SUMMARY
Store-operated Ca2+ channels activated by the depletion of Ca2+ from the endoplasmic reticulum (ER) are a major Ca2+ entry pathway in non-excitable cells and are essential for T cell activation and adaptive immunity. Following store depletion, the ER Ca2+ sensor STIM1 and the CRAC channel protein Orai1 redistribute to ER-plasma membrane (PM) junctions, but the fundamental issue of how STIM1 activates the CRAC channel at these sites is unresolved. Here we identify a minimal, highly conserved 107-aa CRAC activation domain (CAD) of STIM1 that binds directly to the N- and C-termini of Orai1 to open the CRAC channel. Purified CAD forms a tetramer that clusters CRAC channels, but analysis of STIM1 mutants reveals that channel clustering is not sufficient for channel activation. These studies establish a molecular mechanism for store-operated Ca2+ entry in which the direct binding of STIM1 to Orai1 drives the accumulation and the activation of CRAC channels at ER-PM junctions.
INTRODUCTION
Store-operated Ca2+ channels, or SOCs, comprise the major receptor-activated Ca2+ entry pathway in non-excitable cells and play important roles in the control of gene expression, cell differentiation, secretion and Ca2+ homeostasis (Parekh and Putney, 2005). In their native environment, SOCs are activated by the stimulation of phospholipase C (PLC)-coupled receptors that generate inositol 1,4,5-trisphosphate (IP3) and release Ca2+ from the endoplasmic reticulum (ER). The defining feature of SOCs is that they are activated by the reduction of [Ca2+]ER rather than by receptor-associated signaling molecules, such as G proteins, PLC, or IP3. The best characterized store-operated channel is the Ca2+ release-activated Ca2+ (CRAC) channel, whose activation is a steep function of [Ca2+]ER (Luik et al., 2008; Prakriya and Lewis, 2004). CRAC channels play essential roles in T lymphocytes and mast cells, where they provide the pathway for Ca2+ entry triggered by antigen recognition or allergens, respectively, and are required for T cell activation and mast cell degranulation (Feske et al., 2001; Feske et al., 2005; Partiseti et al., 1994; Vig et al., 2008).
The molecular mechanism by which ER Ca2+ depletion activates the CRAC channel has been a mystery since the original proposal of the store-operated Ca2+ entry (SOCE) hypothesis over 20 years ago (Prakriya and Lewis, 2004; Putney, 1986). However, remarkable progress has been made in the past several years following the identification of STIM1 as the ER Ca2+ sensor (Liou et al., 2005; Roos et al., 2005; Zhang et al., 2005) and Orai1 as the pore-forming subunit of the CRAC channel (Prakriya et al., 2006; Vig et al., 2006; Yeromin et al., 2006). Recent studies show that the loss of ER Ca2+ triggers the oligomerization of STIM1 (Liou et al., 2007; Muik et al., 2008; Stathopulos et al., 2006) and its accumulation in regions of the ER located within 10–25 nm of the plasma membrane (Wu et al., 2006), commonly referred to as “puncta.” Orai1 accumulates in overlying regions of the plasma membrane (PM) in register with STIM1 (Luik et al., 2006; Xu et al., 2006), culminating in the local entry of Ca2+ through CRAC channels (Luik et al., 2006). A recent study shows that STIM1 oligomerization is the key event that triggers the redistribution of STIM1 and Orai1, translating changes in [Ca2+]ER into graded activation of the CRAC channel (Luik et al., 2008).
While these studies demonstrate that STIM1 and Orai1 redistribute to ER-PM junctions following depletion of the internal stores it is still not clear how this occurs. STIM1 forms puncta in response to store depletion even when expressed in the nominal absence of Orai1 (Xu et al., 2006), suggesting that its initial target may be independent of Orai1. In contrast, Orai1 only forms puncta in store-depleted cells when co-expressed with STIM1, suggesting that it becomes trapped at ER-PM junctions by binding to STIM1 or an associated protein (Xu et al., 2006). Several parts of the cytosolic domain of STIM1, including the C-terminal polybasic domain, an ERM-like domain, and a serine-proline-rich domain, have been implicated in the activation of Orai1, but their specific roles and interactions in these localization events are not understood (Baba et al., 2006; Huang et al., 2006; Li et al., 2007; Liou et al., 2007).
The molecular mechanism by which STIM1 activates the CRAC channel has also been controversial. A widely considered ‘diffusible messenger’ model posits that STIM1 oligomerization promotes the synthesis of a ‘Ca2+ influx factor’ (CIF), which is delivered locally at ER-PM junctions to stimulate iPLA2β to produce lysolipids that activate ICRAC (Bolotina, 2008). An alternative conformational coupling hypothesis (Berridge, 1995) proposes that STIM1 binds physically to the CRAC channel or to an associated protein to activate Ca2+ entry. Precisely how this binding event might activate the channel is unclear; one recent study has proposed that STIM1 links Orai1 dimers to form active tetrameric channels (Penna et al., 2008), while another study concludes that Orai1 is a tetramer at rest, suggesting instead that STIM1 activates Orai1 by an allosteric mechanism (Ji et al., 2008). The conformational coupling model gains indirect support from the close proximity of STIM1, Orai1 and open CRAC channels at ER-PM junctions (Luik et al., 2006; Wu et al., 2006), and FRET between labeled STIM1 and Orai1 following store depletion (Muik et al., 2008; Navarro-Borelly et al., 2008). In addition, the cytosolic domain of STIM1 (CT-STIM1) expressed in soluble form localizes to the PM in an Orai1-dependent manner and activates ICRAC (Huang et al., 2006; Muik et al., 2008; Zhang et al., 2008) providing indirect evidence that Orai and STIM1 form a protein complex. Co-immunoprecipitation studies have been somewhat equivocal with STIM1 and Orai1 reported to co-immunoprecipitate after store depletion (Yeromin et al., 2006), before store depletion (Vig et al., 2006), or not at all (Gwack et al., 2007). However, co-immunoprecipitation experiments are limited in that they cannot distinguish direct binding of STIM1 to Orai1 from the formation of multi-protein complexes. There is currently no definitive evidence demonstrating a direct interaction between STIM1 and Orai1.
In this study we show that the C-terminal polybasic domain of STIM1 is required for STIM1 targeting to ER-PM junctions in the nominal absence of Orai1. We show that STIM1 binds directly to Orai1 via a 107-residue CRAC activation domain (CAD) that associates with the N- and C-termini of Orai1. Mutant STIM1 proteins that lack the CAD or contain mutations in the CAD show that this domain is necessary and sufficient to cluster and activate CRAC channels, and mutant STIM1 proteins also demonstrate that CRAC-channel clustering and activation are functionally separable events. These results establish that the direct binding of STIM1 to Orai1 is the driving mechanism for both the redistribution and activation of CRAC channels in response to Ca2+ store depletion.
RESULTS
STIM1 Accumulates at ER-PM Junctions by Orai1-dependent and -independent Mechanisms
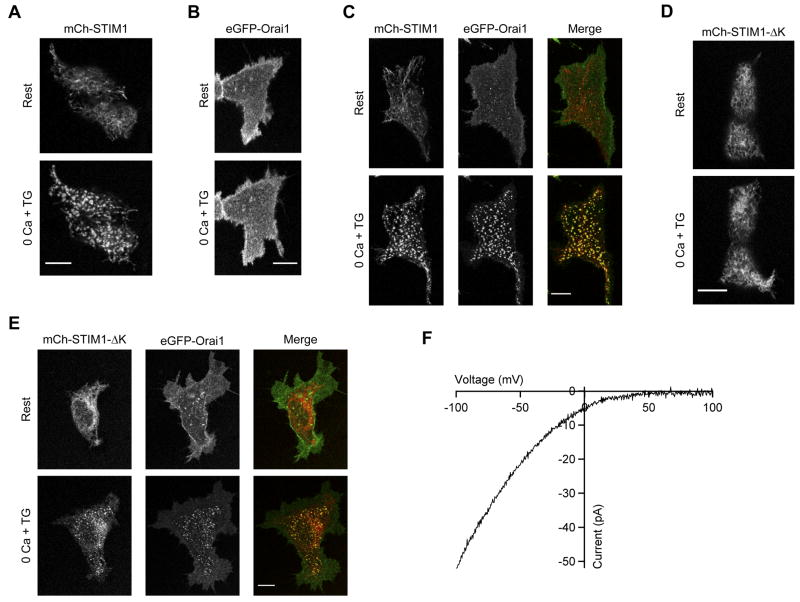
We initially investigated whether the formation of puncta by STIM1 and Orai1 following depletion of stores depends on co-expression of both proteins. When expressed by itself in HEK 293 cells, mCherry-labeled STIM1 (mCh-STIM1) formed distinct puncta after depletion of Ca2+ stores with thapsigargin (TG) (Fig. 1A). In contrast, GFP-myc-Orai1 expressed alone did not form puncta in response to TG (Fig. 1B), but coexpression of mCh-STIM1 restored its ability to form puncta (Fig. 1C). These results suggest that STIM1 recruitment to ER-PM junctions is independent of Orai1, whereas Orai1 recruitment to these sites depends on binding to STIM1 or a STIM1-associated protein as has been previously suggested (Xu et al., 2006).
Figure 1. Deletion of the STIM1 Polybasic Domain Distinguishes Orai1-dependent and independent Mechanisms of STIM1 Accumulation at ER-PM Junctions.
(A) Wild-type mCh-STIM1 expressed alone in HEK 293 cells redistributes from a diffuse ER distribution (Rest) to the cell periphery after store depletion with TG (0 Ca + TG). After depletion, puncta are visible at the cell footprint. (B) eGFP-myc-Orai1 when expressed alone does not redistribute into puncta after depletion. (C) When expressed together, STIM1 and Orai1 form colocalized puncta after store depletion. (D) Unlike wild-type STIM1, STIM1-ΔK expressed alone fails to form puncta; however, when coexpressed with Orai1 both proteins form colocalized puncta after store depletion (E). (F) Store depletion activates ICRAC in HEK 293 cells expressing STIM1-ΔK and Orai1 demonstrating that the polybasic region is not essential for ICRAC activation. All images were taken of cell footprints using confocal microscopy. Scale bars, 10 μm.
To identify the regions of STIM1 that are necessary for puncta formation we investigated the role of the polybasic C-terminal domain (aa 672–685). Deletion of this region (STIM1-ΔK) has been reported to prevent puncta formation and SOC activation in some studies (Huang et al., 2006; Liou et al., 2007) but not in others (Li et al., 2007). We found that STIM1-ΔK failed to form puncta following store depletion when expressed alone in HEK 293 cells (Fig. 1D); however, when expressed together with Orai1, both proteins colocalized in puncta and activated ICRAC after store depletion (Fig. 1E, F). These data suggest that the polybasic domain is required to localize STIM1 to ER-PM junctions in the absence of Orai1 but that a second domain recruits and activates Orai1 at these sites.
Identifying a Minimal Cytosolic Region of STIM1 that Opens the CRAC Channel
To identify the CRAC-activating domain of STIM1, we first tested a series of soluble cytosolic STIM1 fragments for their ability to activate an NFAT-dependent luciferase reporter gene (NFAT-luc). A series of constructs were generated by progressive truncation of the full-length cytosolic region of STIM1 (Fig 2A; STIM1234–685; CT-STIM1) and were transiently expressed in a HEK 293T cell line containing NFAT-luc. Because NFAT-dependent transcription requires the sustained elevation of intracellular Ca2+ ([Ca2+]i) combined with a phorbol ester to activate protein kinase C (PKC), treatment of cells bearing only NFAT-luc with phorbol 12-myristate 13-acetate (PMA; 1 μM) does not stimulate luciferase production. However, PMA in conjunction with 1 μM TG, which activates Ca2+ entry through endogenous CRAC channels, activates NFAT-luc significantly (Fig. 2B). Therefore we compared luciferase production in the presence of PMA with that in PMA + TG to assess the ability of STIM1 fragments to activate endogenous CRAC channels.
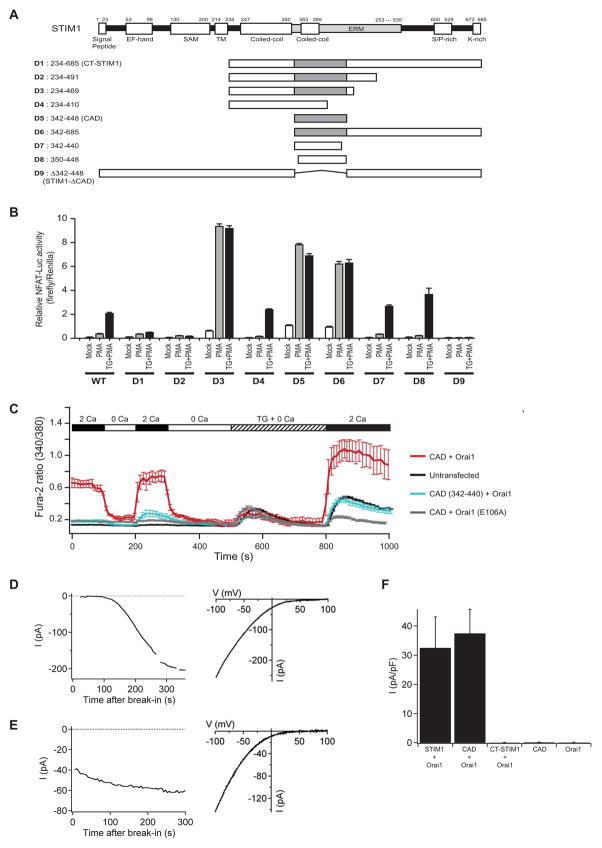
Figure 2. Identification of CAD as a Potent CRAC Channel Activator.
(A) Full-length STIM1 with its putative functional domains (top) and truncated versions of STIM1 (D1-D9; bottom) with the CAD shown in gray. (B) NFAT-dependent luciferase activity in HEK 293T (NFAT-Luc) cells transfected with wild-type (WT) or truncated (D1-D9) STIM1 constructs. Cells were treated with 1 μM PMA or 1 μM TG+PMA as shown. D5 (CAD) is the minimal region that is necessary and sufficient to activate NFAT. Data are shown as mean ± sem (n=4). (C) CAD activates SOCE without depleting intracellular Ca2+ stores. Mean [Ca2+]i (± sem) in untransfected HEK 293 cells (black; n=28) and cells expressing CAD + Orai1 (red; n=15), CAD + Orai1E106A (gray; n=17), or CAD (342–440) + Orai1 (blue; n=12). (D) (left) ICRAC develops slowly in a representative cell cotransfected with WT GFP-STIM1 + myc-Orai1. Current recorded during brief pulses to −100 mV in 2 mM Ca2+o is plotted against time after break-in. (right) Characteristic I–V relationship for ICRAC recorded in 20 mM Ca2+o from the same cell. (E) (left) ICRAC is constitutively active in a representative cell cotransfected with YFP-CAD + myc-Orai1. Current is plotted as in D. (right) I–V relationship for the CAD-induced current recorded in 20 mM Ca2+o from the same cell. (F) Current densities in cells transfected with CAD or Orai1 alone, or cotransfected with myc-Orai1 and GFP-STIM1, YFP-CAD, or YFP-CT-STIM1. Current measured at −100 mV during voltage ramps in 20 mM Ca2+o was normalized to the cell capacitance. Mean values ± sem from 4–5 cells are shown.
While CT-STIM1 did not activate the NFAT reporter gene with PMA alone, truncations of either the C- or N-terminus of this protein generated several active STIM1 peptides (D3, D5, D6). By making additional truncations we identified STIM1342–448 (D5) as the minimal peptide that was sufficient to activate NFAT-luc; we will refer to this domain hereafter as the CRAC activation domain (CAD). Western analysis showed that the inactivity of peptides D1, D2, D4, and D7–9 was not due to inadequate expression (Supplemental Fig. 1A). The CAD encompasses a putative coiled-coil and part of the ERM domain of STIM1, is highly conserved among vertebrates and invertebrates from C. elegans to H. sapiens and is virtually identical to a sequence in STIM2, another ER Ca2+ sensor that controls CRAC channel activation (Brandman et al., 2007) (Supplemental Fig. 2).
To determine if CAD activates store operated Ca2+ influx in cells, we measured [Ca2+]i in HEK 293 cells expressing CAD and Orai1. CAD evoked a sustained [Ca2+]i elevation that was dependent on extracellular Ca2+ (Fig. 2C), was suppressed by CRAC channel nhibitors like 2-APB and 10 μM La3+ (Supplemental Fig. 3A, B), and was not observed in cells coexpressing a dominant-negative nonconductive Orai1 mutant (Orai1E106A; Fig. 2C). Importantly, CAD activated Ca2+ entry without depleting intracellular stores, because the Ca2+ released by TG in Ca2+-free media was similar in CAD-expressing and untransfected cells (Fig. 2C). These results suggest that CAD elevates [Ca2+]i by activating CRAC channels independently of store depletion.
As a definitive test for CRAC channel activation, we conducted whole-cell patch clamp recordings from HEK 293 cells transiently transfected with myc-Orai1 and either YFP-STIM1 or YFP-CAD. In cells expressing full length YFP-STIM1, ICRAC appeared over several minutes following break-in, consistent with the typical slow activation seen in response to passive store depletion (Fig. 2D). ICRAC showed the characteristic inwardly-rectifying current-voltage relation (Fig. 2D, right), inhibition by La3+ and modulation by 2-APB. In contrast, inward current was present from the moment of break-in in cells expressing YFP-CAD (Fig. 2E). This current was identified as ICRAC based on its dependence on extracellular Ca2+, inward rectification, sequential enhancement and inhibition by 2-APB, and inhibition by La3+ (Fig. 2E and Supplemental Fig. 3C, D). ICRAC was not observed at break-in in HEK 293 cells transfected with YFP or YFP-CAD in the absence of Orai1 (Fig. 2F), consistent with the low level of endogenous STIM1 and Orai1 in these cells. Interestingly, the current activated by CAD differed from native ICRAC in that it lacked fast Ca2+-dependent inactivation (Zweifach and Lewis, 1995). Inactivation was restored by addition of STIM1 residues carboxy-terminal to CAD (Mullins, Park, Dolmetsch and Lewis, manuscript in preparation).
Transfection of HEK 293 cells with YFP-CT-STIM1 + myc-Orai1 failed to activate constitutive ICRAC (Fig. 2F) or elevate resting [Ca2+]i (data not shown), consistent with the lack of activity in the NFAT-luciferase assay. CT-STIM1 was inactive even though it was expressed at comparable levels to CAD or wild-type STIM1 (WT-STIM1; Supplemental Fig. 1B). While coexpression of CT-STIM1 + Orai1 did not elevate resting [Ca2+]i in HEK 293 cells, it did so in HEK 293T cells, which express large T antigen and express proteins at substantially higher levels than HEK 293 cells (data not shown). Our studies demonstrate that CAD is a much more potent activator of Orai1 than CT-STIM1, based on its ability to activate ICRAC comparably to WT-STIM1 at expression levels at which CT-STIM1 activity is undetectable (Fig. 2F).
CAD Associates with Orai1 in vivo and in vitro
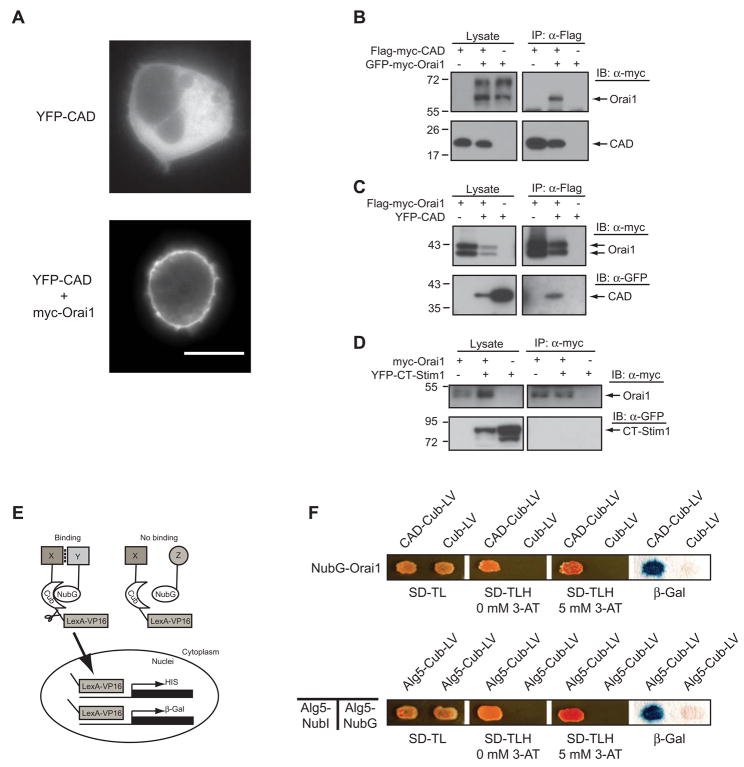
Because cytosolic CAD is a potent activator of Orai1 channels and is not associated with the ER, we hypothesized that CAD might bind to Orai1. To test this idea, we first expressed YFP-CAD with or without myc-Orai1 in HEK 293 cells and examined its intracellular localization. In the absence of Orai1, YFP-CAD was localized diffusely throughout the cytoplasm, but the introduction of Orai1 led to a dramatic recruitment of YFP-CAD to the plasma membrane, suggesting that the two proteins form a complex (Fig. 3A). To provide additional evidence that CAD and Orai1 are part of the same protein complex we expressed Flag-tagged CAD and GFP-tagged Orai1 in HEK 293T cells and immunoprecipitated CAD with anti-Flag antibodies. GFP-myc-Orai1 was detected in the immunoprecipitates from cells co-expressing CAD and Orai1 but not from cells expressing CAD or Orai1 alone (Fig. 3B). The reciprocal experiment, in which Flag-tagged Orai1 was immunoprecipitated using anti-Flag antibodies and YFP-CAD was detected by Western blotting, confirmed that CAD co-immunoprecipitates with Orai1 only when the two proteins are coexpressed (Fig. 3C). Thus, CAD and Orai1 form a protein complex in mammalian cells. Under the same conditions (150 mM salt, 1% Triton X-100) we were unable to detect any interaction between CT-STIM1 and Orai1 (Fig. 3D), suggesting that the affinity of the full-length cytoplasmic domain of STIM1 for Orai1 is much weaker than that of the isolated CAD. This result may explain why CT-STIM1 is a weak activator of CRAC channels and suggests that CAD is not exposed in CT-STIM1.
Figure 3. CAD Associates with Orai1.
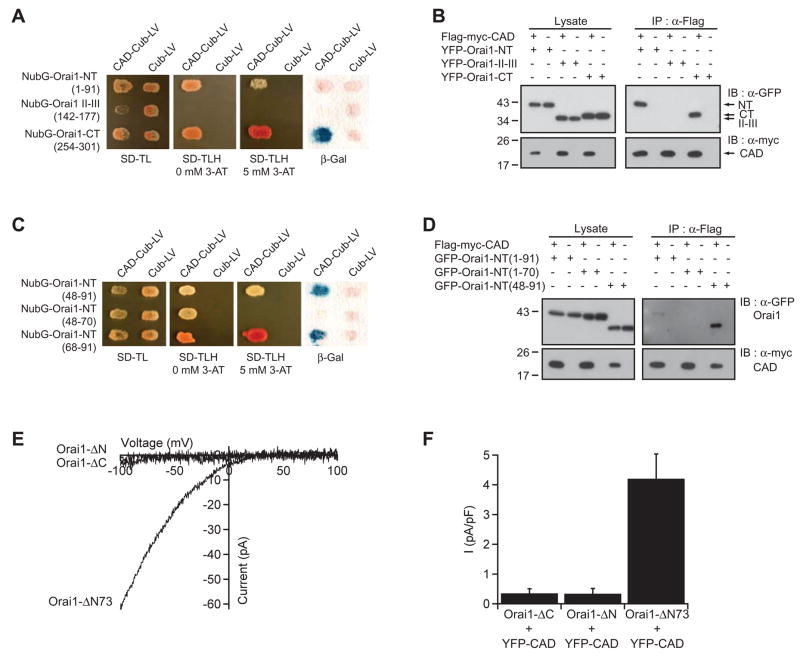
(A) YFP-CAD is cytosolic when expressed by itself in a HEK 293 cell (top), but accumulates at the cell perimeter when coexpressed with Orai1 (bottom). Scale bar, 10 μm. (B, C) Western blots of cell lysates (left) or immunoprecipitated material (right) from cells expressing CAD, Orai1, or CAD + Orai1. Anti-Flag antibodies co-immunoprecipitate GFP-myc-Orai1 with Flag-myc-CAD (B), and co-immunoprecipitate YFP-CAD with Flag-myc-Orai1 (C). (D) CT-STIM1 does not co-immunoprecipitate with myc-Orai1 (representative of 4 experiments). (E) Schematic depiction of the yeast split ubiquitin assay. (F) CAD associates with Orai1 in the split ubiquitin assay. Yeast containing NubG-Orai1 and either Cub-LV alone or CAD-Cub-LV, grow well on plates lacking tryptophan and leucine but containing histidine (SD-TL). Only yeast expressing NubG-Orai1 and CAD-Cub-LV grow on plates lacking all three amino acids (SD-TLH) in the absence or presence of 5 mM 3-aminotriazole (3AT), a competitive HIS3 inhibitor that increases the stringency of selection. Yeast containing CAD-Cub-LV and NubG-Orai1 also activate LacZ whereas cells expressing NubG-Orai1 and Cub do not. The homomeric interaction of Alg5 is shown as a positive control.
As a third test for association of CAD and Orai1 in vivo, and to map the interaction domains of Orai1 we introduced both proteins into a yeast split ubiquitin interaction system (Thaminy et al., 2004). In this system, interaction of the two test proteins reunites the N- and C-terminal fragments of ubiquitin, releasing a LexA-VP16 transcriptional activator that enters the nucleus and activates reporter genes (Fig. 3E). Fusion proteins of Orai1 and the N-terminal fragment of ubiquitin (NubG-Orai1), and of CAD and the C-terminal fragment of ubiquitin fused to LexA-VP16 (CAD-Cub-LV) were introduced into a yeast strain containing LexA His and β-galactosidase reporter genes. Yeast containing both the NubG-Orai1 and CAD-Cub-LV survived on His selection plates and produced significant levels of β-gal, whereas yeast expressing NubG-Orai1 and the Cub-LV domain alone did not (Fig. 3F), indicating that CAD and Orai1 interact with each other in a heterologous system.
CAD Binds Directly to Orai1
To test for direct binding between the CAD and Orai1 we generated a GST-tagged CAD peptide in E. coli and an Orai1 protein containing C-terminal octa-histidine and N-terminal EEYMPME (“EE”) tags in insect Hi5 cells. We incubated purified GST-CAD or GST alone with purified EE-Orai1-His8 and used glutathione beads to precipitate the complexes. GST-CAD co-precipitated Orai1, while GST alone did not, indicating that CAD binds directly to Orai1 in vitro (Fig. 4A).
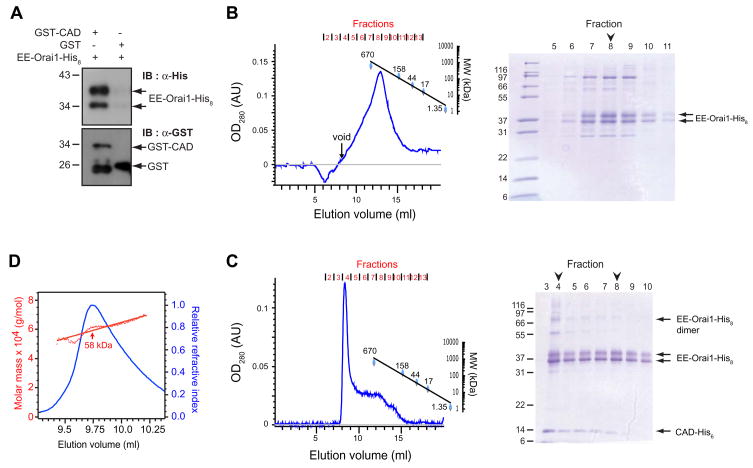
Figure 4. CAD Binds Directly to Orai1.
(A) Glutathione beads precipitate EE-Orai1-His8 with GST-CAD but not GST alone. (B) (Left) Size-exclusion profile of purified EE-Orai1-His8. On SDS-PAGE (right), peak fractions contain EE-Orai1-His8 as a pair of glycosylated and unglycosylated bands at ~37 kDa. (C) (Left) Size-exclusion profile of EE-Orai1-His8 and CAD-His6 coexpressed in Hi5 cells and affinity purified in 0.5 M NaCl. Much of the CAD and Orai1 co-elute in the void volume (MW > 10 MDa) of the Superose 6 gel filtration column (fraction 4). The band corresponding to the dimer of EE-Orai1-His8 (arrow) was identified by Western blotting. (D) Multi-angle light scattering (MALS) indicates a molecular weight of ~58 kDa for purified CAD-His6, approximately four times the predicted mass of the monomer.
To examine the size of the protein complexes generated by the interaction of Orai1 and CAD, we prepared extracts from Hi5 cells expressing EE-Orai1-His8 alone or in combination with CAD-His6 and analyzed them by size exclusion chromatography. Orai1 and CAD proteins were first affinity-purified with Ni2+-NTA and Orai1 was then immunoprecipitated with anti-EE antibodies. Size exclusion chromatography of Orai1 alone revealed a monodisperse peak with an apparent mass of ~ 290 kD (Fig. 4B, left) consistent with Orai1 forming multimers in the absence of CAD. Analysis of the eluate fractions by SDS-PAGE revealed a doublet of ~37 kD (Fig. 4B, right) representing glycosylated and unglycosylated forms of EE-Orai1-His8 as determined by the ability of tunicamycin to collapse the top band to the lower band (data not shown). In contrast to Orai1 alone, EE-Orai1-His8 purified from cells coexpressing CAD-His6 eluted largely in the void volume, consistent with the formation of a large protein complex of molecular weight > 10 MDa (Fig. 4C). The complex was stable in 0.5 M NaCl, suggesting a high-affinity hydrophobic interaction of multiple CAD and Orai1 proteins. Importantly, complex formation did not involve significant amounts of additional proteins, indicating that the interaction between CAD and Orai1 is direct (Fig. 4C). The excess free CAD-His6, isolated by gel filtration after Ni2+-NTA purification of the complex, was analyzed by multi-angle light scattering (MALS) which indicated a molecular weight of ~58 kD, or 4.4 times the predicted weight of 13.2 kD for the CAD-His6 monomer (Fig. 4D). Together, these results show that Orai1 exists as a multimer in the absence of CAD, and CAD forms a tetramer in free solution that links together multiple Orai1 multimers.
STIM1-CAD Binds to the N- and C-termini of Orai1
To identify regions of Orai1 important for activation by CAD, we characterized the binding of CAD to Orai1 using two approaches. First we expressed fusions of NubG with the N-terminus, the II–III cytoplasmic loop and the C-terminus of Orai1 in yeast together with different CAD-Cub-LV constructs. Survival of colonies in selection media and production of β-gal revealed that CAD binds to the N- and C-terminus of Orai1 but not to the II–III loop or to the C ubiquitin fragment alone (Fig. 5A). Based on the level of β-gal activity, CAD appears to interact with the Orai1 C-terminus with a higher affinity than with the N-terminus. We next expressed fusions of YFP with the N-terminus, the II–III loop and the C-terminus of Orai1 in HEK 293T cells together with Flag-myc-CAD. Immunoprecipitation of Flag-myc-CAD followed by Western blotting revealed that CAD interacts with the N- and C-terminus of Orai1 but not with the II–III loop (Fig. 5B).
Figure 5. CAD Binds to Both the N- and C-termini of Orai1.
(A) In this split ubiquitin assay,β-gal production and growth of transformants on plates lacking histidine indicate a strong interaction between CAD and the C-terminus of Orai1, a weaker interaction with the N-terminus, and lack of interaction with the II–III loop of Orai1. (B) CAD in HEK 293T cells co-immunoprecipitates with YFP-tagged N-terminal and C-terminal fragments of Orai1 but not with the II–III loop. (C) Split ubiquitin assays showing that CAD interacts with aa 48–91 of the Orai1 N-terminus; this appears to be due to binding to aa 68–91 rather than aa 48–70. (D) CAD co-immunoprecipitates with Orai1-NT(48–91) but only very weakly with the entire N-terminus of Orai1 (aa 1–91). (E) Whole-cell recordings in 20 mM Ca2+o from HEK 293 cells co-expressing truncated Orai1-GFP proteins and YFP-CAD. Deletion of the N- or C-terminus of Orai1 abrogates function, but a substantial level of ICRAC is generated by Orai1-ΔN73. (F) Summary of ICRAC measurements (current at −100 mV, 20 mM Ca2+o, normalized to cell capacitance) with the indicated constructs (4 cells each, mean ± sem).
To explore the interaction between the N-terminus of Orai1 and CAD in more detail we mapped the subregion within the N-terminus of Orai1 that is responsible for CAD binding, using both the split ubiquitin and HEK 293T co-immunoprecipitation assays. In the yeast assay, CAD interacted strongly with Orai148–91 and Orai168–91 but not with Orai148–70. Similarly, in the HEK cell assay CAD co-immunoprecipitated with Orai148–91 but not with Orai11–70, suggesting that CAD binds to the region of aa 70–91 (Fig. 5D). The interaction between CAD and Orai148–91 was significantly stronger than that between CAD and the full-length N-terminus of Orai1 (Fig. 5D, right), suggesting that aa 1–48 reduce the affinity between CAD and the isolated Orai1 N-terminus.
We next tested the function of the CAD-binding regions of Orai1 using whole-cell recording. We introduced CAD into HEK 293 cells together with Orai1 lacking the full N- (Orai1-ΔN) or C-terminus (Orai1-ΔC) or the initial 73 residues of the N-terminus (Orai1-ΔN73) preceding the CAD binding site (Li et al., 2007). Western blotting and immunostaining of Orai1 containing an extracellular HA epitope confirmed that these mutations do not alter the expression or cell surface localization of the channels (Supplemental Fig. 4). CAD constitutively activated ICRAC in cells expressing Orai1-ΔN73 but not in cells expressing Orai1-ΔN or Orai1-ΔC (Fig. 5E, F), showing that the N- and C-termini of Orai1 are both necessary for activation by CAD but that aa 1–73 are not absolutely required (see also (Li et al., 2007). Furthermore, deletion of aa 73–84 from Orai1 suppressed CAD-induced Ca2+ influx (Supplemental Fig. 5). Taken together with the results of Fig. 5C and D, these findings suggest that CAD binding to the C-terminus of Orai1 and the membrane-proximal region of the Orai1 N-terminus is required to activate the CRAC channel.
CAD Clusters CRAC Channels
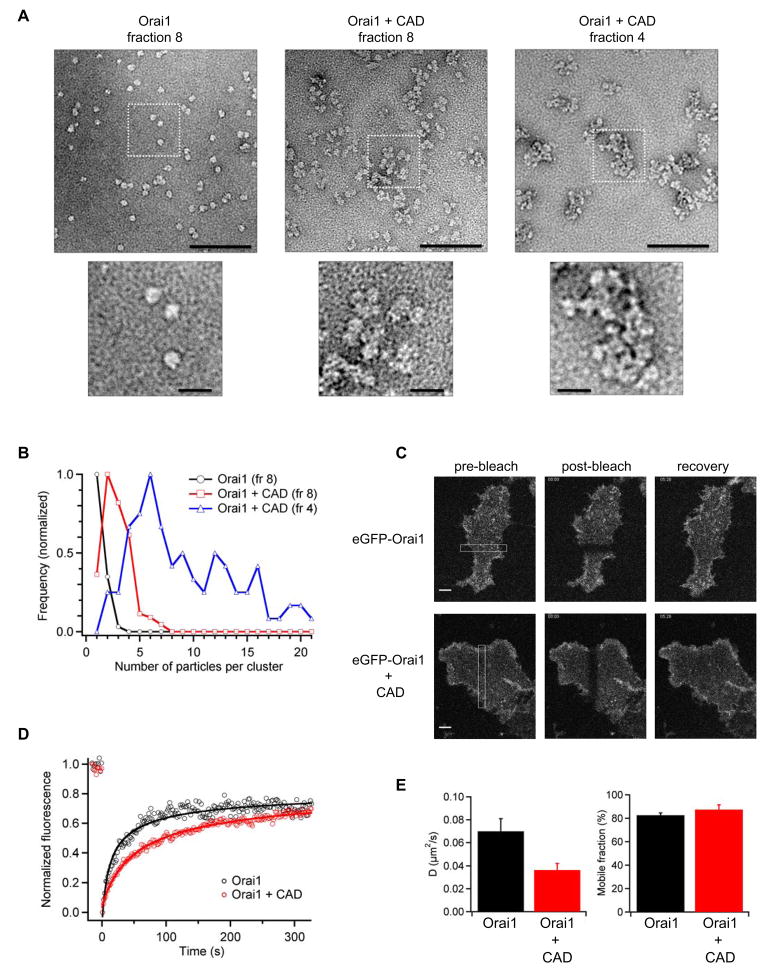
The large size of the CAD/Orai1 complex indicates that CAD clusters Orai1. To investigate the nature of these complexes and test for non-specific aggregation, we examined purified material from the gel filtration column by negative stain single-particle electron microscopy (Fig. 6A, B). Analysis of purified Orai1 alone revealed primarily particles of 8–10 nm diameter, presumably representing single CRAC channels, with a low frequency of pairs and triplets. In contrast, in the presence of CAD we observed clusters of Orai1 unitary particles that increased in frequency and size with increasing molecular weight of the column eluates. Taken together with the MALS results these images suggest that tetramers of CAD bind to multiple sites on CRAC channels to create these clusters.
Figure 6. CAD Links Multiple CRAC Channels to Form Clusters.
(A) Negative stain electron microscopy of purified EE-Orai1-His8 (left; fraction 8 from Fig. 4B), or complexes of EE-Orai1-His8 and CAD-His6 (right panels; fractions 4 and 8 from Fig. 4C). Scale bars, 100 nm (top), 20 nm (bottom, enlargements from dashed boxes). (B) Quantitation of cluster sizes for each condition shown in A; Orai1 (n=174), Orai1 + CAD (fr 8, n=134; fr 4, n=90). Each histogram is normalized to its maximum bin value. (C) FRAP of HEK 293 cells expressing eGFP-Orai1 (top row) or eGFP-Orai1 + CAD (bottom row). Images are shown at the indicated times after bleaching a bar across the cell footprint (left). Scale bars, 5 μm. (D) Time course of FRAP from single cells expressing eGFP-Orai1 alone (black) or with CAD (red). The superimposed fits indicate diffusion coefficients of 0.12 μm2/s (Orai1) and 0.026 μm2/s (Orai1 + CAD). (E) Mean diffusion coefficients and mobile fractions of GFP-Orai1 expressed alone (n=9) or with CAD (n=6; means ± sem).
To test whether CAD also clusters CRAC channels in intact cells, we conducted fluorescence recovery after photobleaching (FRAP) experiments of GFP-Orai1 expressed alone or with CAD in HEK 293 cells. Our FRAP measurements of GFP-Orai1 alone suggest that 83 ± 2% of the channels are mobile in the membrane with an effective diffusion coefficient (D) of 0.070 ± 0.011 μm2/s (n=9, Fig. 6C–E). In cells co-expressing GFP-Orai1 and CAD, the mobile fraction was unchanged (87 ± 4%) but Orai1 diffusion was slowed by a factor of two (D = 0.036 ± 0.006 μm2/s, n=6). Thus, CAD does not appear to anchor CRAC channels to an immobile substrate, but does slow their diffusion significantly, consistent with CAD-induced clustering of Orai1 in the cell membrane.
The Role of CAD in Orai1 Binding and Activation by STIM1
A key question is whether CAD is responsible for the clustering and activation of CRAC channels by full-length STIM1 following store depletion. To address this, we measured the ability of mCherry-tagged STIM1 variants containing CAD mutations to co-cluster with eGFP-Orai1 and activate Ca2+ entry in response to TG (Fig. 7). STIM1-Orai1 colocalization was quantified as the fraction of total STIM1 and Orai1 fluorescence that was recruited to regions of high fluorescence covariance (Supplemental Fig. 6), while Ca2+ influx was measured with single cell Ca2+ imaging. For each set of experiments we measured mCherry and GFP fluorescence to confirm that differences in the activity of the various STIM1 constructs were not due to differences in expression level (Supplemental Fig. 7).
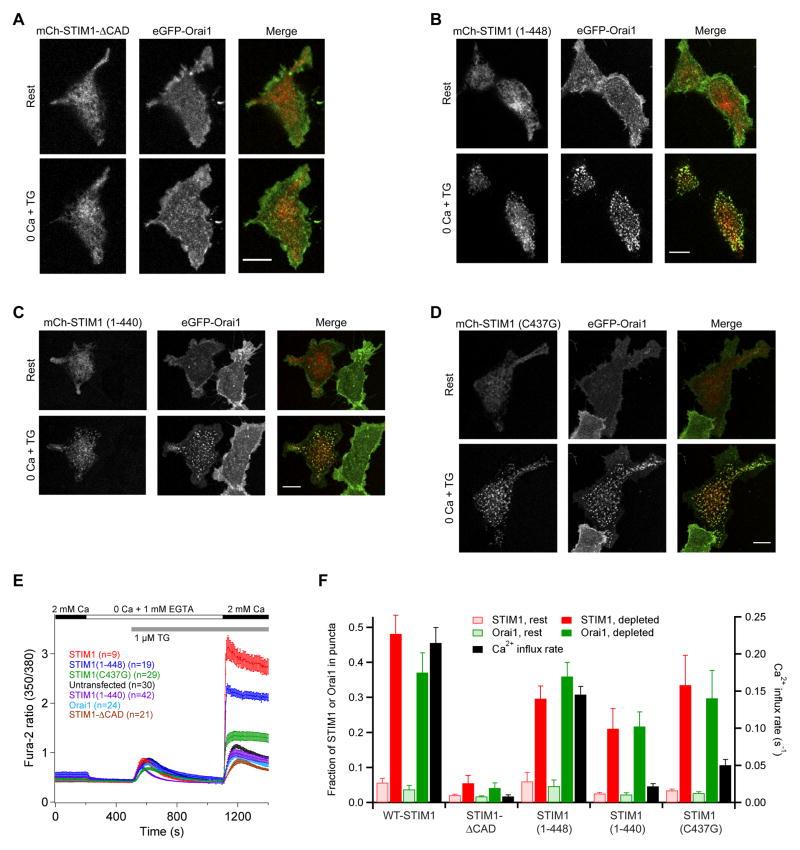
Figure 7. CRAC Channel Clustering and Activation by STIM1 are Separable and Require the CAD Region.
(A) When coexpressed with Orai1, STIM1-ΔCAD fails to form puncta and cluster Orai1 after store depletion. In contrast, STIM11–448 (B), STIM11–440 (C), and STIM1 C437G (D) co-accumulate with Orai1 in puncta after store depletion. In A–D, images are confocal micrographs of the HEK 293 cell footprint. Scale bars, 10 μm. (E) Ca2+ measurements (mean ± sem) in HEK 293 cells expressing the indicated constructs. In all cases, Orai1 was tagged with eGFP and the STIM1 or its variant was tagged with mCherry. (F) Puncta formation and Ca2+ influx in cells expressing Orai1 and STIM1 variants. Each set of bars shows the fraction of the cell s STIM1 or Orai1 fluorescence that is colocalized to puncta before and after store depletion (mean ± sem, n=4 cells for each) and the initial rate of Ca2+ entry (dR/dt, where R is the fura-2 350/380 fluorescence ratio) measured 10–20 s after readdition of Ca2+ in E.
We first deleted the CAD from WT-STIM1 (STIM1- CAD) and found that when expressed with eGFP-Orai1, mCh-STIM1-ΔCAD failed to form puncta, cluster Orai1 or activate Ca2+ entry in response to TG (Fig. 7A, E, F). Moreover, in the NFAT-luciferase assay, STIM1-ΔCAD inhibited NFAT activation by endogenous channels following treatment with TG + PMA (Fig. 2B), suggesting that it forms non-functional oligomers with endogenous STIM1. These results show that the CAD is required for STIM1 to form puncta and to recruit and activate Orai1 at ER-PM junctions.
To test whether the CAD in STIM1 can function independently of the residues C-terminal to it, we truncated STIM1 from the C-terminus to the end of the CAD (STIM11–448). When coexpressed with Orai1, STIM11–448 behaved much like WT-STIM1; it was distributed throughout the ER of resting cells, and following store depletion it formed puncta with Orai1 and activated Ca2+ entry (Fig. 7B, E), though both puncta and Ca2+ entry were somewhat less pronounced than with WT-STIM1 (Fig. 7F). Therefore the region of STIM1 carboxy-terminal to CAD is not absolutely required for CRAC channel binding or activation, although it may make a minor contribution to both.
Deleting the last 8 aa from the CAD (342–440) peptide eliminates its ability to activate CRAC channels (Fig. 2B, C). We therefore tested whether this mutation also prevents STIM11–448 from activating Orai1 by deleting the final 8 aa to generate STIM11–440. STIM11–440 failed to activate SOCE after store depletion (Fig. 7E, F), supporting the idea that CAD function is required for CRAC channel activation by STIM1. Surprisingly, however, STIM11–440 retained the ability to form puncta and cluster Orai1, though to a lesser extent than WT-STIM1 (Fig. 7C, F). Consistent with this result, co-immunoprecipitation experiments revealed that the CAD 342–440 retained the ability to bind to Orai1 (Supplemental Fig. 8) even though it cannot activate it. We obtained a similar result with a STIM1 C437G mutant, which formed pronounced puncta with Orai1 but only marginally activated SOCE (Fig. 7D–F). Together, these data indicate that clustering of CRAC channels by itself is not sufficient to cause channel opening.
DISCUSSION
The STIM1 Polybasic Domain Directs Orai1-independent Formation of STIM1 Puncta
Our results with STIM1-ΔK reveal a new role for the highly conserved lysine-rich domain in STIM1 trafficking and resolve the current debate about its necessity for CRAC channel activation. We found that while STIM1 can form puncta when expressed alone, STIM1-ΔK requires co-expression of Orai1, showing that the polybasic domain enables STIM1 binding to an Orai1-independent target at ER-PM junctions. These results explain discrepancies in prior studies, where STIM1-ΔK failed to redistribute when expressed alone in HEK 293 or HeLa cells (Huang et al., 2006; Liou et al., 2007) but did form puncta and activate ICRAC when coexpressed with Orai1 in HEK 293 cells (Li et al., 2007). The latter finding is now explained by the direct binding of the CAD domain in STIM1-ΔK to Orai1.
The existence of multiple mechanisms for targeting STIM1 to ER-PM junctions is is expected to enhance productive interactions between STIM1 and Orai1 following Ca2+ store depletion. Because endogenous levels of STIM1 and Orai1 are generally modest and their interaction is restricted to ER-PM junctions which cover only ~5% of the cell surface (Wu et al., 2006), the probability that freely diffusing STIM1 and Orai1 proteins will encounter and bind to each other is expected to be low under physiological conditions. However, the initial recruitment of STIM1 to the ER-PM junctions via the polybasic domain will generate a high local concentration of CAD sites beneath the plasma membrane, thereby increasing the likelihood that a diffusing CRAC channel will bind to STIM1 and enhancing the speed and extent of CRAC channel activation.
CAD is the Domain of STIM1 that Activates the CRAC Channel
In previous studies the STIM1 cytosolic region was truncated or mutated to probe the functions of putative domains that had been identified through bioinformatic analysis. Significantly, all of the truncations that completely inhibit CRAC channel activation and prevent the formation of puncta eliminate all or part of CAD (aa 342–448). These include the “delta-ERM” (deleted aa 251–535) (Huang et al., 2006), “delta-ST” (aa 1–390) (Baba et al., 2006), and “delta-C2” (aa 1–424) (Li et al., 2007) variants. The inhibitory effects of these deletions can now all be ascribed to a loss of CAD function rather than suggesting roles for the ERM, serine-proline-rich, and polybasic domains in SOC activation. The CAD s central role is further underscored by the potent ability of CAD peptide and STIM11–448 to activate SOCE and by the loss of STIM1 activity following deletion of CAD or the introduction of mutations that inhibit CAD function (Fig. 7).
Heterologous expression of the full-length cytosolic STIM1 domain, CT-STIM1 has been reported to activate CRAC channels (Huang et al., 2006; Muik et al., 2008; Zhang et al., 2008). In our studies, CT-STIM1 failed to bind to Orai1 or evoke detectable CRAC channel activity when expressed at moderate levels at which CAD bound to Orai1 and strongly activated CRAC channels. We only observed an effect of CT-STIM1 when it was overexpressed in HEK 293T cells along with Orai1, suggesting that it binds to Orai1 with very low affinity. The much-reduced activity of CT-STIM1 relative to CAD suggests that the CAD may be hidden within CT-STIM1 and that a conformational change (perhaps caused by STIM1 oligomerization) is needed to expose the CAD after store depletion.
CAD Binds Directly to Orai1 to Activate the CRAC Channel
Since the discovery of STIM1 and Orai1 and the demonstration that they co-cluster at ER-PM junctions in response to store depletion, there has been considerable interest and uncertainty about how STIM1 activates the CRAC channel across the narrow 10–25 nm ER-PM gap. Several possible mechanisms have been proposed, including local release of a diffusible messenger (Bolotina, 2008), or a physical interaction with Orai1 mediated either directly or through unidentified auxiliary proteins (Muik et al., 2008; Varnai et al., 2007; Yeromin et al., 2006). Our GST-pulldown and coelution results with purified proteins provide the first definitive evidence that CAD binds directly to Orai1 without the necessity for auxiliary partners. Furthermore, the ability of CAD to activate CRAC channels throughout the cell independently of ER Ca2+ depletion argues strongly against the necessity for a diffusible messenger like CIF to be released by depletion-induced changes in the STIM1 luminal domain (Bolotina, 2008). Our results provide strong evidence for a conformational coupling mechanism in which binding of the STIM1 CAD to Orai1 induces conformational changes that lead directly to opening of the CRAC channel.
CAD Interacts with Multiple Regions of Orai1
The N- and C-terminal domains of Orai1 have previously been implicated in CRAC channel clustering and activation. Deletion of the Orai1 C-terminus or an L273S mutation in the C-terminus prevents STIM1 from clustering Orai1 and activating ICRAC whereas deletion of the N-terminus does not affect clustering but precludes channel activation (Li et al., 2007; Muik et al., 2008)but see (Takahashi et al., 2007). In addition, CT-STIM1 was shown to bind weakly to the isolated Orai1 C-terminus but not the N-terminus in vitro (Muik et al., 2008). These studies concluded that STIM1 interacts physically only with the Orai1 C-terminus, with the N-terminus being required for channel gating. Our results using yeast split ubiquitin and mammalian cell co-immunoprecipitation assays confirm the strong binding of CAD to the Orai1 C-terminus but also demonstrate an interaction with the membrane proximal part of the N-terminus (aa 73–91), a region that is required for ICRAC activation (Li et al., 2007) (Fig. 5F, Supplemental Fig. 5). Because CAD binds to two regions of Orai1 that are essential for channel activation, we hypothesize that CAD may provide the energy for CRAC channel gating by bridging the N- and C-termini of the channel.
CAD-induced Clustering of CRAC Channels is Independent of Activation
Purified CAD forms a tetramer in solution (Fig. 4D) and can cluster Orai1/CRAC channel particles into extended arrays both in vivo and in vitro (Fig. 6). This implies that each CAD tetramer has at least two binding sites for Orai1 and each CRAC channel at least two binding sites for CAD. A key question is, does CRAC channel clustering itself deliver a signal for channel activation? A recent study suggests that STIM1 dimerizes resting dimers of Orai1 in the plasma membrane to create functional homotetrameric CRAC channels (Penna et al., 2008). While we have not addressed this issue in detail, our results do not support the conclusion of Penna et al. Negative stain EM data show that CAD induces the clustering of Orai1 particles that are similar in size to pure Orai1 particles, which is inconsistent with the merging of two Orai1 dimers to form a tetramer (Fig. 6A). Instead, these data support the idea that the CRAC channel has a constant subunit stoichiometry (Ji et al., 2008) and that CAD links multiple channels together. Furthermore, the ability of the STIM1 truncation mutant (1–440) (and to a lesser extent the C437G mutant) to cluster Orai1 at ER-PM junctions without activating the channel (Fig. 7F) shows that clustering and activation are separable processes. Thus, CRAC channel clustering itself is not sufficient to induce opening, implying that CAD operates through an allosteric mechanism to gate the CRAC channel.
A Molecular Mechanism for CRAC Channel Activation
The results of this study together with previous data suggest the following model for the activation of Orai1 by STIM1. Depletion of ER Ca2+ stores causes a conformational change in the luminal EF-hand/SAM domain of STIM1 that leads to its oligomerization. Oligomerization has two functional effects. First, it increases the avidity of the polybasic region, enabling it to target STIM1 to ER-PM junctions; second, it causes a conformational change in STIM1 to expose the CAD. The locally high concentration of exposed STIM1 CAD at these sites promotes binding to Orai1, creating a trap to accumulate diffusing CRAC channels and leading to the formation of tightly colocalized STIM1-Orai1 clusters. Finally, the binding of CAD alters the conformation of Orai1 to drive the opening of CRAC channels and evoke Ca2+ entry. More direct evidence for key steps, e.g., the structural basis of STIM1 oligomerization and conformational changes accompanying the exposure of the CAD and CRAC channel opening, will be needed to test this model definitively.
EXPERIMENTAL PROCEDURES
Cells and transfection
HEK 293 and HEK 293T cells (ATCC) were cultured in DMEM with GlutaMax (GIBCO, Carlsbad, CA), 10% FBS (Hyclone, Logan, UT), and 1% penicillin/streptomycin (Mediatech, Hargrave, VA). A HEK 293 cell line with an inducible eGFP-myc-Orai1 was generated using the Flp-In T-REx system (Invitrogen) and was maintained with 50 μg/ml hygromycin and 15 μg/ml blasticidin. Cells were transfected at 90% confluency with 0.2–0.5 μg DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer s instructions.
NFAT-luciferase Assays
HEK 293T cells were co-transfected with the indicated constructs and an NFAT reporter gene (firefly luciferase gene C-terminal to 4X-NFAT site from the IL-2 gene). Cotransfection with the Renilla luciferase (pRLTK) gene driven by the TK promoter was used to control for cell number and transfection efficiency. After 12–18 h, cells were treated with a control DMSO solution (mock), PMA (1 μM), or PMA plus TG (1 μM) for 8 h. Assays were performed using the Dual Luciferase Reporter assay system (Promega). For each condition, luciferase activity was measured with 4 samples taken from duplicate wells using a 96-well automated luminometer (Turner Biosystems). Results are represented as the ratio of firefly to Renilla luciferase activity.
Immunoprecipitation and Immunoblot Analysis
Transfected HEK 293T cells (12–24 h) were washed with PBS and lysed in 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, and protease inhibitors. Lysates were spun at 12,000 rpm for 10 min, and the supernatant was incubated with anti-Flag M2 agarose beads (Sigma), or anti-myc or -HA antibody followed by IgG agarose beads (Pierce). Lysates and immunoprecipitates were subjected to SDS-PAGE, probed with HRP-conjugated secondary antibody and detected by enhanced chemiluminescence (Pierce).
Split-Ubiquitin Yeast Two Hybrid Assay
Screening was performed according to the manufacturer s instructions (Dualsystems Biotech). Transformed yeast were selected on media lacking Trp and Leu (-TL). Interaction was observed by cell growth on plates lacking Trp, Leu, and His (-TLH) in the presence of 3-aminotriazole (3-AT). Protein interactions were also assessed by measuring lacZ activity using the chromogenic substrate X-gal (5-bromo-4-chloro-3-indolyl-β-galactopyranoside).
GST Pull-down Assays
For GST pull-down assays, either GST-CAD (0.16 μM) or GST (0.15 μM) was incubated with EE-Orai1-His8 (0.1 μM) for 1 h in buffer containing (in mM): 20 Tris-HCl (pH 8.0), 150 NaCl, 20 imidazole, and 1 DTT with protease inhibitors. After addition of glutathione sepharose the beads were centrifuged, washed with the above buffer, and the precipitated proteins were eluted with boiling SDS and analyzed by SDS-PAGE and Western blotting.
Purification of EE-Orai1-His8
EE-Orai1-His8 and His6-CAD were integrated into baculoviruses and expressed alone (Orai1) or together (Orai1 + CAD) in Hi5 cells (Invitrogen, Carlsbad, CA) for 48 h at 28 °C. After cell lysis, membranes were solubilized by resuspension in 1% DDM (n-dodecyl-β-d-maltoside; Anatrace, IL, USA). Protein was purified using Ni-NTA beads, and the eluted protein was then incubated with γ-bind Protein G Sepharose beads and anti-EE antibody (Covance, CA, USA) overnight at 4 °C, and eluted with 1 mg/ml EE peptide (Anaspec, CA, USA). DDM was maintained at 0.1%, and the NaCl concentration was 0.5 M up to this purification step. The protein was then passed over a Superose 6 size exclusion column equilibrated in 20 mM Tris pH 8, 150 mM NaCl, 10% glycerol, 0.02% DDM and 0.004% CHS to remove aggregated material and EE peptide. Orai1 was > 98% pure as judged by SDS-PAGE analysis.
MALS analysis of His6-CAD
His6-CAD was isolated from cells expressing His6-CAD and EE-Orai1-His8 and analyzed by MALS using a DAWN EOS light scattering system (Wyatt Technology, Santa Barbara, CA). The detector responses were normalized against monomeric bovine serum albumin.
Electron microscopy
EE-Orai1-His8 alone or copurified with His6-CAD was diluted to a final concentration of ~0.01 mg/ml in 20 mM Tris, 150 mM NaCl, and 0.02% DDM buffer and was negatively stained with uranyl formate as described (Ohi et al., 2004). Images were recorded with a Phillips CM-10 electron microscope equipped with a tungsten filament operated at 100 kV. Images were taken at a nominal magnification of 39,000× and a defocus of −1.5 μm on a Gatan 1k × 1k CCD camera.
Confocal Microscopy
6 h after transfection, HEK 293 cells were plated onto sterilized coverslips coated with poly-D-lysine and maintained in complete DMEM for an additional 12–18 h before imaging in Ringer s solution containing (in mM): 155 NaCl, 4.5 KCl, 2 CaCl2, 1 MgCl2, 10 D-glucose, and 5 Na-Hepes, pH 7.4. To deplete stores, cells were treated with 1 μM TG in Ca2+-free Ringer s (prepared by substituting 2 mM MgCl2 and 1 mM EGTA for CaCl2) for 10 min. eGFP and mCherry were excited simultaneously at 488 and 594 nm, respectively, on a Leica SP2 AOBS inverted confocal microscope equipped with a PL APO 100x/NA 1.4 oil immersion objective. Fluorescence emissions were collected at 615–840 nm (mCherry) and 510–570 nm (eGFP). All experiments were performed at 22–25°C.
Fluorescence Recovery After Photobleaching (FRAP)
Inducible HEK 293 cells were transiently transfected with mCh-CAD and maintained with 10 μM LaCl3 to suppress chronic Ca2+ entry. After 15–16 h, eGFP-myc-Orai1 expression was induced with 1 μg/ml tetracycline in medium containing LaCl3. Cells were rinsed with 2 mM Ca2+ Ringer s solution at 24 h post-transfection prior to imaging on the confocal system described above. Prebleach and recovery images were scanned at 488 nm at low power (20 mW Ar laser, 50% power, 6% transmission). A 3-μm strip was bleached at 50% laser power with full transmission for ~2 s. Bleaching during the recovery period was negligible (<5%). FRAP recovery curves were analyzed as described (see Supplemental Methods).
Ca2+ Imaging
For Fig. 2, cells were loaded at 37 C in DMEM with 1μM fura-2/AM for 30 min. Ratiometric Ca2+ imaging was performed at 340 and 380 nm in 2 mM Ca2+ Ringer s solution with a Nikon Eclipse 2000-U inverted microscope equipped with a fluorescent arc lamp, excitation filter wheel, and a Hamamatsu Orca CCD camera. Images were collected using Openlab (Improvision) and analyzed using Igor Pro.
For Fig. 7, cells were loaded as above with 2 μM fura-2/AM for 25 min. Ratiometric Ca2+ imaging was performed with 350 and 380 nm excitation in 2 mM Ca2+ Ringer s solution on an Axiovert 35 inverted microscope using a VideoProbe imaging system as described (Bautista et al., 2002). mCherry-positive cells were identified using a 540 ± 12 nm excitation and a 580LP emission filter (Chroma).
Electrophysiology
HEK 293 cells were transfected 8–24 h prior to electrophysiology experiments with STIM1- and Orai1- derived constructs in a 1:1 mass ratio using Lipofectamine 2000. Cells transfected with CAD + Orai1 were cultured in 10 μM LaCl3 to avoid the toxicity of constitutively active ICRAC, and LaCl3 was washed out immediately before seal formation. ICRAC in cells cultured without LaCl3 was similar to that in cells cultured with LaCl3, but most cells without LaCl3 died soon after break-in.
Currents were recorded using standard whole-cell patch clamp techniques (Prakriya and Lewis, 2001). Pipettes of resistance 2–5 MΩ were filled with an internal solution containing (in mM) 150 Cs aspartate, 8 MgCl2, 10 EGTA, and 10 HEPES (pH 7.2 with CsOH). Currents were sampled at 5 kHz and filtered at 2 kHz, and all voltages were corrected for the junction potential of the pipette solution relative to Ringer s in the bath (−13 mV).
Supplementary Material
Acknowledgments
The authors thank M. Wu for early Ca2+ imaging experiments on CT-STIM1, D. Oh for helpful comments, and M. Cahalan, S. Feske, T. Meyer, S. Muallem, C. Romanin, and T. Xu for cDNA constructs. This work was supported by grants from the NIH (R.E.D. and R.S.L.), the Mathers Charitable Foundation (R.S.L.), graduate fellowships from the NSF and Stanford U. (E.D.C.), an NIH Medical Scientist Training Program Grant (P.J.H.), and the Korea Research Foundation (KRF-2005-214-C00222) and the SPARK program (C.Y.P.). T.W. and K.C.G. are Investigators of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baba Y, Hayashi K, Fujii Y, Mizushima A, Watarai H, Wakamori M, Numaga T, Mori Y, Iino M, Hikida M, Kurosaki T. Coupling of STIM1 to store-operated Ca2+ entry through its constitutive and inducible movement in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2006;103:16704–16709. doi: 10.1073/pnas.0608358103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista DM, Hoth M, Lewis RS. Enhancement of calcium signalling dynamics and stability by delayed modulation of the plasma-membrane calcium-ATPase in human T cells. J Physiol. 2002;541:877–894. doi: 10.1113/jphysiol.2001.016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ. Capacitative calcium entry. Biochem J. 1995;312:1–11. doi: 10.1042/bj3120001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotina VM. Orai, STIM1 and iPLA2beta: a view from a different perspective. J Physiol. 2008;586:3035–3042. doi: 10.1113/jphysiol.2008.154997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Liou J, Park WS, Meyer T. STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ levels. Cell. 2007;131:1327–1339. doi: 10.1016/j.cell.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feske S, Giltnane J, Dolmetsch R, Staudt LM, Rao A. Gene regulation mediated by calcium signals in T lymphocytes. Nat Immunol. 2001;2:316–324. doi: 10.1038/86318. [DOI] [PubMed] [Google Scholar]
- Feske S, Prakriya M, Rao A, Lewis RS. A severe defect in CRAC Ca2+ channel activation and altered K+ channel gating in T cells from immunodeficient patients. J Exp Med. 2005;202:651–662. doi: 10.1084/jem.20050687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwack Y, Srikanth S, Feske S, Cruz-Guilloty F, Oh-hora M, Neems DS, Hogan PG, Rao A. Biochemical and functional characterization of Orai proteins. J Biol Chem. 2007;282:16232–16243. doi: 10.1074/jbc.M609630200. [DOI] [PubMed] [Google Scholar]
- Huang GN, Zeng W, Kim JY, Yuan JP, Han L, Muallem S, Worley PF. STIM1 carboxyl-terminus activates native SOC, Icrac and TRPC1 channels. Nat Cell Biol. 2006;8:1003–1010. doi: 10.1038/ncb1454. [DOI] [PubMed] [Google Scholar]
- Ji W, Xu P, Li Z, Lu J, Liu L, Zhan Y, Chen Y, Hille B, Xu T, Chen L. Functional stoichiometry of the unitary calcium-release-activated calcium channel. Proc Natl Acad Sci U S A. 2008;105:13668–13673. doi: 10.1073/pnas.0806499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lu J, Xu P, Xie X, Chen L, Xu T. Mapping the interacting domains of STIM1 and Orai1 in Ca2+ release-activated Ca2+ channel activation. J Biol Chem. 2007;282:29448–29456. doi: 10.1074/jbc.M703573200. [DOI] [PubMed] [Google Scholar]
- Liou J, Fivaz M, Inoue T, Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci U S A. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, Jr, Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luik RM, Wang B, Prakriya M, Wu MM, Lewis RS. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature. 2008;454:538–542. doi: 10.1038/nature07065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luik RM, Wu MM, Buchanan J, Lewis RS. The elementary unit of store-operated Ca2+ entry: local activation of CRAC channels by STIM1 at ER-plasma membrane junctions. J Cell Biol. 2006;174:815–825. doi: 10.1083/jcb.200604015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik M, Frischauf I, Derler I, Fahrner M, Bergsmann J, Eder P, Schindl R, Hesch C, Polzinger B, Fritsch R, et al. Dynamic Coupling of the Putative Coiled-coil Domain of ORAI1 with STIM1 Mediates ORAI1 Channel Activation. J Biol Chem. 2008;283:8014–8022. doi: 10.1074/jbc.M708898200. [DOI] [PubMed] [Google Scholar]
- Navarro-Borelly L, Somasundaram A, Yamashita M, Ren D, Miller RJ, Prakriya M. STIM1-Orai1 interactions and Orai1 conformational changes revealed by live-cell FRET microscopy. J Physiol. 2008;586:5383–5401. doi: 10.1113/jphysiol.2008.162503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh AB, Putney JW., Jr Store-operated calcium channels. Physiol Rev. 2005;85:757–810. doi: 10.1152/physrev.00057.2003. [DOI] [PubMed] [Google Scholar]
- Partiseti M, Le Deist F, Hivroz C, Fischer A, Korn H, Choquet D. The calcium current activated by T cell receptor and store depletion in human lymphocytes is absent in a primary immunodeficiency. J Biol Chem. 1994;269:32327–32335. [PubMed] [Google Scholar]
- Penna A, Demuro A, Yeromin AV, Zhang SL, Safrina O, Parker I, Cahalan MD. The CRAC channel consists of a tetramer formed by Stim-induced dimerization of Orai dimers. Nature. 2008;456:116–120. doi: 10.1038/nature07338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M, Feske S, Gwack Y, Srikanth S, Rao A, Hogan PG. Orai1 is an essential pore subunit of the CRAC channel. Nature. 2006;443:230–233. doi: 10.1038/nature05122. [DOI] [PubMed] [Google Scholar]
- Prakriya M, Lewis RS. Potentiation and inhibition of Ca2+ release-activated Ca2+ channels by 2-aminoethyldiphenyl borate (2-APB) occurs independently of IP3 receptors. J Physiol. 2001;536:3–19. doi: 10.1111/j.1469-7793.2001.t01-1-00003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M, Lewis RS. Store-operated calcium channels: properties, functions and the search for a molecular mechanism. Adv Molec Cell Biol. 2004;32:121–140. [Google Scholar]
- Putney JW., Jr A model for receptor-regulated calcium entry. Cell Calcium. 1986;7:1–12. doi: 10.1016/0143-4160(86)90026-6. [DOI] [PubMed] [Google Scholar]
- Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopulos PB, Li GY, Plevin MJ, Ames JB, Ikura M. Stored Ca2+ depletion-induced oligomerization of STIM1 via the EF-SAM region: An initiation mechanism for capacitive Ca2+ entry. J Biol Chem. 2006;281:35855–35862. doi: 10.1074/jbc.M608247200. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Murakami M, Watanabe H, Hasegawa H, Ohba T, Munehisa Y, Nobori K, Ono K, Iijima T, Ito H. Essential role of the N-terminus of murine Orai1 in store-operated Ca2+ entry. Biochem Biophys Res Commun. 2007;356:45–52. doi: 10.1016/j.bbrc.2007.02.107. [DOI] [PubMed] [Google Scholar]
- Thaminy S, Miller J, Stagljar I. The split-ubiquitin membrane-based yeast two-hybrid system. Meth Mol Biol. 2004;261:297–312. doi: 10.1385/1-59259-762-9:297. [DOI] [PubMed] [Google Scholar]
- Varnai P, Toth B, Toth DJ, Hunyady L, Balla T. Visualization and manipulation of plasma membrane-endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 Complex. J Biol Chem. 2007;282:29678–29690. doi: 10.1074/jbc.M704339200. [DOI] [PubMed] [Google Scholar]
- Vig M, Beck A, Billingsley JM, Lis A, Parvez S, Peinelt C, Koomoa DL, Soboloff J, Gill DL, Fleig A, et al. CRACM1 multimers form the ion-selective pore of the CRAC channel. Curr Biol. 2006;16:2073–2079. doi: 10.1016/j.cub.2006.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vig M, DeHaven WI, Bird GS, Billingsley JM, Wang H, Rao PE, Hutchings AB, Jouvin MH, Putney JW, Kinet JP. Defective mast cell effector functions in mice lacking the CRACM1 pore subunit of store-operated calcium release-activated calcium channels. Nat Immunol. 2008;9:89–96. doi: 10.1038/ni1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MM, Buchanan J, Luik RM, Lewis RS. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Lu J, Li Z, Yu X, Chen L, Xu T. Aggregation of STIM1 underneath the plasma membrane induces clustering of Orai1. Biochem Biophys Res Commun. 2006;350:969–976. doi: 10.1016/j.bbrc.2006.09.134. [DOI] [PubMed] [Google Scholar]
- Yeromin AV, Zhang SL, Jiang W, Yu Y, Safrina O, Cahalan MD. Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature. 2006;443:226–229. doi: 10.1038/nature05108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SL, Kozak JA, Jiang W, Yeromin AV, Chen J, Yu Y, Penna A, Shen W, Chi V, Cahalan MD. Store-dependent and -independent modes regulating Ca2+ release-activated Ca2+ channel activity of human Orai1 and Orai3. J Biol Chem. 2008;283:17662–17671. doi: 10.1074/jbc.M801536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SL, Yu Y, Roos J, Kozak JA, Deerinck TJ, Ellisman MH, Stauderman KA, Cahalan MD. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifach A, Lewis RS. Rapid inactivation of depletion-activated calcium current (ICRAC) due to local calcium feedback. J Gen Physiol. 1995;105:209–226. doi: 10.1085/jgp.105.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.