10.1073/pnas.0807752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bae JH, Lew ED, Yuzawa S, Schlessinger J. Structural basis underlying a novel mechanism for control of receptor tyrosine kinase signal selectivity. Cell. 2009 in press. [Google Scholar]
- 20.Donaldson LW, Gish G, Pawson T, Kay LE, Forman-Kay JD. Structure of a regulatory complex involving the Abl SH3 domain, the Crk SH2 domain, and a Crk-derived phosphopeptide. Proc Natl Acad Sci U S A. 2002;99:14053–14058. doi: 10.1073/pnas.212518799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan B, Lanyi A, Song HK, Griesbach J, Simarro-Grande M, Poy F, Howie D, Sumegi J, Terhorst C, Eck MJ. SAP couples Fyn to SLAM immune receptors. Nat Cell Biol. 2003;5:155–160. doi: 10.1038/ncb920. [DOI] [PubMed] [Google Scholar]
- *22.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026.. This study provides an example of the power of phosphoproteomics in identifying global intracellular phosphorylation events, and describes 103 tyrosine phosphorylation sites in HeLa cells, 53 of which are induced by EGF treatment.
- 23.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci U S A. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *24.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046.. This paper describes the development of technology to profile tyrosine phosphorylation sites selectively by using mass spectrometry to analyze P.Tyr-containing tryptic peptides isolated by affinity purification with anti-P.Tyr antibodies from digests of whole cell proteins.
- 25.Luo W, Slebos RJ, Hill S, Li M, Brabek J, Amanchy R, Chaerkady R, Pandey A, Ham AJ, Hanks SK. Global impact of oncogenic Src on a phosphotyrosine proteome. J Proteome Res. 2008;7:3447–3360. doi: 10.1021/pr800187n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amanchy R, Kalume DE, Iwahori A, Zhong J, Pandey A. Phosphoproteome analysis of HeLa cells using stable isotope labeling with amino acids in cell culture (SILAC) J Proteome Res. 2005;4:1661–1671. doi: 10.1021/pr050134h. [DOI] [PubMed] [Google Scholar]
- *27.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225.. This review catalogues Ser/Thr and Tyr kinases that are implicated in cancer through consistent mutations or altered expression.
- 28.Hunter T. Treatment for chronic myelogenous leukemia: the long road to imatinib. J Clin Invest. 2007;117:2036–2043. doi: 10.1172/JCI31691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levitzki A, Mishani E. Tyrphostins and other tyrosine kinase inhibitors. Annu Rev Biochem. 2006;75:93–109. doi: 10.1146/annurev.biochem.75.103004.142657. [DOI] [PubMed] [Google Scholar]
- *30.Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–4817. doi: 10.1182/blood-2008-07-077958.. A compelling narrative telling the story behind the development of imatininb for the treatment of chronic myelogenous leukemia written by the person primarily responsible.
- 31.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 32.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- *34.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946.. Studies showing that several RTKs are activated in most glioblastoma cells, and that combined inhibition of more than one these activated RTK is required to inhibit the proliferation of these cancer cells.
- 35.Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K, Possemato A, Nardone J, Innocenti G, Wetzel R, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci U S A. 2008;105:692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDermott U, Sharma SV, Dowell L, Greninger P, Montagut C, Lamb J, Archibald H, Raudales R, Tam A, Lee D, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104:19936–19941. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *37.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766.. This is the first report of the use of cancer genome resequencing to identify causal mutations in a protein kinase that had not previously been implicated in human cancer.
- *38.Bardelli A, Parsons DW, Silliman N, Ptak J, Szabo S, Saha S, Markowitz S, Willson JK, Parmigiani G, Kinzler KW, et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300:949. doi: 10.1126/science.1082596.. This describes the first kinome-wide analysis for cancer-associated mutations in tyrosine kinases in human cancer.
- *39.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610.. A comprehensive sequence of analysis of the coding regions of the entire kinome in 210 diverse human cancers reveals a significant number of protein kinases with putative “driver” mutations that may be causal in cancer.
- 40.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025.. This study describes the use of P.Tyr peptide profiling by mass spectrometry to identify new tyrosine kinases activated in non-small cell lung carcinoma.
- 41.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 42.Baldwin A, Li W, Grace M, Pearlberg J, Harlow E, Munger K, Grueneberg DA. Kinase requirements in human cells: II. Genetic interaction screens identify kinase requirements following HPV16 E7 expression in cancer cells. Proc Natl Acad Sci U S A. 2008;105:16478–16483. doi: 10.1073/pnas.0806195105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bommi-Reddy A, Almeciga I, Sawyer J, Geisen C, Li W, Harlow E, Kaelin WG, Jr, Grueneberg DA. Kinase requirements in human cells: III. Altered kinase requirements in VHL-/- cancer cells detected in a pilot synthetic lethal screen. Proc Natl Acad Sci U S A. 2008;105:16484–16489. doi: 10.1073/pnas.0806574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grueneberg DA, Degot S, Pearlberg J, Li W, Davies JE, Baldwin A, Endege W, Doench J, Sawyer J, Hu Y, et al. Kinase requirements in human cells: I. Comparing kinase requirements across various cell types. Proc Natl Acad Sci U S A. 2008;105:16472–16477. doi: 10.1073/pnas.0808019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grueneberg DA, Li W, Davies JE, Sawyer J, Pearlberg J, Harlow E. Kinase requirements in human cells: IV. Differential kinase requirements in cervical and renal human tumor cell lines. Proc Natl Acad Sci U S A. 2008;105:16490–16495. doi: 10.1073/pnas.0806578105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 47.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 48.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]